Abstract
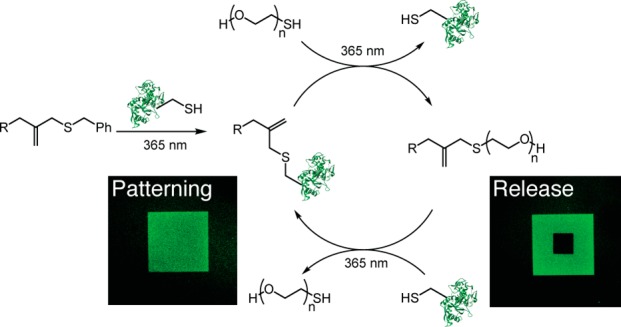
Biomolecule-functionalized hydrogels have emerged as valuable cell culture platforms to recapitulate the mechanical and biochemical properties of the extracellular niche. The typical strategy to functionalize hydrogels with biomolecules involves directly tethering them to the hydrogel backbone resulting in a static material. Thus, this approach fails to capture the dynamic changes in biomolecule composition that occur during biological processes or that may be required for regenerative medicine applications. Moreover, it also limits the scope of biomolecules to simple peptides, as signaling proteins generally have poor stability under cell culture conditions and lose their bioactivity over time. To that end, we sought to develop a bioconjugation reaction that would enable reversible and repeatable tethering of signaling proteins to hydrogels, so that spent protein could be released on-demand and replaced with fresh protein as needed. Specifically, we designed an allyl sulfide chain-transfer agent that enables a reversible, photomediated, thiol–ene bioconjugation of signaling proteins to hydrogels. Upon addition of a thiolated protein to the allyl sulfide moiety, the previously tethered protein is released, and the “ene” functionality is regenerated. Using this approach, we demonstrate that protein patterning can be achieved in hydrogels through a thiol–ene reaction, and the patterned protein can then be released through a subsequent thiol–ene reaction of a PEG thiol. Importantly, this process is repeatable through multiple iterations and proceeds at physiologically relevant signaling protein concentrations. Finally, we demonstrate that whole signaling proteins can be patterned and released in the presence of cells, and that cells respond to their presentation with spatial fidelity. Combined, these data represent the first example of a methodology that enables fully reversible and repeatable patterning and release of signaling proteins from hydrogels.
Short abstract
Protein-functionalized hydrogels provide more accurate extracellular matrix mimics. Herein, we describe fully reversible and repeatable patterning and release of signaling proteins from hydrogels.
Introduction
Synthetic hydrogel materials seek to recapitulate critical aspects of the extracellular matrix for modeling cellular microenvironments.1 Hydrogels synthesized from poly(ethylene glycol) (PEG) polymers act as a “blank slate,” enabling complete user control over the mechanical and biochemical signals presented to cells.2 Manipulating cellular phenotypes in vitro often requires the presentation of biochemical signals to achieve desired alterations in cellular adhesion,3 migration,4 and differentiation.5,6 To that end, biomolecules, including peptides and proteins, are grafted on to the scaffold via direct conjugation to the polymer backbone to facilitate their presentation to cells.7
While any bioconjugation reaction can be adapted to achieve biomolecule tethering,8,9 photomediated bioconjugations,10,11 including the thiol–ene photoclick reaction, are particularly attractive as they enable spatial presentation, or patterning, of cues.10−19 Biomolecule patterning via the thiol–ene reaction involves the addition of a thiolated biomolecule to alkene functionalities on the polymer backbone to afford a thioether aduct.20 The reaction is radical mediated and initiated by light to enable spatial control over where in the hydrogel the reaction proceeds. Moreover, the process is cytocompatible and can be performed in the presence of cells.21 Performing the thiol–ene reaction in conjunction with photolithographic methods enables the assembly of biomolecule gradients and complex three-dimensional patterns with high precision to design materials capable of mimicking the inherently heterogeneous nature of the cellular niche.22−24 Because these approaches generally afford a covalent tether between the biomolecule and polymer backbone, the resulting materials are static and do not allow for user control over dynamic changes that occur during many biological processes. Indeed, biomolecules must be presented over time, as cells interpret signals from biomolecules on the order of seconds to days to change their phenotypes.25 Introducing biochemical cues into a hydrogel network in a dynamic fashion would greatly improve extracellular matrix mimicry to enable specialized cell culture platforms.26 For example, platforms for stem cell expansion and differentiation could be improved through the sequential presentation of multiple different biochemical cues over time.27 Additionally, spatiotemporal patterning of multiple different cues could be used to engineer more accurate in vitro disease models where the composition of extracellular matrix proteins changes over the course of disease progression.28
To address the limitations associated with traditional covalent bioconjugation, new photomediated strategies have emerged that enable the patterning and subsequent release of biomolecules in hydrogels through the inclusion of a photolabile linkage.29,30 Upon irradiation, the photolabile linker is cleaved, releasing the tethered biomolecule from the hydrogel. Because the bioconjugation handle is cleaved and thus consumed, patterning and release can only be performed once. From an experimental design standpoint, it may be necessary to sequentially pattern and release multiple different biomolecules of interest. Perhaps more importantly, the patterning of whole signaling proteins has remained a significant challenge due to their low stability under cell culture condition. For protein patterning to be practical, it is necessary that spent protein can be released from the hydrogel and replaced with fresh protein as needed.
Given these considerations, we sought to develop a methodology that would not only enable reversible patterning and release of proteins, but also be repeatable so that protein patterning and release can be performed through multiple iterations. We drew inspiration from the allyl sulfide chain-transfer agent which has been used extensively in the design of stress relaxing polymer systems.31−33 We hypothesized that incorporating pendant allyl sulfide moieties onto the hydrogel backbone would enable full reversible and repeatable tethering of whole signaling proteins from a hydrogel through a photomediated thiol–ene click reaction (Scheme 1). Due to the nature of the allyl sulfide as a chain-transfer agent, upon addition of protein to the allyl sulfide handle, any previously tethered protein would be released, and the alkene would be regenerated enabling subsequent thiol–ene reactions. We previously demonstrated that the allyl sulfide chain-transfer agent can be used to reversibly pattern peptides in hydrogels.34 However, the patterning of whole signaling proteins poses a significant challenge over peptides—proteins are employed under dilute concentrations (picomolar to nanomolar) compared to peptides (millimolar) and are significantly bulkier, both inhibiting their reaction rates toward addition and release. Herein, we describe the development of a new allyl sulfide chemical handle that enables reversible and repeatable patterning of signaling proteins in hydrogels. Using this approach, we demonstrate that multiple different proteins can be patterned in and released from hydrogels through sequential thiol–ene reactions. We also demonstrate that this process is cytocompatible, and signaling proteins can be patterned and released in the presence of cells to control cell phenotype.
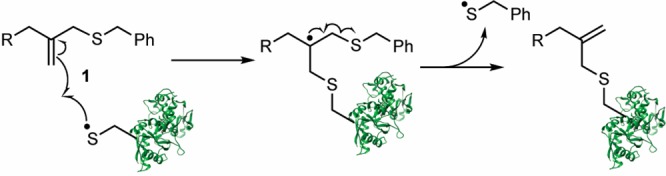
Scheme 1. Mechanism of the Thiol–ene Reaction between a Thiyl Radical-Containing Protein and the Allyl Sulfide Moiety 1 (R Represents the Hydrogel Backbone).
Results and Discussion
Drawing inspiration from the regenerative nature of chain-transfer agents employed in reversible addition–fragmentation chain-transfer (RAFT) polymerizations, we postulated the allyl sulfide chain-transfer agent 1 would enable reversible tethering and release of thiol-containing proteins (Scheme 1).31−34 While we drew inspiration from RAFT polymerizations, which typically cannot be performed in the presence of oxygen, a key advantage of the thiol–ene reaction is it is not prone to oxygen inhibition and thus can be performed under cell culture conditions.35 We hypothesized that photoinitiation will lead to the generation of thiyl radicals on the protein of interest. Attack of the thiyl radical at the allyl sulfide moiety results in the formation of an unstable intermediate that rapidly undergoes β-scission resulting in covalent tethering of the thiolated protein, release of benzyl mercaptan, and regeneration of the alkene. Since the radical intermediate is asymmetrical, the more stable radical will be favored for release upon β-scission. In our system, both the benzyl mercaptan radical and the protein thiyl radical have similar stabilities.36 Thus, we hypothesized that initial product formation will favor the protein adduct, since protein concentrations is higher relative to released benzyl mercaptan.
The overall strategy to achieve sequential protein tethering and release is outlined in Scheme 2. Photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate 2 (LAP) was selected due to its rapid rate of initiation upon irradiation with 365 nm light and cytocompatibility.37 After protein tethering to the hydrogel backbone, its release can be achieved through a subsequent thiol–ene reaction of a PEG thiol. This process results in tethering of PEG, regeneration of the allyl sulfide alkene, and release of the previously tethered protein. A third thiol–ene reaction can then be performed to tether a new protein of interest, while releasing the PEG thiol. Since the alkene is regenerated during each reaction, this process should be repeatable, thus enabling a methodology to reversibly pattern and release signaling proteins of interest with spatial fidelity within a hydrogel.
Scheme 2. Proposed Strategy To Reversibly Tether Signaling Proteins to Hydrogels through the Allyl Sulfide Moiety.
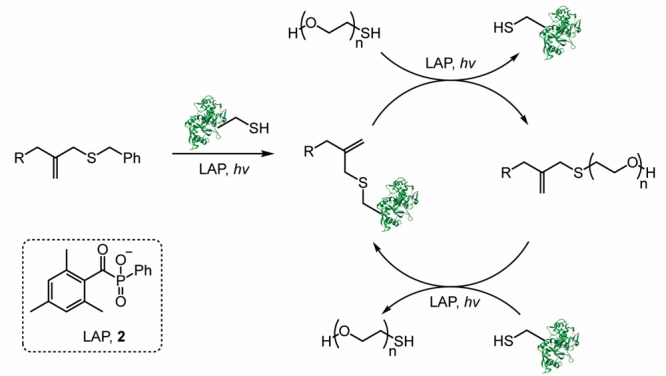
Thiolated signaling proteins are tethered through a thiol–ene reaction. To release the protein, a subsequent thiol–ene reaction is performed with mPEG-SH. This process of protein tethering followed by release should be repeatable. R represents the hydrogel backbone.
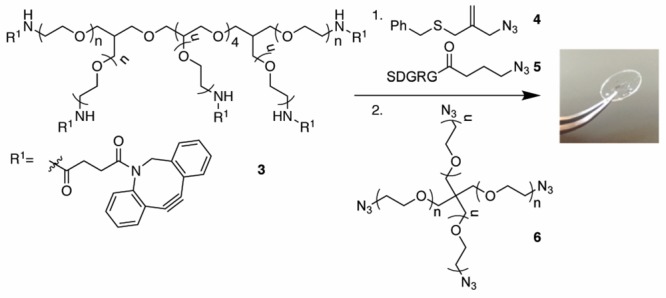
To synthesize allyl sulfide hydrogels (Scheme 3), we employed strain-promoted azide–alkyne cycloaddition (SPAAC) polymerization.38 Eight-armed PEG40K-dibenzylcyclooctyne (DBCO) 3 was functionalized with azido allyl sulfide 4 and the cell adhesive azido-RGDS peptide 5 to give a final gel concentration of 1 and 2 mM, respectively. Polymerization was performed between eight-armed PEG40K-DBCO and four-armed PEG20K azide 6 on an azide-functionalized coverslip to give a final gel composition of 10 wt % PEG. Gels were swelled in the presence of nPEG7-N3 to quench any unreacted DBCO that could also participate in the thiol–ene bioconjugation during protein tethering. The inclusion of a large excess of allyl sulfide (1 mM) in the hydrogels relative to the concentration of signaling proteins that will be employed during tethering (<100 μM) ensures that any nonspecific addition of cysteine-containing serum proteins should not interfere with the overall tethering reaction efficiency.
Scheme 3. Synthesis of Allyl Sulfide SPAAC Hydrogels on an Azide-Functionalized Glass Coverslip.
With allyl sulfide gels in hand, we first sought to optimize protein tethering to the allyl sulfide hydrogels. We selected human transferrin, an 80 kDa iron transporting protein, as a model protein for kinetic studies.39 Transferrin is larger than most signaling proteins, so its optimization of tethering and release should be translatable to other signaling proteins of interest. To ensure reactive thiols on transferrin are available to undergo the thiol–ene reaction, transferrin was treated with N-hydroxysuccinimidyl-PEG1K thiol to functionalize lysine residues nonspecifically.
A key advantage of the thiol–ene reaction is its high efficiency, whereby a single initiation event mediates hundreds or thousands of subsequent reactions.20 Thus, under idealized conditions, the thiol–ene bioconjugation proceeds with catalytic photoinitiator with respect to the biomolecule. Signaling proteins, however, are often employed under dilute conditions (<100 μM). At these concentrations, we hypothesized that radical propagation may be inhibited, and thus the reaction would proceed with dependence upon LAP concentration. To that end, we first sought to determine the effect of photoinitiator stoichiometry on protein tethering efficiency. Allyl sulfide hydrogels were swollen with 100 μM thiolated transferrin and a range of concentrations of LAP (0, 10, 20, or 100 μM). The hydrogels were irradiated with 5 mW/cm2 365 nm light for 180 s through a chrome photomask with a repeating 250 μm wide striped pattern. After washing away unreacted transferrin, protein tethering was visualized via immunostaining for transferrin. Normalized fluorescence intensity was measured as a readout of transferrin immobilization. After quantifying pixel intensity, a 1:1 stoichiometric ratio of LAP to transferrin yielded the most intense transferrin patterns (Figure 1A). Immobilization of transferrin scaled linearly with increasing LAP concentration (R = 0.998), suggesting radical propagation is indeed inhibited, and stoichiometric LAP relative to transferrin is required for efficient protein immobilization (Figure 1B).
Figure 1.
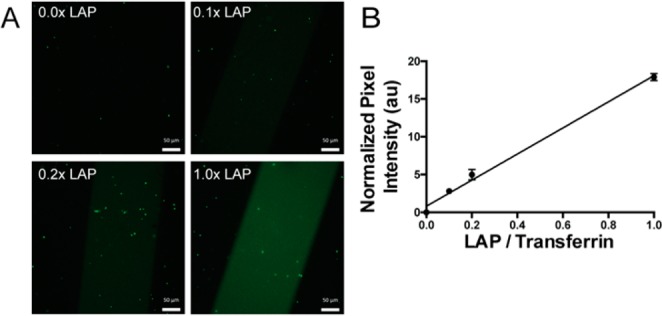
Protein patterning efficiency is a function of LAP concentration. Thiolated transferrin (100 μM) and LAP (0, 10, 20, 100 μM) were swelled into allyl sulfide hydrogels and irradiated through a chrome photomask with 5 mW/cm2 365 nm light for 180 s. Hydrogels were washed with PBS and immunostained for transferrin for visualization (A). Pixel intensity was averaged for the patterned region and subtracted from the average pixel intensity of the nonpatterned region to obtain normalized pixel intensity (B). Scale bars represent 50 μm.
Continuing with stoichiometric LAP, we next explored the effect of light intensity (Io) and light dose on protein patterning. Hydrogels were swelled with 100 μM thiolated transferrin and 100 μM LAP and irradiated through a chrome photomask with 365 nm light (0.5, 5, 50 mW/cm2) for a range of light doses (0–600 s). After immunostaining for transferrin, normalized pixel intensity was measured for each gel. Protein patterning increased with increasing light intensity and dose (Figure 2A). For both Io = 5 and 50 mW/cm2, protein tethering plateaued at similar intensities, but the rate at which protein tethering plateaued increased with higher light intensity. Since we expected that propagation would be inhibited under these conditions, we hypothesized that this plateau in the protein signal may correlate with LAP consumption. Indeed, the theoretical rate of photolysis of LAP (eq S5) followed the same trend as protein patterning, further supporting our observations that protein patterning is dependent upon LAP initiation and that propagation is inhibited under these conditions (Figure 2B). With these data combined, we selected Io = 5 mW/cm2 for 180 s with stoichiometric LAP as the optimal conditions to achieve maximal protein tethering to allyl sulfide hydrogels (Figure 2C).
Figure 2.
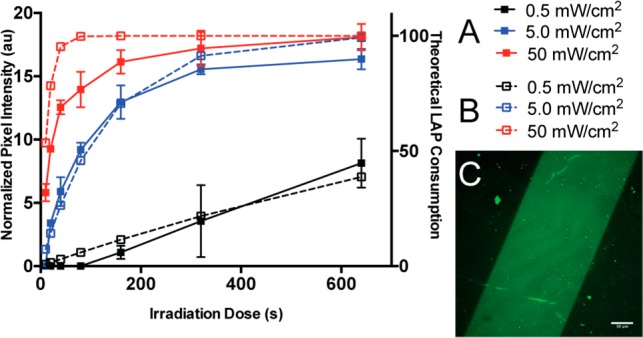
Protein patterning increases with light intensity and dose. Hydrogels were swelled with 100 μM thiolated transferrin and 100 μM LAP and irradiated through a chrome photomask with 365 nm light at specified intensities and doses. Gels were immunostained for transferrin, and normalized pixel intensity was measured (A, solid line). The rate of consumption of LAP was calculated with respect to Io and dose (B, dashed line). Maximal protein patterning was achieved with 5 mW/cm2 365 nm light for 180 s (C).
A key advantage of the allyl sulfide moiety is the regeneration of the alkene upon tethering of the protein of interest, thus enabling a subsequent thiol–ene reaction to release the bound protein. Importantly, this process should be repeatable to afford a methodology to immobilize and release signaling proteins of interest in demand. We chose to release previously patterned protein from the hydrogels through a thiol–ene reaction of PEG thiol resulting in tethering of an inert PEG moiety to the hydrogel. To optimize the release conditions, we explored the effect of LAP concentration and Io on the reaction. Hydrogels patterned with transferrin in 250 μm stripes were swelled with a release solution consisting of 50 mM PEG1K thiol and a range of catalytic LAP concentrations (0.1, 1.0, and 10 mM). By employing a large excess of PEG thiol relative to allyl sulfide concentration in the hydrogel (1 mM), reactivity with all allyl sulfide moieties should occur to ensure release of all tethered protein. Hydrogels were placed on a chrome photomask with a repeating 250 × 250 μm square pattern and irradiated with 365 nm light of varying intensity (0.5, 5.0, and 50 mW/cm2) for 180 s. Thus, the release reaction occurs in 250 × 250 μm regions within the striped transferrin patterns. The hydrogels were then washed and immunostained, and normalized pixel intensity was measured (Figure 3A). Transferrin release increased with increasing LAP concentration and light intensity, as indicated by a decrease in fluorescence within the irradiated regions. Remarkably, when hydrogels were irradiated with Io = 50 mW/cm2 365 nm light with 10 mM LAP, a >97% decrease in fluorescence signal intensity was observed relative to the original pattern signal intensity (Figure 3B). These results suggest near-complete release of protein can be achieved via this methodology. Furthermore, LAP concentration and light intensity may be tuned to achieve protein patterns of varied concentrations within the patterned regions.
Figure 3.
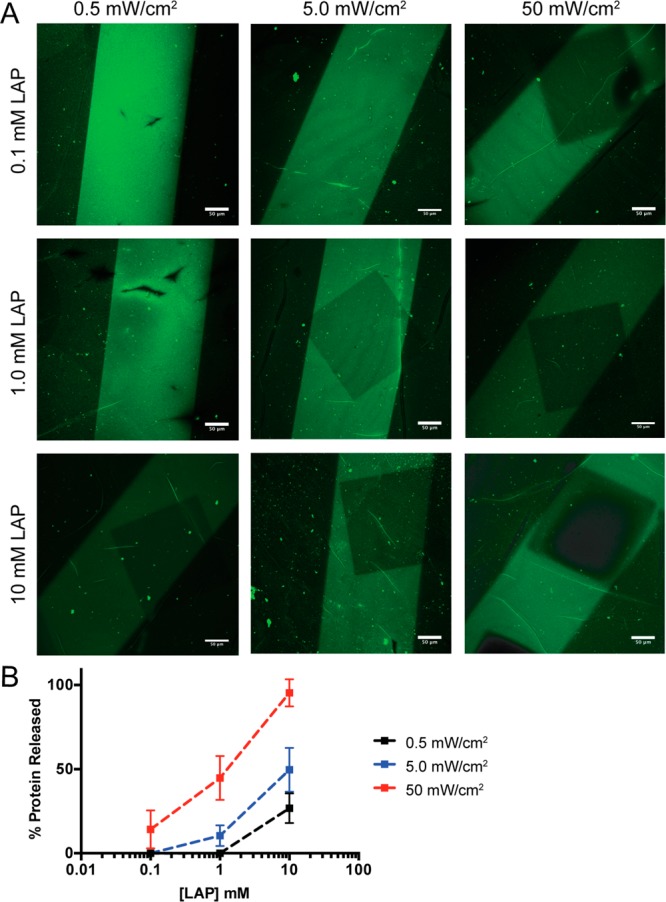
Protein release depends on LAP concentration and light intensity. Hydrogels with patterned transferrin were swelled with 50 mM mPEG1K-SH and LAP (0.1, 1.0, 10 mM), placed over a chrome photomask with a repeating 250 μm × 250 μm square pattern, and irradiated with 365 nm light at specific Io (0.5, 5.0, 50 mW/cm2). Immunostaining for transferrin was used to visualize patterning (A). Percent transferrin release was calculated by measuring the pixel intensity of the released region and pixel intensity of the original patterned region, normalized to the pixel intensity of unpatterned regions (B). Scale bar represents 50 μm.
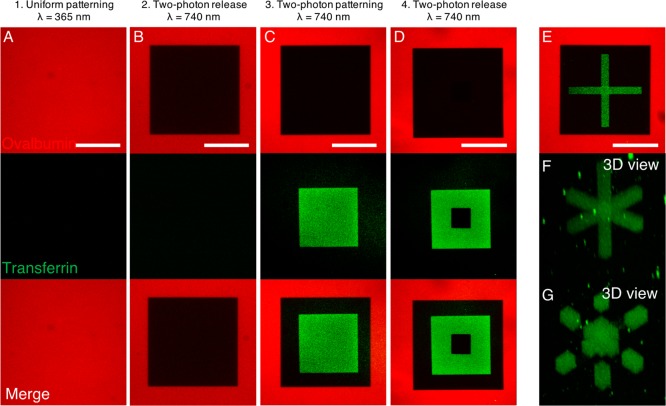
Having optimized protein tethering and release from allyl sulfide hydrogels, we next sought to demonstrate that the process of protein patterning and release can also be controlled in three-dimensions and is repeatable through multiple iterations with multiple different proteins (Figure 4). To control spatial patterning in three-dimensions, multiphoton photolithography can be employed. Multiphoton patterning techniques rely on near-simultaneous absorption of two or more photons and therefore restrict initiation to the focal volume, imparting precise three-dimensional control over the protein tethering reaction.40 Further, the nonlinear absorption increases the penetration depth for multiphoton strategies. We first patterned the thiolated and fluorescently labeled immunogenic protein ovalbumin. Ovalbumin was tethered uniformly to the hydrogel using the optimized exposure conditions (365 nm, 5 mW/cm2, 5 min; Figure 4A). Two-photon photolithography (λ = 740 nm) was then employed to release a 200 μm × 200 μm square within the original transferrin pattern through a subsequent thiol–ene reaction of PEG thiol (Figure 4B). To demonstrate protein patterning could be performed a subsequent time, thiolated and fluorescently labeled transferrin was patterned through two-photon photolithography in a 133 μm × 133 μm square within the released region (Figure 4C). Finally, a 44 μm × 44 μm square pattern of transferrin was released through another thiol–ene reaction of PEG thiol (Figure 4D). Additionally, to leverage the 3D capabilities provided by two-photon patterning, a “coordinate axes” shape was patterned in the released area (Figure 4E,F). Selected 3D regions of this shape were then replaced with PEG1K-thiol (Figure 4G). These data demonstrate not only that the regenerative nature of the allyl sulfide handle as the same region of a hydrogel can be patterned multiple times, but also that multiple different proteins can be patterned sequentially.
Figure 4.
Sequential tethering and release of multiple proteins with 3D control. Thiolated ovalbumin (AlexaFluor 555 conjugate) was uniformly tethered to the hydrogel (A), then selectively released using PEG1K-thiol (B), followed by tethering (C) and release (D) of thiolated transferrin (AlexaFluor 488 conjugate). Two-photon photolithography is used to pattern proteins with 3D control. A “coordinate axes” shape of transferrin is patterned within the volume of released ovalbumin (E, middle plane; F, 3D view), and selected regions of the 3D shape can be removed (G). Scale bars 100 μm.
Finally, we also characterized the concentration of transferrin tethered to the hydrogel through an enzyme-linked immunosorbent assay (ELISA) analysis (Figure S3). Transferrin patterning in the hydrogel proceeded through a broad range of physiologically relevant concentrations (10–10 000 pg/mL). Importantly, a linear relationship (R2 = 0.98) between the amount of transferrin swelled into the hydrogel and the amount patterned on the hydrogel was identified, suggesting the resultant protein concentration patterned in the hydrogel can easily be predicted a priori.
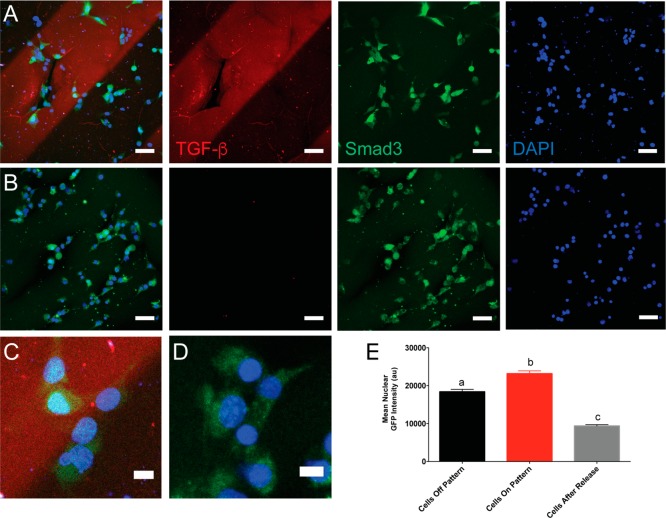
A key benefit of the thiol–ene reaction is that it is cytocompatible, so tethering of biomolecules can be performed in the presence of cells with minimal cytotoxicity. Moreover, since the thiol–ene reaction is light mediated, complex three-dimensional patterns of biomolecules can be assembled within the gel on-demand, vastly expanding the traditional experimental space for evaluating how cells respond to spatiotemporal protein cues. We sought to use this strategy of protein patterning followed by release with a signaling protein to verify that cells can respond to dynamic changes in protein patterning. We selected transforming growth factor-β1 (TGF-β1) for use as a signaling protein of interest. TGF-β1 is a growth factor principally implicated in many fibrotic diseases and is a target of interest for therapeutic intervention.41 Moreover, TGF-β1 exists bound to the extracellular matrix in vivo, and cells interact with TGF-β1 through direct interactions with the matrix.42,43 Thus, tethering TGF-β1 to hydrogels may function as a physiologically relevant presentation platform. When TGF-β1 binds to the TGF-β1 receptor on the surface of cells, a signaling cascade proceeds culminating in translocation of the transcription factor Smad3 from the cytosol to the nucleus. We postulated that the nuclear translocation of Smad3 could be used as a real-time readout of TGF-β1 signaling for our experiments. Thus, we modified mouse embryonic fibroblasts (MEFs) via retroviral transfection with a GFP-Smad3 fusion, expressed at endogenous levels.44,45 Importantly, Smad3 translocation is transient—cells treated with TGF-β1 exhibit rapid nuclear localization, and removal of TGF-β1 results in localization of Smad3 to the cytosol within minutes.44
Thiolated TGF-β1 (10 ng/mL) was patterned into 250 μm stripes through a chrome photomask. To visualize TGF-β1 patterns during microscopy, gels were simultaneously patterned with 10 μM fluorescently labeled thiolated transferrin. MEF GFP-Smad3 cells were seeded on top of the gels and allowed to spread for 3 h prior to imaging (Figure 5A). GFP signal was measured within the nucleus for cells both on and off the TGF-β1 patterns (Figure 5C), and we observed a ∼1.2-fold increase in nuclear GFP signal for cells on top of the TGF-β1 patterns relative to those not on the pattern (Figure 5E). These data demonstrate that cells can signal through tethered TGF-β1 and that the TGF-β1 cellular response can be controlled through its spatial presentation. To remove the TGF-β1 signal from the hydrogels, the cell-laden gels were swelled with 50 mM PEG thiol and 10 mM LAP for 1 h. The gels were then flood irradiated with 50 mW/cm2 365 nm light for 180 s to release all patterned TGF-β1. After washing the gels with media for 90 min, cells remained attached and were subsequently imaged (Figure 5B). As expected, mean nuclear GFP intensity was ∼2.0-fold lower after release of TGF-β1 (Figure 5D,E) compared to cells that were on the patterned regions initially. Finally, cell viability was also assessed before and after irradiation, and no significant decrease in viability was observed (Figure S4). Although we did not observe cells on patterns and off patterns influencing each other via paracrine signaling, open questions remain regarding how cells on patterns and off patterns communicate with each other and how patterning of signals may be used to influence entire cell populations or define the length scales over which paracrine signaling occurs. Combined, these data demonstrate how the allyl sulfide functional handle can be used to pattern and release signaling proteins within hydrogel matrices in the presence of cells. Our patterning strategy also enables user control to manipulate cellular phenotype with spatial fidelity.
Figure 5.
GFP-Smad3-expressing mouse embryonic fibroblasts respond to TGF-β1 patterned hydrogels. MEFs were seeded on hydrogels patterned with TGF-β1 (10 ng/mL) and cultured for 3 h (A). TGF-β1 was released in situ via a subsequent thiol–ene reaction with 50 mM PEG thiol, and cells were cultured for an additional 90 min (B). Inset of merged image shows cells within the pattern contain the Smad3 reporter in the nucleus (C, D). Mean nuclear GFP signal was measured for cells within the patterned region, outside the patterned region, and for cells after release of TGF-β1 (E) . At least 260 cells were analyzed per condition. Letters a, b, and c denote groups that are statistically distinct (p < 0.001) according to two-way ANOVA with Bonferroni testing for multiple comparisons. Data are shown as mean ± standard error (SEM). TGF-β1 (red), DAPI (blue), Smad3 (green).
Conclusions
In conclusion, we have designed an allyl sulfide handle that allows reversible thiol–ene protein patterning within cell-laden hydrogels. Due to the ability of the allyl sulfide to undergo chain transfer, the “ene” functionality is regenerated, and any tethered protein is released during each subsequent thiol–ene reaction. Moreover, this process of patterning and releasing proteins on-demand is repeatable, which enables facile replenishment of active proteins in hydrogel matrices. Importantly, we demonstrate that signaling proteins maintain their bioactivity and can be employed to control cell phenotype in a user-defined manner. TGF-β1 patterning induced localized cellular responses, and upon release of TGF-β1, cells returned to an unstimulated phenotype. We anticipate our protein patterning strategy will enable unique platforms to maintain and alter cell phenotype for precision medicine applications, where biomolecules can be tethered and released from a hydrogel to control cell phenotype for a wide array of applications, including immunoengineering and stem cell expansion.46 Combined, the allyl sulfide moiety is a powerful bioconjugation handle enabling real-time user control over the presentation of signaling proteins in hydrogels.
Acknowledgments
This work was supported by the NSF DMR 1408955, the NIH HL132353-02, the Howard Hughes Medical Institute, the NSF GRFP (T.E.B.), the NIH F32 HL137256-01 (B.A.A.), and the Burroughs Wellcome Fund Postdoctoral Enrichment Program (B.A.A.).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00325.
Synthetic procedures and molecular characterization for all new compounds, hydrogel fabrication, protein thiolation and fluorescent labeling, protein swelling characterization by fluorescence recovery after photobleaching analysis, photokinetic calculations, procedures for single-photon and two-photon protein patterning and release, immunostaining protocol, ELISA protocol, and generation of GFP-Smad3 cell line and cell culture (PDF)
The authors declare no competing financial interest.
Notes
No unexpected or unusually high safety hazards were encountered.
Supplementary Material
References
- Tibbitt M. W.; Anseth K. S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103 (4), 655–663. 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-C.; Anseth K. S. PEG hydrogels for the controlled release of biomolecules in regenerative medicine. Pharm. Res. 2009, 26 (3), 631–643. 10.1007/s11095-008-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hern D. L.; Hubbell J. A. Incorporation of adhesion peptides into nonadhesive hydrogels useful for tissue resurfacing. J. Biomed. Mater. Res. 1998, 39 (2), 266–276. . [DOI] [PubMed] [Google Scholar]
- DeLong S. A.; Moon J. J.; West J. L. Covalently immobilized gradients of bFGF on hydrogel scaffolds for directed cell migration. Biomaterials 2005, 26 (16), 3227–3234. 10.1016/j.biomaterials.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Wen J. H.; Vincent L. G.; Fuhrmann A.; Choi Y. S.; Hribar K. C.; Taylor-Weiner H.; Chen S.; Engler A. J. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 2014, 13 (10), 979–987. 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musah S.; Morin S. A.; Wrighton P. J.; Zwick D. B.; Jin S.; Kiessling L. L. Glycosaminoglycan-binding hydrogels enable mechanical control of human pluripotent stem cell self-renewal. ACS Nano 2012, 6 (11), 10168–10177. 10.1021/nn3039148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf M. P.; Gilbert P. M.; Blau H. M. Designing materials to direct stem-cell fate. Nature 2009, 462 (7272), 433–441. 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari E. Bioconjugation of hydrogels for tissue engineering. Curr. Opin. Biotechnol. 2011, 22 (5), 655–660. 10.1016/j.copbio.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S. A.; Baker A. E. G.; Shoichet M. S. Designing peptide and protein modified hydrogels: selecting the optimal conjugation strategy. J. Am. Chem. Soc. 2017, 139 (22), 7416–7427. 10.1021/jacs.7b00513. [DOI] [PubMed] [Google Scholar]
- Fisher S. A.; Tam R. Y.; Fokina A.; Mahmoodi M. M.; Distefano M. D.; Shoichet M. S.. Photo-immobilized EGF chemical gradients differentially impact breast cancer cell invasion and drug response in defined 3D hydrogels. Biomaterials 2018, in press; 10.1016/j.biomaterials.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam R. Y.; Smith L. J.; Shoichet M. S. Engineering cellular microenvironments with photo- and enzymatically responsive hydrogels: toward biomimetic 3D cell culture models. Acc. Chem. Res. 2017, 50 (4), 703–713. 10.1021/acs.accounts.6b00543. [DOI] [PubMed] [Google Scholar]
- Lee T. T.; Garcia J. R.; Paez J. I.; Singh A.; Phelps E. A.; Weis S.; Shafiq Z.; Shekaran A.; del Campo A.; García A. J. Light-triggered in vivo activation of adhesive peptides regulates cell adhesion, inflammation and vascularization of biomaterials. Nat. Mater. 2015, 14 (3), 352–360. 10.1038/nmat4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim J. C.; Marozas I. A.; Anseth K. S. Thiol-ene and photo-cleavage chemistry for controlled presentation of biomolecules in hydrogels. J. Controlled Release 2015, 219 (C), 95–106. 10.1016/j.jconrel.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramlich W. M.; Kim I. L.; Burdick J. A. Synthesis and orthogonal photopatterning of hyaluronic acid hydrogels with thiol-norbornene chemistry. Biomaterials 2013, 34 (38), 9803–9811. 10.1016/j.biomaterials.2013.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubko C. A.; Basak A.; Majumdar S.; Cao X. Dynamic cell patterning of photoresponsive hyaluronic acid hydrogels. J. Biomed. Mater. Res., Part A 2014, 102 (2), 381–391. 10.1002/jbm.a.34712. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Liu C.; Yang C.; Wu Y.; Yu F.; Chen Y.; Du J. Aptamer-patterned hydrogel films for spatiotemporally programmable capture and release of multiple proteins. ACS Appl. Mater. Interfaces 2018, 10 (10), 8546–8554. 10.1021/acsami.8b00191. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Trout W. S.; Liu S.; Andrade G. A.; Hudson D. A.; Scinto S. L.; Dicker K. T.; Li Y.; Lazouski N.; Rosenthal J.; Thorpe C.; Jia X.; Fox J. M. Rapid bioorthogonal chemistry turn-on through enzymatic or long wavelength photocatalytic activation of tetrazine ligation. J. Am. Chem. Soc. 2016, 138 (18), 5978–5983. 10.1021/jacs.6b02168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming Z.; Fan J.; Bao C.; Xue Y.; Lin Q.; Zhu L. Photogenerated aldehydes for protein patterns on hydrogels and guidance of cell behavior. Adv. Funct. Mater. 2018, 28, 1706918. 10.1002/adfm.201706918. [DOI] [Google Scholar]
- Adzima B. J.; Tao Y.; Kloxin C. J.; DeForest C. A.; Anseth K. S.; Bowman C. N. Spatial and temporal control of the alkyne–azide cycloaddition by photoinitiated Cu(II) reduction. Nat. Chem. 2011, 3 (3), 256–259. 10.1038/nchem.980. [DOI] [PubMed] [Google Scholar]
- Hoyle C. E.; Bowman C. N. Thiol-ene click chemistry. Angew. Chem., Int. Ed. 2010, 49 (9), 1540–1573. 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- Nguyen K. T.; West J. L. Photopolymerizable hydrogels for tissue engineering applications. Biomaterials 2002, 23 (22), 4307–4314. 10.1016/S0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- DeForest C. A.; Polizzotti B. D.; Anseth K. S. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater. 2009, 8 (8), 659–664. 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzotti B. D.; Fairbanks B. D.; Anseth K. S. Three-dimensional biochemical patterning of click-based composite hydrogels via thiolene photopolymerization. Biomacromolecules 2008, 9 (4), 1084–1087. 10.1021/bm7012636. [DOI] [PubMed] [Google Scholar]
- Vega S. L.; Kwon M. Y.; Song K. H.; Wang C.; Mauck R. L.; Han L.; Burdick J. A. Combinatorial hydrogels with biochemical gradients for screening 3D cellular microenvironments. Nat. Commun. 2018, 9 (1), 614. 10.1038/s41467-018-03021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng G.; Bhalla U. S.; Iyengar R. Complexity in biological signaling systems. Science 1999, 284 (5411), 92–96. 10.1126/science.284.5411.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. E.; Anseth K. S. Spatiotemporal hydrogel biomaterials for regenerative medicine. Chem. Soc. Rev. 2017, 46, 6532–6552. 10.1039/C7CS00445A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick J. A.; Mauck R. L.; Gerecht S. To serve and protect: hydrogels to improve stem cell-based therapies. Cell Stem Cell 2016, 18 (1), 13–15. 10.1016/j.stem.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Gill B. J.; West J. L. Modeling the tumor extracellular matrix: tissue engineering tools repurposed towards new frontiers in cancer biology. J. Biomech. 2014, 47 (9), 1969–1978. 10.1016/j.jbiomech.2013.09.029. [DOI] [PubMed] [Google Scholar]
- DeForest C. A.; Anseth K. S. Photoreversible patterning of biomolecules within click-based hydrogels. Angew. Chem., Int. Ed. 2012, 51 (8), 1816–1819. 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest C. A.; Tirrell D. A. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat. Mater. 2015, 14 (5), 523–531. 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- Kloxin C. J.; Scott T. F.; Bowman C. N. Stress relaxation via addition-fragmentation chain transfer in a thiol-ene photopolymerization. Macromolecules 2009, 42 (7), 2551–2556. 10.1021/ma802771b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. E.; Marozas I. A.; Anseth K. S. Amplified photodegradation of cell-laden hydrogels via an addition-fragmentation chain transfer reaction. Adv. Mater. 2017, 29 (11), 1605001. 10.1002/adma.201605001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. Y.; Kloxin C. J.; Abuelyaman A. S.; Oxman J. D.; Bowman C. N. Novel dental restorative materials having low polymerization shrinkage stress via stress relaxation by addition-fragmentation chain transfer. Dent. Mater. 2012, 28 (11), 1113–1119. 10.1016/j.dental.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandavarapu N. R.; Azagarsamy M. A.; Anseth K. S. Photo-click living strategy for controlled, reversible exchange of biochemical ligands. Adv. Mater. 2014, 26 (16), 2521–2526. 10.1002/adma.201304847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle C. E.; Lee T. Y.; Roper T. Thiol-enes: chemistry of the past with promise for the future. J. Polym. Sci., Part A: Polym. Chem. 2004, 42 (21), 5301–5338. 10.1002/pola.20366. [DOI] [Google Scholar]
- Hioe J.; Zipse H. Radical stability and its role in synthesis and catalysis. Org. Biomol. Chem. 2010, 8 (16), 3609–3617. 10.1039/c004166a. [DOI] [PubMed] [Google Scholar]
- Fairbanks B. D.; Schwartz M. P.; Bowman C. N.; Anseth K. S. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials 2009, 30 (35), 6702–6707. 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.; Smith Callahan L. A.; Hao J.; Guo K.; Wesdemiotis C.; Weiss R. A.; Becker M. L. Strain-promoted crosslinking of PEG-based hydrogels via copper-free cycloaddition. ACS Macro Lett. 2012, 1 (8), 1071–1073. 10.1021/mz3003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautry-Varsat A.; Ciechanover A.; Lodish H. F. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc. Natl. Acad. Sci. U. S. A. 1983, 80 (8), 2258–2262. 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbitt M. W.; Shadish J. A.; DeForest C. A.. Photopolymers for multiphoton lithography in biomaterials and hydrogels. In Multiphoton lithography: techniques, materials, and applications; Stampfl J., Liska R., Ovsianikov A., Eds.; Wiley Publishing: Weinheim, 2016; pp 183–220. [Google Scholar]
- Györfi A. H.; Matei A. E.; Distler J. H. W. Targeting TGF-β signaling for the treatment of fibrosis. Matrix Biol. 2018, 68–69, 8–27. 10.1016/j.matbio.2017.12.016. [DOI] [PubMed] [Google Scholar]
- Schultz G. S.; Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair. Regen. 2009, 17 (2), 153–162. 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- Wipff P.-J.; Rifkin D. B.; Meister J.-J.; Hinz B. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J. Cell Biol. 2007, 179 (6), 1311–1323. 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi Z.; Feng Z.; Chapnick D. A.; Dahl M.; Deng D.; Klipp E.; Moustakas A.; Liu X. Quantitative analysis of transient and sustained transforming growth factor-β signaling dynamics. Mol. Syst. Biol. 2011, 7, 492-1–492-12. 10.1038/msb.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson R. A.; Liu X. Association of v-ErbA with Smad4 disrupts TGF-beta signaling. Mol. Biol. Cell 2009, 20 (5), 1509–1519. 10.1091/mbc.e08-08-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguado B. A.; Grim J. C.; Rosales A. M.; Watson-Capps J. J.; Anseth K. S. Engineering precision biomaterials for personalized medicine. Sci. Transl. Med. 2018, 10 (424), eaam8645. 10.1126/scitranslmed.aam8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.