Abstract
Previous reports have indicated that the abundance of specific microRNAs (miRNA) contained within the exosome/microvesicle compartment of patient biofluids may be useful in diagnosing specific types of cancer. In the present study, the 786-O cell line, which is derived from a clear cell renal cell carcinoma (ccRCC), was used as an in vitro ccRCC tumor model and the human renal proximal tubule cell line HK-2 was used as its normal renal tissue control to investigate the similarities of exosomal content of selected ccRCC miRNA biomarkers in the supernatant with the content of those markers in the cells themselves. A PCR array identified miRNA biomarkers of solid RCC tumors (miR-210, MiR-34a, miR-155-5p and miR-150-5p) that were increased by 2–8 fold in 786-O exosomes compared with the control. These were subsequently chosen for further investigation using TaqMan RT-qPCR in addition to miR-15a and miR-205, which were selected based on prior interest as RCC biomarkers. MiR-15a, −34a, −210 and −155 levels were significantly lower in exosomes when compared with that in whole cells but did not differ between the HK-2 and 786-O cells in either the cytoplasmic, exosome or exosome-free supernatant fractions. By contrast, cytoplasmic miR-150 and miR-205 exhibited significant differences in concentration between the two cell lines. In addition, the cytoplasmic content of miR-150 and miR-205 was mirrored in the exosomal content of these miRNAs. Furthermore, the difference in exosomal miR-205 content was statistically significant. The present study indicated that measurements of the exosomal content of miR-205 and possibly miR-150, but not those of the other examined miRNAs, are proportional to their respective contents in the cells that secreted them. These findings suggest that in vitro RCC systems may be useful in identifying miRNAs with sufficiently high levels of exportation into exosomes; and with sufficiently different expression levels between tumor and normal cells to serve as ccRCC biomarkers in vivo.
Keywords: exosome, microRNA, renal cell carcinoma, biomarker, 786-O cells, HK-2 cells
Introduction
Renal cell carcinoma (RCC) accounts for 2–3% of solid tumors occurring annually and has a relatively high rate of cure if detected early while still confined to the kidney. However, overall mortality is high once the tumor has spread to secondary sites (1,2). Thus, early detection is key to long-time survival. Recent studies have focused on microRNAs (miRNAs) as potential diagnostic and prognostic indicators of several types of cancer since these small non-coding RNAs often act as either oncogenes or tumor suppressors and also since the content of a specific miRNA can vary greatly between a tumor and its tissue of origin (3–5). Such miRNAs can be measured in several readily accessible biofluids (serum, saliva, and urine) where they exist either in a ‘free’ state, bound to specific proteins such as Argonaute, or enclosed within membrane-delimited vesicles (exosomes and other microvesicles) released by normal tissues and tumor cells (5).
To date, several miRNA species have been proposed as having diagnostic and prognostic value in renal cell carcinoma, where either as solitary biomarkers or as part of a diagnostic panel, they appear able to distinguish clear cell renal cell carcinoma (ccRCC) patients from healthy controls and benign renal tumors from malignant ones (6). Most of these panels have been developed from serum samples (7–11), but two recent studies have focused on urine specimens as a source of diagnostic information (12,13). Urinary miRNAs are particularly attractive as RCC biomarkers since urine can be sampled frequently by non-invasive means.
The 786-O cell line originated from a human ccRCC of proximal tubule origin, and is frequently employed in in vitro studies of ccRCC cell behavior (14). Among the characteristics distinguishing the 786-O line from several other ccRCC cultures is a stable Von Hippel Lindau (VHL) mutation that results in overexpression of vascular endothelial growth factor (VEGF) (14). HK-2 cells are an immortalized cell line originating from normal human proximal tubule (15).
In the present study we employed 786-O and HK-2 cell monolayers as the in vitro equivalents of ccRCC tumor and surrounding renal cortical tissue, respectively, to assess the correlation between the content of selected miRNAs extracted from secreted exosomes with miRNA derived from the cell monolayers. Our results suggest that those miRNAs (e.g., miR-150 and miR-205) that are of high concentration in exosomes relative to cytoplasmic concentration and that have the highest levels of differential expression between tumor and non-tumor tissue could be particularly useful as biomarkers of RCC in urinary samples.
Materials and methods
Cell culture
786-O cells (ATCC®CRL-1932™) and HK-2 cells (ATCC®CRL-2190™) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and subcultured in a growth medium containing RPMI-1640 medium, fetal bovine serum (FBS) at a final concentration of 10%, and penicillin (100 U/ml)-streptomycin (100 µg/ml). For cell and exosome collection, cultures of HK-2 and 786-O cells grown to confluence in growth medium in 162 cm2 flasks were switched to 20 ml serum-free RPMI-1640 for 48 h after which exosomes were collected and miRNA extracted from those exosome pellets and from the cell monolayer using the methods described below.
Exosome isolation
For all studies, purified exosomes were collected from medium using two steps of ultracentrifugation as described previously (16). Briefly, conditioned medium was removed from 786-O and HK-2 cultures and the supernatant was then frozen at −80°C until exosome isolation. The cellular monolayer was then washed with PBS, scraped from the flask and frozen at −80°C until miRNA extraction. To isolate exosomes, the frozen medium was thawed and centrifuged at 17,000 × g for 18 min to pellet larger organelles and other membrane structures out, followed by a final centrifugation at 200,000 × g for 1 h and 15 min, and collection of the pellet (exosomal fraction) and supernatant for miRNA extraction.
miRNA extraction
MiRNA was extracted from both exosome and whole cell pellets using the miRNeasy Micro kit (Qiagen Inc., Germantown, MD, USA). Briefly, exosome pellets in polycarbonate ultracentrifuge tubes were suspended in 25 ul phosphate buffered saline (PBS) and transferred to 1.5 ml Eppendorf tubes to which 700 ul QIAzol lysis reagent was added and homogenized by pulling repeatedly through a hypodermic needle. Following the addition of 140 ul chloroform to the homogenate and centrifugation (12,000 × g, 15 min), 1.5 volumes of 100% ethanol was added to the supernatant and the sample was added to an RNAeasy minElute spin column and centrifuged (8,000 × g, 30 sec). The column was then washed with RWT buffer, followed by RPE buffer, and 80% ethanol, and purified miRNA was eluted with 15 µl RNAse-free dH2O and centrifugation at full speed for 2 min. Total RNA content was determined using the nano-drop method.
miRNA was extracted from whole cell pellets using a similar technique with the following modifications: After dissolving the entire cell pellet in 700 ul lysis buffer, 35 ul of the homogenate was removed and added to 630 ul lysis buffer (i.e., 1:20 dilution), and the diluted homogentate was then processed as described for exosome miRNA extraction. MiRNA was extracted from a 1 ml sample of the supernatant and designated as the ‘exosome-free supernatant’ (EFS).
miRNA PCR Array
An miRNA PCR array (Cancer PathwayFinder miRNA PCR Array; Qiagen Inc.) containing a total of 84 miRNA primers that had previously been shown as differentially expressed in a variety of tumor types was used to make an initial selection of miRNAs differently expressed in 786-O cells compared to HK-2 cells.
MiRNA was extracted from collected exosomes as described above, and cDNA was produced from a total of 300 ng RNA from each sample in a reaction volume of 20 ul using the miScript II miRNA RT kit (Qiagen Inc.). A total of 19 ul of each reaction product was added to 200 ul H2O and 100 µl of each diluted cDNA template was added to a common reaction mix containing 2×QuantiTect SYBR-Green PCR Master Mix, 10× miScript Universal Primer, RNase-free water, and 25 ul of the total mix was pipetted into each well of a 96-well Cancer PathwayFinder miRNA PCR Array plate.
RT-qPCR
Reverse transcription (RT) of extracted miRNA was performed to confirm the array results using a TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc., Waltham, MA) and stem-loop oligonucleotides specific for the following miRNAs (mature miRNA sequences in parentheses): miR-15a-5p (UAGCAGCACAUAAUGGUUUGUG), miR-15a-3p (CAGGCCAUAUUGUGCUGCCUCA), miR-16-5p (UAGCAGCACGUAAAUAUUGGCG), miR-150-5p (UCUCCCAACCCUUGUACCAGUG), miR-34a-5p (UGGCAGUGUCUUAGCUGGUUGU), miR-210-3p (CUGUGCGUGUGACAGCGGCUGA), miR-155-5p (UUAAUGCUAAUCGUGAUAGGGGU), and miR-205-5p (UCCUUCAUUCCACCGGAGUCUG) purchased from Thermo Fisher Scientific, Inc. Briefly, RT reactions contained 20 ng of purified miRNA in a volume of 5 ul, and 0.15 ul 100 M dNTP mix w/dTT, 1.5 ul 10X RT buffer, 0.19 µl RNase inhibitor (20 U/ul), 1 µl MultiScribe RT enzyme (50 U/µl), and 3 µl 5X specific primer in a total volume of 15 ul. Reverse transcription was performed in a programmable thermal cycler as follows: 16°C-30 min, 42°C 30 min, and 85°C-5 min, after which 1.33 ul of RT reaction product was added to 10 ul master mix, 7.7 ul H2O, and 1 µl 20X specific primer in a total volume of 20 ul that was amplified by 50 cycles of 15 sec denature/60 sec anneal/extend in a real-time thermal cycler.
MiR-16 was selected as a control due to the relative constancy of its expression in various cultured cell lines in the literature (17), and RT-qPCR for this miRNA was performed in tandem with target miRNAs to determine the optimal normalization procedure.
Data analysis
Statistical analysis was performed on studies repeated at least three times (with separate cultures of confluent cells) using GraphPad Prizm version 7 (GraphPad Software, La Jolla, CA, USA). One-way ANOVA was performed, followed by Tukey's multiple comparisons test, and P<0.05 was considered to indicate a statistically significant difference.
Results
miRNA PCR Array
Although not comprehensive, this array contained primarily miRNAs that have demonstrated an association with human cancers in the literature, many of which have been identified as being up-or downregulated in RCC, specifically. As of this writing, most of the 84 miRNAs in this assay have been identified in published studies as playing a role in RCC, either as oncogenes or tumor suppressors in cultured cells, or as biomarkers in vivo (18–23). Therefore, the probability of finding differences in expression levels of miRNA in the two cell lines was assumed to be high. Since the PCR array consisted of 84 miRNAs that were roughly evenly distributed between oncogenes (upregulated in cancer) and tumor suppressors (downregulated in cancer), expression levels of individual miRNAs were determined by normalization of specific Ct values to the mean Ct for all 84 miRNAs.
We selected for further study several miRNAs that were increased by >2-fold in 786-O exosomes compared with HK-2 exosomes in the array, and whose difference in expression from control was consistent with previous citations of upregulation in either RCC tumors, RCC patient serum/plasma, and/or RCC patient urine compared with control. Using both criteria, four miRNAs were chosen for further study using Taqman RT-qPCR: miR-150-5p, miR-34a-5p, miR-155-5p, and miR-210, increased by at 8-fold, 7-fold, 5-fold and 2-fold, respectively, in the 786-O cells compared with HK-2 in the PCR array. In addition, miR-15a-5p was examined due to its prior identification as a urinary marker of RCC (12). MiR-205 was also of interest because of its previous citations as a markedly up- or downregulated miRNA in urogenital cancers (24), and RCC cell lines (25) compared to controls.
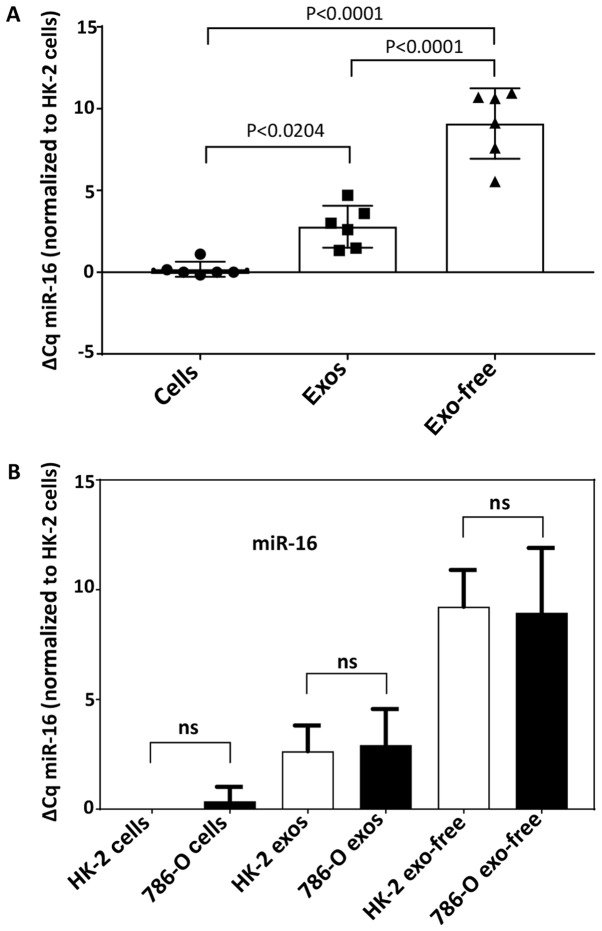
Normalization of the target genes in the PCR array was accomplished by subtracting the mean Ct of all genes in each plate from the Cq value of individual target miRNAs in the respective plate (17). However, this normalization method was not feasible to use with the limited number of miRNAs selected for TaqMan RT-qPCR. Therefore, endogenous miR-16, a gene frequently used as a housekeeping gene in miRNA studies (17), was assessed for differences in expression between the two cell types, and among the three pools (exosome, exosome-free, and cytoplasmic) from which miRNA was extracted (Fig. 1). The results showed significant differences in miR-16 Cq value among the three pools, with whole cells exhibiting the lowest mean Cq (i.e., the highest concentration) and EFS exhibiting the highest Cq. Mean miR-16 Cq, however, did not differ between HK-2 and 786-O cells for any of the three pools (Fig. 1). Since variability was least in the whole cell fraction, the miR-16 content of whole cells chosen for normalization in the studies described below.
Figure 1.
Levels of miR-16, the reference gene used in these studies, varies significantly with sample source (cell cytoplasm, exosomes, exosome-free supernatant) (A) but not between HK-2 and 786-0 cells (B). Bars represent the ΔCq ± SD of miR-16 measured in each sample and normalized to the value for HK-2 cells in three separate experiments. Open bars are HK-2; filled bars are 786-O. miR, microRNA; ns, not significant.
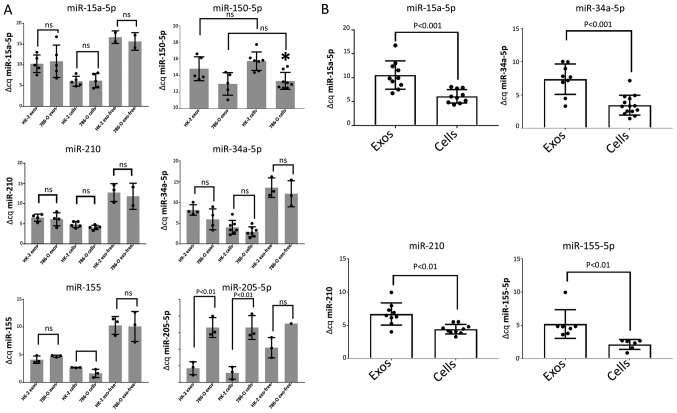
Fig. 2A shows the results of RT-qPCR for selected mature miRNAs obtained from the exosomes, exosome-free medium, or whole cells obtained from confluent cultures of either HK-2 cells or 786-O cells, with all results expressed as Δ Cq normalized to the miR-16 expression level in whole cell pellets. MiR-15a-5p levels exhibited no difference between the HK-2 and 786-O cell lines in either the exosomes, exosome-free medium, or in the whole cells (Fig. 2A). Of the four miRNAs selected from the array based on upregulation in 786-O cells and on previous literature citations, miR-34a-5p, miR-155 and miR-210, showed elevations in 786-O cells using TaqMan RT-qPCR consistent with the array results, but the differences did not attain significance (Fig. 2A). However, significant differences were obtained between the exosomal concentration of each of these miRNAs and its respective intracellular content when results for HK-2 and 786-O cells were pooled (Fig. 2B), indicating that the secreted exosomes contained only between 5 and 37% of the miRNA concentration in the parent cells. The content of the target miRNAs in the exosome and cell free fraction was even lower than this, with miRNA levels ranging between undetectable (miR-150-5p) and only 0.5% of the content of the whole cell.
Figure 2.
(A) ΔCq of six different target miRNAs measured using miRNA extracted from the cell cytoplasm, exosomes and exosome-free supernatant obtained from HK-2 and 786-O cells and normalized to Cq for miR-16 in HK-2 whole cell miRNA. Individual data points representing extractions from different cell cultures and conditioned media are represented by individual filled circles, with the mean value shown as a light gray bar with standard deviation brackets. ΔCq between selected groups were compared to indicate any significant differences. *P<0.05 vs. HK-2 cells. (B) Exosome content of miR-15a-5p, miR-34a-5p, miR-210 and miR-155 is significantly lower than in whole cells. Data from HK-2 cells and 786-O cells pooled from values shown in Fig. 2A are shown as filled circles with the means ± SD of those values shown as unfilled bars. miR, microRNA.
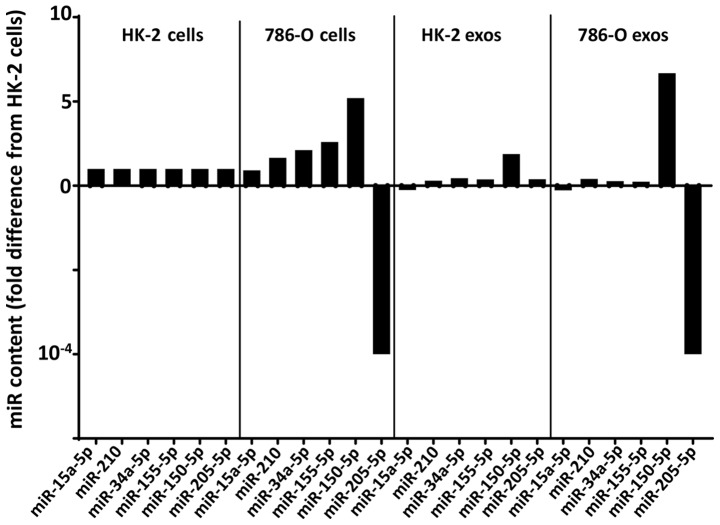
In contrast to these four miRNA species, both miR-150 and miR-205 exhibited a significant difference in intracellular expression levels between the two cell lines, with miR-150 showing a 5.2-fold upregulation and miR-205 showing a 10,000-fold downregulation in 786-O cells relative to HK-2 (Figs. 2A and 3). Also distinguishing miR-150 from the others was its higher content in the exosome fraction than in the whole cell fraction in both HK-2 and 786-O cells. These two factors may have contributed to the 3.6-fold increase of miR-150 in 786-O exosomes compared to HK-2 cells, although the P-value for this difference (P=0.10) did not reach significance (Fig. 3).
Figure 3.
Fold difference in exosome and cellular miRNA in HK-2 cells and 786-O cells normalized to intracellular HK-2 miRNA content. miR, microRNA.
miR-205 was unlike all other miRNAs used in this study in showing a significantly lower (10,000-fold) expression level in 786-O cells than in HK-2 (P=0.0043). Most importantly, the reduction in cellular miR-205 content was reflected in the exosomal miR-205 content of the two cell lines with a 10,000-fold (P=0.0040) lower level in 786-O cells than in HK-2.
Discussion
In the absence of a single unique biomarker for early stage ccRCC, attention has recently focused on changes in serum or urinary levels of multiple miRNA species (i.e., a miRNA panel) to provide an early indication of the presence of this cancer (7–13). There is a considerable lack of uniformity, however, in the specific miRNAs that make up those diagnostic panels. For example, of three such panels consisting of 2–5 miRNAs each (9–11), only a single miRNA (miR-378) was present in more than one panel. Similarly, in the few studies performed thus far of urinary miRNAs as potential biomarkers for ccRCC, one has pointed to miR-15a as a particularly effective indicator of disease (12), and the other (13) has noted that miR-150-5p is significantly upregulated in the urine of ccRCC patients.
To gain a better understanding of how secreted miRNAs measured in readily accessible biofluids reflect the status of those miRNAs in the tumor of origin, we have used in vitro culture models of ccRCC and of normal renal proximal tubule (786-O and HK-2 cells, respectively) to measure selected miRNAs in secreted exosomes and EFS and compare their expression levels with those in whole cells.
Our results show that even in a simplified in vitro system where miRNA in secreted exosomes might be expected to have a higher correlation with cellular miRNA content than serum or urinary exosomes have with tumors in vivo, only exosomal miR-150 and miR-205 were strongly correlated with cellular levels, and of those two, only miR-205 reached significance.
In addition, miR-205 was the only miRNA that was significantly reduced (~10,000-fold in both cells and exosomes) in 786-O cells compared to control HK-2 cells (Fig. 3). MiR-205 in the exosome-free fraction was almost undetectable in either cell line. This result indicates that collection of the exosome fraction and measurement of the miR-205 content of that fraction can distinguish 786-O cells from HK-2 cells in culture. However, whether exosomal miR-205 is a potentially useful biomarker for detecting the presence of ccRCC in vivo remains unclear. The relationship between the amount of exosomal miR-205 secreted from a tumor in vivo and its concentration in a particular biofluid is not as direct as the relationship between the miR-205 content of cultured cells and the overlying supernatant, and the detection of changes in biofluid miR-205 from a growing tumor mass that possesses very little of that miRNA could pose a challenge, especially in plasma or serum samples. A recent large-scale study of plasma miRNAs in ccRCC patients and controls did not list miR-205 as being among the most differentially expressed, however the assays were performed on plasma rather than exosome extracts as in the present study (26).
In regard to the functional significance of this finding, Hirata et al (25) had observed the downregulation of miR-205 in both 786-O cells, and in A-498 RCC cells compared to levels in HK-2 cells. Since both RCC lines possess a mutated VHL gene (14), it is possible that the mutation could be related to the reduced miR-205 expression both in the cancer lines and in RCC tumors bearing the VHL mutation in vivo. The biological relevance of miR-205 in RCC remains unclear (27,28), but the targeting of several oncogenic genes including VEGF-A (27) and MALAT1 (25) by miR-205 suggests that it normally acts as a tumor suppressor that is inactivated in RCC.
MiR-150, the miRNA that was identified in the PCR microarray assay as displaying the greatest increase (8-fold) in 786-O cells was the only miRNA to show significant upregulation (5.2-fold) in 786-O cells using RT-qPCR. Moreover, the 3.3-fold difference in exosomal miRNA content between HK-2 and 786-O cells, although not reaching significance, reflected the difference in whole cell miR-150 content between the two cell types. Another distinctive feature of miR-150 observed in this study was its approximately two-fold higher concentration in exosomes than in whole cell cytoplasm. This feature, shared with miR-451 and several other miRNAs, is its preferential import into microvesicles compared with other miRNAs (29), a feature that might favor them as ccRCC biomarkers. The failure to show a significant difference in exosomal miR-150 in the present study could be due to the relatively small amounts of miRNA extractable from exosomes compared to whole cells, and also to the relatively high Cq value for miR-150-5p compared to the other miRNAs. Future studies with additional ccRCC cell lines will address these issues.
Like miR-150, miR-15a had also shown promise as a urinary biomarker for RCC in a recent clinical study (12). However, our in vitro results could discern no difference in miR-15a-5p levels between 786-O cells and HK-2, either in the secreted exosomes or in the parent cells, suggesting that the reported clinical changes in urinary miR-15a-5p are unlikely to be due to increased expression in RCC tumor cells.
In contrast to miR-150, a preferentially exported miRNA, miR-15a was determined by Guduric-Fuchs et al (29) to be preferentially retained within cells rather than exported (29). This observation is supported by our findings that the miR-15a-5p content of both HK-2 and 786-0 cells is about 16-fold higher than its content in the exosomes secreted from the corresponding cell line. MiR-34a, miR-210, and miR-155 also exhibited significantly lower levels in exosomes than in whole cells (Fig. 3), and none of the four was demonstrably different in concentration in exosomes between the two cell lines.
In conclusion, the present study, limited initially to 84 miRNA species important in cancer pathways and six miRNAs selected for further study, shows that only two of the miRNAs (miR-205 and miR-150) were sufficiently altered from control in 786-O cells to serve as exosomal markers of ccRCC, even in the simplified in vitro model that we use here. One of these, miRNA-150, is increased in both 786-O cells and its secreted exosomes, in close agreement with differences in the exosomal miR-150 content of patient and control urine (13), but opposite to differences in circulating levels of miR-150 that were observed in earlier clinical studies (10,26). In vitro observations of cancer-associated differences in miR-150 (and other miRNA) content in assays such as the present one could therefore be especially relevant to the design of urinary biomarker assays. Furthermore, urinary assays of exosomal miRNA may be more amenable to the selection of downregulated ccRCC miRNAs as biomarkers owing to the close contact of the intial tumor to the pathway to final urine. Many of these downregulated miRNAs, like miR-205, could have immense differences in expression levels between cancer and non-cancer cells that would help make RCC easier to detect.
We suggest that comparison of miRNA content of exosomes with miRNA content of whole cells in homogenous cultures of specific strains of RCC could help to identify specific miRNAs that are particularly useful urinary biomarkers for monitoring changes in primary ccRCC and metastases.
Acknowledgements
Not applicable.
Funding
The present study was performed with the support of USUHS (grant no. R083406616).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
VCC conceived the project idea and executed experiments; HL designed and executed experiments; DFS aided in the design of the project and wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 2.Greef B, Eisen T. Medical treatment of renal cancer: New horizons. Br J Cancer. 2016;115:505–516. doi: 10.1038/bjc.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med. 2006;79:131–140. [PMC free article] [PubMed] [Google Scholar]
- 4.Wuchty S, Arjona D, Bozdag S, Bauer PO. Involvement of microRNA families in cancer. Nuc Acids Res. 2012;40:8219–8226. doi: 10.1093/nar/gks627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Zhang KY, Liu SM, Sen S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules. 2014;19:1912–1938. doi: 10.3390/molecules19021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sellitti DF, Doi SQ. MicroRNAs in renal cell carcinoma. Microrna. 2015;4:26–35. doi: 10.2174/2211536604666150713105247. [DOI] [PubMed] [Google Scholar]
- 7.Iwamoto H, Kanda Y, Sejima T, Osaki M, Okada F, Takenaka A. Serum miR-210 as a potential biomarker of early clear cell renal cell carcinoma. Int J Oncol. 2014;44:53–58. doi: 10.3892/ijo.2013.2169. [DOI] [PubMed] [Google Scholar]
- 8.Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brünagel G, von Ruecker A, Müller SC, Ellinger J. MicroRNAs in renal cell carcinoma: Diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6:e25787. doi: 10.1371/journal.pone.0025787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Hu J, Lu M, Gu H, Zhou X, Chen X, Zen K, Zhang CY, Zhang T, et al. A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Sci Rep. 2015;5:7610. doi: 10.1038/srep07610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R, Slaby O. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Transl Med. 2012;10:55. doi: 10.1186/1479-5876-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauser S, Wulfken LM, Holdenrieder S, Moritz R, Ohlmann CH, Jung V, Becker F, Herrmann E, Walgenbach-Brünagel G, von Ruecker A, et al. Analysis of serum microRNAs (miR-26a-2*, miR-191, miR-337-3p and miR-378) as potential biomarkers in renal cell carcinoma. Canc Epidemiol. 2012;36:391–394. doi: 10.1016/j.canep.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 12.von Brandenstein M, Pandarakalum JJ, Kroon L, Loeser H, Herden J, Braun G, Wendland K, Dienes HP, Engelmann U, Fries JW. MicroRNA 15a, inversely correlated to PKCα, is a potential marker to differentiate between benign and malignant renal tumors in biopsy and urine samples. Am J Pathol. 2012;180:1787–1797. doi: 10.1016/j.ajpath.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Butz H, Nofech-Mozes R, Ding Q, Khella HWZ, Szabó PM, Jewett M, Finelli A, Lee J, Ordon M, Stewart R.et alExosomal microRNAs are diagnostic biomarkers and can mediate cell-cell communication in renal cell carcinoma Eur Urol Focus 2210–218.2016 10.1016/j.euf.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 14.Brodaczewska KK, Szczylik C, Fiedorowicz M, Porta C, Czarnecka AM. Choosing the right cell line for renal cancer research. Mol Canc. 2016;15:83. doi: 10.1186/s12943-016-0565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan MJ, Johnson G, Kirk J, Fuerstenberg SM, Zager RA, Torok-Storb B. HK-2: An immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994;45:48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- 16.Vlasov P, Doi SQ, Sellitti DF. FRTL-5 rat thyroid cells release thyroglobulin sequestered in exosomes: A possible novel mechanism for thyroglobulin processing in the thyroid. J Thyroid Res. 2016;2016:9276402. doi: 10.1155/2016/9276402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data normalization strategies for microRNA quantification. Clin Chem. 2015;61:1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Dai Y, Yang J, Chen T, Yin Y, Tang M, Hu C, Zhang L. Microarray analysis of microRNA expression in renal clear cell carcinoma. Eur J Surg Oncol. 2009;35:1119–1123. doi: 10.1016/j.ejso.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, Ganesan S, Bhanot G, Liou LS. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75:835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 20.Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach H, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13:3918–3928. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakada C, Matsuura K, Tsukamoto Y, Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida T, Sato F, et al. Genome-wide microRNA expression profiling in renal cell carcinoma: Significant down-regulation of miR-141 and miR-200c. J Pathol. 2008;216:418–427. doi: 10.1002/path.2437. [DOI] [PubMed] [Google Scholar]
- 22.Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, The BT. MicroRNA profiling of human kidney cancer subtypes. Int J Oncol. 2009;35:109–114. doi: 10.3892/ijo_00000318. [DOI] [PubMed] [Google Scholar]
- 23.Weng L, Wu X, Gao H, Mu B, Li X, Wang JH, Guo C, Jin JM, Chen Z, Covarrubias M, et al. MicroRNA profiling of clear cell renal cell carcinoma by whole-genome small RNA deep sequencing of paired frozen and formalin-fixed, paraffin-embedded tissue specimens. J Pathol. 2010;222:41–51. doi: 10.1002/path.2736. [DOI] [PubMed] [Google Scholar]
- 24.Huang X, Liang M, Dittmar R, Wang L. Extracellular microRNAs in urologic malignancies: Chances and challenges. Int J Mol Sci. 2013;14:14785–14799. doi: 10.3390/ijms140714785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Canc Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chanudet E, Wozniak MB, Bouaoun L, Byrnes G, Mukeriya A, Zaridze D, Brennan P, Muller DC, Scelo G. Large-scale genome-wide screening of circulating microRNAs in clear cell renal cell carcinoma reveals specific signatures in late-stage disease. Int J Cancer. 2017;141:1730–1740. doi: 10.1002/ijc.30845. [DOI] [PubMed] [Google Scholar]
- 27.Orang AV, Safaralizaden R, Hosseinpour Feizi MA. Insights into the diverse roles of miR-205 in human cancers. Asian Pac J Cancer Prev. 2014;15:577–583. doi: 10.7314/APJCP.2014.15.2.577. [DOI] [PubMed] [Google Scholar]
- 28.Qin AY, Zhang XW, Liu L, Yu JP, Li H, Wang SZE, Ren XB, Cao S. Mir-205 in cancer: An angel or a devil? Eur J Cell Biol. 2013;92:54–60. doi: 10.1016/j.ejcb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Guduric-Fuchs J, O'Conner A, Camp B, O'Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.