Abstract
Background
High-altitude polycythemia (HAPC) is a chronic high-altitude disease that can lead to an increase in the production of red blood cells in the people who live in the plateau, a hypoxia environment, for a long time. The most frequent symptoms of HAPC include headache, dizziness, breathlessness, sleep disorders, and dilation of veins. Although chronic hypoxia is the main cause of HAPC, the fundamental pathophysiologic process and related molecular mechanisms responsible for its development remain largely unclear yet.
Aim/methods
This study aimed to explore the related hereditary factors of HAPC in the Chinese Han and Tibetan populations. A total of 140 patients (70 Han and 70 Tibetan) with HAPC and 60 healthy control subjects (30 Han and 30 Tibetan) were recruited for a case-control association study. To explore the genetic basis of HAPC, we investigated the association between HAPC and both phosphatidylinositol-4,5-bisphosphonate 3-kinase, catalytic subunit delta gene (PIK3CD) and collagen type IV α3 chain gene (COL4A3) in Chinese Han and Tibetan populations.
Results/conclusion
Using the unconditional logistic regression analysis and the false discovery rate (FDR) calculation, we found that eight SNPs in PIK3CD and one SNP in COL4A3 were associated with HAPC in the Tibetan population. However, in the Han population, we did not find any significant association. Our study suggested that polymorphisms in the PIK3CD and COL4A3 were correlated with susceptibility to HAPC in the Tibetan population.
Keywords: High-altitude polycythemia, PIK3CD, COL4A3
Introduction
High-altitude polycythemia (HAPC) is a chronic high-altitude disease, characterized by excessive erythrocytosis. The clinical HAPC is diagnosed by a hemoglobin concentration ≥ 19 g/dL for females and ≥ 21 g/dL for males, according to the criteria established in the VI World Congress on Mountain Medicine and High-altitude Physiology in 2004 [1]. More than 140 million people are living at high altitudes above 2500 m worldwide, majorly in the Andes, Ethiopian Highlands, and Qinghai-Tibet Plateau [2]. The Qinghai-Tibet Plateau is the highest plateau in the world, which covers a large area with low oxygen in natural environment, and millions of people are living and working in this region. It is well known that the body’s hemoglobin concentration increases due to the hypoxic environment of high altitude, and therefore, this response is crucial for people who adapt to live at high altitudes. Some studies show that a number of populations suffer from chronic mountain sickness because they stay long at high altitudes [3]. HAPC mainly leads to a significant increase in blood viscosity, causing damage to microcirculatory and immune response disturbances such as vascular thrombosis, extensive organ damage, and sleep disorders [4, 5]. It is reported that the prevalence of HAPC in the Qinghai-Tibet Plateau is around 5 to 18% [1], and the prevalence of HAPC increases with the altitude. As the construction of the Qinghai-Tibet Railway has been completed, a number of Han populations migrate to Tibet. The incidence of HAPC among immigrants is significantly higher than the high-altitude natives [6]. As the Tibetan population keeps genetic adaptations, they can easily adapt to the high-altitude hypoxia environment, for example, showing lower hemoglobin levels and lower hematocrit. Many studies have noted that there are some significant differences in the genomes between immigrants and high-altitude natives, which indicates that genetic factors may contribute to the development of HAPC, although the molecular mechanisms and pathogenesis are still under study. In our study, we aimed to investigate the associations between susceptibility to HAPC and two new candidate genes that are related to the oxygen metabolism in red blood cells but have not been reported before.
The first candidate, PIK3CD, encodes the p110δ catalytic subunit of phosphoinositide 3-kinaseδ (PI3Kδ), a member of a big family of metalloenzymes. PI3Kδ is a heterodimer comprising the p110δ and p85 family regulatory subunit and expressed predominantly in leukocytes. Therefore, it plays an important role in the proliferation, survival, and activation of leukocytes [7–9]. The expression pattern and functions of PIK3CD are very important in PI3K/Akt pathway. Recently, research studies revealed that PI3K/Akt mediated the stabilization of HIF-1α (hypoxia-inducible factors-1α) [10], and it was also involved in the increase of HIF-1α protein level [11]. Meanwhile, HIF-1α plays an important role in transcriptionally upregulating erythropoietin (EPO) in hypoxia and affecting the amount of red blood cells [12].
The second candidate, COL4A3, encodes a subunit of type IV collagen that is a structural protein of the alveolar extracellular matrix (ECM) and mostly found in the kidney, lung, and basement membranes. It is located at 2q35-q3 and mainly contains 51 exons [13]. Type IV collagen is involved in various physiological conditions, including aging, diabetes, kidney disease, scarring, and pulmonary fibrosis [14]. The ECM is important to the structure and function of cell types. It contributes to many processes, such as cellular proliferation, differentiation, migration, and apoptosis [15].
Results
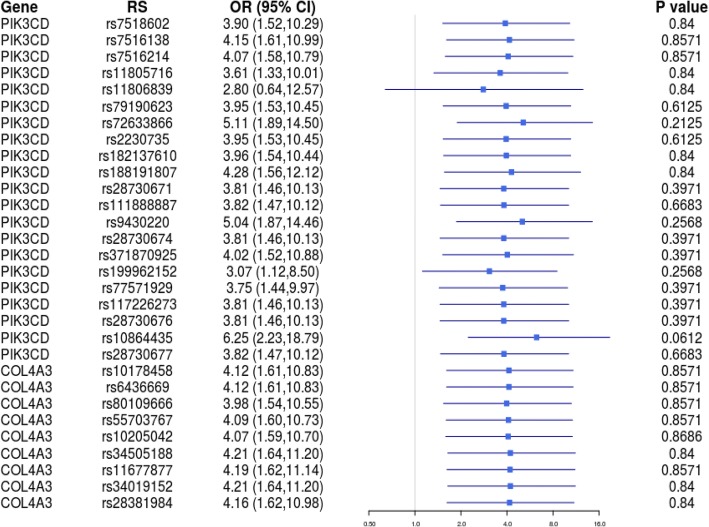
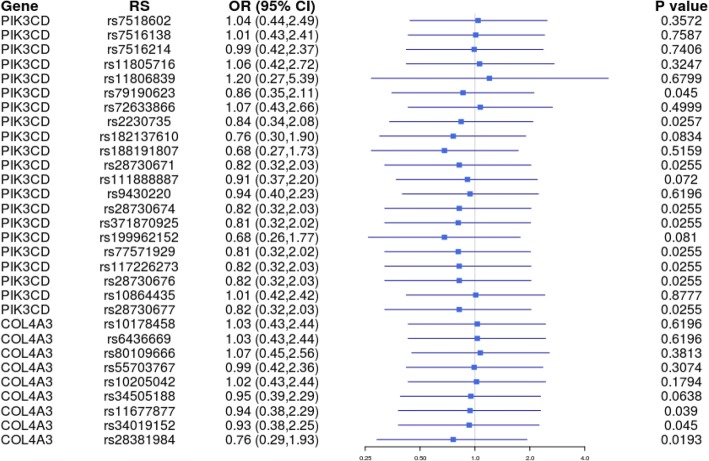
The demographics of HAPC patients and controls are shown in Table 1. The basic characteristics of candidate SNPs in the Han and Tibetan subjects are summarized in Table 2 (Fig. 1) and Table 3 (Fig. 2). We analyzed the associations between SNPs and HAPC using unconditional logistic regression analysis. In the Han population, rs72633866 (P1 = 0.033 before adjustment and P2 = 0.014 after adjustment for age), rs9430220 (P1 = 0.081 and P2 = 0.029), rs199962152 (P1 = 0.024 and P2 = 0.034), and rs10864435 (P1 = 0.013 and P2 = 0.002) in PIK3CD were significantly associated with HAPC. In the Tibetan subjects, rs2230735 (P1 = 0.008 and P2 = 0.008), rs28730671 (P1 = 0.007 and P2 = 0.007), rs111888887 (P1 = 0.034 and P2 = 0.034), rs28730674 (P1 = 0.007 and P2 = 0.007), rs371870925 (P1 = 0.007 and P2 = 0.007), rs199962152 (P1 = 0.045 and P2 = 0.040), rs77571929 (P1 = 0.005 and P2 = 0.005), rs117226273 (P1 = 0.007 and P2 = 0.007), rs28730676 (P1 = 0.007 and P2 = 0.007), and rs28730677 (P1 = 0.007 and P2 = 0.007) in PIK3CD were significantly associated with HAPC. Furthermore, rs34505188 (P1 = 0.028 and P2 = 0.028), rs11677877 (P1 = 0.013 and P2 = 0.013), rs34019152 (P1 = 0.018 and P2 = 0.018), and rs28381984 (P1 = 0.001 and P2 = 0.001) in COL4A3 were associated with HAPC.
Table 1.
Demographics of the control individuals and patients with high-altitude polycythemia
| Variables | Han | Tibetan | ||
|---|---|---|---|---|
| Case (n = 70) | Control (n = 30) | Case (n = 70) | Control (n = 30) | |
| Male | 35 | 15 | 35 | 15 |
| Female | 35 | 15 | 35 | 15 |
Table 2.
Basic information of candidate SNPs in Han subjects
| SNP_ID | Gene | Alleles A/B | Case (N) | Control (N) | OR (95% CI) | P | P1 | P2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | AA | AB | BB | |||||||
| rs7518602 | PIK3CD | C/T | 4 | 16 | 50 | 0 | 6 | 23 | 3.899 (1.515–10.290) | 0.961 | 0.261 | 0.425 |
| rs7516138 | PIK3CD | G/A | 4 | 19 | 47 | 0 | 11 | 18 | 4.150 (1.615–10.989) | 0.993 | 0.959 | 0.800 |
| rs7516214 | PIK3CD | G/A | 4 | 19 | 46 | 0 | 11 | 18 | 4.070 (1.582–10.786) | 0.993 | 0.924 | 0.829 |
| rs11805716 | PIK3CD | T/C | 13 | 17 | 32 | 3 | 9 | 14 | 3.610 (1.335–10.014) | 0.961 | 0.516 | 0.488 |
| rs11806839 | PIK3CD | G/C | 7 | 11 | 12 | 2 | 6 | 6 | 2.800 (0.640–12.570) | 0.993 | 0.627 | 0.616 |
| rs79190623 | PIK3CD | C/T | 63 | 7 | 0 | 23 | 6 | 0 | 3.952 (1.533–10.448) | 0.766 | 0.160 | 0.245 |
| rs72633866 | PIK3CD | G/A | 64 | 4 | 0 | 22 | 6 | 0 | 5.107 (1.892–14.498) | 0.236 | 0.033 | 0.014 |
| rs2230735 | PIK3CD | A/G | 63 | 7 | 0 | 23 | 6 | 0 | 3.952 (1.533–10.448) | 0.766 | 0.160 | 0.245 |
| rs182137610 | PIK3CD | A/C | 63 | 7 | 0 | 24 | 5 | 0 | 3.961 (1.540–10.444) | 0.961 | 0.321 | 0.510 |
| rs188191807 | PIK3CD | G/A | 57 | 3 | 0 | 23 | 3 | 0 | 4.280 (1.564–12.125) | 0.961 | 0.287 | 0.486 |
| rs28730671 | PIK3CD | C/T | 65 | 5 | 0 | 23 | 6 | 0 | 3.808 (1.463–10.129) | 0.509 | 0.061 | 0.129 |
| rs111888887 | PIK3CD | T/C | 65 | 5 | 0 | 24 | 5 | 0 | 3.816 (1.471–10.117) | 0.821 | 0.141 | 0.312 |
| rs9430220 | PIK3CD | T/C | 37 | 28 | 4 | 9 | 18 | 2 | 5.044 (1.871–14.464) | 0.245 | 0.081 | 0.029 |
| rs28730674 | PIK3CD | A/G | 65 | 5 | 0 | 23 | 6 | 0 | 3.808 (1.463–10.129) | 0.509 | 0.061 | 0.129 |
| rs371870925 | PIK3CD | T/C | 65 | 3 | 0 | 24 | 5 | 0 | 4.016 (1.520–10.884) | 0.509 | 0.050 | 0.112 |
| rs199962152 | PIK3CD | A/G | 63 | 2 | 0 | 22 | 5 | 0 | 3.065 (1.118–8.502) | 0.245 | 0.024 | 0.034 |
| rs77571929 | PIK3CD | T/C | 64 | 5 | 0 | 23 | 6 | 0 | 3.737 (1.439–9.970) | 0.509 | 0.065 | 0.132 |
| rs117226273 | PIK3CD | G/T | 65 | 5 | 0 | 23 | 6 | 0 | 3.808 (1.439–10.129) | 0.509 | 0.061 | 0.129 |
| rs28730676 | PIK3CD | T/C | 65 | 5 | 0 | 23 | 6 | 0 | 3.808 (1.436–10.129) | 0.509 | 0.061 | 0.129 |
| rs10864435 | PIK3CD | C/T | 63 | 7 | 0 | 20 | 9 | 0 | 6.247 (2.235–18.787) | 0.051 | 0.013 | 0.002 |
| rs28730677 | PIK3CD | G/A | 65 | 5 | 0 | 24 | 5 | 0 | 3.816 (1.471–10.117) | 0.821 | 0.141 | 0.312 |
| rs10178458 | COL4A3 | T/C | 0 | 9 | 61 | 0 | 4 | 25 | 4.116 (1.069–10.826) | 0.993 | 0.900 | 0.803 |
| rs6436669 | COL4A3 | A/G | 0 | 9 | 61 | 0 | 4 | 25 | 4.116 (1.069–10.826) | 0.993 | 0.900 | 0.803 |
| rs80109666 | COL4A3 | G/A | 49 | 19 | 2 | 22 | 7 | 0 | 3.984 (1.545–10.552) | 0.993 | 0.433 | 0.707 |
| rs55703767 | COL4A3 | G/T | 53 | 15 | 2 | 21 | 7 | 1 | 4.088 (1.600–10.732) | 0.993 | 0.732 | 0.762 |
| rs10205042 | COL4A3 | C/T | 2 | 15 | 53 | 1 | 7 | 21 | 4.073 (1.592–10.704) | 0.993 | 0.732 | 0.869 |
| rs34505188 | COL4A3 | G/A | 46 | 19 | 5 | 18 | 11 | 0 | 4.214 (1.637–11.199) | 0.993 | 0.787 | 0.587 |
| rs11677877 | COL4A3 | A/G | 46 | 19 | 5 | 17 | 12 | 0 | 4.187 (1.625–11.138) | 0.993 | 0.997 | 0.725 |
| rs34019152 | COL4A3 | G/A | 46 | 19 | 5 | 18 | 11 | 0 | 4.214 (1.637–11.199) | 0.993 | 0.787 | 0.587 |
| rs28381984 | COL4A3 | C/T | 27 | 32 | 11 | 9 | 16 | 4 | 4.160 (1.623–10.984) | 0.993 | 0.711 | 0.599 |
SNP single-nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval, P value FDR-calculated P value, P1 P value calculated by unconditional logistic regression analysis, P2 P value adjusted for age
Fig. 1.
Forest plots for the ORs in the Han population
Table 3.
Basic information of candidate SNPs in Tibetan subjects
| SNP_ID | Gene | Alleles A/B | Case (N) | Control (N) | OR (95% CI) | P | P1 | P2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | AA | AB | BB | |||||||
| rs7518602 | PIK3CD | C/T | 5 | 29 | 36 | 1 | 10 | 19 | 1.043 (0.438–2.490) | 0.612 | 0.240 | 0.238 |
| rs7516138 | PIK3CD | G/A | 11 | 26 | 33 | 3 | 13 | 14 | 1.015 (0.428–2.411) | 0.992 | 0.735 | 0.733 |
| rs7516214 | PIK3CD | G/A | 11 | 26 | 32 | 3 | 13 | 14 | 0.992 (0.416–2.366) | 0.982 | 0.688 | 0.691 |
| rs11805716 | PIK3CD | T/C | 15 | 27 | 17 | 6 | 8 | 13 | 1.061 (0.416–2.718) | 0.555 | 0.207 | 0.206 |
| rs11806839 | PIK3CD | G/C | 5 | 7 | 4 | 2 | 9 | 3 | 1.205 (0.270–5.393) | 0.893 | 0.593 | 0.612 |
| rs79190623 | PIK3CD | C/T | 58 | 10 | 1 | 18 | 11 | 1 | 0.862 (0.349–2.106) | 0.081 | 0.018 | 0.018 |
| rs72633866 | PIK3CD | G/A | 57 | 8 | 1 | 25 | 2 | 0 | 1.071 (0.434–2.656) | 0.791 | 0.366 | 0.367 |
| rs2230735 | PIK3CD | A/G | 59 | 10 | 1 | 17 | 12 | 1 | 0.844 (0.337–2.079) | 0.046 | 0.008 | 0.008 |
| rs182137610 | PIK3CD | A/C | 57 | 10 | 1 | 18 | 9 | 1 | 0.764 (0.301–1.897) | 0.150 | 0.050 | 0.045 |
| rs188191807 | PIK3CD | G/A | 53 | 5 | 1 | 22 | 5 | 0 | 0.685 (0.266–1.726) | 0.791 | 0.454 | 0.396 |
| rs28730671 | PIK3CD | C/T | 63 | 6 | 1 | 19 | 10 | 1 | 0.821 (0.324–2.035) | 0.046 | 0.007 | 0.007 |
| rs111888887 | PIK3CD | T/C | 61 | 6 | 1 | 21 | 8 | 1 | 0.906 (0.368–2.205) | 0.130 | 0.034 | 0.034 |
| rs9430220 | PIK3CD | T/C | 29 | 34 | 5 | 12 | 14 | 4 | 0.940 (0.395–2.234) | 0.851 | 0.536 | 0.535 |
| rs28730674 | PIK3CD | A/G | 63 | 6 | 1 | 19 | 10 | 1 | 0.821 (0.324–2.035) | 0.046 | 0.007 | 0.007 |
| rs371870925 | PIK3CD | T/C | 64 | 4 | 1 | 20 | 9 | 1 | 0.812 (0.320–2.017) | 0.046 | 0.007 | 0.007 |
| rs199962152 | PIK3CD | A/G | 58 | 4 | 1 | 19 | 6 | 1 | 0.685 (0.256–1.766) | 0.146 | 0.045 | 0.040 |
| rs77571929 | PIK3CD | T/C | 63 | 5 | 1 | 19 | 10 | 1 | 0.814 (0.320–2.024) | 0.046 | 0.005 | 0.005 |
| rs117226273 | PIK3CD | G/T | 63 | 6 | 1 | 19 | 10 | 1 | 0.821 (0.324–2.035) | 0.046 | 0.007 | 0.007 |
| rs28730676 | PIK3CD | T/C | 63 | 6 | 1 | 19 | 10 | 1 | 0.821 (0.324–2.035) | 0.046 | 0.007 | 0.007 |
| rs10864435 | PIK3CD | C/T | 55 | 15 | 0 | 25 | 4 | 1 | 1.011 (0.423–2.417) | 0.992 | 0.879 | 0.878 |
| rs28730677 | PIK3CD | G/A | 63 | 6 | 1 | 19 | 10 | 1 | 0.821 (0.324–2.035) | 0.046 | 0.007 | 0.007 |
| rs10178458 | COL4A3 | T/C | 4 | 21 | 45 | 4 | 7 | 19 | 1.026 (0.432–2.442) | 0.851 | 0.539 | 0.537 |
| rs6436669 | COL4A3 | A/G | 4 | 21 | 45 | 4 | 7 | 19 | 1.026 (0.432–2.442) | 0.851 | 0.539 | 0.537 |
| rs80109666 | COL4A3 | G/A | 60 | 9 | 1 | 28 | 2 | 0 | 1.070 (0.449–2.559) | 0.655 | 0.271 | 0.267 |
| rs55703767 | COL4A3 | G/T | 47 | 21 | 2 | 18 | 8 | 4 | 0.990 (0.415–2.360) | 0.524 | 0.184 | 0.184 |
| rs10205042 | COL4A3 | C/T | 0 | 23 | 47 | 4 | 8 | 18 | 1.017 (0.425–2.437) | 0.305 | 0.102 | 0.102 |
| rs34505188 | COL4A3 | G/A | 42 | 20 | 8 | 10 | 14 | 6 | 0.948 (0.390–2.292) | 0.115 | 0.028 | 0.028 |
| rs11677877 | COL4A3 | A/G | 42 | 21 | 7 | 10 | 13 | 7 | 0.941 (0.385–2.290) | 0.070 | 0.013 | 0.013 |
| rs34019152 | COL4A3 | G/A | 42 | 20 | 8 | 10 | 13 | 7 | 0.928 (0.380–2.254) | 0.081 | 0.018 | 0.018 |
| rs28381984 | COL4A3 | C/T | 24 | 39 | 7 | 23 | 6 | 1 | 0.761 (0.294–1.928) | 0.035 | 0.001 | 0.001 |
The abbreviations were the same as Table 2
Fig. 2.
Forest plots for the ORs in the Tibetan population
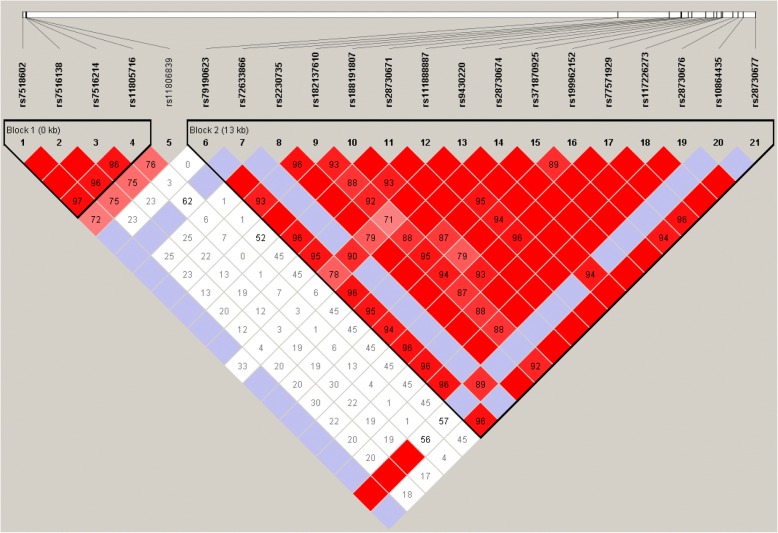
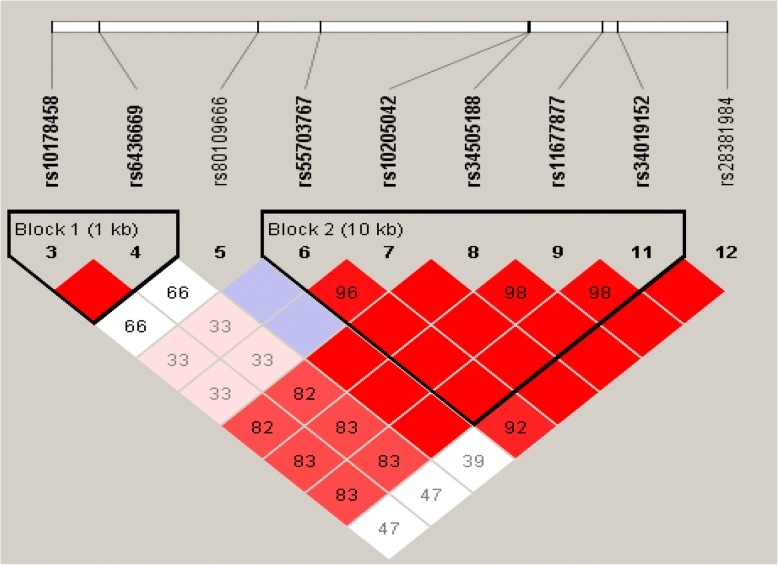
After using FDR to correct for multiple comparisons, in the Tibetan subjects, we found that rs2230735 (OR = 0.844, 95% CI = 0.337–2.079, P = 0.046), rs28730671 (OR = 0.821, 95% CI = 0.324–2.035, P = 0.046), rs28730674 (OR = 0.821, 95% CI = 0.324–2.035, P = 0.046), rs371870925 (OR = 0.812, 95% CI = 0.320–2.017, P = 0.046), rs77571929 (OR = 0.814, 95% CI = 0.320–2.024, P = 0.046), rs117226273 (OR = 0.821, 95% CI = 0.324–2.035, P = 0.046), rs28730676 (OR = 0.821, 95% CI = 0.324–2.035, P = 0.046), rs28730676 (OR = 0.821, 95% CI = 0.324–2.035, P = 0.046), and rs28730677 (OR = 0.821, 95% CI = 0.324–2.035, P = 0.046) in PIK3CD were significantly associated with HAPC. Furthermore, rs28381984 (OR = 0.761, 95% CI = 0.294–1.928, P = 0.035) in COL4A3 was associated with HAPC in the Tibetan population. But in the Han population, we did not find any significant association. In addition, using haplotype analysis, two blocks were detected among the PIK3CD SNPs (Fig. 3): block 1 contains rs7518602, rs7516138, rs7516214, and rs11805716 and block 2 contains rs79190623, rs72633866, rs2230735, rs182137610, rs188191807, rs28730671, rs111888887, rs9430220, rs28730674, rs371870925, rs199962152, rs77571929, rs117226273, rs28730676, rs10864435, and rs28730677. Two blocks were detected among the COL4A3 SNPs too (Fig. 4): block 1 contains rs10178458 and rs6436669 and block 2 contains rs55703767, rs10205042, rs34505188, rs11677877, and rs34019152. These SNPs within the same genes showed strong linkage in-between.
Fig. 3.
Haplotype block map for the 15 PIK3CD SNPs
Fig. 4.
Haplotype block map for the 21 COL4A3 SNPs
Discussion
Tibet covers a vast area with a harsh hypoxic natural environment. According to a report in 2006, approximately 12 million people permanently settled down in this region. This number constantly increases every year; the increase mainly comes from the Han population that are emigrating from plain areas [16]. HAPC is a serious disease that threatens the health of people in the plateau area, especially those who have emigrated from a low-altitude area. In the past, a large number of patients with HAPC have been investigated with a focus on the pathophysiologic mechanisms of this disease. Nevertheless, a lot of questions remain to be elucidated. With the completion of the human genome project, much research has been shifted to human genetic variation, which was one of NIH’s Roadmap Initiatives for 2008 [17]. As we all know, in Tibet, in order to adapt to altitude hypoxia, the body increases the hemoglobin concentration to increase the efficiency of carrying oxygen, and this response is crucial for the Han population who adapt to live at high altitudes. Compared with the Han people, the Tibetan population keeps genetic adaptations; they can easily adapt to the high-altitude hypoxia environment. Several studies have indicated that natural selection associated with high-altitude adaptation appears to act on genes in the hypoxic response pathway to regulate erythrocyte production, possibly to prevent or reduce erythrocyte growth [18–22]. Recently, significant progress has been made in the study of the genetic basis of HAPC in Tibetans and Han, and some studies have confirmed that many genes are associated with HAPC. Namely, integrin subunit alpha 6 (ITGA6), erb-b2 receptor tyrosine kinase 4 (ERBB4), EPH receptor A2 (EPHA2), angiotensinogen (AGT), and endothelial PAS domain protein 1 (EPAS1) have been reported to play important roles in HAPC in Tibetans and Han [23–26]. Of these genes, EPHA2 can affect erythrocyte production by regulating EPO production and EPAS1 has been implicated as making the greatest contribution to genetic adaptation to high altitude and to the low Hb concentrations observed in the Tibetan population [18, 21]. Some research have shown that genetic variants selected for adaptation at extreme environmental conditions not only increase cancer risk later on age but may also be the downregulation of erythropoiesis in Tibetans in high altitude [22, 27, 28]. In our study, we found rs2230735, rs28730671, rs28730674, rs371870925, rs77571929, rs117226273, rs28730676, rs28730677, and rs28381984 in PIK3CD and COL4A3 were significantly associated with decreased HAPC risk in the Tibetan population. However, in the Han population, we did not find any significant association. Inspired by the development of genome research and the genetic findings of high-altitude natives, we consider that genetic factors may be involved in the formation of such kind of disease. Our study revealed an association of HAPC with SNPs in PIK3CD and COL4A3 in the Tibetan population.
PIK3CD gene encodes P110δ catalytic subunit that is expressed predominantly in leukocytes and plays a vital role in the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway. According to recent reports, p110δ contributes to the activation of Akt and cell proliferation in primary AML (acute myeloid leukemia) cells [29, 30]. PI3K signaling contributes to many processes, including cell cycle progression, proliferation and differentiation, survival, and migration [8, 9, 31]. PI3K/Akt pathways are critical to HIF-1α transcriptional activity in hypoxia. HIF-1, which is basically a heterodimer transcription factor, composed of HIF-1α and HIF-1β subunits, serves as a central regulator of metabolic adaptation to low oxygen [32]. The HIF-1α subunit is stabilized under hypoxia, translocating to the nucleus, forming a heterodimer with HIF-1β, and transactivating its target genes including EPO. HIF-1α is a factor that was originally thought to be bound to the 3′ enhancer region of the EPO genes, controlling for 100–200 genes that are involved in angiogenesis, glycolysis, and erythropoiesis [33]. The main organ for EPO production is the liver during the fetus stage, whereas it becomes the kidney after birth. However, there is small amount of expression in the other organs of the body, such as the brain, spleen, lungs, testis, and placenta. Further, it is a necessary glycoprotein, which does not only promote the maturation of red blood cell from erythroid progenitors but also mediates erythropoiesis. It is identified as an inducer of erythropoiesis and can promote excessive cell production. In addition, previous studies showed that inhibited PI3K/Akt signaling pathway led to decreased hematopoietic stem cell (HSC) proliferation. This suggests that such kind of pathway is important for HSC proliferation. HSC is involved in the formation of HAPC, expansion of the population, and enforcement of erythroid lineage-committed differentiation [34]. Therefore, we speculate that PIKCD may affect the generation of EPO and the decrease of HSC appreciation through the PI3K/Akt signaling pathway. In this study, we show here for the first time that the PIK3CD gene plays a crucial role in the production of erythrocyte, so PIK3CD has a significant influence on the formation of HAPC.
COL4A3 is an important risk gene for HAPC that is also linked to many diseases such as the Alport syndrome, focal segmental glomerulosclerosis, and type 2 diabetes [35–37]. Furthermore, it is important to the structure and function of various cell types and contributes to a variety of processes. Although the functional effects of the polymorphisms have not yet been elucidated fully, our current results show that the variants may have an effect on COL4A3 expression or activity. Therefore, it may play an important role in modulating the susceptibility to HAPC. By searching the KEGG pathway database, we found that COL4A3 can bind to receptors on cell surface and promote the activation of PI3K/Akt. Under hypoxic conditions, this pathway can promote the production of hypoxia-inducible factor and increase the cell cycle, and thereby promote the increase of EPO and the amount of red blood cells. Therefore, it is speculated that COL4A3 may affect the production of EPO through the PI3K/Akt signaling pathway, thus affecting the production of red blood cells. Consequently, COL4A3 gene may be a useful marker for the formation of HAPC. We also show here for the first time that the COL4A3 gene plays a crucial role in the production of erythrocyte. Meanwhile, based on the results of our research, COL4A3 was significantly associated with erythropoiesis in hypoxia. It is suggested that gene polymorphisms may be relevant to the susceptibility to HAPC.
The genome research era has also opened the road to studying the basis of susceptibility to chronic mountain sickness (CMS) [38]. Gene polymorphisms have set the platform for the analysis of the molecular mechanisms of adaptation to life at high altitudes [39]. Tibet, an average elevation above 4000 m, is commonly regarded as the “Roof of the World” and has a unique genetic background and dietary and lifestyle habits. In this study, we have suggested that several genetic polymorphisms are associated with susceptibility to HAPC and each polymorphism may contribute to only a small relative risk of HAPC. It shows a complex interplay between exposure to hypoxic environmental stimuli and genetic background. There are important discoveries revealed by the studies, but there are still a lot of limitations. Due to these limitations, the study power of this paper is limited. On the other hand, the functions of the genetic variants and their mechanisms have not been evaluated in this study. In a following study, we will use animal models to verify the experimental results, to more clearly illustrate how the two genes affect erythrocytosis, which signaling pathways are involved in the formation of the disease, and to try to elucidate the functions of the genetic variants and mechanisms with HAPC.
Conclusion
We analyzed SNPs in PIK3CD and COL4A3 and identified a relationship between genetic polymorphisms and HAPC in the Tibetan people. This study sets out to improve the quality of life of people living in the Qinghai-Tibet Plateau, determines paramount insights into the etiology of HAPC, and may provide more guidance for such people with regard to prolonged and healthy living. However, additional genetic risk factors and functional investigations should be identified in order to further confirm our results.
Materials and methods
Study populations
A total of 140 patients (70 Han and 70 Tibetan) with HAPC and 60 healthy control subjects (30 Han and 30 Tibetan) were recruited for a case-control association study. The 200 subjects who participated in this research had resided at an altitude of above 4000 m, and these samples were collected from the General Hospital in Tibet Military Region and the second People’s Hospital of Tibet Autonomous Region. Written informed consent was obtained from each individual. Patients met the diagnostic criteria for HAPC, i.e., males with hemoglobin ≥ 21 g/dL or females with hemoglobin ≥ 19 g/dL, and had no high-altitude cerebral edema and chronic respiratory disorders or secondary polycythemia due to hypoxemia caused by certain chronic diseases. Moreover, subjects have no endocrinological, nutritional, and metabolic diseases. Healthy individuals were randomly selected as controls. The experimental protocol was established by the Ethics Committee of the Xizang Minzu University.
Epidemiological and clinical data
We used a standardized epidemiological questionnaire to collect demographic and clinical data, including information on gender, age, residential region, ethnicity, family history of cancer, and education status. Furthermore, the patient information was collected through physicians or from medical chart review. All participants signed informed consent, and 5 ml of peripheral blood was taken from each participant in this study.
Selection of SNPs and methods of genotyping
Thirty SNPs from PIK3CD and COL4A3 were chosen for analysis in this study, including 21 SNPs in PIK3CD and 9 SNPs in COL4A3 with minor allele frequency (MAF) > 0.05 in the Asian population HapMap database, and SNP genotyping was performed utilizing Illumina sequencing platform for exon sequencing of PIKCD and COL4A3. Because the genetic background of Han and Tibetan populations has not been compared yet, we selected these two candidate genes based on their relations to the oxygen metabolism in red blood cells, which were related to high-altitude adaptation in the Chinese Han and Tibetan populations.
Statistical analysis
The data were analyzed using an R program, Haploview, and Excel. Unconditional logistic regression analysis was used to calculate odds ratios (ORs), 95% confidence intervals (CIs), and P values for comparisons between cases and controls. Multiple comparisons were corrected using FDR, and FDR-corrected P < 0.05 was considered to indicate a significant difference.
Acknowledgements
We are grateful to those who collected samples in the Tibetan Plateau, and we thank those who participated in the study and worked hard on the research. This work was supported by the National Natural Science Foundation of China (No. 31460286; 31660307; 31260252; 31330038), the Natural Science Foundation of Xizang (Tibet) Autonomous Region (No. Z2014A09G2-3), the Innovation Support Program for Young Teachers of Tibet Autonomous Region (No. QCZ2016-27; QCZ2016-29; QCZ2016-34), and the Science and Technology Department Project of Tibet Autonomous Region (No. 2016ZR-MQ-06; 2015ZR- 13-19).
Funding
This work was supported by the National Natural Science Foundation of China (No. 31460286; 31660307; 31260252; 31330038), the Natural Science Foundation of Xizang (Tibet) Autonomous Region (No. Z2014A09G2-3), the Innovation Support Program for Young Teachers of Tibet Autonomous Region (No. QCZ2016-27; QCZ2016-29; QCZ2016-34), and the Science and Technology Department Project of Tibet Autonomous Region (No. 2016ZR-MQ-06; 2015ZR- 13-19).
Abbreviations
- 95%CI
95% confidence intervals
- AGT
Angiotensinogen
- AML
Acute myeloid leukemia
- CMS
Chronic mountain sickness
- COL4A3
Collagen type IV α3 chain gene
- ECM
Extracellular matrix
- EPAS1
Endothelial PAS domain protein 1
- EPHA2
EPH receptor A2
- ERBB4
Erb-b2 receptor tyrosine kinase 4
- FDR
False discovery rate
- HAPC
High-altitude polycythemia
- HIF-1α
Hypoxia-inducible factors-1α
- HSC
Hematopoietic stem cell
- ITGA6
Integrin subunit alpha 6
- OR
Odds ratio
- PIK3CD
Catalytic subunit delta gene
Authors’ contributions
LK conceived and designed the study, supervised the project, and drafted the manuscript. XF participated in the design of study and data analysis and helped to draft the manuscript. LM, ZZhang, YL, MH, LL, YZ, FL, PC, and YL contributed to the sample collection and experiments. XL helped in the English language editing. ZZhao contributed to sample collection and experiments. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Ministry of Medicine of Xizang Minzu University, ethical approval of biomedical ethics research project number 201801.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Xiaowei Fan, Lifeng Ma and Zhiying Zhang contributed equally to this work.
Contributor Information
Xiaowei Fan, Email: 13582173987@163.com.
Lifeng Ma, Email: 316454760@qq.com.
Zhiying Zhang, Email: 630352985@qq.com.
Yi Li, Email: liyistat@gmail.com.
Meng Hao, Email: Haombio@gmail.com.
Zhipeng Zhao, Email: 15110700046@fudan.edu.cn.
Yiduo Zhao, Email: 1330550039@qq.com.
Fang Liu, Email: 3916309@qq.com.
Lijun Liu, Email: 29354889@qq.com.
Xingguang Luo, Email: 563647380@qq.com.
Peng Cai, Email: 499535455@qq.com.
Yansong Li, Email: 411463682@qq.com.
Longli Kang, Email: longli_kang@163.com.
References
- 1.Leon-Velarde F, et al. Consensus statement on chronic and subacute high altitude diseases. High Alt Med Biol. 2005;6(2):147–157. doi: 10.1089/ham.2005.6.147. [DOI] [PubMed] [Google Scholar]
- 2.Otten, E.J., High altitude: an exploration of human adaptation.: edited by Hornbein TF and Schoene RB. New York, Marcel Dekker, Inc. 2001, 982 pages, $235. J Emerg Med, 2003. 25(3): p. 345-346.
- 3.Reeves JT, Leon-Velarde F. Chronic mountain sickness: recent studies of the relationship between hemoglobin concentration and oxygen transport. High Altitude Medicine & Biology. 2004;5(2):147. doi: 10.1089/1527029041352090. [DOI] [PubMed] [Google Scholar]
- 4.Guan W, et al. Sleep disturbances in long-term immigrants with chronic mountain sickness: a comparison with healthy immigrants at high altitude. Respir Physiol Neurobiol. 2015;206:4–10. doi: 10.1016/j.resp.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Jiang C, et al. Gene expression profiling of high altitude polycythemia in Han Chinese migrating to the Qinghai-Tibetan plateau. Mol Med Rep. 2012;5(1):287–293. doi: 10.3892/mmr.2011.632. [DOI] [PubMed] [Google Scholar]
- 6.Wu TY. Chronic mountain sickness on the Qinghai-Tibetan plateau. Chin Med. 2005;118(2):161–168. [PubMed] [Google Scholar]
- 7.Vanhaesebroeck B, et al. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc Natl Acad Sci U S A. 1997;94(9):4330. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chantry D, et al. p110delta, a novel phosphatidylinositol 3-kinase catalytic subunit that associates with p85 and is expressed predominantly in leukocytes. J Biol Chem. 1997;272(31):19236. doi: 10.1074/jbc.272.31.19236. [DOI] [PubMed] [Google Scholar]
- 9.Kok K, Geering B, Vanhaesebroeck B. Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem Sci. 2009;34(3):115. doi: 10.1016/j.tibs.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Gaber T, et al. Hypoxia inducible factor (HIF) in rheumatology: low O2! See what HIF can do! Ann Rheum Dis. 2005;64(7):971. doi: 10.1136/ard.2004.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dayan F, et al. A dialogue between the hypoxia-inducible factor and the tumor microenvironment. Cancer Microenvironmen. 2008;1(1):53–68. doi: 10.1007/s12307-008-0006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka T, Nangaku M. Recent advances and clinical application of erythropoietin and erythropoiesis-stimulating agents. Exp Cell Res. 2012;318(9):1068–1073. doi: 10.1016/j.yexcr.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 13.Stabuc-Silih M, et al. Polymorphisms in COL4A3 and COL4A4 genes associated with keratoconus. Mol Vis. 2009;15(300-01):2848–2860. [PMC free article] [PubMed] [Google Scholar]
- 14.Osman OS, et al. A novel method to assess collagen architecture in skin. Bmc Bioinformatics. 2013;14(1):1–10. doi: 10.1186/1471-2105-14-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan T, et al. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol Cell Biol. 2009;29(6):1575–1591. doi: 10.1128/MCB.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu T, Kayser B. High altitude adaptation in Tibetans. High Alt Med Biol. 2006;7(3):193. doi: 10.1089/ham.2006.7.193. [DOI] [PubMed] [Google Scholar]
- 17.Pennisi, E., Research funding. Are epigeneticists ready for big science? Science, 2008. 319(5867): p. 1177. [DOI] [PubMed]
- 18.Yi X, et al. Sequencing of fifty human exomes reveals adaptation to high altitude. Science. 2010;329(5987):75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford JE, et al. Natural selection on genes related to cardiovascular health in high-altitude adapted Andeans. Am J Hum Genet. 2017;101(5):752–767. doi: 10.1016/j.ajhg.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329(5987):72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 21.Beall CM, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107(25):11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigham A, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6(9):e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, et al. EPAS1 gene polymorphisms are associated with high altitude polycythemia in Tibetans at the Qinghai-Tibetan Plateau. Wilderness Environ Med. 2015;26(3):288–294. doi: 10.1016/j.wem.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, et al. An EPAS1 haplotype is associated with high altitude polycythemia in male Han Chinese at the Qinghai-Tibetan plateau. Wilderness Environ Med. 2014;25(4):392–400. doi: 10.1016/j.wem.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, et al. Associations of high altitude polycythemia with polymorphisms in EPAS1, ITGA6 and ERBB4 in Chinese Han and Tibetan populations. Oncotarget. 2017;8(49):86736. doi: 10.18632/oncotarget.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, et al. Associations of high altitude polycythemia with polymorphisms in EPHA2 and AGT in Chinese Han and Tibetan populations. Oncotarget. 2017;8(32):53234–53243. doi: 10.18632/oncotarget.18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macinnis MJ, Koehle MS, Rupert JL. Evidence for a genetic basis for altitude illness: 2010 update. High Alt Med Biol. 2010;11(4):349–368. doi: 10.1089/ham.2010.1030. [DOI] [PubMed] [Google Scholar]
- 28.Voskarides K. Combination of 247 genome-wide association studies reveals high cancer risk as a result of evolutionary adaptation. Mol Biol Evol. 2017;35(2):473–485. doi: 10.1093/molbev/msx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Billottet C, et al. A selective inhibitor of the p110|[delta]| isoform of PI 3-kinase inhibits AML cell proliferation and survival and increases the cytotoxic effects of VP16. Oncogene. 2006;25(50):6648. doi: 10.1038/sj.onc.1209670. [DOI] [PubMed] [Google Scholar]
- 30.Sujobert P, et al. Essential role for the p110delta isoform in phosphoinositide 3-kinase activation and cell proliferation in acute myeloid leukemia. Blood. 2005;106(3):1063. doi: 10.1182/blood-2004-08-3225. [DOI] [PubMed] [Google Scholar]
- 31.Vanhaesebroeck B, et al. p110δ, a novel phosphoinositide 3-kinase in leukocytes. Proc Natl Acad Sci U S A. 1997;94(9):4330. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, et al. PI3K/Akt is required for heat shock proteins to protect hypoxia-inducible factor 1α from pVHL-independent degradation. J Biol Chem. 2004;279(14):13506–13513. doi: 10.1074/jbc.M310164200. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka T, Nangaku M. Recent advances and clinical application of erythropoietin and erythropoiesis-stimulating agents. Exp Cell Res. 2012;318(9):1068. doi: 10.1016/j.yexcr.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 34.Li P, et al. Regulation of bone marrow hematopoietic stem cell is involved in high-altitude erythrocytosis. Exp Hematol. 2011;39(1):37–46. doi: 10.1016/j.exphem.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Jingyuan X, et al. COL4A3 mutations cause focal segmental glomerulosclerosis. J Mol Cell Biol. 2014;6(6):498–505. doi: 10.1093/jmcb/mju040. [DOI] [PubMed] [Google Scholar]
- 36.Guo L, et al. Mutation analysis of COL4A3 and COL4A4 genes in a Chinese autosomal-dominant Alport syndrome family. J Genet. 2017;96(2):389. doi: 10.1007/s12041-017-0786-7. [DOI] [PubMed] [Google Scholar]
- 37.Saravani S, et al. Association of COL4A3 (rs55703767), MMP-9 (rs17576) and TIMP-1 (rs6609533) gene polymorphisms with susceptibility to type 2 diabetes. Biomed Rep. 2017;6(3):329–334. doi: 10.3892/br.2017.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas PK, et al. Neurological manifestations in chronic mountain sickness: the burning feet-burning hands syndrome. J Neurol Neurosurg Psychiatry. 2000;69(4):447–452. doi: 10.1136/jnnp.69.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen J. DNA duplications and deletions help determine health. Science. 2007;317(5843):1315–1317. doi: 10.1126/science.317.5843.1315. [DOI] [PubMed] [Google Scholar]