Abstract
The Chinese century egg, or pidan, is a traditional preparation of duck eggs that can be stored for months at room temperature without degradation. Raw eggs are soaked in a strong alkaline and salt solution, and the albumin gradually forms a stable, transparent gel. Here, we show that pidan gels belong to the class of materials formed from “patchy particles”. We found that the β-sheet structure of ovalbumin, the major protein constituent of egg white, is preserved during gelation, while α-helical regions undergo a degree of unfolding into unstructured random coils that may form attractive patches. Upon dilution in additional strong base, the phase behavior of pidan gels is consistent with patchy-particle thermodynamics. This protein gel is also physically and structurally similar to the protein gels that form the squid lens. Both systems exhibit patchy thermodynamics, and the constituent proteins share physical features including a structured, charged core, and polar, unstructured “arms” that form attractive patches. Our work provides a path toward rational design of proteins for precisely structured, volume-spanning materials.
Short abstract
Similarities between proteins in Chinese century eggs and squid lenses show that partial disorder and high, uniform surface charge are correlated with low-valence, equilibrium gelation.
Introduction
It was recorded 400 years ago in the Ming dynasty in China that duck egg white, a protein-dense liquid, forms a transparent, edible gel when exposed to a high base and high salt material (eggs were traditionally covered with clay and wood ash)1 (Figure 1a). The Chinese word is transliterated as pidan, where “pi” means tough or rubber, and “dan” means egg. Pidan can be stored and used for years without rotting or noticeable degradation (hence one translation of the word pidan is “century egg” in English). In fact, pidan is so structurally stable that the preparation is a main ingredient in a traditional soup (Figure 1b), in which the gelled egg structure remains intact and transparent after reaching boiling temperature. We recently showed that proteins in the squid lens form similarly thermostable, transparent equilibrium gels throughout a wide range of packing fractions via patchy-colloidal physics.2 We hypothesize that, similar to the material found in squid lens, the gel formed from egg albumin in pidan may be at a thermodynamic minimum, and therefore the result of patchy-colloidal self-assembly physics similar to the processes evolved in the squid lens.
Figure 1.
Images of pidan and ovalbumin gelation. (a) Photograph of lab-made pidan, a traditional method for preserving eggs in alkaline solution. This treatment results in the egg white forming a transparent gel so stable that pidan is known as “century eggs” in English. (b) Traditional rice porridge with pidan. The pidan is cooked in the rice, and the egg white gel remains intact and transparent. (c) Gels formed from ovalbumin, the major constituent of egg white, in strong base and strong acid. Ovalbumin in 0.25 M NaOH solution immediately forms a colorless, transparent gel (left vial), whereas ovalbumin in 0.25 M HCl solution gradually forms an opaque gel (right vial). The inset of part c shows an imaging target behind the ovalbumin in 0.25 M NaOH, demonstrating the gel’s high transparency.
“Patchy colloid” physics is a general physical framework for understanding the thermodynamics and self-assembly of colloidal particles with anisotropic interactions.3−7 Applying patchy-particle physics to proteins provides an appealingly simple and physical picture in which the rich landscape of protein–protein interactions arises simply from the presence of anisotropic, attractive patches on the surface of proteins that otherwise experience repulsive interactions.8 As a loose analogy, the polymers formed by actin proteins can be conceptualized as arising from monomers with attractive patches comprised of hydrophobic surfaces containing salt bridges, and flat, polar surfaces that interact repulsively, with the interplay between the two surface chemistries then assembling monomers into chainlike structures with flat surfaces.9 If these familiar sorts of protein–protein attractive interactions are few in number and small in area on a single protein, but general between proteins in a system, “patchy” thermodynamics results. Patchy particles may therefore be a useful paradigm for gaining insight into protein-based self-assembly, since the theory describing the thermodynamics of these systems is relatively complete and advanced.4,6,8 To date, patchy particles that assemble into self-limiting designed patterns have been designed and fabricated.10,11 However, macroscopic, homogeneous, volume-spanning materials have not yet been prepared by these methods.
Theory predicts that patchy-particle systems exhibit novel physical phase behaviors, especially novel gelation properties.12−15 “Patchy” thermodynamics is distinct and distinguishable from isotropic colloidal interactions in the following ways. Thermodynamic interactions of particles with isotropic potentials may only form rigid, volume-spanning materials at extremely high packing fractions near the so-called jamming transition, where translational movement of particles becomes inhibited due to the high particle density of the system. At these high densities, a crystalline or glassy phase occurs. Crystalline phases are at or near true energy minima of a system like this, while glassy phases may be very far from equilibrium, but further particle movement is blocked due to mechanical entanglement. Therefore, noncrystalline organizations of isotropic colloids may only be rigid and volume-spanning if they are “arrested”; i.e., the particle organization may be very far from an equilibrium state, but it is impossible to move closer to equilibrium due to jamming. The qualitative sense that this degree of entanglement only occurs at very high particle densities is also reflected in previous physical descriptions.16,17 To the extent that many mono- or oligodisperse protein solutions behave effectively like isotropic colloids in bulk,18 the ability to engineer the bulk material properties of protein-based materials is limited by these thermodynamic phenomena; under these conditions, volume-spanning materials only form at high densities and with a limited number of specific geometries. For the characteristic energy of isotropic protein–protein interactions at room temperature, reducing the density of the system via dilution will result either in a complete solubilization of the original colloids, if the density is low enough, or in liquid–liquid phase separation, with some colloids becoming isolated in a sparse phase and some arrested clusters remaining in the dense phase. Because the clusters are arrested, it may be possible to mechanically break them into smaller pieces, but their density will not readily change.
In contrast, patchy-particle theory predicts how stable colloidal gels of very low packing fraction can form at equilibrium: Bianchi and colleagues showed that in the patchy-particle phase diagram, the spinodal line systematically moves toward a lower solute density with decreasing ⟨M⟩, or average coordination number of a system, leaving most of the region outside the spinodal line in a stable, liquid/gelled phase.8 Previous work has shown that squid eye lens proteins, S-Crystallins, harness this patchy-particle phase diagram to form optically transparent gels over a large span of protein packing fractions (from 5% to nearly 100%), resulting in gradient index optics.2 Our experimental work with the S-Crystallin system shows that reconstituted version of these materials will span macroscopic volumes, though in the animal, the gels are formed in cells of volume ∼10–16 L. Since these gels arising from anisotropic interactions form at equilibrium, and the area of the phase space that is unstable can be small, these systems can exist flexibly and stably at a wide range of densities. This is in contrast to the isotropic case where only very high densities can form rigid or gelled materials. Similarly, because thermodynamic properties of the system are determined by the average valence, many different particles comprising different bonding geometries can be employed in a single, stable composite, allowing the properties of the resulting material to be tuned via network topology.
Previous work on pidan showed that it is a transparent, elastic gel formed from a fine-stranded protein network.19,20 Ovalbumin (molecular weight of 45 kDa, pI ≈ 4.59) is the most abundant protein in the egg white, composing about 55% of the total egg white proteins by weight,21 and seems to be responsible for most of the structure observed in pidan gels. Eiser and colleagues found that, when treated with high pH, the egg albumen proteins lose structure to an unknown degree and aggregate into a network with local order.19
Here, we used small-angle X-ray scattering (SAXS), dilution experiments, sequence analysis, and two biochemical assays of protein secondary structure (tyrosine absorbance and a dye-binding assay) to test the hypothesis that the Chinese century egg system is an additional natural example of proteins behaving as low-valence patchy particles that are therefore able to form volume-spanning equilibrium gels. Our results suggest that, for proteins in pidan, the partial loss of structure induced by strong alkaline conditions is associated with the formation of low-valence gels (average coordination number of about 2.2). We also found structural commonalities between the pidan system and the previously described squid lens system that allow us to generalize a set of structural features that seem to be important to realize general patchy-particle physics in protein-based systems. In particular, the anisotropic attractive patches seem to be realized by unstructured, unfolded polar loops in the protein structure, while the repulsive, hard-sphere-like interactions are encoded by a highly charged and folded protein core.
Results
Chinese Century Egg and Ovalbumin Gel
After 2 weeks of treatment in 0.9 M sodium hydroxide and 0.5 M sodium chloride, or in 0.9 M sodium hydroxide alone, the albumen of fresh quail eggs became a transparent, gelled material, with a slight yellow pigmentation (Figure 1a); there was no observable difference between the material that resulted from the preparation with or without sodium chloride. The appearance and texture was consistent with commercial pidan, which is traditionally prepared from duck eggs. These preparations were stable after 18 months of storage at 4 °C. The only apparent change in the material after this period was that the transparent yellow pigmentation darkened to transparent brown.
We also observed that solutions of purified ovalbumin protein (62 mg/mL) formed transparent, colorless gels when sodium hydroxide was added (Figure 1c). The gelation behavior of this solution depended on the concentrations of both purified ovalbumin and sodium hydroxide in the preparation (Table 1). At low concentrations of sodium hydroxide (0.05 M), gelation never occurred, and at high concentrations (between 0.15 and 0.25 M), the preparation initially gelled but then reliquefied after a period of hours or days. At intermediate concentrations (0.05–0.15 M), the resulting gels were stable for up to 1 day on the bench, at which time they were discarded. In contrast to gels made from whole eggs, these pure ovalbumin gels were all transparent and colorless, and did not show any yellow pigmentation (Figure 1c). We also explored the influence of sodium chloride on the gelation procedure, both to better understand the nature of protein–protein interactions during gelation, and to probe the purpose of sodium chloride in the original pidan formulation. Sodium chloride concentrations greater than 0.25 M prevented gelation, with the resulting solution remaining liquid after 1 day. With sodium chloride less than 0.25 M, gelation kinetics were slowed, but gels formed after a few minutes.
Table 1. Gelation of 62 mg/mL (Φ = 5%) Solutions of Ovalbumin.
| acid/base | acid/base concn (M) | NaCl concn (M) | pH | result | 2 h later | 1 day later |
|---|---|---|---|---|---|---|
| NaOH | 0.05 | 0 | 12.55a | liquid | liquid | liquid |
| NaOH | 0.075 | 0 | 12.85a | gel slowly | gel | gel |
| NaOH | 0.1 | 0 | 13.00 | gel slowly | gel | gel |
| NaOH | 0.15 | 0 | 13.26a | gel | gel | liquid |
| NaOH | 0.25 | 0 | 13.40 | gel | gel | liquid |
| NaOH | 0.5 | 0 | 13.70 | gel | liquid | liquid |
| NaOH | 0.75 | 0 | 13.88 | gel | liquid | liquid |
| NaOH | 0.1 | 0.05 | 13.00 | gel | gel | |
| NaOH | 0.1 | 0.1 | 13.00 | gel | gel | |
| NaOH | 0.1 | 0.25 | 13.00 | gel | gel | |
| NaOH | 0.1 | 0.3 | 13.00 | liquid | liquid | |
| NaOH | 0.1 | 0.4 | 13.00 | liquid | liquid | |
| HCl | 0.25 | 0 | 0.60 | white gel |
measured pH.
Given that the original pidan preparation solution contains 0.5 M sodium chloride, but this salt concentration inhibits gel formation in a solution of pure ovalbumin, we measured the chloride ion concentration of pidan prepared from whole eggs in 0.9 M NaOH and 0.5 M NaCl. We found the chloride ion concentration to be 0.01–0.02 M, an order of magnitude lower than the concentration that causes gelation to fail. Therefore, while high salt concentrations do inhibit the protein–protein interactions that lead to gelation, in the traditional preparation of whole eggs in a bath, the salt concentration within the eggshell remains too low to inhibit the gelation process. Given that apparently identical gels also form when salt is excluded from the bath, the inclusion of salt in the traditional recipe for pidan seems to have more to do with salt being a nearly universal ingredient in food preservation processes rather than with salt’s influence on the material behavior of the eggs.
In contrast, adding a similar concentration of hydrochloric acid into the same aqueous ovalbumin solution immediately resulted in formation of a white precipitate. This precipitate became increasingly viscous for the first few minutes after the addition of the acid. After 1/2 h, an opaque, white gel appeared in the tube (Figure 1c).
Phase Behavior of Diluted Pidan Tissues
We measured the density of pidan and determined that it contains a protein concentration of 170 mg/mL, for an estimated ovalbumin packing fraction of 12%. We systematically diluted this original material in additional sodium hydroxide solution (pH = 12.90), resulting in a range of lower protein packing fractions from 0.8% to 6.4%. We then centrifuged this diluted material. Upon centrifugation, we observed the reformation of homogeneous, volume-spanning, transparent gels, but only in preparations in which the total protein packing fractions were greater than 4.5%. It is only possible to reform homogeneous materials at lower densities via dilution in the stable regime of a phase space; for colloidal materials, patchy-colloidal thermodynamics is the only regime in which this is possible at densities far from the jamming transition. Therefore, this result demonstrates that individual ovalbumin proteins in this preparation undergo highly anisotropic, structured attractive interactions in addition to a hard-sphere-like repulsion, and can be rationalized via the patchy-particle thermodynamic regime.
When protein packing fraction was less than 3.3%, we observed liquid–liquid phase separation of the system (Figure 2). In the dilute preparations where liquid–liquid phase separation was observed, the packing fraction of protein in the resulting dense phase increased with the increase of the packing fraction of the system as a whole. The overall behavior of these ovalbumin gel preparations that are diluted past 3.3% is very similar to that of a colloidal clay system that was also shown to be patchy-colloidal in nature.13 In contrast, as expected, dissolving ovalbumin in water at neutral pH resulted in a homogeneous, transparent solution of soluble protein monomers for all protein packing fractions <3.3% (data not shown).
Figure 2.
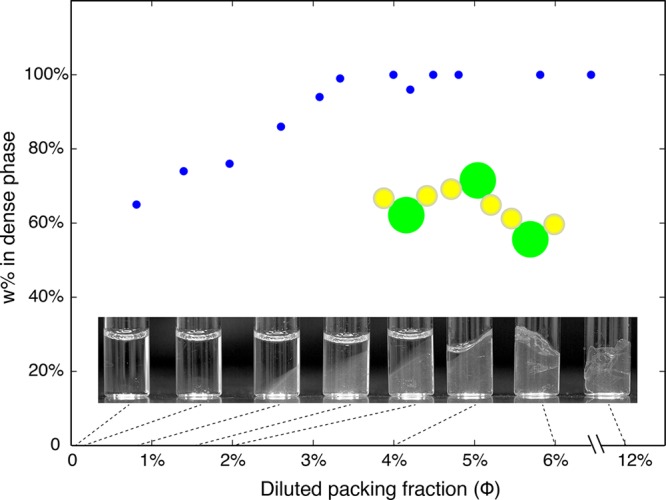
Phase separation of pidan with decreasing protein packing fraction. We diluted pidan material from an initial packing fraction of 12% in increasing volumes of NaOH solution (pH 12.90). Photographs show the system through a range of packing fractions, with volume-spanning gels forming at packing fractions greater than 3.3% and liquid–liquid phase separation occurring at all packing fractions less than about 3.3%. Blue circles show the fraction of protein in the gelled or dense phase of the system as a function of packing fraction; the fraction of total protein forming the dense phase decreased with dilution of the system. This phase behavior is consistent with a patchy-particle system13 but not with an isotropic colloidal system. A patchy particle (green circles) interacts with others via certain patches (yellow circles) on the surface.
Small-Angle X-ray Scattering
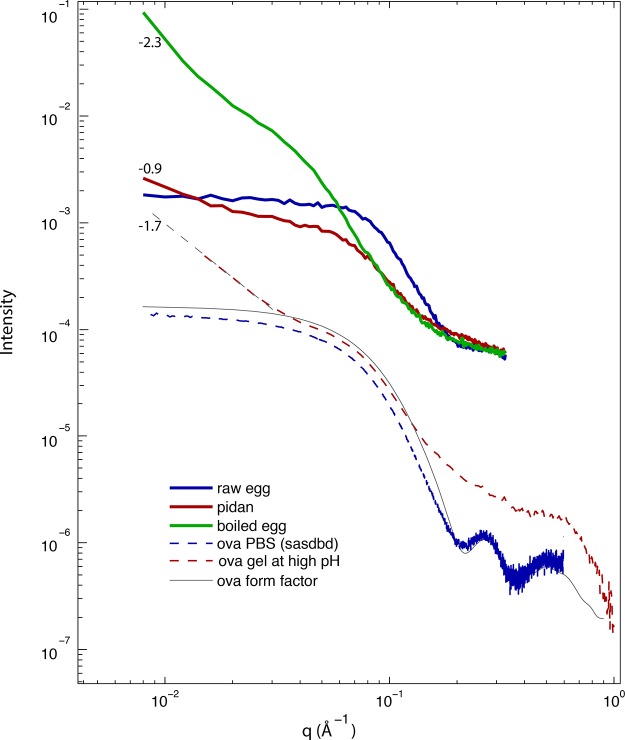
The literature data for X-ray scattering from a solution of ovalbumin in PBS as a function of wave vector (q) (https://www.sasbdb.org/data/SASDAL2) were very similar to those of a calculated form factor considering the atomic positions of the crystal structure 1OVA22 (Figure 3). Both the literature data and an independent calculation of form factor from a crystal structure showed a flat intensity at q < 0.08 Å–1. There was also a sharp decrease with increasing q at 0.08 < q < 0.2 Å–1, followed by peaks at q = 0.27 Å–1 and q = 0.5 Å–1 in both characterizations.
Figure 3.
Small- and wide-angle X-ray scattering (SAXS/WAXS) from egg white and ovalbumin, I(q) for all liquid and gel materials described in this study. Curves are as follows: blue curve, raw egg white; red curve, traditional pidan; green curve, boiled egg; dashed red curve, alkaline ovalbumin gel; dashed blue curve, ovalbumin from SASBDB; gray curve, ovalbumin form factor calculated from crystal structure 1OVA. Numeric labels show the log–log slope of those data for q < 3 × 10–2 Å–1, providing an estimate of the fractal dimension of a material at length scales larger than a protein monomer. In pidan, the absence of the peak at q ∼ 0.25 Å–1 that is present in native ovalbumin suggests that the high-base treatment induces partial loss of protein secondary structure. The low-q log–log slopes of pidan and high-base ovalbumin at q < 0.04 Å–1 (slope from −0.6 to −1.7) are intermediate between those of liquid egg white and an ovalbumin at neutral pH (|slope| ≪ 1) and boiled egg (slope ∼−2.3), indicating that pidan protein forms a network with large-scale structure with a lower fractal dimension than the gelled boiled egg material.
We also measured scattering intensity as a function of q for two related protein solutions: raw egg white and ovalbumin in solution at neutral pH. Both of these solutions had very similar X-ray scattering to the ovalbumin form factor described above (Figure 3). Each sample showed a very flat region at q < 0.08 Å–1, and a sharp decrease as a function of q from 0.08 < q < 0.2 Å–1. At q = 0.27 Å–1, we observed local maxima for both the raw egg white and neutral ovalbumin solution that were similar to the prominent peak in the form factor at this value of q. These local maxima correspond to a D-spacing of 23 Å and result from the internal structure of ovalbumin protein. For neutral ovalbumin solution, there was an additional peak at q = 0.5 Å–1 that also corresponded to a major feature in the form factor.
When treated with strong base, both pidan and alkaline ovalbumin gels showed patterns in scattering intensity that were similar to each other but distinct from the protein solutions described above. Unlike the protein solutions, at low q, both pidan and alkaline ovalbumin gels showed a decrease in the X-ray scattering intensity with increasing q (Figure 3). The slope in a log–log plot of I(q) for pidan was about −0.9 between 0.008 < q < 0.02 Å–1, while that of alkaline ovalbumin gel was −1.7. Between 0.03 < q < 0.15 Å–1, the scattering patterns of both pidan and alkaline ovalbumin exhibited shoulders at the D-spacing consistent with the size of an ovalbumin monomer (50 Å). At q > 0.15 Å–1, neither pidan nor high-pH ovalbumin showed any prominent local maxima, as was also the case for the neutral protein solutions.
In contrast, I(q) for boiled egg white was distinct from both neutral protein solutions and the alkaline pidan and ovalbumin gels described above. At low q, the slope in the log–log plot for boiled egg between 0.008 < q < 0.02 Å–1 was −2.3. There was a shoulder in the range 0.03 < q < 0.15 Å–1, but the shoulder was much less pronounced than for pidan or high-base ovalbumin. At large q, there was no peak or local maximum in scattering intensity at q = 0.27 Å–1 (Figure 3).
UV Absorption at Various Alkaline pH Values
As a general assay of changes in protein structure of ovalbumin upon gelation, we characterized the oxidation, and therefore solvent exposure of tyrosine residues using UV–vis absorbance. The UV absorbance spectrum of ovalbumin solutions changed dramatically with increasing sodium hydroxide concentration (Figure 4). When pH was between 8 and 10, we observed three peaks in the UV absorbance spectrum at 220, 230, and 280 nm, and a local minimum at 250 nm. The overall shape of the spectrum changed little throughout this pH range, and was also similar to the absorbance spectrum at neutral pH. However, as the pH of the system increased from 10 to 13, the absorbance peak at 220 nm disappeared; the peak at 280 nm shifted to 290 nm, and features in the spectra appeared at 242 and 250 nm.
Figure 4.
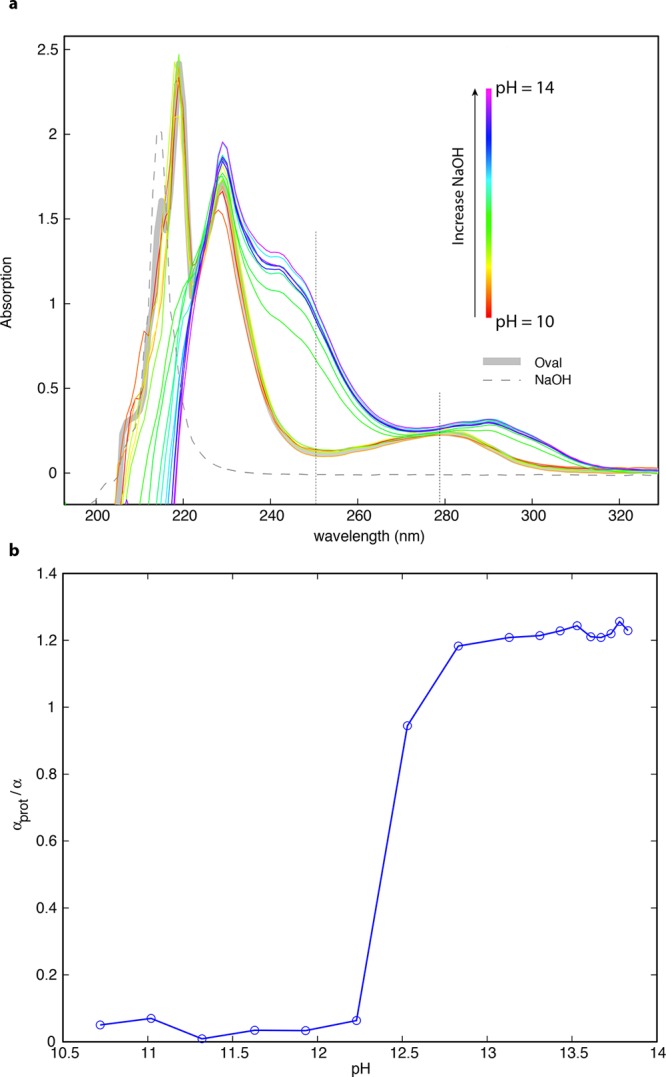
Tyrosine oxidation assay. (a) UV absorbance spectra of ovalbumin solutions in the pH range 10–14. Gray solid line shows the absorption spectrum of ovalbumin at neural pH. Gray dashed line shows the absorption spectrum of 2 M NaOH solution alone. Remaining spectra are color-coded in rainbow order with red lines indicating ovalbumin solution with pH ∼ 10 and violet lines indicating ovalbumin solution with pH ∼ 14. The absorbance at 250 nm is associated with tyrosine, and increases with tyrosine oxidation; the absorbance at 280 nm indicates tryptophan absorbance. A dramatic, nonlinear change in ovalbumin structure, as indicated by tyrosine oxidation, occurs between pH 12.3 and pH 13.0. (b) Parameter αprot/α (tyrosine oxidation per protein molecular weight) as a function of pH; αprot/α indicates the number of tyrosines exposed relative to the average molecular weight of the protein. Tyrosine oxidation is unchanged from the native state, but undergoes a sharp increase between pH 12.25 and 12.5. Pidan formation occurs between pH 12.5 and pH 13. There is a slower increase in tyrosine oxidation from pH 13 to pH 13.5, where it appears to reach a maximum. At pH greater than 13.5, ovalbumin gels slowly degraded, perhaps due to cleavage of the protein backbone.
Using the method of Melo and colleagues,23 we used these changes in the absorbance spectrum with pH to estimate the fraction of tyrosines in the protein structure that become deprotonated and therefore exposed to solvent as a function of pH; this parameter is (αprot/α) (Figure 4b). When the pH of the system was less than 11, αprot/α was < 0.1. When pH increased from 12.3 to 13, αprot/α increased dramatically from 0.1 to a maximum of 1.2, observed when the pH was greater than 13.
Estimates of Ovalbumin Structure
We used DAMMIF to estimate a real-space configuration of amino acids consistent with the inverse-space data in the function I(q) (Figure 5). This prediction showed a 4 × 4 × 6 nm cluster of amino-acid-size particles, which is roughly the size of one ovalbumin molecule. This cluster had two parts, a cylinder of about 4.5 × 3 nm and a cone-shaped part of about 3.5 × 5 nm, with an angle of about 90° separating these domains.
Figure 5.
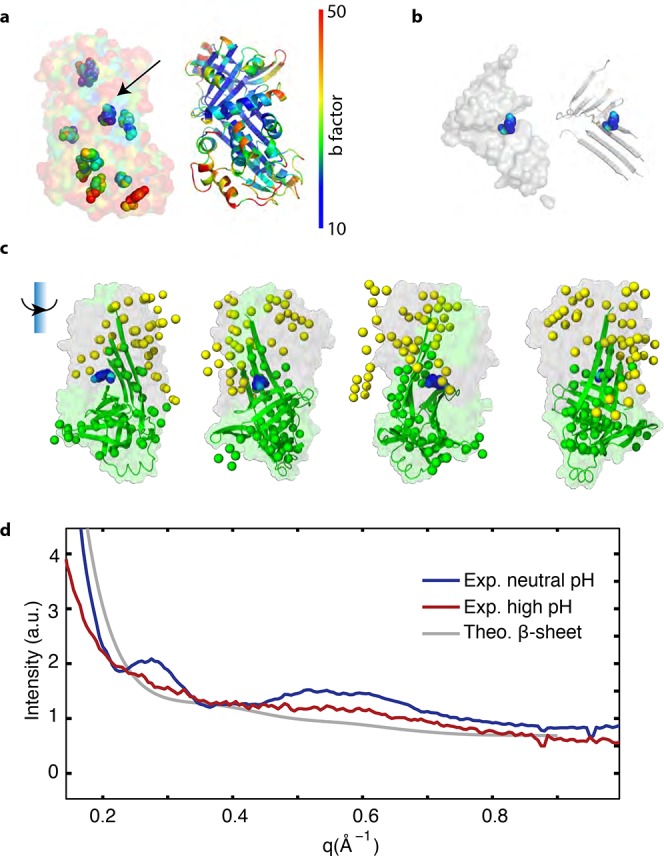
Estimation of the secondary structure of ovalbumin in pidan. (a) b-factors of 1OVA (transparent surface), highlighting tyrosines (filled spheres). Arrow indicates the tyrosine with a b-factor one standard deviation lower than the average tyrosine in 1OVA. This residue is likely the one most buried from solvent exposure. The ribbon structure of 1OVA is shown to the right. (b) Isolated β-sheet structure of ovalbumin. The left image shows the surface, and the right shows the ribbon structure. 1OVA contains eight β-strands forming two β-sheets. The most stable tyrosine (shown as spheres colored by b-factor) is located at the closest point of contact between these two sheets. (c) Multiple views of the β-sheet structure of ovalbumin (surface) superimposed with a numerical prediction of the protein structure of pidan gels (green and yellow spheres). Each rotation represents 90° around the axis indicated. Green spheres show regions that overlap well with the β-sheet structure of ovalbumin, which may serve as the hard core of a patchy particle. Green and gray surfaces are the regions of the native protein structure predicted to remain folded, and to unfold, respectively. Regions of numerical prediction that do not overlap with crystal structure are shown in yellow; these areas are also predicted to be disordered and potentially sites of general polar, attractive interactions. (d) WAXS I(q), characterizing structure internal to a protein, of raw egg white (blue curve), pidan (red curve), and a calculated form factor of only the β-sheet structure from native ovalbumin (1OVA, gray curve). WAXS of pidan is intermediate between that of the native protein and the isolated β-sheet of the native protein, suggesting that the native protein undergoes a partial unfolding leaving the β-sheets and little additional structure in the pidan conformation.
The native ovalbumin structure consists of several α-helical segments folded around a β-sheet core.22 We considered the hypothesis that only the core β-sheet structure of the ovalbumin fold is maintained after high-base treatment, a result that is also consistent with previous experimental analyses of this system for which the degree of unfolding upon gelation was ambiguous19 (Figure 5). We therefore visualized the β-sheet-only structure in the 1OVA structural model of the protein, and observed an “L” shape with a size of about 3 × 4.5 × 4.5 nm. The dimension of one leg of the “L” was about 2 × 3 × 4.5 nm, and the other leg was about 3 × 3 × 4 nm, with the angle between them ∼90°.
We calculated a form factor for the isolated β-sheet structure at the core of native ovalbumin in order to predict how this structure, in the absence of other defined structure, might scatter at high values of q in a SAXS experiment (Figure 5d). At q > 0.2 Å–1, both this calculated β-sheet form factor and the experimental scattering data for ovalbumin at high pH are nearly featureless. In contrast, the X-ray scattering of native ovalbumin shows two prominent peaks at q = 0.25 and 0.6 Å–1.
Congo Red Binding Assay of Secondary Structure
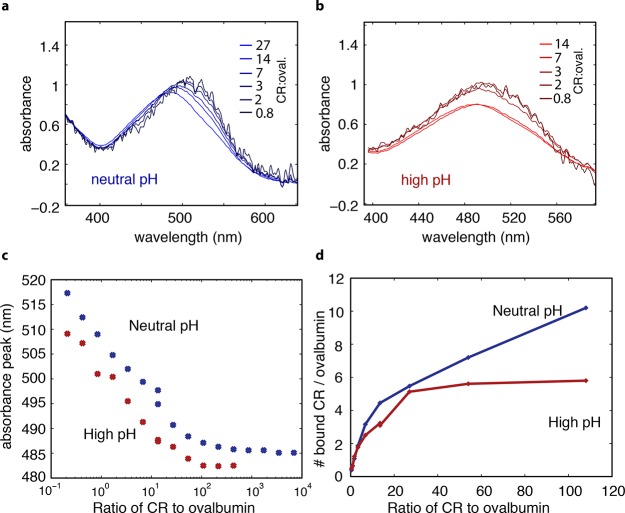
Utilizing shifts in the absorption peak of the dye Congo red (CR) when bound to protein secondary structure, we assayed the relative proportion of protein surface area with secondary structure to surface area composed of random coil in these systems. The absorbance of CR undergoes a red-shift when it is bound to protein secondary structure, but there is no peak shift in the presence of random-coil structure.24,25 Therefore, comparing the relative peak shifts of a given CR to ovalbumin ratio in neutral and alkaline systems allows us to determine the relative amount of secondary structure present at each pH. When a system contained 0.2 CR molecules per ovalbumin, such that most CR molecules were bound to a protein, the absorbance peak occurred at 517 nm in neutral pH and 510 nm in alkaline pH, suggesting that there was more secondary structure present in neutral pH (Figure 6a,b). When we increased the relative number of CR present per ovalbumin, such that relatively fewer CR would be bound to ovalbumin, the absorbance peak of the dye shifted to shorter wavelengths in both systems. Because we were able to observe a change in both systems, we conclude that there was some secondary structure present in both systems. This shift in wavelength began to saturate at a ratio of about 100 CR:ovalbumin. In alkaline solution, a complete saturation was observed, while, in neutral solution, the rate of peak shift markedly decreased, but the shift continued to a concentration ratio of ∼1000, similarly suggesting that there was somewhat more secondary structure present in the neutral system (Figure 6c). In neutral solution, the peak shifted from 517 to 485 nm (change of −32 nm), while in alkaline solution the peak shifted from 510 to 483 nm (change of −27 nm); the greater magnitude of the peak shift in neutral solution also suggests that more secondary structure was present compared to alkaline solution, while the presence of a peak shift in both systems again suggests that secondary structure is present in both systems. In both systems, when the molar ratio of CR to ovalbumin was less than 100, the position of the CR absorption peak decreased linearly with the log of the molar ratio CR:ovalbumin. In neutral solution, this rate was −10.6, and in alkaline solution the rate was −11.6 nm per ten-fold change in concentration ratio.
Figure 6.
Surface area of folded regions of ovalbumin (Ova) estimated by Congo red (CR) binding assay. (a) Visible absorbance of CR over a range of CR:ovalbumin concentration from 0.8 to 27, neutral pH. CR bound to ovalbumin secondary structure absorbs at a longer wavelength than CR free in solution, so as ovalbumin is added to the solution relative to CR, thereby increasing opportunities for CR to bind ovalbumin, the overall absorption of the system shifts to the right. (b) Same as part a, but in 0.5 M NaOH with the CR:Ova concentration from 0.8 to 14. A similar peak shift, of a smaller overall magnitude, was observed at high pH. (c) CR absorbance peak as a function of CR:Ova concentration ratio. Blue symbols label CR:Ova at neutral pH, and red symbols label CR:Ova in high pH consistent with pidan formation. (d) Calculated number of CR bound to one ovalbumin monomer as a function of CR:Ova concentration ratios for both neural pH (blue) and high pH (red). Because CR only binds to secondary structure but not random coil, this assay allows us to estimate the surface area of secondary structure in the native protein compared to that of the conformation found in pidan. The results suggest that the pidan conformation consists of some secondary structure, and that the surface area of secondary structure is roughly 60% that of the native conformation.
We used these data in combination with the definition of the binding constant to estimate the number of CR molecules bound to an ovalbumin molecule in a given state, and thereby estimate the surface area of secondary structure present in each system. As the number of CR per ovalbumin increased from 0.4 to 100, the average number bound to an ovalbumin increased from 0.4 to 10 in the neutral system, and 0.4 to 6 in the alkaline system (Figure 6d). Therefore, if we assume that the number of CR molecules bound to the protein is proportional to the surface area of protein secondary structure, then there is about 40% less secondary structure surface area in the high-base condition compared to neutral pH. These results are not consistent with the complete loss of protein structure in the alkaline system.
Temperature Dependence and the Structure Factor of Squid Lens and Pidan
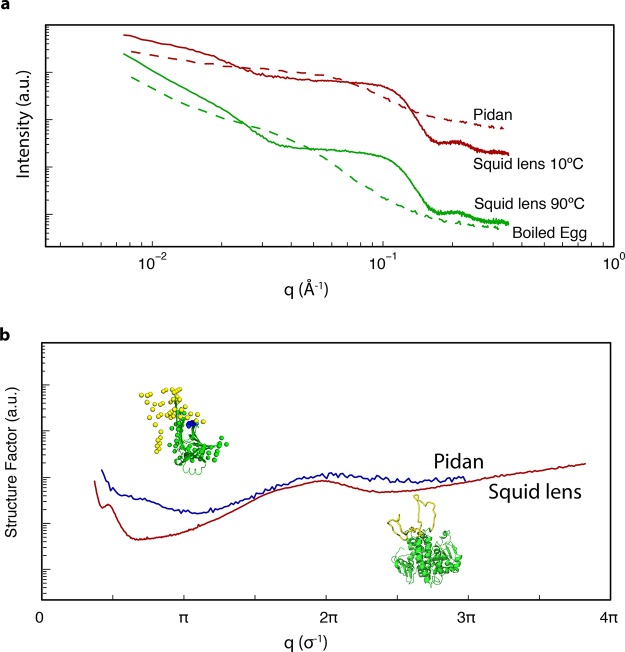
We compared the structure of pidan to that of the previously described protein gels in squid lens. The SAXS pattern of pidan and the outer, most sparse layer (100% layer) of the squid eye lens were similar (Figure 7a). At q < 0.03 Å–1, I(q) decreased slowly until a shoulder between 0.03 < q < 0.2 Å–1, corresponding to the size of a single protein in both systems.
Figure 7.
Hypothetical structural origins of patchy thermodynamics in S-Crystallin and pidan proteins. (a) I(q) from SAXS for pidan (red dashed line) and boiled egg white (green dashed line, this study), and squid lens at 10 °C (red solid line) and 90 °C (green solid line, lens data from ref (2)). In both the squid and the egg system, the slope at low values of q becomes sharper upon heating to near-boiling, while the overall shape of the curve does not change at high values of q. (b) Structure factors of pidan (blue curve) and squid lens (red curve), normalized to the diameter of an individual monomer (σ). Both structure factors show a peak at the diameter of the protein (2π/σ), indicating frequent pairwise interactions; a minimum at π/σ, consistent with reduced density fluctuation at distances of a few σ, and a log−log slope of ∼−2.5 at q ≤ 0.5π/σ. Insets show predicted structures of squid S-Crystallin proteins, and ovalbumin in the high-base state that induces patchy-particle thermodynamics. Our hypothesis about the regions of protein structure responsible in both systems for hard-sphere interaction is shown in green, and the regions inferred to be responsible for patchy attractive interactions are shown in yellow.
We also compared the structure of boiled egg and squid lens heated to 90 °C; these materials were also similar to each other. Their SAXS intensities showed a sharp decrease at low q, with the slope of −2.3 in the log–log plot, greater than that of pidan and squid lens at 10 °C (Figure 7a).
We also analyzed these SAXS data as a function of wave vector normalized to the diameter of the single proteins in a given system, or σ (σ = 30 Å for pidan monomers and σ = 40 Å for lens proteins) (Figure 7b). At a large spatial scale with qσ < 2, there was a decrease in the intensity, which was sharper in the lens layers than pidan. In the intermediate range where 2 < qσ < 6, there was a trough in both cases. For pidan, the minimum was at one-third the intensity of the nearby plateau. In contrast, the lens minimum was 20 times smaller compared to the peak at q = 0.13 Å–1. At qσ = 6, there was a peak at the real size position of protein–protein interaction for both cases.
Intrinsic Disorder of Ovalbumin in High pH
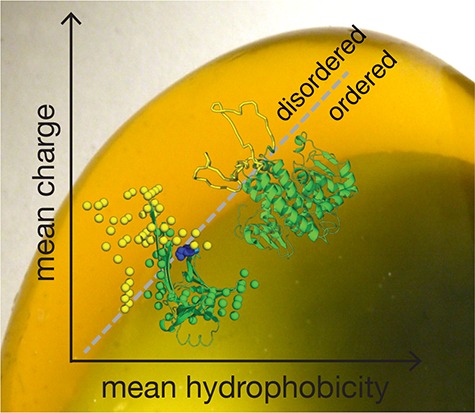
It is possible to assay the tendency to folded order vs intrinsic disorder in a protein by comparing the average mean hydropathy of residues to their average mean charge. In this space, there is a sharp boundary between order and disorder described by the line ⟨R⟩ = 2.785⟨H⟩ – 1.151, where ⟨R⟩ and ⟨H⟩ are the mean hydrophobicity and the mean net charge, respectively.26 To better understand the order–disorder transition upon gelation of ovalbumin, we analyzed the ovalbumin sequence using an 11-amino-acid sliding window, and calculated the displacement within this order–disorder space for each calculation. We found that the whole-sequence location of ovalbumin shifted from the ordered side of this space to the disordered side when the pH shifted from neutral to pH 12 where gelation begins (Figure 8). However, when we analyzed the regions of the sequence responsible for this shift, we found the shift toward intrinsic disorder was concentrated in the region from the N-terminus to around position 130 in the sequence (windows describing positions 39–42, 79–88, and 91–130 shifted to the disordered region of the space). This region of the sequence primarily forms the non-β-sheet regions of the protein, corroborating our hypothesis that most of the structure other than the β-sheet regions unfolds in the gel-forming structure of the protein that forms at high pH (Figure 5). The tyrosine residue that could remain unoxidized at high pH is located at the interface between the β-sheet region and the region that is prone to disorder during gelation, suggesting that its remaining protected from oxidation is necessary for ordered and disordered regions of the protein to maintain structural integrity with respect to each other. Our results above show that gelation is no longer possible once that final tyrosine is oxidized, suggesting that the ordered and intrinsically disordered regions lose structural coherence, shifting the system into a regime of polymeric random-coil-like interactions rather than patchy-colloidal interactions. Similarly, the “loop” regions of S-Crystallins that are responsible for the linkages in patchy-colloidal assembly are located in the disordered region of this space, while the “body” regions of the same proteins are located well within the ordered region of the space (Figure 8).
Figure 8.
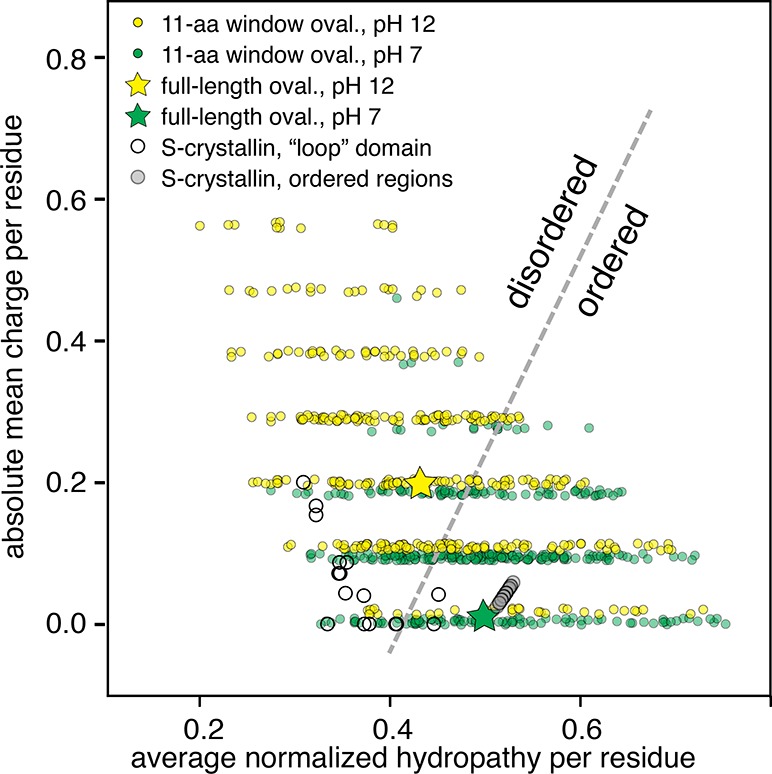
Protein order vs intrinsic disorder is determined by the location of a sequence within the space describing average net charge of a residue vs average normalized hydrophobicity of a residue in a protein. The yellow star shows the location of the entire ovalbumin sequence at pH 12; the green star shows the location of ovalbumin at pH 7. Yellow dots represent a single 11-residue window of ovalbumin at pH 12, while green dots represent the same windows at pH 7. The high-pH ovalbumin is predicted to be in the disordered region. The S-Crystallin loop structure (empty dots) and the rest (gray dots) are also plotted. The loops are mostly locating at the disordered region, whereas the rest of the S-Crystallin form a line in the ordered region.
Discussion
Pidan as a Gelled Protein Network
We observed the formation of transparent gels when both raw eggs and solutions of pure ovalbumin protein were treated with strong base such that the pH of the system was >12.3. Therefore, in both the whole-egg pidan and the gels of purified protein, the constituent proteins form a volume-spanning structure with minimal density fluctuation at the scale of visible wavelengths of light (∼400–800 nm, or 100s of protein diameters). In contrast, when the same concentration of ovalbumin protein was treated with a similar concentration of strong acid, an opaque, white gel resulted, such that the same polymer in acid forms gels characterized by large density fluctuation at visible light length scales.
Since it was possible to dilute this system with additional strong base solution and recover a homogeneous volume-spanning material at the new average density, down to a final volume fraction of 3.3%, these materials must exist in the stable regime of the governing phase diagram. For colloidal materials, the only known phase diagram that predicts this behavior is that of patchy particles, or colloids that simultaneously exhibit anisotropic attractive interactions and a general hard-sphere-like repulsion. Liquid–liquid phase separation occurred when the ovalbumin protein packing fraction was lower than 3.3%. When this result is placed in the context of the patchy-colloid phase diagram, we estimate that the average coordinate number ⟨M⟩ of the constituent pidan protein is ∼2.2. We did this by inferring that, after phase separation, the denser of the two resulting materials must re-equilibrate immediately to the right of the spinodal line that governs the total system, and interpolating the known spinodal lines accordingly.8 Since the pidan protein packing fraction is 3.7 times higher than the packing fraction where liquid–liquid phase separation starts to appear for ⟨M⟩ = 2.2, the pidan material formed from whole eggs is therefore located outside of the relevant spinodal line in the phase diagram, and is also likely at an equilibrium state. This observation may explain why this transparent gel and traditional food is stable for many years, and also after extensive boiling.
Stable gels (defined as no change after >24 h on the bench) occur spontaneously at the pH at which tyrosine ionization, or αprot/α, was ∼90% of the maximum possible given the number of tyrosines in the protein sequence. Therefore, it is likely that as gel formation and protein unfolding occurs, one tyrosine out of 10 total in the protein remains protected within residual secondary structure. This in turn suggests that the one tyrosine that is especially stable in the crystal structure and embedded within the core β-sheets of the structure remains buried. This result is consistent with an overall picture in which most of the ovalbumin structure unfolds in high-base treatment other than the β-sheet core of the native fold. A CR dye-binding assay also indicates that roughly 60% of the secondary structure surface area of the native fold is still present in the alkaline condition.
These results, together with our SAXS data, may provide some insight into the protein structural origin of the patchy-colloidal behavior, and how this system differs from the opaque materials formed from the same proteins. I(q) of the pidan and ovalbumin high-base gels had intermediate slopes at low q of −0.9 and −1.7, in contrast to the boiled system, where this slope was −2.3. This result implies that pidan and high-base ovalbumin form networks with large-scale structure, but this structure has lower density fluctuation at length scales of 100s of nm than the material that forms when the egg white is boiled.
Pidan Protein Structure
How does high-base treatment, but not neutral pH or acid treatment, alter ovalbumin protein structure to form monomeric particles of low average valence (⟨M⟩ ∼ 2.2)? The structure factor of pidan shows a peak at qσ = 2π, where σ = 3 nm, indicating that the unit particle of the larger gelled protein network is an individual globular protein monomer that has radius of gyration ∼2.5 nm in folded state (https://www.sasbdb.org/data/SASDAL2) (Figure 7). Simultaneously, the peak at q ∼ 0.25 Å–1, indicating structure internal to a monomer, is diminished relative to the measurement of the system at neutral pH, suggesting that the high-base treatment induces at least partial loss of protein secondary structure. Previous work on this system using circular dichroism (CD) to characterize protein structure has suggested that the proteins in pidan may be completely unfolded.19 However, CD is not especially sensitive to distinguishing random-coil structure alone from mixtures of random-coil and β-sheet structure. Our CR binding assay, in which a dye absorbance peak shift still occurs in the high-base treatment (consistent with the presence of some secondary structure), combined with the SAXS result that there is a particle the size of a monomer present, suggests that the loss of ovalbumin secondary structure is partial rather than complete in pidan. Our experiment calculating αprot/α, indicative of the fraction of tyrosines exposed to solvent, may allow us to infer a bit more about the nature of this structural change.
Given our result that, as pidan gels form, one of 10 tyrosines in the protein sequence remains buried from solvent, we were able to predict which tyrosine may contribute to the folded state. The average b-factor of atoms in tyrosines in the structure was 25.1; however, the most N-terminal tyrosine had an average atomic b-factor of 10.6, or more than a standard deviation less than the residue average in the protein. This tyrosine is located near the core of the protein, at the end of the first β-strand in the structure and near the hinge between the two regions of β-sheet. Our data taken together make it possible to make a low-resolution estimate of the protein structure within the pidan gel network. Together, the data generate a picture in which high-base treatment induces a partial loss of ovalbumin secondary structure by disrupting some of the α-helical structures, but leaving intact most of the β-sheet core, including one tyrosine of the original protein fold.
In order to understand how a partial unfolding of the ovalbumin structure gives rise to specific patchy interactions, we studied the intrinsic disorder of the sequence as a function of sequence position; ultimately the disorder of a region of protein can be predicted by its position in a space comparing net charge per residue to net hydrophobicity per residue. We found that, given the changes in charge and hydrophobicity that occur in a high-base environment, the overall position of ovalbumin in this space shifts from the ordered to the disordered region of the space (Figure 8). However, the regions of the protein responsible for this shift were concentrated in the N-terminal region of the protein sequence, which gives rise to most of the α-helical structure of the folded protein. Therefore, the novel attractive interactions that emerge and cause gelation are correlated with the introduction of intrinsic disorder into the N-terminal half of the protein sequence and structure.
There are similarities between our observations about low-valence ovalbumin structure and the structure of the protein patchy particles (S-Crystallins) we recently described in squid lens. In the low-valence squid lens proteins/particles, we showed that a pair of unstructured loops protrudes from the spherical, globular region particle. Pairwise interactions between these random-coil loops constitute the low-valence patch–patch interactions in this system, while the high charge on the folded, globular region of the protein apparently forms the hard-sphere aspect required by patchy colloid theory.2 In the ovalbumin particles that form pidan gels, there are similarly unstructured regions, and a high net surface charge, relative to the same proteins at neutral pH. We speculate that, like in the squid lens, the low-valence patch–patch interactions in pidan gels form via polar linkages between unfolded regions of the protein, while the stable β-sheet structure gives rise to the charged, hard-sphere region required by the theory.
Our data do not allow us to specify exactly how the introduction of partial intrinsic disorder to the ovalbumin structure at high pH structurally encodes the angularly small attractive interactions required to generate the gelation behaviors we observe. However, it will be possible in the future to test two structural hypotheses. In the first, there are two distinct coils that form from the intrinsically disordered region, perhaps resulting from the unfolding of two regions connecting two β-strands into random coil. In this hypothesis, there are two disordered loops that directly form two distinct linkages for attractive interactions, directly analogous to the structural origin of low-valence interactions in the S-Crystallin system. In a second structural hypothesis, the unfolding of the protein on one end could generate a Janus-like particle able to undergo nonspecific attractive interactions with the still-ordered regions of a neighboring particle. In either hypothesis, higher-valence interactions could still be formed from less-frequent nonspecific polar or hydrophobic interactions between protein surfaces.
Previous work showed that ovalbumin heated to 80 °C forms opaque gels (similar to our boiled egg experiment); however, if the protein is covalently decorated with succinic acid first, heating the system to 80 °C results in a transparent gel.27 Succinylation of ovalbumin also induces a stable, partially unfolded ovalbumin structure with double the aspect ratio of the folded protein,28 consistent with our observation of partial unfolding and a high aspect ratio in high-base treatment.29 Given that the particles that form transparent gels are more rodlike than globular, these uncoiled regions seem to be located on either end of the β-sheet structure, forming a rodlike particle with sticky ends. We speculate that the degree of disorder in these random-coil loops and their interactions may be important for a system to exhibit patchy-colloidal thermodynamics rather than crystallization or uncontrolled filament or amyloid formation.30,31 Since SAXS data suggest that ovalbumin proteins apparently do completely unfold in the boiled egg case (the shoulder in I(q) at the D-spacing of the monomer diameter disappears), and then form a higher average valence network, the effective ⟨M⟩ of these protein systems may be related to the degree of protein unfolding present. That is, partial unfolding seems to lead to low-valence behavior, while total unfolding leads to high-valence behavior of the same protein polymer. Similarly, at lower temperatures, the oxidation of the final tyrosine in the ovalbumin sequence causes a loss of the ability to gel, also suggesting that unfolding into near-random-coil results in a loss of the anisotropic interactions required for equilibrium gelation.
High surface charge also seems to be important for achieving patchy rather than isotropic interactions in both systems (high protein surface charge occurs at physiological pH for S-Crystallins and high pH for pidan). The surface charge of the low-valence S-Crystallins is around +8, while the total charge of ovalbumin at pH 12 is estimated to be −56 (http://protcalc.sourceforge.net). We speculate that this similar, high surface charge generates the repulsive, hard-sphere-like interaction between the folded regions of the protein, which is required for “patchy” thermodynamics and prevents random attractive aggregation.
DNA-nanostars have also been used as an experimental realization of patchy thermodynamics.5,32−35 A typical DNA-nanostar molecule has hydrodynamic radius ∼4.5–4.7 nm, similar to the size of ovalbumin and S-Crystallin protein. The valences of these DNA-nanostar systems can be engineered to be 3 or 4,5 higher than the predicted pidan ⟨M⟩ of 2.2, or the minimum observed in the squid lens of 2.1. The multivalence DNA-nanostars form equilibrium gels at room temperature33,36 and can avoid crystallization,35 both of which are similar to the protein systems. However, in contrast to the two protein-based systems outlined here, the DNA-nanostar system showed liquid–liquid phase separation at low temperature (<10 °C for M = 4), while, at high temperature (>65 °C), double-stranded regions of DNA denature into single strands, fundamentally changing the nature of the particle.5 Also, the reported DNA-nanostar systems are volume-spanning over very small volumes (50 μL5,36) compared to pidan (∼50 mL for a duck egg), and pidan gel is stable while boiling.19 Given that it is potentially more cost-effective to generate gram-scale quantities of proteins via expression than gram-scale quantities of engineered DNA via synthesis, proteins may provide advantages over DNA-based systems as attempts to engineer nanoscale self-assembled systems move forward.
The similarities between all these protein systems, and their differences from chemically simpler DNA systems, suggests that, in order to make nanoparticles into low-valence patchy networks, it is important to control the degree of partial unfolding of the protein polymer, and thereby control the relative amount of random coil relative to the surface area of a hard-core-like region of folded protein. High surface charge of the folded region may in turn enforce hard-sphere-like interactions between the central bodies of the particles. These structural states of controlled random coil and high surface charge are less accessible in DNA-based systems made of polymers of four, uncharged chemical bases. It is also possible for low-valence proteins to be stable past the boiling point of water, while DNA melts at around 60 °C. The greater chemical complexity of 20 amino acids in a polymer may then be required for achieving true volume-spanning materials of controlled valence that have not so far been accessed using a DNA strategy. The similarities between two disparate protein systems able to form low-density equilibrium gels suggest that a generalized strategy for achieving these self-assembling materials is to generate regions of polar disorder in the number and geometry of desired attractive patches, and to maintain a high, uniform surface charge throughout other regions of the protein to generate hard-sphere-like repulsion there. In the case of pidan and ovalbumin, this structure occurs at pH 12, while, in the case of the squid lens, proteins exhibit these structural properties at physiological pH. These insights may help with the rational design of proteins and nanoparticles for volume-spanning self-assembly. By lowering ⟨M⟩ of the protein system via either partial unfolding or the introduction of hydrophilic, disordered loops, it may be possible to design nanoscaled systems that avoid liquid–liquid phase separation at any concentration.
Materials and Methods
Chinese Century Egg Preparation
Traditional Chinese century eggs (pidan) are made from duck eggs. We used quail eggs in our experiment because the volume is smaller to allow many more discrete samples with the same amount of reagent, while the protein composition is similar. Raw quail eggs purchased from a local Asian supermarket were treated as follows to make pidan. These intact quail eggs were soaked in a solution of excess 0.9 M sodium hydroxide (Sigma-Aldrich ACS reagent, >97.0% pellets, PO 221465) and 0.5 M sodium chloride (Sigma-Aldrich SLBB9000V). We also prepared eggs in 0.9 M sodium hydroxide alone. After two days of this high-base treatment, we rinsed the eggshells, discarded any broken eggs, and replaced the high-base solution with fresh solution. After 13–16 days at room temperature, the eggs were removed from the solution and stored in airtight containers at 4 °C for future use. A new egg was peeled right before each experiment described below. In this work, no unexpected or unusually high safety hazards were encountered.
Ovalbumin Solution and Ovalbumin Gel
To study the gelation of purified ovalbumin protein, 25 mg of ovalbumin was dissolved into a series of 250–390 μL of deionized water. To this solution another 10–150 μL of 2 M NaOH was added such that the total volume of each sample was 400 μL. The ovalbumin final concentration was fixed at 62 mg/mL, but the concentration of NaOH varied from 0.05 to 0.75 M. We also made a similar set of samples but with strong acid instead of strong base: protein water solution was made by dissolving 125 mg of ovalbumin into 1.5 mL of deionized water, and then 0.5 mL of 1 M HCl solution for a final concentration of 62 mg/mL ovalbumin and 0.25 M HCl.
To prepare ovalbumin solutions/gels for spectral measurements and SAXS experiments, the ovalbumin proteins were first dissolved into PBS (100 tablets, MP Biomedicals, catalog no. 2810305) with the ovalbumin concentration of 85 mg/mL; then the solution was centrifuged at 14 800g at 10 °C for 10 min, and the pellet was discarded. For ovalbumin solutions at high pH, 0.5 M NaOH (Sigma Aldrich PO 306576-25G) was added 1:3 and mixed with the protein solution. The protein concentrations were determined using a UV–vis spectrophotometer (NanoDrop2000). The ovalbumin used in this paper was either Sigma-Aldrich, lyophilized powder, ≥98%, agarose gel electrophoresis, PO A5503-10G); or Sigma-Aldrich, powder, 62–88%, agarose gel electrophoresis, PO A5253-250G.
Phase Behavior of Diluted Pidan Tissues
Pidan material was homogenized in NaOH solution (pH = 12.90) with different pidan weight percentages. The solutions were then centrifuged at 14800g at 10 °C for 10 min. After centrifugation, some solutions have liquid–liquid phase separation, and the volumes of sparse phase solutions were measured using a pipet. The weights of egg protein in both dense and spare phase were measured after lyophilization overnight with the weight of NaOH subtracted. The packing fractions of the egg protein at different solutions were estimated using the density of an ovalbumin protein (45 kDa, volume = 7 × 2 × 4 nm).
SAXS
Small-angle X-ray scattering (SAXS) measurements were performed both at the University of Pennsylvania (Penn) and the X9 beam line of National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL).
Measurements at Penn use a rotating-anode system with Cu Kα radiation (λ = 1.542 Å) from a Bruker-Nonius FR591 generator operated at 3.4 kW. Collimation was performed using circular pinholes and Osmic Max-Flux confocal optics. The scattered X-rays were collected using a Bruker Hi-Star area detector. Data were collected at sample–detector distances of 54 and 150 cm, and the data merged to create a single profile over the range q ∈ [0.008, 0.35] Å–1. To minimize background, an integral vacuum with a pressure of < 0.3 mbar was maintained along the entire flight path. The sample temperature was either at room temperature or maintained at 10 °C by a TMS 94 and LNP 94/2 temperature control unit (Linkam Scientific Instruments Ltd., Waterfield, Tadworth, United Kingdom).
The measurements at the NSLS were made at an incident X-ray energy of 8.00 keV. These measurements were done in air due to constraints of the sample holder geometry. Each measurement was made at a single sample position with a MAR-CCD detector at the wave vector range of q ∈ [0.08, 1] Å–1.
All sample tissues were mounted in custom-built O-ring-sealed sample holders with clear ruby mica windows (7.5 and 12 mm in diameter, Attwater Group, Preston, England). Raw egg white liquid was loaded to completely fill in the sample chamber (approximately 45 μL) using a glass pipet so that there were no air bubbles observed. The sample chambers were carefully sealed to prevent leakage in the vacuum of the sample chamber, and no leaks were observed during the scans.
On the Penn apparatus, exposure times for each angle range were 30 min for raw egg, pidan, and boiled egg, and 4 h for ovalbumin solutions and gels. At NSLS, exposure times were 2–4 min for all experiments. We did not observe time dependence in the scattering patterns for any sample. Primary data reduction was performed using a field-specific software Datasqueeze (https://www.sas.upenn.edu/~heiney/html-physics/datasqueeze/index.html).
We performed buffer subtraction from the solution samples as follows: for ovalbumin solutions measured at Penn, the scattering intensities were first normalized to the product of I(q = 0) and exposure time, and then a buffer subtraction was performed. For the wide-angle data taken at BNL, the normalization was performed based on a background peak at q = 0.39 Å–1 with the width of 0.04 Å–1. A linear regression was performed by fitting the scattering from the buffer using the scattering from the ovalbumin solution. The coefficients were used to adjust the relative position of these two sets of data, and then the buffer signal was subtracted from the solution intensity. Finally, the buffer subtracted intensity was vertically adjusted by subtracting the minimum of this intensity to avoid producing negative intensities.
Form Factor and Structure Factor
The calculated form factors of ovalbumin, β-sheet structure from ovalbumin, and S-Crystallin were estimated using the FOXS online server (https://modbase.compbio.ucsf.edu/foxs/). This calculation used the atom positions of the protein and the Debye formula as follows:
| 1 |
Here, fi(q) (fj(q)) is the form factor of the ith (jth) atom, which is approximated by the number of electrons in the atom. The variable d is the pair distance between atom i and j. The protein structure of the ovalbumin (1OVA) was obtained from the Protein Data Bank (doi.org/10.2210/pdb1ova/pdb).22 The β-sheet structure was obtained by manually selecting the atoms in the ovalbumin that corresponds to the β-sheet structure. The form factor of the squid eye lens was averaged over the compositions of S-Crystallins, as described by Cai and colleagues.2 Then, the structure factor of the ovalbumin at high pH was determined by dividing the scattering intensity as a function of angle by this form factor. The structure factor of pidan was calculated from pidan SAXS intensity divided by the raw quail egg SAXS intensity because little interaction was observed between the proteins in the raw quail egg white in our SAXS data.
DAMMIF
The most likely real-space configuration of amino acids consistent with the SAXS result was obtained using DAMMIF.37 Here we used GNOM38 program parameters of nonzero condition at r = rmin and nonzero condition at r = rmax, with rmax = 70 Å. For DAMMIF, we used a simulation sphere diameter of 70 Å and particle diameter 3.2 Å, consistent with a single spherical particle representing a single amino-acid residue. No symmetry was presumed, we used the setting consistent with unknown particle anisometry, and all other settings were set to the program defaults.
UV Absorption at Various Alkaline pH Values
The structural change of ovalbumin at high pH was studied using the UV absorption of proteins dissolved at various pH solutions. Ovalbumin was diluted in 20 mM Tris-HCl with 50 mM sodium chloride solution to a final concentration of 3 mg/mL. Various amounts of NaOH were added, and the final pH of these solutions ranged from 10.7 to 13.8. The UV–vis absorption was measured using a NanoDrop spectrophotometer (NanoDrop2000, ThermoFisher Scientific). Because the absorbance at 250 nm depends on the average ionization state of tyrosine, and the red-shift of the ∼280 nm peak shift is due to the ionization of the phenolic hydrogen, we estimated the degree of tyrosine residue exposure (αprop/α) at the protein surface using the absorbance at 250 and 278 nm following the method described by Melo and colleagues.23
Congo Red Binding Experiment
We prepared a series of dilutions of purified ovalbumin protein (Sigma-Aldrich A5503-10G) in both neutral (phosphate-buffered saline, pH 7.4) and alkaline (0.5 M NaOH, pH 13.7) solution. To remove any particulates, the solutions were centrifuged at 14.8Kg at 10 °C for 10 min; little precipitate was observed, and we isolated the supernatant by pipetting. A 300 μM stock solution of the dye Congo red (CR, Sigma Aldrich C6277-25G) was also prepared in PBS and similarly centrifuged at 14.8Kg for 10 min with the supernatant isolated by pipetting. We then performed two sets of serial 2-fold dilutions of this CR stock solution using the neutral and alkaline ovalbumin solutions. This procedure had the effect of reducing the number of CR molecules relative to a fixed number of ovalbumin molecules in a fixed volume. In this dilution series, the ovalbumin concentration was fixed at 22.2 μM, while the CR concentration decreased in 2-fold steps from 300 to 4.69 μM, such that the molar ratio of CR to ovalbumin decreased from 13.5 to 0.21. We prepared another two sets of 2-fold dilutions in which we held the concentration of CR fixed while decreasing the concentration of either neutral or alkaline ovalbumin, all in a fixed volume. In this second dilution series, the CR concentration was fixed at 300 μM, and ovalbumin concentration decreased from 22.2 to 0.04 μM, such that the molar ratio of CR to ovalbumin increased from 13.5 to 6912.
The UV–vis absorbance of these solutions was similarly measured using a NanoDrop spectrophotometer (NanoDrop2000, ThermoFisher Scientific). In neutral solution without protein, CR has a strong absorbance peak at 484 nm. However, when CR interacts with protein secondary structure, this absorbance peak shifts to longer wavelengths.24 Notably, there seems to be little or no structured interaction of CR with random-coil protein structure.25 We systematically located the peak in each absorbance assay using the MATLAB Gaussian fit function cftool. Because ovalbumin has a second, fixed absorbance peak in the UV between 330 and 340 nm, we used this algorithm to fit the two Gaussian peaks of each spectrum, one between 330 and 340 and the second between 400 and 600 nm, with no other constraints.
Next, we inferred the average number of CR molecules bound to each ovalbumin molecule using the following logic. In this section, all concentrations are in the unit of μM. When the CR concentration is low compared to the ovalbumin concentration, we assumed that the binding of a single CR to ovalbumin will not influence the binding of other molecules of CR to ovalbumin. Therefore, the chemical reaction will be:
| 2 |
where O is unbound ovalbumin, CR is unbound Congo red, and OCR is Congo red bound to ovalbumin.
The equilibrium constant for this reaction is then
| 3 |
where [Cb]f is the concentration of bonded CR at the final state, and [Cub] is the concentration of unbonded CR at the final state. The concentration O can then be estimated using the initial ovalbumin concentration, [O]i, and the concentration of the bound CR ([Cb]f) using
| 4 |
where B is the average number of CR binding to an ovalbumin. The ratio of the number of bound CR ([Cb]) to the total number of CR ([Cb] + [Cub]) present can then be estimated using the peak shift from the absorption measurement:31
| 5 |
Therefore, the number of CR bound to each ovalbumin can be estimated as
| 6 |
Given this logic, we then performed a manual numeric fit to find reasonable values for the binding constant K by minimizing the number of binding events in the system while avoiding negative values of the CR to ovalbumin ratio. A value of K less than 0.6 μM–1 resulted in negative values of CR to ovalbumin ratio, while a value of K greater than 5 μM–1 resulted in saturation of the number of CR bound to a given ovalbumin. We therefore estimate K to be approximately 3 μM–1.
Protein Order vs Disorder
To predict whether protein regions were ordered or disordered, we calculated the net charge and the hydrophobicity following the method described by Uversky and colleagues.26 We used ovalbumin amino acid (a.a) sequence from chain A (protein data bank, 1OVA), and 19 previously characterized of S-Crystallin sequences from L. opalescens.39 To predict residue charges in high pH, we determined the likely charge given the pKa of the side chains at both neutral pH and pH = 12. We used the Kyte–Doolittle hydropathy scale, normalized from 0 to 1, as described by Uversky and colleagues.26,40 Since hydropathy is a function of aliphatic surface area, hydropathy values will not fundamentally shift at high pH, unless the overall charge of a residue changes. Residues that were predicted to gain a charge at high pH were assigned a hydropathy value of −3.72, or the mean hydropathy of the charged amino acids at neutral pH. For ovalbumin, moving averages with window size 11 were calculated for both the net charge per residue and hydropathy per residue. S-Crystallin sequences were divided into “loop” residues and “body” residues by determining the boundaries of the third exon that encodes the “loop” or disordered region responsible for low-valence linkages.2,41 At both neutral and high pH, the average distances from each amino acid to the order–disorder separation line were calculated either in an 11-residue moving window for ovalbumin, or for the loop vs body domains for S-Crystallins.
Squid Species and Dissection
Specimens of the squid Doryteuthis pealeii were obtained from the Marine Biological Laboratory at Woods Hole, MA. Lenses were excised from the suspending tissue and either used fresh or stored at −80 °C either alone for use in SAXS. Before each experiment, a lens was peeled into four concentric layers, and the outermost layer, whose outermost edge was at 100% of the total radius, was used in the temperature dependence SAXS measurement. The SAXS experimental details of the squid lens follow the method by Cai and colleagues.2
Acknowledgments
The authors thank E. Eiser for helpful discussions, P. A. Heiney for discussions and expert assistance with SAXS, and D. R. Vann for expert assistance with ion chromatography. Financial support was provided by a Packard Foundation Fellowship for Science and Engineering, Sloan Foundation, NSF-1351935, Kaufman Foundation, and University of Pennsylvania to A.M.S. SAXS data are archived at Small Angle Scattering Biological Data Bank under SASDDV8–Y8.
The authors declare no competing financial interest.
References
- Fang Y.Wu Li Xiao Shi; Shanghai Gu Ji Chu Ban She, 1987; Vol. 867; pp 863–864. [Google Scholar]
- Cai J.; Townsend J. P.; Dodson T. C.; Heiney P. A.; Sweeney A. M. Eye Patches: Protein Assembly of Index-Gradient Squid Lenses. Science 2017, 357, 564–569. 10.1126/science.aal2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E.; Blaak R.; Likos C. N. Patchy Colloids: State of the Art and Perspectives. Phys. Chem. Chem. Phys. 2011, 13, 6397–6410. 10.1039/c0cp02296a. [DOI] [PubMed] [Google Scholar]
- Russo J.; Tartaglia P.; Sciortino F. Reversible Gels of Patchy Particles: Role of the Valence. J. Chem. Phys. 2009, 131, 014504. 10.1063/1.3153843. [DOI] [PubMed] [Google Scholar]
- Biffi S.; Cerbino R.; Bomboi F.; Paraboschi E. M.; Asselta R.; Sciortino F.; Bellini T. Phase Behavior and Critical Activated Dynamics of Limited-Valence DNA Nanostars. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 15633–15637. 10.1073/pnas.1304632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Lu Z.-Y.; Sun Z.-Y. Self-assembly of Amphiphilic Patchy Particles with Different Cross-linking Densities. Soft Matter 2012, 8, 7073–7080. 10.1039/c2sm25078k. [DOI] [Google Scholar]
- Glotzer S. C.; Solomon M. J. Anisotropy of Building Blocks and Their Assembly into Complex Structures. Nat. Mater. 2007, 6, 557–562. 10.1038/nmat1949. [DOI] [PubMed] [Google Scholar]
- Bianchi E.; Largo J.; Tartaglia P.; Zaccarelli E.; Sciortino F. Phase Diagram of Patchy Colloids: Towards Empty Liquids. Phys. Rev. Lett. 2006, 97, 168301. 10.1103/PhysRevLett.97.168301. [DOI] [PubMed] [Google Scholar]
- Miguel M.; Manso M.; Lopez-Fandino R.; Ramos M. Comparative Study of Egg White Proteins from Different Species by Chromatographic and Electrophoretic Methods. Eur. Food Res. Technol. 2005, 221, 542–546. 10.1007/s00217-005-1182-8. [DOI] [Google Scholar]
- Mohanty P. S.; Yethiraj A.; Schurtenberger P. Deformable Particles with Anisotropic Interactions: Unusual Field-Induced Structural Transitions in Ultrasoft Ionic Microgel Colloids. Soft Matter 2012, 8, 10819–10822. 10.1039/c2sm26729b. [DOI] [Google Scholar]
- Zhang Z.; Keys A.; Chen T.; Glotzer S. Self-assembly of Patchy Particles into Diamond Structures Through Molecular Mimicry. Langmuir 2005, 21, 11547–11551. 10.1021/la0513611. [DOI] [PubMed] [Google Scholar]
- Corezzi S.; De Michele C.; Zaccarelli E.; Fioretto D.; Sciortino F. A Molecular Dynamics Study of Chemical Gelation in a Patchy Particle Model. Soft Matter 2008, 4, 1173–1177. 10.1039/b719872h. [DOI] [PubMed] [Google Scholar]
- Ruzicka B.; Zaccarelli E.; Zulian L.; Angelini R.; Sztucki M.; Moussaid A.; Narayanan T.; Sciortino F. Observation of Empty Liquids and Equilibrium Gels in a Colloidal Clay. Nat. Mater. 2011, 10, 56–60. 10.1038/nmat2921. [DOI] [PubMed] [Google Scholar]
- Romano F.; Sanz E.; Sciortino F. Phase Diagram of a Tetrahedral Patchy Particle Model for Different Interaction Ranges. J. Chem. Phys. 2010, 132, 184501. 10.1063/1.3393777. [DOI] [Google Scholar]
- Romano F.; Tartaglia P.; Sciortino F. Gas-liquid Phase Coexistence in a Tetrahedral Patchy Particle Model. J. Phys.: Condens. Matter 2007, 19, 322101. 10.1088/0953-8984/19/32/322101. [DOI] [Google Scholar]
- Liu A. J.; Nagel S. R. Nonlinear dynamics: Jamming is not just cool any more. Nature 1998, 396, 21. 10.1038/23819. [DOI] [Google Scholar]
- Lubchenko V. Theory of the structural glass transition: A pedagogical review. Adv. Phys. 2015, 64, 283–443. 10.1080/00018732.2015.1057979. [DOI] [Google Scholar]
- Cardinaux F.; Zaccarelli E.; Stradner A.; Bucciarelli S.; Farago B.; Egelhaaf S. U.; Sciortino F.; Schurtenberger P. Cluster-driven dynamical arrest in concentrated lysozyme solutions. J. Phys. Chem. B 2011, 115, 7227–7237. 10.1021/jp112180p. [DOI] [PubMed] [Google Scholar]
- Eiser E.; Miles C. S.; Geerts N.; Verschuren P.; MacPhee C. E. Molecular Cooking: Physical Transformations in Chinese ‘Century’ Eggs. Soft Matter 2009, 5, 2725–2730. 10.1039/b902575h. [DOI] [Google Scholar]
- DOI E. Gels and Gelling of Globular-Proteins. Trends Food Sci. Technol. 1993, 4, 1–5. 10.1016/S0924-2244(05)80003-2. [DOI] [Google Scholar]
- Sugimoto Y.; Sanuki S.; Ohsako S.; Higashimoto Y.; Kondo M.; Kurawaki J.; Ibrahim H.; Aoki T.; Kusakabe T.; Koga K. Ovalbumin in Developing Chicken Eggs Migrates from Egg White to Embryonic Organs While Changing its Conformation and Thermal Stability. J. Biol. Chem. 1999, 274, 11030–11037. 10.1074/jbc.274.16.11030. [DOI] [PubMed] [Google Scholar]
- Stein P.; Leslie A.; Finch J.; Carrell R. Crystal-Structure of Uncleaved Ovalbumin at 1.95 ÅResolution. J. Mol. Biol. 1991, 221, 941–959. 10.1016/0022-2836(91)80185-W. [DOI] [PubMed] [Google Scholar]
- Pinho Melo E.; AiresBarros M.; Costa S.; Cabral J. Thermal Unfolding of Proteins at High pH Range Studied by UV Absorbance. J. Biochem. Biophys. Methods 1997, 34, 45–59. 10.1016/S0165-022X(97)01202-5. [DOI] [PubMed] [Google Scholar]
- Chiti F.; Webster P.; Taddei N.; Clark A.; Stefani M.; Ramponi G.; Dobson C. Designing Conditions for in vitro Formation of Amyloid Protofilaments and Fibrils. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 3590–3594. 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana R.; Uversky V.; Nielsen L.; Fink A. Is Congo Red an Amyloid-specific Dye?. J. Biol. Chem. 2001, 276, 22715–22721. 10.1074/jbc.M011499200. [DOI] [PubMed] [Google Scholar]
- Uversky V. N.; Gillespie J. R.; Fink A. L. Why are “natively unfolded” proteins unstructured under physiologic conditions?. Proteins: Struct., Funct., Genet. 2000, 41, 415–427. . [DOI] [PubMed] [Google Scholar]
- Mine Y. Laser Light Scattering Study on the Heat-Induced Ovalbumin Aggregates Related to its Gelling Property. J. Agric. Food Chem. 1996, 44, 2086–2090. 10.1021/jf960153z. [DOI] [Google Scholar]
- Kidwai S.; Ansari A.; Salahuddin A. Effect of Succinylation (3-Carboxypropionylation) on Conformation and Immunological Activity of Ovalbumin. Biochem. J. 1976, 155, 171–180. 10.1042/bj1550171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers M.; Broersen K.; Barneveld P. A.; Stuart M. A. C.; Hamer R. J.; De Jongh H. J.; Visschers R. W. Net Charge Affects Morphology and Visual Properties of Ovalbumin Aggregates. Biomacromolecules 2008, 9, 3165–3172. 10.1021/bm800751e. [DOI] [PubMed] [Google Scholar]
- Pusey P. N.; Zaccarelli E.; Valeriani C.; Sanz E.; Poon W. C. K.; Cates M. E. Hard Spheres: Crystallization and Glass Formation. Philos. Trans. R. Soc., A 2009, 367, 4993–5011. 10.1098/rsta.2009.0181. [DOI] [PubMed] [Google Scholar]
- Sabate R.; Estelrich J. Aggregation Characteristics of Ovalbumin in Beta-sheet Conformation Determined by Spectroscopy. Biopolymers 2002, 67, 113–120. 10.1002/bip.10061. [DOI] [PubMed] [Google Scholar]
- Bomboi F.; Romano F.; Leo M.; Fernandez-Castanon J.; Cerbino R.; Bellini T.; Bordi F.; Filetici P.; Sciortino F. Re-Entrant DNA Gels. Nat. Commun. 2016, 7, 13191. 10.1038/ncomms13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciortino F.; Zaccarelli E. Equilibrium Gels of Limited Valence Colloids. Curr. Opin. Colloid Interface Sci. 2017, 30, 90–96. 10.1016/j.cocis.2017.06.001. [DOI] [Google Scholar]
- Rovigatti L.; Bomboi F.; Sciortino F. Accurate Phase Diagram of Tetravalent DNA Nanostars. J. Chem. Phys. 2014, 140, 154903. 10.1063/1.4870467. [DOI] [Google Scholar]
- Rovigatti L.; Smallenburg F.; Romano F.; Sciortino F. Gels of DNA Nanostars Never Crystallize. ACS Nano 2014, 8, 3567–3574. 10.1021/nn501138w. [DOI] [PubMed] [Google Scholar]
- Biffi S.; Cerbino R.; Nava G.; Bomboi F.; Sciortino F.; Bellini T. Equilibrium Gels of Low-Valence DNA Nanostars: a Colloidal Model for Strong Glass Formers. Soft Matter 2015, 11, 3132–3138. 10.1039/C4SM02144D. [DOI] [PubMed] [Google Scholar]
- Franke D.; Svergun D. I. DAMMIF, a Program for Rapid ab-initio Shape Determination in Small-angle Scattering. J. Appl. Crystallogr. 2009, 42, 342–346. 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J. Appl. Crystallogr. 1992, 25, 495–503. 10.1107/S0021889892001663. [DOI] [Google Scholar]
- Sweeney A. M.; Des Marais D. L.; Ban Y.-E. A.; Johnsen S. Evolution of graded refractive index in squid lenses. J. R. Soc., Interface 2007, 4, 685–698. 10.1098/rsif.2006.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J.; Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Tomarev S. I.; Zinovieva R.; Piatigorsky J. Characterization of squid Crystallin genes. Comparison with mammalian glutathione S-transferase genes. J. Biol. Chem. 1992, 267, 8604–8612. [PubMed] [Google Scholar]