Abstract
Background
Persons living with human immunodeficiency virus (PLWH) face an increased burden of chronic obstructive pulmonary disease (COPD). Repeated pulmonary infections, antibiotic exposures, and immunosuppression may contribute to an altered small airway epithelium (SAE) microbiome.
Methods
SAE cells were collected from 28 PLWH and 48 HIV- controls through bronchoscopic cytologic brushings. DNA extracted from SAE cells was subjected to 16S rRNA amplification and sequencing. Comparisons of alpha and beta diversity between HIV+ and HIV- groups were performed and key operational taxonomic units (OTUs) distinguishing the two groups were identified using the Boruta feature selection after Random Forest Analysis.
Results
PLWH demonstrated significantly reduced Shannon diversity compared with HIV- volunteers (1.82 ± 0.10 vs. 2.20 ± 0.073, p = 0.0024). This was primarily driven by a reduction in bacterial richness (23.29 ± 2.75 for PLWH and 46.04 ± 3.716 for HIV-, p < 0.0001). Phyla distribution was significantly altered among PLWH, with an increase in relative abundance of Proteobacteria (p = 0.0003) and a decrease in Bacteroidetes (p = 0.0068) and Firmicutes (p = 0.0002). Six discriminative OTUs were found to distinguish PLWH from HIV- volunteers, aligning to Veillonellaceae, Fusobacterium, Verrucomicrobiaceae, Prevotella, Veillonella, and Campylobacter.
Conclusions
Compared to HIV- controls, PLWH’s SAE microbiome is marked by reduced bacterial diversity and richness with significant differences in community composition.
Electronic supplementary material
The online version of this article (10.1186/s12931-018-0835-7) contains supplementary material, which is available to authorized users.
Keywords: HIV, Microbiome, Epithelium, COPD
Background
For the 35 million people living with human immunodeficiency virus (PLWH), antiretroviral therapy (ART) has been a lifesaving measure. As a result, AIDS-related morbidity and mortality have decreased markedly; however, aging with HIV has brought other challenges [1]. For instance, PLWH are more likely to develop chronic obstructive pulmonary disease (COPD) [2] and are also more likely to suffer from severe respiratory symptom burdens even after adjustment for smoking habits [3]. Although the pathogenesis of accelerated COPD in PLWH is poorly understood, the unique risk for pulmonary infections in this setting suggests that shifts in the lung microbiome might account for this phenomenon.
Investigations into the HIV lung microbiome have yielded interesting insights but no clear consensus. Lozupone et al. found that the abundance of Tropheryma whipplei was significantly increased in bronchoalveolar lavage (BAL) samples of PLWH compared with HIV- control subjects [4], while a second study by Beck et al. showed no differences between the two groups [5]. A third study also evaluating BAL demonstrated that PLWH who had advanced disease (CD4 cell counts < 500 cells/mm3) had significantly reduced microbiome diversity when compared to HIV- controls, with diversity starting to return to normal levels once ART was initiated [6]. While these studies have offered the first insights into the HIV lung microbiome, the reliance on BAL fluid may fail to identify important changes at the specific initial site of injury in the pathogenesis of COPD, namely the small airway [7]. The small airway epithelium (SAE) is the first line of defense against toxins such as cigarette smoke and microbial pathogens. In COPD, remodeling of this layer with squamous metaplasia, goblet cell hyperplasia, and breakdown of the epithelial barrier junction are critical to injury development [8]. Moreover, evidence that endotoxins produced by Staphylococcus aureus and Haemophilus influenza can damage epithelial barrier function suggests that an important relationship between the microbiome, epithelial injury, and COPD may exist [9]. Previous work by our group identified that within PLWH, the absence of Pasteurellaceae and Brachybacterium and the presence of Yersinia species in the SAE could help identify those with COPD [10]. Our study explores whether significant differences exist between the SAE microbiomes of PLWH and uninfected controls.
Methods
Study cohort
PLWH were drawn from the patient population at St. Paul’s Hospital in Vancouver, Canada, a tertiary care setting with an active bronchoscopy program and HIV outpatient clinic. Eligible PLWH were patients who were undergoing bronchoscopies for clinical purposes (i.e. for lung masses or nodules or to rule out infection) and were consented for additional research specimen collection during the procedure. All subjects were ≥ 19 years old and provided written informed consent under the University of British Columbia (UBC) Providence Health Care ethics protocol H14–03267. HIV- controls were recruited from patients undergoing lung cancer screening bronchoscopies at the British Columbia Cancer Agency in Vancouver, Canada. With the exception of seven HIV-infected patients who were lost to follow-up, all subjects underwent pre-bronchodilator spirometry according to guidelines provided by the American Thoracic Society and European Respiratory Society [11]. COPD was defined according to criteria outlined by the Global Initiative for Chronic Obstructive Lung Disease (GOLD) [12].
Sample collection
SAE cells were obtained via bronchoscopic cytologic brushings. Samples were obtained prior to the collection of clinical specimens and away from sites of disease as detected by chest computed tomography (CT) imaging performed within a month of the bronchoscopy. The bronchoscope was inserted in the oral cavity into the trachea and bronchi with minimal use of the suction channel to avoid contamination. Cytologic brushes were then directed in the subsegment of interest until resistance was encountered (in the 5th and 6th generation airways). Brushings were taken at that site and collected in Cytolyt (Cytyc, Marlborough, MA) for DNA preservation.
DNA extraction and PCR amplification
DNA was extracted using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Toronto, Ontario). Samples were eluted with 50 ul of distilled water and the DNA concentration was measured by NanoDrop (ThermoFisher Scientific, Waltham, MA). All samples were normalized to 12 ng/ul for downstream experiments. To quantify total bacteria load in each sample, primers specifying the 293 bp amplicon of the 16S rRNA gene were designed using the protocol outlined by Sze et a.l [13]. A pooled library consisting of all the samples with individually labeled indices was generated using the protocol adopted from a dual-index sequencing strategy published by Kozich et al. [14]. One exception to this protocol was that touchdown PCR was used to amplify the 16S rRNA gene fragments spanning the V4 region. PCR products were purified with Agencourt AMPure XP system (Beckman Coulter, Catalog #A63880). Sequencing was performed on the Illumina MiSeq™ platform (Illumina, Redwood City, CA, USA) with 2 × 250 paired end-read chemistry at the UBC Sequencing and Bioinformatics Consortium. Further details regarding the PCR amplification are provided in the supplement.
Microbiome profiling
Sequencing reads were merged, filtered for quality, and processed using the software mothur v.1.35.1 [15] according to the Standard Operating Procedure for MiSeq data (http://www.mothur.org). The accepted sequences were clustered into operational taxonomic units (OTUs) using the 97% identity threshold, and classified using the Ribosomal Database Project (RDP) Classifier [16] and the RDP taxonomy training set (http://rdp.cme.msu.edu). To account for potential sources of contamination, OTUs observed in the negative extraction controls (sterile water processed along with samples) were considered contaminants and removed from downstream analysis.
Statistical analysis
Alpha diversity measures (Richness, Shannon diversity index, and Evenness) [17, 18] were obtained using the Vegan package (V2.3.0) [19] in R (V3.2.0), available at https://www.r-project.org, and compared according to COPD or HIV status using the Mann-Whitney U test [18]. Beta diversity analyses based on Bray-Curtis distance matrix were performed, and differences in microbial communities between groups were tested by Permutational Multivariate Analysis of Variance (PERMANOVA) [20]. Nonmetric Multidimensional Scaling [NMDS] was employed as ordination method for visualization of Bray-Curtis dissimilarity values in both groups [21]. Comparisons of relative taxa abundance (at the phylum level) between groups were performed using the Mann-Whitney U test [18]. To identify the most discriminating OTUs between groups, we employed a random forest algorithm with Boruta feature selection [22, 23], and the Vegan package was used to construct abundance heatmaps.
Results
Study cohort
Demographic details of the study cohort are provided in Table 1. 28 PLWH (7 with COPD and 14 without COPD, 7 with unknown status) and 48 HIV- controls (24 with COPD and 24 without COPD) were enrolled. Males accounted for the majority of the HIV+ cohort (86%) compared to only half of the HIV- cohort. There were no statistically significant differences in lung function and in smoking status between the two cohorts. Nine (32%) of the PLWH had a detectable plasma HIV viral load and the mean CD4 count for this group was 419 cells/mm3. Only four (14%) of the PLWH cohort were not currently on ART.
Table 1.
Study Cohort Characteristics
| PLWH (n = 28) | HIV- (n = 48) | p-value | |
|---|---|---|---|
| Sex | |||
| Male (%) | 24 (86%) | 25 (52%) | 0.003 |
| Female (%) | 4 (14%) | 23 (48%) | |
| Age ± SDa (years) | 57.54 ± 11.85 | 63.00 ± 7.65 | 0.038 |
| FEV1/FVC (%) ± SDa | 70.01 ± 12.62 | 68.39 ± 15.24 | 0.498 |
| Smoking Status | |||
| Current (%) | 12 (43%) | 21 (44%) | 0.988 |
| Past (%) | 14 (50%) | 24 (50%) | |
| Never (%) | 2 (7%) | 3 (6%) | |
| HIV Plasma Viral Load > 50 copies/mL (%) | 9 (32%) | N/A | N/A |
| CD4 cell count ± SDa (cells/mm3) | 419 ± 295 | N/A | N/A |
aSD Standard deviation
Total bacterial load
Total 16S rRNA (reflective of total bacterial loads) was significantly reduced in PLWH compared to the HIV- cohort (0.27 ± 1.10 copies/ng DNA vs. 3.25 ± 8.44 copies/ng DNA, p = 0.0076). However, there were no differences in total 16S rRNA between HIV+ patients with and without COPD (p = 0.653) or between HIV- controls with and without COPD (p = 0.719).
Bacterial diversity
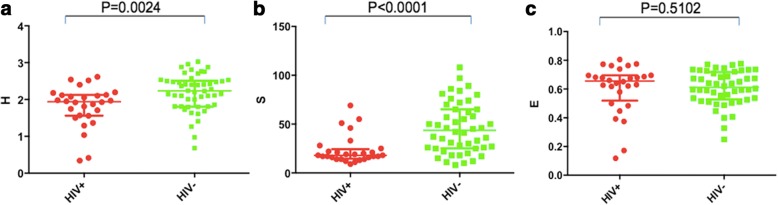
Alpha diversity was assessed using the Shannon diversity index, richness, and evenness (Fig. 1). There was a significant decrease in Shannon diversity in the HIV+ compared to the HIV- groups (1.82 ± 0.10 vs. 2.20 ± 0.073, p = 0.0024), driven mainly by a significant reduction in bacterial richness (23.29 ± 2.75 for HIV+ and 46.04 ± 3.716 for HIV-, p < 0.0001). The difference in bacterial richness (i.e. overall species count) was confirmed in a rarefaction curve in which more species were found in the HIV- group for each random sample compared with the HIV+ group (Additional file 1: Figure S1). There was no significant difference in evenness between the two groups (0.60 ± 0.032 for HIV+, 0.61 ± 0.017 for HIV-, p = 0.5102). Across the four groups (HIV + COPD+, HIV + COPD-, HIV-COPD+, and HIV-COPD-), there were significant differences in Shannon diversity (p = 0.0195) and richness (p = 0.0006) by Kruskal-Wallis tests. However, this was entirely driven by differences between the HIV+ and HIV- groups as there were no significant differences observed between the COPD+ and COPD- groups. There was also no significant difference in evenness between the four groups (p = 0.38). There was a significant direct correlation between FEV1/FVC and Shannon diversity (p = 0.0095, R = 0.39) in the HIV- group but not within the HIV+ group (p = 0.1427, R = 0.34). To ensure that the differences between HIV+ and HIV- groups was not confounded by the clinical indication for bronchoscopy within the HIV+ group, we compared Shannon diversity and beta diversity by indication for bronchoscopy, use of inhaled corticosteroids, cancer diagnosis, and treatment or prophylactic antibiotics. No significant differences were found in any of these categories (Additional file 1: Figures S2–S6). There was no correlation between Shannon diversity and CD4 count, nor was there a difference between those with detectable and undetectable viral loads.
Fig. 1.
Shannon diversity a, richness b, and evenness c are shown for PLWH (red) and HIV- subjects (green). Shannon diversity and richness were both significantly lower in PLWH compared with HIV- subjects. There was no significant difference in species evenness between the two groups
Phyla distribution
A significant increase was observed in the relative abundance of Proteobacteria in PLWH (0.38 ± 0.25 in HIV+ vs. 0.19 ± 0.19 in HIV-, p = 0.0003) (Fig. 2). Decreased abundance of Bacteroidetes (0.23 ± 0.19 in HIV+ vs. 0.35 ± 0.16 in HIV-, p = 0.0068) and Firmicutes (0.18 ± 0.17 in HIV+ vs. 0.33 ± 0.16 in HIV-, p = 0.0002) were found in the HIV+ group compared to the HIV- group. Figure 3 shows the distribution of phyla across all four patient groups. According to a Kruskal-Wallis test, there was a significant difference in Bacteroidetes (p = 0.0031), Proteobacteria (p = 0.0013) and Firmicutes (p = 0.0017) across the four groups. Post-hoc tests were performed using Dunn’s Multiple Comparison method; the differences observed in Bacteroidetes and Proteobacteria were driven by the difference between the HIV+ and HIV- groups in the COPD- population. The difference observed in Firmicutes was driven by the difference between the HIV+ and HIV- groups in the COPD+ population. There was no significant difference between COPD+ patients and COPD- patients, irrespective of HIV status.
Fig. 2.
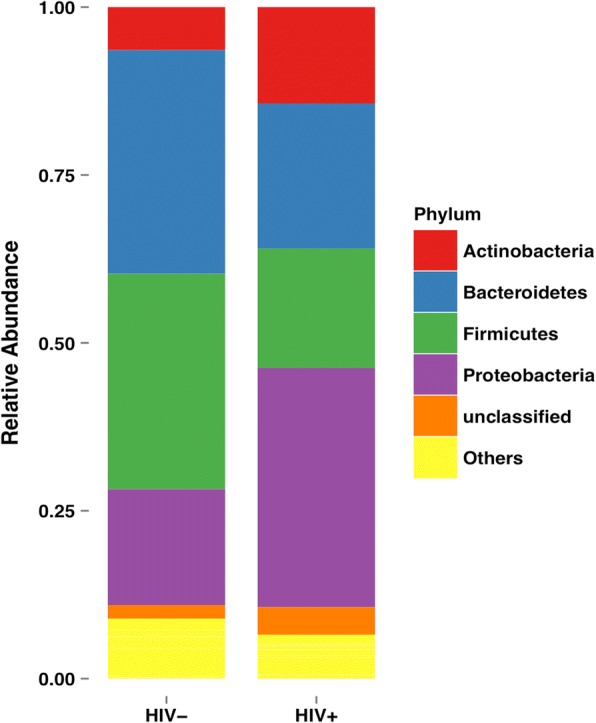
The relative distribution of phyla between PLWH and HIV- patients is shown. There was a significant increase in Proteobacteria (p = 0.0003) and decrease in Bacteroidetes (p = 0.0068) and Firmicutes (p = 0.0002) in PLWH compared to HIV- patients
Fig. 3.
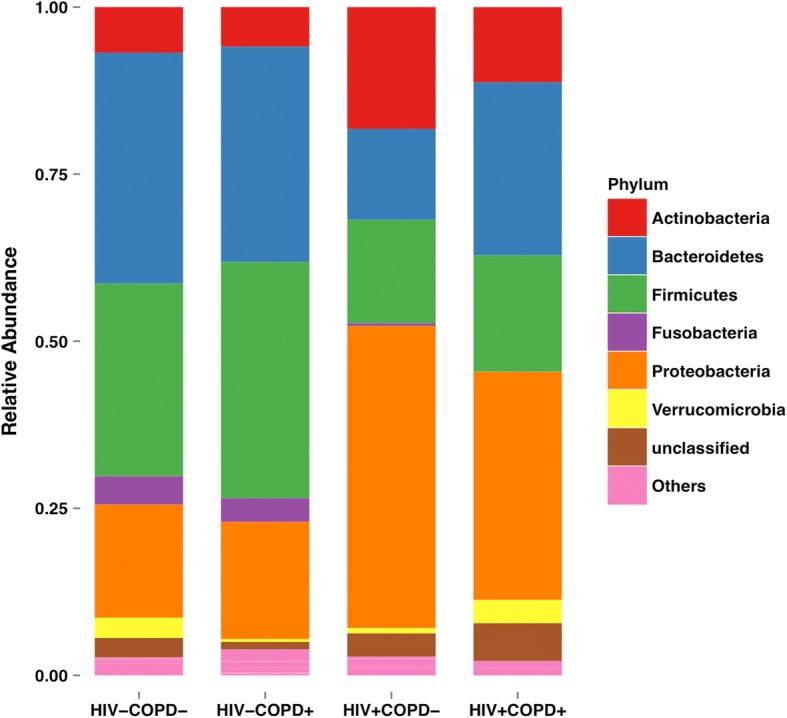
The distribution of phyla across all four groups is shown. By Kruskal-Wallis tests, there were significant differences between the groups in Bacteroidetes (p = 0.0031), Proteobacteria (p = 0.0013), and Firmicutes (p = 0.0017). Post-hoc tests by Dunn’s Multiple Comparison method demonstrated that the differences observed in Bacteroidetes and Proteobacteria were driven entirely by the difference between the HIV + COPD- and HIV-COPD- groups. However, the difference observed in Firmicutes was driven by the difference between the HIV + COPD+ and HIV-COPD+ groups. There were no significant differences in phyla distribution between the COPD+ and COPD- groups
Microbial compositional analysis
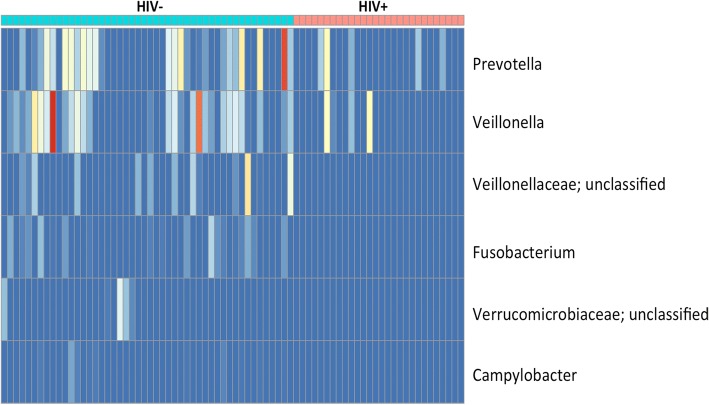
Because there was no difference in alpha diversity between the COPD+ and COPD- groups, beta diversity analysis was restricted to comparing the study cohort by HIV status. Using non-metric multidimensional scaling analysis, a significant difference in community composition was detected between the HIV+ and HIV- groups (PERMANOVA = 0.001) (Fig. 4). Using Boruta feature selection with Random Forest analysis, six OTUs were found to be able to discriminate PLWH from the HIV- group. Figure 5 is a heatmap with yellow representing a lower relative abundance, red representing a higher relative abundance and blue representing samples that did not contain such particular OTUs. OTUs that aligned to Veillonellaceae, Fusobacterium, Verrucomicrobiaceae and Campylobacter were not found in PLWH. Moreover, Prevotella and Veillonella were mainly present in HIV- group. These findings suggest that these six discriminative OTUs were able to separate PLWH from the HIV- group.
Fig. 4.
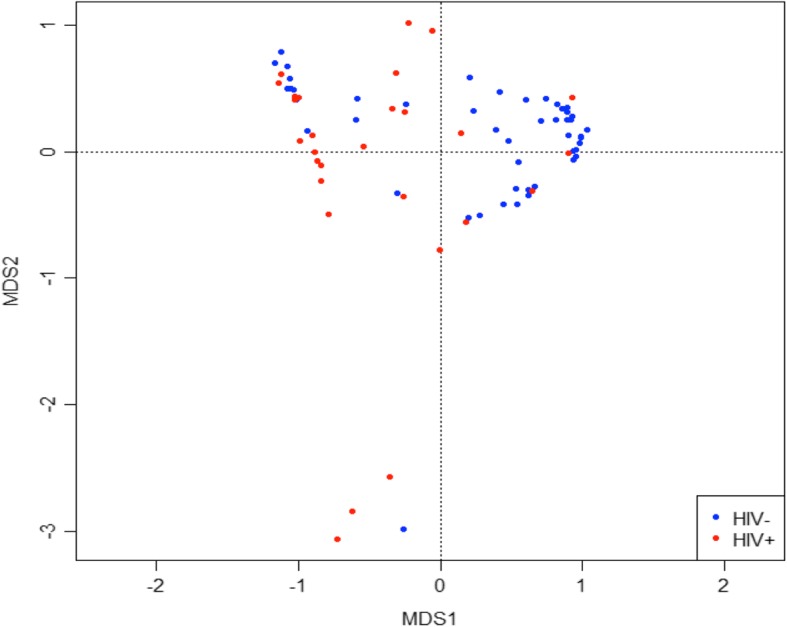
Non-metric multidimensional scaling analysis demonstrates a significant difference in bacterial community composition between PLWH (red) and HIV- subjects (blue) (PERMANOVA = 0.001)
Fig. 5.
In this heat map of OTU abundance, blue indicates a complete absence of the OTU, yellow a lower relative abundance, and red a higher relative abundance. Six OTUs were found to help distinguish between PLWH and HIV- subjects
Discussion
In this first comparison of the HIV and non-HIV SAE microbiome, we discovered that PLWH, had significantly lower bacterial loads, microbial diversity, and species richness compared to HIV- controls. While this observation may seem contrary to what one would expect in a disease associated with immunosuppression and frequent pulmonary infections, it is in fact consistent with numerous microbiome studies comparing disease and non-disease states. Specifically, regardless of the organ examined, disease states are often associated with lower microbial diversity, suggesting that a certain degree of diversity is a hallmark of healthy tissue [24–27]. In the lung, for instance, chronic respiratory conditions such as cystic fibrosis [28, 29] and COPD [30] are associated with lower alpha diversity in sputum and lung samples. Disease severity in the lung also appears to track inversely with diversity [30–32]. In similar fashion, HIV and/or the repeated antibiotic exposures these patients may have experienced due to frequent infections may lower SAE diversity. Supporting this theory are the recent findings by Twigg et al. that patients with uncontrolled HIV have significantly decreased BAL diversity compared with HIV-uninfected controls [6].
The connection between decreased diversity and dysbiosis to COPD pathogenesis is likely multifactorial. In fact, this relationship may be different between HIV+ and HIV- groups, with only the latter showing a correlation between decreased diversity and reduced FEV1/FVC. We were unable to demonstrate significant microbiome differences between patients with and without COPD, regardless of their HIV status. One possible explanation may be that our study was underpowered to detect any difference. Larger studies examining the relationship between airflow obstruction, HIV, and the microbiome are warranted. Targeted brushings in areas of advanced emphysema compared with brushings taken from normal lung within the same PLWH may also help us understand what role the microbiome may play in COPD pathogenesis. Linking the microbiome with metabolomic, transcriptomic, and epigenetic modifications will provide additional clues as to how dysbiosis can set the stage for progressive airflow obstruction. In a previous study by our group looking specifically at the SAE microbiome and its associated transcriptome in HIV [10], we demonstrated that the abundance of Firmicutes was negatively correlated with the expression of cilia-related genes and positively correlated with the expression of immune response genes. Haemophilus species were also negatively correlated with the expression of cilia-related genes. These genetic pathways could be critical in the pathogenesis of accelerated COPD in HIV; however, direct comparisons of these relationships to those observed in HIV- subjects are further required.
Interestingly, the SAE phyla distribution in PLWH was markedly different from that in HIV- controls in a pattern that was reminiscent of the differences previously noted between COPD and non-COPD lungs in an HIV- cohort. We found that PLWH had an increase in Proteobacteria and decreases in both Bacteroidetes and Firmicutes compared with HIV- controls. Similarly, Sze et al. found that lung tissue from GOLD Stage 4 COPD patients had increased Proteobacteria and decreased Bacteroidetes and Firmicutes compared to control lungs [30]. For PLWH, changes in the abundance of these three phyla may represent an early stage in the COPD development. Longitudinal studies evaluating the progression of phyla distribution from healthy to diseased lungs may help to clarify this association.
A novel aspect of this study was the use of SAE cells to investigate the unique lung microbiome in HIV. Previous studies have largely focused on BAL fluid, a useful compartment with which to identify generalized inflammation in the lung but not one that necessarily provides specific information on the pathogenesis of COPD. Profound structural changes occur in the SAE in COPD, including squamous metaplasia, ciliary dysfunction, mucous cell hyperplasia, and the breakdown of apical junctional barriers [8]. Even prior to the onset of overt COPD, smoking-related changes in the airway epithelium can be observed. These include senescent signatures determined by telomere length and growth differential factor 15 production [33, 34] and gene expression alterations along immunity and oxidative stress pathways [35]. SAE changes in HIV have not yet been fully characterized, although two studies have recently shed greater light. In one study, the presence of X4 tropic HIV increased both epithelial cell layer permeability and the expression of pro-inflammatory cytokines [36]. In another study, HIV was found to bind to airway epithelial basal cells, resulting in a tissue-destructive phenotype [37]. Whether or not the distinct microbiome of the HIV SAE plays a role in these processes is certainly a question worth pursuing in future experiments.
There are a number of limitations noted in our study. First, contamination by oropharyngeal and environmental elements is always a concern in a lung microbiome study in which specimens are obtained via bronchoscopy. This is particularly true for organs with relatively low microbial biomass such as the lung. Ideally, reagent samples and oral and bronchoscope channel washes prior to the procedure would have helped to identify potential contaminants in bronchial brushings [38]. Nonetheless, all bronchoscopies were performed with no suction used upon insertion of the bronchoscope to avoid oral and large airway contamination. Second, patients enrolled had other pulmonary concerns, including lung masses, nodules, and pneumonia. While these could conceivably pose as confounders, we did not find significant diversity differences between those with and without these conditions. This was likely due to the fact that sample acquisition took place specifically in lobes of the lung away from clinically important lesions. Further studies evaluating the microbiome in asymptomatic PLWH will be necessary moving forward to confirm our findings.
Conclusions
As the demographics of PLWH shift towards older ages, the spectre of chronic lung diseases looms large and improved understanding of their pathogenesis in the setting of HIV will become imperative. Dysbiosis in the HIV SAE marked by significantly reduced microbial diversity may be one important hallmark or instigator of chronic lung diseases such as COPD and further explorations of the microbiome’s role in this process are warranted.
Additional file
Figure S1. A rarefaction curve is shown for HIV- subjects (blue) and HIV+ subjects (red). Each line represents a study subject. Figure S2. Shannon diversity is shown for the HIV+ group divided by indication for bronchoscopy (n = 18 for lung nodule, n = 10 for pneumonia). Figure S3. Shannon diversity is shown for the HIV+ group comparing those were diagnosed with lung cancer (blue, n = 9) and those without lung cancer (red, n = 21). Figure S4. Shannon diversity is shown for the HIV+ group comparing those who were taking inhaled corticosteroids (blue, n = 4) and those who were not (red, n = 24). Figure S5. Shannon diversity is shown for the HIV+ group comparing those who were taking treatment dose antibiotics (blue, n = 8) and those who were not (red, n = 20). Figure S6. Shannon diversity is shown for the HIV+ group comparing those who were taking treatment prophlactic antibiotics (blue, n = 3) and those who were not (red, n = 25). (DOCX 651 kb)
Acknowledgements
The authors would like to acknowledge Fernando Studart for his assistance in editing the manuscript.
An abstract version of this manuscript was presented at the American Thoracic Society Conference in San Francisco, CA, in May of 2016.
Funding
This study was funded by the Canadian Institutes of Health Research and the BC Lung Association. JML is supported by the Canadian Institutes of Health Research, the Michael Smith Foundation for Health Research, and the St. Paul’s Hospital Foundation.
Availability of data and materials
Data are available from the corresponding author with reasonable request.
Abbreviations
- ART
Antiretroviral therapy
- BAL
Bronchoalveolar lavage
- COPD
Chronic obstructive pulmonary disease
- CT
Computed tomography
- FEV1
Forced expiratory volume in 1 s
- FVC
Forced vital capacity
- GOLD
Global Initiative for Chronic Obstructive Lung Disease
- HIV
Human immunodeficiency virus
- OTU
Operational taxonomic unit
- PCR
Polymerase chain reaction
- PLWH
Persons living with HIV
- SAE
Small airway epithelium
- UBC
University of British Columbia
Authors’ contributions
Data acquisition: SX, MAS, EAV, TS, MH, SG, SS, CN, WL, SL, DDS, SFPM, JML, AT, JM. Data analysis: SX, MAS, JY, JML. Manuscript writing: SX, AT, JML. Editing: SX, AT, MAS, EAV, MH, JY, JM, DDS, SFPM, JML. All authors read and approved the final manuscript.
Authors’ information
JML is the guarantor of the manuscript, had full access to all of the data in the study, and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Consent for publication
All subjects provided written informed consent under the University of British Columbia (UBC)-approved Providence Health Care ethics protocol H14–03267. As no individual data are presented, consent for publication is not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12931-018-0835-7) contains supplementary material, which is available to authorized users.
References
- 1.Hasse B, Ledergerber B, Furrer H, Battegay M, Hirschel B, Cavassini M, Bertisch B, Bernasconi E, Weber R. Morbidity and aging in HIV-infected persons: the Swiss HIV cohort study. Clin Infect Dis. 2011;53:1130–1139. doi: 10.1093/cid/cir626. [DOI] [PubMed] [Google Scholar]
- 2.Crothers K, Butt AA, Gibert CL, Rodriguez-Barradas MC, Crystal S, Justice AC. Veterans aging cohort 5 project T: increased COPD among HIV-positive compared to HIV-negative veterans. Chest. 2006;130:1326–1333. doi: 10.1378/chest.130.5.1326. [DOI] [PubMed] [Google Scholar]
- 3.Brown J, Roy A, Harris R, Filson S, Johnson M, Abubakar I, Lipman M. Respiratory symptoms in people living with HIV and the effect of antiretroviral therapy: a systematic review and meta-analysis. Thorax. 2017;72:355–366. doi: 10.1136/thoraxjnl-2016-208657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, Mitreva M, Abubucker S, Martin J, Yao G, et al. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am J Respir Crit Care Med. 2013;187:1110–1117. doi: 10.1164/rccm.201211-2145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck JM, Schloss PD, Venkataraman A, Twigg Iii H, Jablonski KA, Bushman FD, Campbell TB, Charlson ES, Collman RG, Crothers K, et al. Multi-center comparison of lung and oral microbiomes of HIV-infected and HIV-uninfected individuals. Am J Respir Crit Care Med. 2015;192:1335–1344. doi: 10.1164/rccm.201501-0128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Twigg HL, 3rd, Knox KS, Zhou J, Crothers KA, Nelson DE, Toh E, Day RB, Lin H, Gao X, Dong Q, et al. Effect of advanced HIV infection on the respiratory microbiome. Am J Respir Crit Care Med. 2016;194:226–235. doi: 10.1164/rccm.201509-1875OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonough JE, Yuan R, Suzuki M, Seyednejad N, Elliott WM, Sanchez PG, Wright AC, Gefter WB, Litzky L, Coxson HO, et al. Small-airway obstruction and emphysema in chronic obstructive pulmonary disease. N Engl J Med. 2011;365:1567–1575. doi: 10.1056/NEJMoa1106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaykhiev R, Crystal RG. Early events in the pathogenesis of chronic obstructive pulmonary disease. Smoking-induced reprogramming of airway epithelial basal progenitor cells. Ann Am Thorac Soc. 2014;11(5):S252–S258. doi: 10.1513/AnnalsATS.201402-049AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puchelle E, Zahm JM, Tournier JM, Coraux C. Airway epithelial repair, regeneration, and remodeling after injury in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3:726–733. doi: 10.1513/pats.200605-126SF. [DOI] [PubMed] [Google Scholar]
- 10.Sze MA, Xu S, Leung JM, Vucic EA, Shaipanich T, Moghadam A, Harris M, Guillemi S, Sinha S, Nislow C, et al. The bronchial epithelial cell bacterial microbiome and host response in patients infected with human immunodeficiency virus. BMC Pulm Med. 2016;16:142. doi: 10.1186/s12890-016-0303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MR, Crapo R, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, Enright P, van der Grinten CP, Gustafsson P, et al. General considerations for lung function testing. Eur Respir J. 2005;26:153–161. doi: 10.1183/09031936.05.00034505. [DOI] [PubMed] [Google Scholar]
- 12.Vestbo J, Hurd SS, Agusti AG, Jones PW, Vogelmeier C, Anzueto A, Barnes PJ, Fabbri LM, Martinez FJ, Nishimura M, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 13.Sze MA, Abbasi M, Hogg JC, Sin DD. A comparison between droplet digital and quantitative PCR in the analysis of bacterial 16S load in lung tissue samples from control and COPD GOLD 2. PLoS One. 2014;9:e110351. doi: 10.1371/journal.pone.0110351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan XC, Huttenhower C. Chapter 12: human microbiome analysis. PLoS Comput Biol. 2012;8:e1002808. doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia Y, Sun J. Hypothesis testing and statistical analysis of microbiome. Genes & Diseases. 2017;4:138–148. doi: 10.1016/j.gendis.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 20.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 21.Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62:142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 23.Kursa MB, Rudnicki WR. Feature selection with the boruta package. J Stat Softw. 2010;36:1–13. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 24.Alipour M, Zaidi D, Valcheva R, Jovel J, Martinez I, Sergi C, Walter J, Mason AL, Wong GK, Dieleman LA, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in Paediatric ulcerative colitis. J Crohns Colitis. 2016;10:462–471. doi: 10.1093/ecco-jcc/jjv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Paiva CS, Jones DB, Stern ME, Bian F, Moore QL, Corbiere S, Streckfus CF, Hutchinson DS, Ajami NJ, Petrosino JF, Pflugfelder SC. Altered mucosal microbiome diversity and disease severity in Sjogren syndrome. Sci Rep. 2016;6:23561. doi: 10.1038/srep23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers MB, Firek B, Shi M, Yeh A, Brower-Sinning R, Aveson V, Kohl BL, Fabio A, Carcillo JA, Morowitz MJ. Disruption of the microbiota across multiple body sites in critically ill children. Microbiome. 2016;4:66. doi: 10.1186/s40168-016-0211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganju P, Nagpal S, Mohammed MH, Nishal Kumar P, Pandey R, Natarajan VT, Mande SS, Gokhale RS. Microbial community profiling shows dysbiosis in the lesional skin of vitiligo subjects. Sci Rep. 2016;6:18761. doi: 10.1038/srep18761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zemanick ET, Wagner BD, Robertson CE, Ahrens RC, Chmiel JF, Clancy JP, Gibson RL, Harris WT, Kurland G, Laguna TA, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J. 2017;50:1700832. doi: 10.1183/13993003.00832-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flight WG, Smith A, Paisey C, Marchesi JR, Bull MJ, Norville PJ, Mutton KJ, Webb AK, Bright-Thomas RJ, Jones AM, Mahenthiralingam E. Rapid detection of emerging pathogens and loss of microbial diversity associated with severe lung disease in cystic fibrosis. J Clin Microbiol. 2015;53:2022–2029. doi: 10.1128/JCM.00432-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sze MA, Dimitriu PA, Suzuki M, McDonough JE, Campbell JD, Brothers JF, Erb-Downward JR, Huffnagle GB, Hayashi S, Elliott WM, et al. The host response to the lung microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;192:483–445. doi: 10.1164/rccm.201502-0223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers GB, van der Gast CJ, Cuthbertson L, Thomson SK, Bruce KD, Martin ML, Serisier DJ. Clinical measures of disease in adult non-CF bronchiectasis correlate with airway microbiota composition. Thorax. 2013;68:731–737. doi: 10.1136/thoraxjnl-2012-203105. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Nunez M, Millares L, Pomares X, Ferrari R, Perez-Brocal V, Gallego M, Espasa M, Moya A, Monso E. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J Clin Microbiol. 2014;52:4217–4223. doi: 10.1128/JCM.01967-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Q, Jiang D, Matsuda JL, Ternyak K, Zhang B, Chu HW. Cigarette smoke induces human airway epithelial senescence via growth differentiation factor 15 production. Am J Respir Cell Mol Biol. 2016;55:429–438. doi: 10.1165/rcmb.2015-0143OC. [DOI] [PubMed] [Google Scholar]
- 34.Walters MS, De BP, Salit J, Buro-Auriemma LJ, Wilson T, Rogalski AM, Lief L, Hackett NR, Staudt MR, Tilley AE, et al. Smoking accelerates aging of the small airway epithelium. Respir Res. 2014;15:94. doi: 10.1186/s12931-014-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med (Berl) 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 36.Brune KA, Ferreira F, Mandke P, Chau E, Aggarwal NR, D'Alessio FR, Lambert AA, Kirk G, Blankson J, Drummond MB, et al. HIV impairs lung epithelial integrity and enters the epithelium to promote chronic lung inflammation. PLoS One. 2016;11:e0149679. doi: 10.1371/journal.pone.0149679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung NPY, Ou X, Khan KMF, Salit J, Kaner RJ, Crystal RG. HIV reprograms human airway basal stem/progenitor cells to acquire a tissue-destructive phenotype. Cell Rep. 2017;19:1091–1100. doi: 10.1016/j.celrep.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A rarefaction curve is shown for HIV- subjects (blue) and HIV+ subjects (red). Each line represents a study subject. Figure S2. Shannon diversity is shown for the HIV+ group divided by indication for bronchoscopy (n = 18 for lung nodule, n = 10 for pneumonia). Figure S3. Shannon diversity is shown for the HIV+ group comparing those were diagnosed with lung cancer (blue, n = 9) and those without lung cancer (red, n = 21). Figure S4. Shannon diversity is shown for the HIV+ group comparing those who were taking inhaled corticosteroids (blue, n = 4) and those who were not (red, n = 24). Figure S5. Shannon diversity is shown for the HIV+ group comparing those who were taking treatment dose antibiotics (blue, n = 8) and those who were not (red, n = 20). Figure S6. Shannon diversity is shown for the HIV+ group comparing those who were taking treatment prophlactic antibiotics (blue, n = 3) and those who were not (red, n = 25). (DOCX 651 kb)
Data Availability Statement
Data are available from the corresponding author with reasonable request.