In the United States, the gold standard for malaria diagnosis is microscopic blood smear examination. Because malaria is not endemic in the United States, diagnostic capabilities may be limited, causing delays in diagnosis and increased morbidity and mortality.
KEYWORDS: malaria, diagnostic testing, United States, CLSI guidelines
ABSTRACT
In the United States, the gold standard for malaria diagnosis is microscopic blood smear examination. Because malaria is not endemic in the United States, diagnostic capabilities may be limited, causing delays in diagnosis and increased morbidity and mortality. A survey of the malaria diagnostic practices of U.S. laboratories was conducted from June to July 2017; members of the American Society for Microbiology's listserv received a questionnaire inquiring about malaria diagnostic test availability, techniques, and reporting. Results were assessed using the Clinical and Laboratory Standards Institute (CLSI) guidelines for malaria diagnostics. After excluding incomplete and duplicate responses, responses representing 175 laboratories were included. Most labs (99%) received at least one specimen annually for malaria diagnosis, and 31% reported receiving only 1 to 10 specimens. The majority (74%) diagnosed five or fewer cases of malaria per year. Most (90%) performed blood smears on-site. Two-thirds (70%) provided initial blood smear results within 4 h. Although diagnostic testing for malaria was available 24/7 at 74% (141) of responding laboratories, only 12% (17) met criteria for analysis and reporting of malaria testing, significantly more than reported in a similar survey in 2010 (3%; P < 0.05). The majority of laboratories surveyed had the capability for timely diagnosis of malaria; few comply with CLSI guidelines. Inexperience may factor into this noncompliance; many laboratories see few to no cases of malaria per year. Although reported adherence to CLSI guidelines was higher than in 2010, there is a need to further improve laboratory compliance with recommendations.
INTRODUCTION
In 2016, there were an estimated 216 million cases of malaria, with 445,000 deaths globally (1). Malaria is endemic in 91 countries, and it continues to be a leading cause of morbidity and mortality worldwide (1). An estimated 74 million U.S. travelers departed for international destinations in 2015 (2), and up to four million will develop illnesses that are severe enough to prompt care seeking during or after their trip (3). Nearly one-quarter of travelers develop a febrile illness and of those, nearly one-third were diagnosed with malaria (4). In the United States, 1,500 to 1,900 imported cases of malaria are reported to the Centers for Disease Control and Prevention (CDC) annually, with 1,517 cases reported in 2015 (5–11).
Between 2010 and 2015, a total of 46 fatalities due to malaria occurred in the United States (5–11). Diagnostic delays potentially contributed to 18 of these deaths, highlighting the need for prompt and accurate diagnosis and timely treatment (6–12). Up to 92% of patients presenting to physicians without expertise in tropical medicine experience diagnostic and treatment delays due to failure to consider the diagnosis of malaria on initial presentation and to laboratory errors in recognition and species identification (13). In a Canadian study by Kain et al., 45% of laboratories provided incorrect species identification or no species identification at all (13), impeding optimal treatment.
There are significant challenges to diagnosing malaria, especially in an area where it is not endemic. A range of challenges may contribute to these laboratory errors. For example, nonimmune individuals may present with severe illness even with low parasite densities, and parasite morphology may be altered because of the use of chemoprophylaxis. Given the rarity of cases, laboratory technicians may have limited experience identifying malaria parasites (12).
The Clinical and Laboratory Standards Institute (CLSI) produces guidelines for laboratories in an effort to reduce diagnostic delays and improve laboratory competencies (14). The guidelines for malaria diagnosis include:
Availability of diagnostic capabilities 24 h/day, 7 days/week.
Preparation of at least three thick and three thin smears from each sample received.
Use of Giemsa stain for definitive diagnosis.
Examination of at least 300 fields using 100× objective under oil immersion.
Examination of at least 10 fields on a thin smear to determine percent parasitemia (the Centers for Disease Control and Prevention [CDC] recommends counting 500 to 2,000 red blood cells on a thin smear for patients in the United States who may have lower levels of parasitemia).
Immediate reporting of microscopy results.
Kain et al. found that most community laboratories did not perform malaria smears on an urgent basis, nor did they routinely report species identification or percent parasitemia (13). Overall, these laboratories had significant reporting delays, misdiagnoses, and incorrect species identifications (13). A 2010 survey of U.S. laboratories found that most responding laboratories performed thick and thin smears on-site; however, only 3% of laboratories performing malaria diagnostic testing around the clock met all CLSI guidelines and regulatory practices; 28% of these stated that species identification took more than 24 h (15). This study was conducted to provide an updated assessment of laboratory practices related to malaria diagnosis in the United States.
MATERIALS AND METHODS
A survey of United States-based laboratories was conducted from 26 June to 24 July 2017, to determine current malaria diagnostic testing practices. The web-based survey (SurveyMonkey Inc., San Mateo, CA) included questions on testing practices, diagnostic test availability, methodologies, time to reporting, and who performs diagnostic testing (see the supplemental material for full survey). A convenience sample of laboratory personnel was recruited through an introductory email and two reminder e-mails, all of which included a link to the survey, which were distributed to the American Society for Microbiology (ASM) listservs ClinMicroNet and DivC. These include microbiology technologists, clinical laboratory scientists, pathologists, and hematologists worldwide. The survey was limited to individuals residing in the United States.
The survey was based on a prior survey used by Abanyie et al. (15), with edits for clarification and additional questions to enable removal of duplicate responses. Responses were limited to one per physical laboratory using a combination of variables, including internet protocol (IP) address, ZIP code, and institution type/population served. In cases where there appeared to be duplicate responses following review of this combination of variables, we selected one respondent based on the response that was most complete or was from the most senior respondent (e.g., laboratory director or microbiology director). Adherence to CLSI guidelines for malaria diagnosis were assessed using the same questions as in the 2010 survey. Laboratories meeting the following seven criteria were defined as in compliance with recommendations: 24/7 diagnostic capabilities, preliminary report within 4 h, report of percent parasitemia within 6 h, species identification within 24 h, examination of at least 300 fields for a negative smear, counting at least 500 red blood cells (RBCs) on a thin smear to determine percent parasitemia, and use of Giemsa stain for definitive diagnosis. Survey results were analyzed using Excel (Microsoft Office 2013, Seattle, WA) and RStudio (RStudio Team 2016, Boston, MA). Frequencies and averages were computed, and chi-square or Fischer's exact tests were used, as appropriate, to compare between groups. A P value of <0.05 was considered statistically significant.
Ethical review.
The study was determined not to require review by the Emory University Institutional Review Board and was approved as research not involving human subjects by the office of the Associate Director for Science, Center for Global Health at the U.S. Centers for Disease Control and Prevention. Contacted individuals were provided information about the study and informed that participation was voluntary and that they could end the survey at any time.
RESULTS
Study participants.
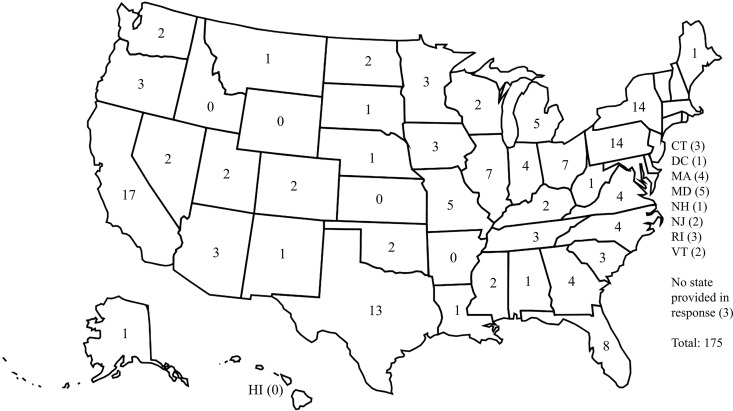
A total of 237 participants initiated the survey, and 190 (80.2%) completed it. After exclusion of 15 duplicates, the final set of responses represented 175 laboratories. Laboratories were located in 45 of the 50 (90%) states, with the greatest number of responses from California (17; 9.7%), followed by New York (14; 8%), Pennsylvania (14; 8%), and Texas (13; 7.4%) (Fig. 1). The majority of respondents self-identified as laboratory directors (92; 52.6%) or microbiologists (48; 27.4%). Responses from university and community hospital-affiliated labs each comprised about 40% of the total, with 70 respondents from university hospitals and 68 from community hospitals (Table 1). The majority (71%) of the laboratories serve both adult and pediatric populations; 21% serve an exclusively adult population, and 8% serve a pediatric population. Of responding laboratories, 30 (17%) report being either a commercial referral laboratory or central laboratory for multiple hospitals.
FIG 1.
Responses included in the analysis by state.
TABLE 1.
Classification of laboratory type with number of responses, on-site testing, and CLSI practices
| Laboratory type | No. (%) of responsesa | No. (%) of laboratories with on-site testing | No. (%) of laboratories meeting CLSI guidelines and regulatory practices |
|---|---|---|---|
| University hospital | 70 (40) | 69 (99) | 7 (10) |
| Community hospital | 68 (39) | 58 (85) | 7 (12) |
| Commercial referral laboratory | 10 (6) | 9 (90) | 1 (11) |
| VAb hospital | 5 (3) | 4 (80) | 0 (0) |
| Primary care center | 2 (1) | 2 (100) | 0 (0) |
| Other | 20 (11) | 18 (90) | 2 (11) |
| All | 175 | 160 (91) | 17 (10) |
Total and percentages based on 175 responses to question. Other laboratories include consolidated regional laboratories (8), public health laboratories (6), other referral laboratories (2), military hospital laboratory (1), consultant (1), and laboratories that did not provide detail on affiliation (2).
VA, Veterans Affairs.
Diagnostic testing availability on-site.
The majority of respondents (91%; 160) reported that their laboratories offered at least one type of diagnostic test for malaria on-site; the remaining 9% (15) offer only send-out testing. Almost all university hospital (69; 99%) and most community hospital (58; 85%) laboratories offered on-site malaria testing. A total of 141 (81%) laboratories report on-site testing with in-house or on-call personnel 24 h a day, 7 days a week. The remaining laboratories provide coverage over the majority of the working day, but with various hours.
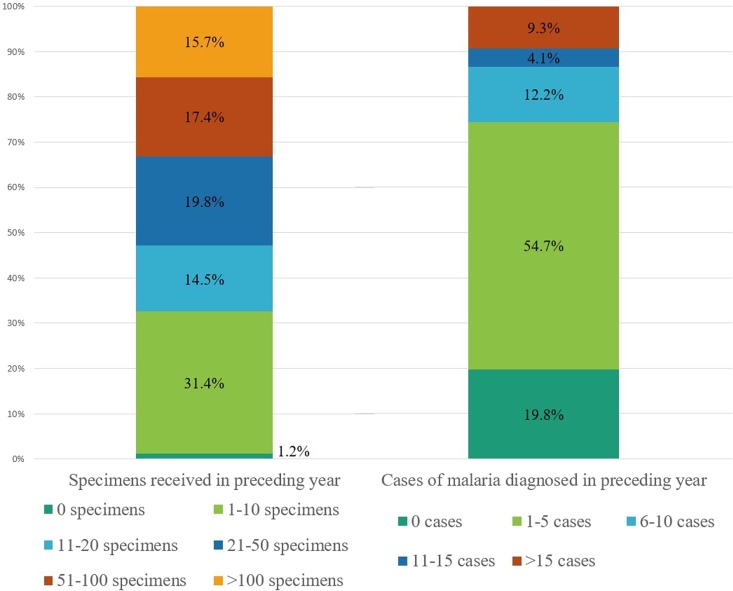
The annual number of specimens received by the laboratories was generally small, with the majority (66%) receiving fewer than 50 specimens per year and two (1%) receiving none at all (Fig. 2). The majority (75%) of labs had five or fewer confirmed cases of malaria in the preceding year, while 16 laboratories (9%) had more than 15 confirmed cases in the preceding year (Fig. 2). Of laboratories that provide any on-site testing, 99% (158) perform microscopy (thick and thin blood smears); the remaining two provide rapid diagnostic testing (RDT) as the only testing method. RDT is performed on-site at a total of 65 laboratories (40%); microscopy is performed either simultaneously or subsequent to RDT at these laboratories. A total of 10 (6%) laboratories perform PCR on-site, while an additional 119 (68%) access PCR as a send-out test.
FIG 2.
Number of specimens received for malaria diagnostic testing and number of confirmed cases of malaria in percentages for the year preceding the survey. (n = 172).
Technique of on-site testing.
Of 158 laboratories that perform on-site microscopy, 143 (91%) reported on the staining technique. Twenty percent of laboratories reported using more than one method of staining; 57% use Giemsa stain in at least one step of microscopic analysis, 46% use Wright-Giemsa, and 17% use Wright stain.
Approximately half of respondents (73; 46%) report a practice of reviewing at least 300 fields under oil immersion prior to considering a blood smear negative, 35% review 100 to 299 oil immersion fields (OIFs), and 5% review <100 OIFs. To determine the percent parasitemia, 20% of laboratories count >2,000 red blood cells (RBCs), 49% count 500 to 2,000 RBCs, and 9% count <500 RBCs.
Reporting.
Of the 158 laboratories that perform microscopy on-site, 142 (90%) responded regarding the length of time required for them to give a preliminary report (presence or absence of parasites). Of those, the majority (100; 70%) provide a preliminary report of the smear within 4 h of specimen receipt, with 9 providing a preliminary report in less than an hour. Twenty-four (17%) laboratories provide a preliminary report in 5 to 12 h, 15 (11%) within 13 to 24 h, and 3 (2%) require more than 24 h to provide a report.
Although less than half (48%) of laboratories that perform microscopy on-site provide a report of parasitemia within 6 h of specimen receipt, the majority do so within 24 h (128; 81%). Nine laboratories (6%) do not report percent parasitemia, reporting only whether the smear is positive or negative.
Different departments, most commonly microbiology (59%), hematology (24%), or pathology (2%), screen the smear prior to releasing an initial report. Approximately 60% of laboratories require review of the smear by a laboratory technician and a supervisor (either laboratory director, microbiologist, pathologist, or hematologist) prior to making a report available; in the remainder, two laboratory technicians review the smear prior to providing a report. In some cases, a preliminary report is released quickly if the specimen is received overnight, with the final report following confirmation provided the next morning.
Overall, 65% (113/175) of surveyed laboratories provide species diagnosis within the recommended 24-hour period; all perform this testing on-site. Fifty-nine (43%) of these laboratories determine species of malaria within 6 h of specimen receipt. Overall, species determination is available on-site at 135 of 159 responding laboratories (85%), including 100% of university hospital laboratories, 80% of Veterans Affairs (VA) hospital laboratories, 69% of community hospital laboratories, and 83% of other laboratories. Fifty-eight (36%) laboratories make the species diagnosis on-site, but subsequently send the specimen to another laboratory (e.g., state health laboratory) for confirmation and 39 (67%) have preliminary results in less than 24 h.
CLSI and regulatory practice compliance summary.
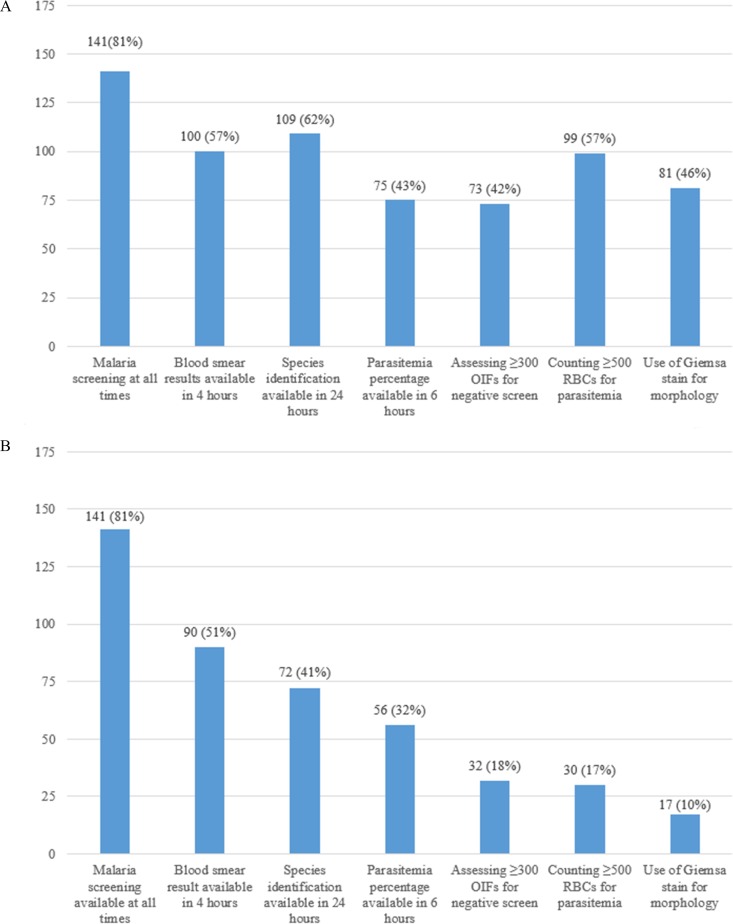
Of 141 laboratories that provide on-site testing 24/7, only 17 (12%) were in compliance with all seven criteria (Fig. 3); this represents 10% of all responding laboratories. These include seven university hospital laboratories, seven community hospital laboratories, two commercial referral laboratories, and one health system core laboratory (Table 1). There was no significant difference in compliance between the laboratories with the highest volume of specimens for malaria diagnostic testing compared to the laboratories with the lowest volume (P = 0.16). An additional 15 laboratories are able to provide a preliminary result within 4 h if RDT results are considered; however, none of these laboratories subsequently meet all of the remaining guidelines.
FIG 3.
(A) Number and proportion of laboratories with any capability for malaria diagnosis (n = 175). (B) Number and proportion of laboratories with 24/7 capability for malaria diagnosis that meet consecutive CLSI guidelines and regulatory practices (n = 175). Malaria screening refers to the ability to perform microscopic examination of a blood smear 24 h a day, 7 days a week. OIF, oil immersion field; RBCs, red blood cells.
Off-site testing.
Sixteen (9%) laboratories send out all specimens for diagnostic testing of malaria, including 11 community hospital laboratories, one university hospital laboratory, one VA laboratory, and three other laboratories. Of these 16 laboratories, the majority receive few specimens for malaria testing; 2 received no specimens, 10 received 1 to 10 specimens, 3 received 11 to 20 specimens, and 1 received >100 specimens in the preceding year. Similarly, only 6 of these laboratories identified any confirmed cases of malaria in the preceding year: 5 diagnosed 1 to 5 cases, 1 diagnosed 11 to 15 cases, and 1 diagnosed more than 15 cases.
Of those laboratories relying on send-out testing for malaria diagnosis, 75% use microscopy as the primary diagnostic methodology. In addition, 56% report sending specimens for RDT and 75% for PCR. A positive or negative result is available within 24 h for two laboratories, with an additional two receiving results within 48 h. Percent parasitemia is reported within 24 h and species identification within 48 h at two of these laboratories; 10 laboratories state that it takes more than 48 h to report out percent parasitemia and species identification.
DISCUSSION
In areas where malaria is nonendemic, multiple factors contribute to difficulties in providing a rapid and accurate diagnosis of malaria, including delays in recognizing the need for testing due to low burden of disease, delays in the availability of test results, lack of diagnostic capabilities, and inexperience of laboratory personnel. CLSI guidelines provide parameters to ensure an accurate and timely diagnosis, and ideally, all laboratories performing smears should follow these recommendations. However, in this survey, only a small number of responding U.S. laboratories met all seven CLSI criteria for accurate and timely malaria diagnosis and reporting.
The results of the malaria smear, including parasitemia percentage and species of malaria, are critical to selecting the appropriate antimalarial for treatment. Diagnostic delays are associated with the majority of morbidity and mortality associated with malaria in returned travelers, and thus it is important that laboratories have the ability to perform accurate malaria testing 24 h/day, 7 days/week, and provide initial results within 4 h (12, 16). Although the majority of laboratories (83%) have the ability to test for malaria at any time, and 70% provide a positive or negative blood smear result within 4 h of obtaining a specimen, there is still room for improvement.
While microscopy remains the preferred option, in laboratories where microscopy is not available 24/7, RDTs may be an option for rapid screening. These are widely used throughout Africa, particularly in lower-level facilities where microscopes or trained microscopists are not available, as well as in community treatment programs. Many brands of RDT are available globally, with various product characteristics. In the United States, only the BinaxNOW Malaria test (Alere, Waltham, MA) is FDA approved; however, it is only approved for testing in a validated laboratory and not at the point of care. This test uses an immunochromatographic assay to detect Plasmodium antigens (both Plasmodium falciparum-specific histidine-rich protein II [HRPII] and pan-Plasmodium lactate dehydrogenase [LDH]) in 15 min, with high sensitivity and specificity for P. falciparum (94 to 100% and 94.2%, respectively), but much lower sensitivity for detecting non-falciparum species (67 to 86%), particularly P. ovale and P. malariae (17–21). This can provide enough information to begin targeted therapy. RDT sensitivity, similarly to microscopy, depends on parasite concentration, and false negatives may occur in the setting of low-density parasitemia, while false positives may be seen in cases of recent treatment, especially with HRPII-based RDTs (16). In one study from a U.S. laboratory, the sensitivity of BinaxNOW RDT was demonstrated to be 100% for parasitemia as low as 0.1%, although it was only 40% (2/5) for patients with <0.1% parasitemia, all of which had non-falciparum malaria (21). In another study from Toronto, BinaxNOW had an overall sensitivity of 86% compared to microscopy. However, compared to PCR, the sensitivities of microscopy and RDT were similar, 86% and 84%, respectively. Similar to the other study, the majority of false negatives were for non-falciparum species (22). Furthermore, RDTs provide a qualitative assessment and give no information on parasite density. For these reasons, all RDT results should be confirmed with microscopy as soon as possible and no later than 24 h, as microscopy provides additional information on species identification and parasitemia in positive cases. While the limitations of using RDTs need to be recognized, in laboratories where there are not trained microscopists available 24/7 to perform microscopic diagnosis, the use of a rapid diagnostic test, such as BinaxNOW, may allow for rapid provision of a preliminary diagnosis until the specimen can be tested by a reference laboratory. In cases where testing is unavailable on-site, microscopy and PCR should be the only tests provided for malaria diagnosis.
The remainder of the CLSI guidelines and regulatory practices define standards for laboratory practices related to species identification and quantification of parasitemia. Species identification is important for selecting the appropriate antimalarial for treatment. The species of malaria and area of acquisition provide information about drug resistance. Additionally, both P. vivax and P. ovale infections require dual therapy, consisting of an antimalarial to treat the acute infection and, in those that are tested and confirmed to have normal glucose-6-phosphate dehydrogenase (G6PD) activity, primaquine to eliminate dormant hypnozoites, which may reside in the liver for years after primary infection with these species and cause recurrences of malaria. Thick and thin blood smears allow for greater sensitivity in malaria screening and species morphology detection, respectively. It should be further noted that thick and thin smears should be prepared as soon as possible following blood collection; if blood is collected in anticoagulants such as EDTA and held for too long (more than 24 to 48 h), morphological changes may occur, making microscopic diagnosis less reliable (16). However, once thick and thin smears have been prepared and fixed, reading can be delayed without compromising quality. CLSI guidelines recommend a Giemsa stain, as opposed to Wright-Giemsa or other rapid stains, to ensure proper visualization of morphological features (malaria pigment, Schüffner's dots, Maurer's clefts, etc.) that can aid in detection and species identification. Giemsa staining is only used in 57% of laboratories surveyed, with 46% reporting use of Wright-Giemsa stain (which is used to differentiate white blood cells and which may have less accuracy in species identification; see reference 23); this may be due to lack of a dedicated parasitology section at these laboratories. Parasite density should be determined by examining a minimum of 500 red blood cells (RBCs) on a thin smear, as examining fewer fields could lead to missing the diagnosis, particularly in cases with low parasite density; 79% of laboratories follow this recommendation (parasite density, or percent parasitemia, refers to the percentage of infected RBCs, calculated as [number of infected RBCs/total number of RBCs counted] × 100, where a multiply infected RBC counts as 1 infected RBC). From a clinical perspective, it is important to identify patients with hyperparasitemia (>5% of RBCs infected), as these patients require urgent and aggressive treatment due to significantly higher chance of death.
Limitations.
Potential participants for the survey were members of a listserv maintained by the ASM. This listserv includes international members and can include multiple individuals from a single laboratory. Thus, it is impossible to calculate an accurate response rate. While we had responses from a wide variety of laboratory types, the overall sample was nonrandom and was small compared to the overall number of laboratories in the U.S. (nationally, 366 laboratories participate in proficiency testing with the College of American Pathologists for BinaxNOW; R. Rolf, College of American Pathologists, personal communication), and the results may not be generalizable. ClinMicroNet and DivC primarily include those with a background in microbiology and may not contain physicians that oversee laboratories; thus, this group may be underrepresented. Additionally, laboratories may have malaria testing managed by hematology. These laboratories, along with their testing methods, may be underrepresented, as the recruitment methods were aimed at microbiologists. There is also a potential for responder or recall bias, as the responses were self-reported and not observed or verified. For example, it is possible that laboratories that have a higher volume of malaria or those that have more sophisticated methods of testing may have been more likely to respond. Still, our results provide information about the range of approaches to malaria diagnosis and indicate that compliance with CLSI guidelines is incomplete.
Conclusions.
The majority of participating laboratories within the United States have the capability to perform diagnostic testing for malaria. However, only a small proportion of the laboratories surveyed were in compliance with all of the CLSI guidelines. Many laboratories see few to no cases of malaria per year, and each member of the laboratory may only work on a fraction of those cases; this inexperience can lead to diagnostic errors and contribute to poor patient outcomes. Given the life-threatening nature of malaria and the rapidity with which it can progress, compliance with CLSI guidelines remains unacceptably low, and work is needed to ensure improvement. It is possible that an update or review of standard operating procedures is needed to improve laboratory compliance with these guidelines and recommendations.
Supplementary Material
ACKNOWLEDGMENTS
We thank the American Society for Microbiology (Washington, DC) for facilitating recruitment using their listservs, ClinMicroNet and DivC.
No financial support was received for this study.
The findings and conclusions presented in this article are those of the authors and do not necessarily reflect the official position of the U.S. Centers for Disease Control and Prevention.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00461-18.
REFERENCES
- 1.World Health Organization. 2017. World malaria report 2017. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.World Tourism Organization (UNWTO). 2017. UNWTO tourism highlights—2017 edition. World Tourism Organization (UNWTO), Madrid, Spain. [Google Scholar]
- 3.Freedman DO, Weld LH, Kozarsky PE, Fisk T, Robins R, von Sonnenburg F, Keystone JS, Pandey P, Cetron MS. 2006. Spectrum of disease and relation to place of exposure among ill returned travelers. N Engl J Med 354:119–130. doi: 10.1056/NEJMoa051331. [DOI] [PubMed] [Google Scholar]
- 4.Leder K, Torresi J, Libman MD, Cramer JP, Castelli F, Schlagenhauf P, Wilder-Smith A, Wilson ME, Keystone JS, Schwartz E, Barnett ED, von Sonnenburg F, Brownstein JS, Cheng AC, Sotir MJ, Esposito DH, Freedman DO, GeoSentinel Surveillance N. 2013. GeoSentinel surveillance of illness in returned travelers, 2007–2011. Ann Intern Med 158:456–468. doi: 10.7326/0003-4819-158-6-201303190-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arguin PM, Tan KR. 2017. CDC yellow book: health information for international travel 2018. Oxford University Press, New York, NY. [Google Scholar]
- 6.Mali S, Kachur SP, Arguin PM. 2012. Malaria surveillance—United States, 2010. MMWR Surveill Summ 61(2):1–17. [PubMed] [Google Scholar]
- 7.Cullen KA, Arguin PM. 2013. Malaria surveillance—United States, 2011. MMWR Surveill Summ 62(5):1–17. [PubMed] [Google Scholar]
- 8.Cullen KA, Arguin PM. 2014. Malaria surveillance—United States, 2012. MMWR Surveill Summ 63(12):1–22. [PubMed] [Google Scholar]
- 9.Cullen KA, Mace KE, Arguin PM. 2016. Malaria surveillance—United States, 2013. MMWR Surveill Summ 65(2):1–22. doi: 10.15585/mmwr.ss6502a1. [DOI] [PubMed] [Google Scholar]
- 10.Mace KE, Arguin PM. 2017. Malaria surveillance—United States, 2014. MMWR Surveill Summ 66(12):1–24. doi: 10.15585/mmwr.ss6612a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mace KE, Arguin P, Tan KR. 2018. Malaria surveillance—United States, 2015. MMWR Surveill Summ 67(7):1–28. doi: 10.15585/mmwr.ss6707a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bronzan RN, McMorrow ML, Kachur SP. 2008. Diagnosis of malaria: challenges for clinicians in endemic and non-endemic regions. Mol Diagn Ther 12:299–306. doi: 10.1007/BF03256295. [DOI] [PubMed] [Google Scholar]
- 13.Kain KC, Harrington MA, Tennyson S, Keystone JS. 1998. Imported malaria: prospective analysis of problems in diagnosis and management. Clin Infect Dis 27:142–149. doi: 10.1086/514616. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2000. Laboratory diagnosis of blood-borne parasitic diseases; approved guideline. CLSI document M15-A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Abanyie FA, Arguin PM, Gutman J. 2011. State of malaria diagnostic testing at clinical laboratories in the United States, 2010: a nationwide survey. Malaria J 10:340. doi: 10.1186/1475-2875-10-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia LS. 2016. Diagnostic medical parasitology, 6th ed ASM Press, Washington, DC. [Google Scholar]
- 17.Alere Inc. 2015. BinaxNOW malaria test kit product instructions. Alere Inc., Scarborough, ME. [Google Scholar]
- 18.Durand F, Crassous B, Fricker-Hidalgo H, Carpentier F, Brion JP, Grillot R, Pelloux H. 2005. Performance of the Now Malaria rapid diagnostic test with returned travellers: a 2-year retrospective study in a French teaching hospital. Clin Microbiol Infect 11:903–907. doi: 10.1111/j.1469-0691.2005.01253.x. [DOI] [PubMed] [Google Scholar]
- 19.Farcas GA, Zhong KJ, Lovegrove FE, Graham CM, Kain KC. 2003. Evaluation of the Binax NOW ICT test versus polymerase chain reaction and microscopy for the detection of malaria in returned travelers. Am J Trop Med Hyg 69:589–592. doi: 10.4269/ajtmh.2003.69.589. [DOI] [PubMed] [Google Scholar]
- 20.Stauffer WM, Cartwright CP, Olson DA, Juni BA, Taylor CM, Bowers SH, Hanson KL, Rosenblatt JE, Boulware DR. 2009. Diagnostic performance of rapid diagnostic tests versus blood smears for malaria in US clinical practice. Clin Infect Dis 49:908–913. doi: 10.1086/605436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimaio MA, Pereira IT, George TI, Banaei N. 2012. Performance of BinaxNOW for diagnosis of malaria in a U.S. hospital. J Clin Microbiol 50:2877–2880. doi: 10.1128/JCM.01013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khairnar K, Martin D, Lau R, Ralevski F, Pillai DR. 2009. Multiplex real-time quantitative PCR, microscopy and rapid diagnostic immuno-chromatographic tests for the detection of Plasmodium spp: performance, limit of detection analysis and quality assurance. Malar J 8:284. doi: 10.1186/1475-2875-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathison BA, Pritt BS. 2017. Update on malaria diagnostics and test utilization. J Clin Microbiol 55:2009–2017. doi: 10.1128/JCM.02562-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.