Abstract
Nasopharyngeal carcinoma (NPC) is prevalent in Africa and East Asia, particularly in the southern areas of China. Previous data has demonstrated that microRNAs (miRNAs/miRs) may be involved in the formation and progression of NPC. The deregulation of miR-342 has been identified in multiple types of cancer. However, to the best of our knowledge, there are no data concerning miR-342 in NPC. The present study aimed to measure miR-342 expression in NPC, and to investigate its roles in NPC initiation and progression, in addition to the underlying molecular mechanisms. miR-342 was significantly downregulated in NPC tissues and cell lines. Low miR-342 expression was associated with distant metastasis and tumor node metastasis stage in patients with NPC. The restoration of the expression of miR-342 suppressed cell proliferation and invasion of NPC in vitro. In addition, ZEB1 was identified as a direct target gene of miR-342 in NPC. Downregulation of ZEB1 mimicked the tumor-suppressive roles of miR-342 in NPC. Taken together, the present study identified that miR-342 directly targeted ZEB1 to inhibit NPC cell growth and invasion, which may provide a novel therapeutic target for the treatments of patients with this malignancy.
Keywords: nasopharyngeal cancer, microRNA-342, invasion, proliferation, zinc finger E-box-binding homeobox 1
Introduction
Head and neck carcinomas, which are derived from the oral and pharyngeal squamous epithelium, are a group of malignancies that includes the tumors of the larynx, hypopharynx, oropharynx, nasopharynx, paranasal sinuses and the oral and nasal cavity (1). Nasopharyngeal carcinoma (NPC) is prevalent in Africa and East Asia, particularly in the southern areas of China, with an estimated morbidity of 20–50 cases per 100,000 individuals per year globally in 2002 (2,3). Epstein-Barr virus infection, tobacco use and genetic susceptibility are considered to contribute to the pathogenesis of NPC (4). At present, radiotherapy is the primary therapeutic approach for patients with early-stage NPC, whereas concomitant radio and chemotherapy is the standard treatment protocol for local advanced NPC (5). In spite of the development of therapeutic strategies, the prognosis of patients with NPC remains poor (6). The primarily reasons for treatment failure in patients with NPC are local recurrence and local or distant metastasis (7). Therefore, clarification of the mechanisms underlying the progression of NPC, which may provide novel therapeutic targets for patients with this disease, is urgent.
MicroRNAs (miRNAs/miRs) are single-stranded, short non-coding RNA molecules, containing 22–25 nucleotides (8); they post-transcriptionally regulate gene expression through base pairing to the 3′ untranslated regions (3′UTRs) of their target genes, causing translational inhibition or mRNA degradation, thereby decreasing protein expression (9). A number of studies have demonstrated that miRNAs serve notable roles in tumorigenesis and tumor development through the regulation of a variety of pathways in physiological and pathological processes, including tumor cell proliferation, cycle, differentiation, angiogenesis, invasion and metastasis (10–12). Aberrant expression of miRNAs has been observed in numerous types of human cancer, including NPC (13). In addition, an increasing number of studies have indicated that miRNAs may be involved in NPC formation and progression. For example, miR-338 is expressed at low levels in NPC, and inhibits tumor cell proliferation and migration by negative regulation of hypoxia-induced factor-1α (14). miR-205, which is upregulated in NPC, enhanced cell growth, metastasis and induced apoptosis through directly targeting the tumor protein p53-inducible nuclear protein (15). Therefore, miRNAs may have potential to facilitate the development of novel therapeutic biomarkers for the diagnosis, therapy and prognosis of human cancer owing to their tissue- and disease-specific expression and regulatory functions (16,17).
The deregulation of miR-342 has been identified in multiple types of cancer (18–23). However, to the best of our knowledge, there is no data concerning miR-342 in NPC. The present study aimed to measure miR-342 expression in NPC and investigate its functions in NPC initiation and progression, in addition to the underlying molecular mechanisms.
Materials and methods
Clinical tissues
A total of 52 NPC tissues (age range, 26–71 years; mean age, 48 years) and 11 normal nasopharyngeal epithelium specimens (age range, 34–62 years; mean age, 41 years) were obtained at Department of Otorhinolaryngology, Jining No. 1 People's Hospital (Jining, China). All tissues were frozen in liquid nitrogen and stored at −70°C. None of these patients were treated with radiotherapy or chemotherapy prior to biopsy. The Tumor Node Metastasis (TNM) staging system was used to classify all patients with NPC (24). The present study was approved by the Ethical Review Committee of Jining No. 1 People's Hospital. Written informed consent was also provided by each patient.
Cell lines and transient transfection
The human NPC SUNE1, 5–8F and 293T cell lines were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). These cells were grown in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin, and 100 U/ml streptomycin. The immortalized normal nasopharynx epithelial NP69 cells was obtained from the ATCC and cultured in keratinocyte-serum-free medium (Thermo Fisher Scientific, Inc.) supplemented with 30 µg/ml bovine pituitary extract (BD Biosciences, San Jose, CA, USA). All cell lines were maintained in an atmosphere of 5% CO2 at 37°C.
miR-342 mimics, negative control (NC) mimics, zinc finger E-box-binding homeobox 1 (ZEB1) and NC small interfering RNA (siRNA) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The miR-342 mimics sequence was 5′-UCUCACACAGAAAUCGCACCCGU-3′, and the NC sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The ZEB1 siRNA sequence was 5′-AACUGAACCUGUGGAUUAU-3′ and the NC siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. SUNE1 cells were seeded into 6-well plates with a density of 8×105 cells per well and incubated in an atmosphere of 5% CO2 at 37°C. When the cells reached 60–70% confluence, transient transfection was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following transfection for 48 h, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to detect miR-342 expression. MTT and Transwell invasion assays were conducted at 24 and 48 h post transfection. Western blotting analysis was performed at 72 h following transfection.
RT-qPCR
Total RNA was extracted from homogenized tissues and cells (SUNE1, 5–8F and NP69) using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A Moloney-Murine Leukemia Virus Reverse Transcription system (cat no. M1701; Promega Corporation, Madison, WI, USA) was used to synthesize the cDNA according to the manufacturer's protocol. For miR-342 expression, qPCR was conducted using a Hairpin-it™ miRNAs qPCR Quantitation kit (Shanghai GenePharma Co., Ltd.), with U6 as an internal control. The thermocycling conditions for qPCR were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 12 sec and 58°C for 40 sec. SYBR Green PCR Master Mix (Takara Biotechnology Co., Ltd., Dalian, China) was employed to measure ZEB1 mRNA expression, with β-actin as an internal control. The thermocycling conditions for qPCR were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primers were designed as follows: miR-342 forward, 5′-GTGCTATCTGTGATTGAGGGA'-3 and reverse, 5′-CGGGTGCGATTTCTGTG'-3; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; ZEB1 forward, 5′-CTCGAGCATTTAGACACAAGCG-3′ and reverse, 5′-TTGCCCTTCCTTTCCTGTGT-3′; β-actin forward, 5′-ATGGGTCAGAAGGATTCCTATGTG-3′ and reverse, 5′-CTTCATGAGGTAGTCAGTCAGGTC-3′. The relative expression of miR-342 and ZEB1 was calculated using the 2−ΔΔCq method (25).
MTT assay
At 24 h following transfection, cells were harvested and then seeded into 96-well plates at a density of 3×103 in each well. Cells were incubated in an atmosphere of 5% CO2 at 37°C for 1, 2, 3 and 4 days. An MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was performed at each time point. Briefly, 20 µl MTT solution (5 mg/ml) was added into the cultured cells and incubated at 37°C for an additional 4 h. The culture medium was removed carefully and 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added into each well. Absorbance at 490 nm was determined using an automatic multi-well spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were performed in triplicate.
Transwell invasion assay
Cell invasion ability was quantified using transwell apparatus (Costar; Corning Incorporated, Corning, NY, USA) coated with 50 ng/ml Matrigel (BD Biosciences, San Jose, CA, USA). At 48 h after transfection, cells were collected and suspended in FBS-free culture RPMI-1640 medium. A total of 1×105 cells were seeded into the upper chamber, and the bottom chamber was filled with 500 µl culture medium containing 10% FBS. Cells were incubated in an atmosphere of 5% CO2 at 37°C for 48 h. Cells remaining on the upper surface of the transwell apparatus were mechanically removed and the invaded cells were fixed in 4% paraformaldehyde at room temperature for 15 min, stained with 0.5% crystal violet at room temperature for 15 min and washed with PBS (Gibco; Thermo Fisher Scientific, Inc.). Values for the invasion ability were obtained by counting 5 random fields of cells/membrane under a light microscope (CKX41; Olympus Corporation, Tokyo, Japan) at a magnification, ×200.
Bioinformatics analysis
Bioinformatics analysis was performed to predicate the potential targets of miR-342 using TargetScan (Release 6.0: November 2011; https://www.targetscan.org) and PicTar (last update: March 26, 2007; http://pictar.mdc-berlin.de/). The search term used was has-miR-342-3p.
Western blotting
Transfected cells were harvested at 72 h post-transfection and lysed with Radioimmunoprecipitation Assay buffer with protease inhibitors (Cell Biolabs, Inc., San Diego, CA, USA). A Bicinchoninic Acid Assay kit (Pierce Biotechnology Inc., Rockford, IL, USA) was used to detect protein concentration. Equal amounts of protein (30 µg) were resolved by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). Following blocking at room temperature for 1 h with 5% non-fat milk in TBS containing 0.1% Tween-20 (TBST), the membranes were incubated with primary antibodies against ZEB1 (1:1,000 dilution; cat. no., sc-81428; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and GADPH (1:1,000 dilution; cat. no., sc-81545; Santa Cruz Biotechnology, Inc.) at 4°C overnight. Following this, the membranes were washed with TBST three times and blotted with horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; cat. no., sc-2005; Santa Cruz Biotechnology, Inc.) at room temperature for 1 h. The protein bands were developed using chemiluminescence detection reagents (Pierce; Thermo Fisher Scientific, Inc.) and analyzed using Quantity One software version 4.62 (Bio-Rad Laboratories, Inc.).
Luciferase reporter assay
A wild-type (Wt) ZEB1 3′-UTR containing the binding sequences of miR-342 (psiCHECK-ZEB1-3′UTR Wt) and a mutant type (Mut) ZEB1 3′-UTR lacking the binding sequences of miR-342 (psiCHECK-ZEB1-3′UTR Mut) were synthesized by Shanghai GenePharma Co., Ltd. 293T cells (ATCC) were seeded into 24-well plates at 40–50% confluence. Subsequent to adherence, 293T cells were transfected with miR-342 mimics or NC, along with psiCHECK-ZEB1-3′UTR WT or psiCHECK-ZEB1-3′UTR MUT, using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, transfected cells were collected and the luciferase activities were determined by a Dual-Luciferase® Reporter Assay system (Promega Corporation) following the manufacturer's protocol. Renilla luciferase activity was used for normalization.
Statistical analysis
Results were expressed as the means ± standard deviation and compared with paired Student's t-tests or one-way analysis of variance using SPSS 20.0 software (IBM Corp., Armonk, NY, USA). Student-Newman-Keuls test was used as a post hoc test. Spearman correlation analysis was used to explore the association between miR-342 and ZEB1 mRNA in NPC tissues. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-342 is downregulated in NPC tissues and cell lines
Firstly, the miR-342 expression level in human NPC tissues and normal nasopharyngeal epithelium specimens was measured using RT-qPCR. As demonstrated in Fig. 1A, miR-342 expression was significantly downregulated in NPC tissues compared with normal nasopharyngeal epithelium specimens (P<0.05). In addition, the expression levels of miR-342 in the NPC SUNE1 and 5–8F cell lines and immortalized normal nasopharynx epithelial NP69 cell line were detected. The results demonstrated that miR-342 expression in the NPC cell lines was reduced in comparison with the NP69 cells (Fig. 1B; P<0.05). In addition, the associations between miR-342 expression and clinicopathological factors of patients with NPC were investigated. As summarized in Table I, low miR-342 expression was markedly associated with distant metastasis (P=0.005) and TNM stage (P=0.010). However, there was no significant association with gender (P=0.815) or age (P=0.250) in NPC.
Figure 1.
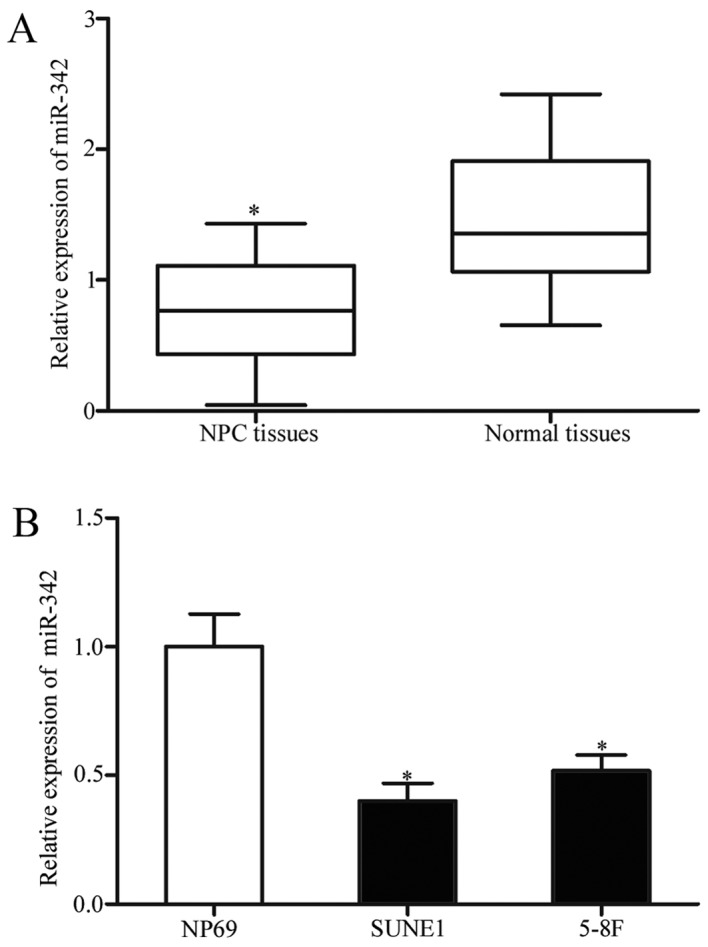
Expression levels of miR-342 in NPC tissues and cell lines. (A) miR-342 was expressed at low levels in NPC tissue compared with normal nasopharyngeal epithelium specimens. Data were presented as box plots. Top of the box indicates the upper quartile. Bottom of the box indicates the lower quartile. The central line in the box indicates the median. The whiskers indicate min to max. *P<0.05 vs. normal nasopharyngeal epithelium specimens. (B) Expression levels of miR-342 in two NPC cell lines were decreased compared with that in the immortalized normal nasopharynx epithelial NP69 cell line. *P<0.05 vs. NP69. miR-342, microRNA-342; NPC, nasopharyngeal cancer.
Table I.
Association between miR-342 expression and clinicopathological parameters in nasopharyngeal carcinoma.
| miR-342 expression | ||||
|---|---|---|---|---|
| Variable | Patients, n | Low | High | P-value |
| Sex | 0.815 | |||
| Male | 33 | 18 | 15 | |
| Female | 19 | 11 | 8 | |
| Age, years | 0.250 | |||
| <45 | 25 | 16 | 9 | |
| ≥45 | 27 | 13 | 14 | |
| Distant metastasis | 0.005 | |||
| No | 32 | 13 | 19 | |
| Yes | 20 | 16 | 4 | |
| TNM stage | 0.010 | |||
| I+II | 28 | 11 | 17 | |
| III+IV | 24 | 18 | 6 | |
TNM, Tumor-Node-Metastasis; miR-320, microRNA-320.
miR-342 suppresses cell proliferation and invasion of NPC
As miR-342 was expressed at low levels in NPC, miR-342 may serve important roles in NPC initiation and progression. To confirm this hypothesis, SUNE1 cells were transfected with miR-342 mimics or NC. Following transfection for 48 h, data from the RT-qPCR assay indicated that the levels of miR-342 were markedly increased in the SUNE1 cells transfected with miR-342 mimics (Fig. 2A; P<0.05). To evaluate the biological functions of miR-342 in NPC cell proliferation and invasion, MTT and transwell invasion assays were performed. The MTT assay demonstrated that the ectopic expression of miR-342 suppressed the proliferation of SUNE1 cells (Fig. 2B; P<0.05). Consistent with this, cell invasion was markedly reduced in SUNE1 cells transfected with miR-342 mimics, compared with those transfected with the NC (Fig. 2C; P<0.05). These results indicated that miR-342 may serve as a tumor suppressor in NPC cell growth and metastasis.
Figure 2.
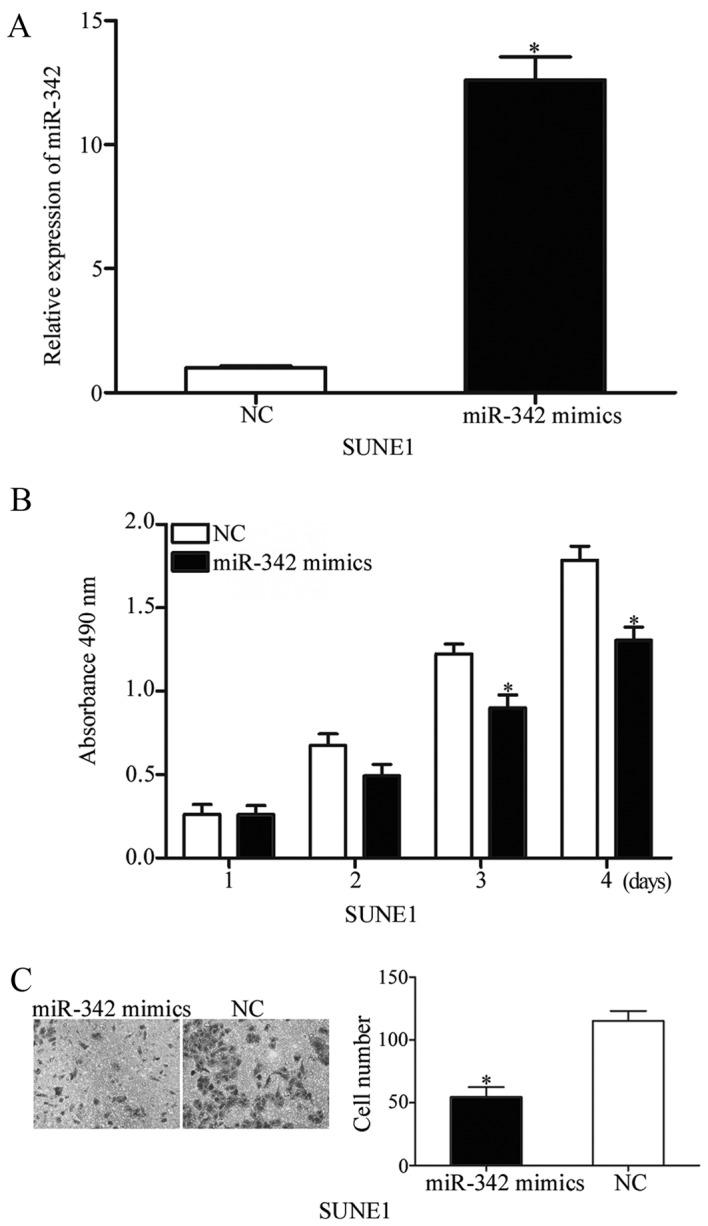
Restoration of miR-342 expression inhibits cell proliferation and invasion of nasopharyngeal cancer in vitro. (A) miR-342 expression was significantly higher in SUNE1 cells transfected with miR-342 mimics compared with the NC group. (B) miR-342 overexpression significantly suppressed SUNE1 cells proliferation. (C) Inhibition of cell invasion ability by miR-342 in SUNE1 cells, as determined by Transwell invasion assay. *P<0.05 vs. NC. miR-342, microRNA-342; NC, negative control.
ZEB1 is a direct target of miR-342 in NPC
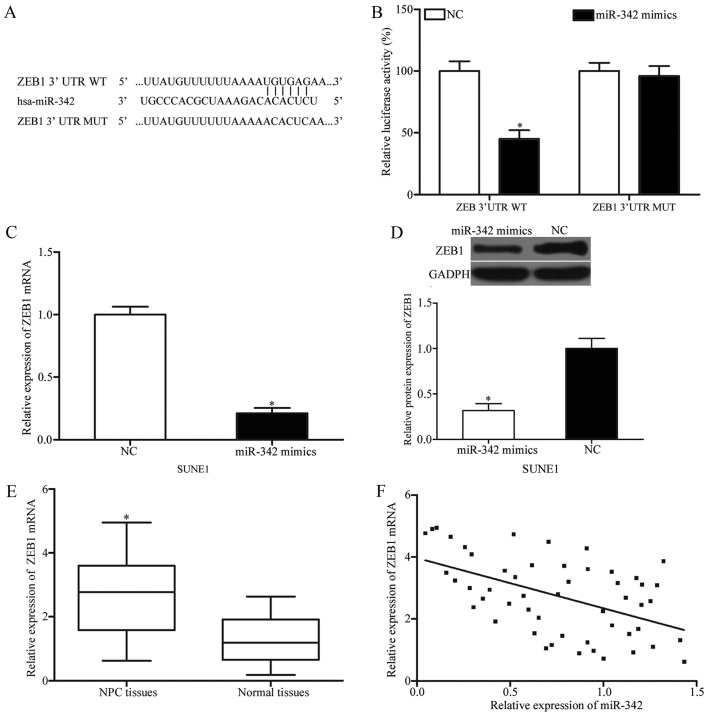
To investigate the mechanisms by which miR-342 inhibits the cell proliferation and invasion of NPC further, bioinformatics analysis was performed to identify the potential targets of miR-342. Among these putative targets, ZEB1 was selected for additional analysis (Fig. 3A) as it had previously been demonstrated to contribute to multiple cellular processes, including cell growth, apoptosis, migration, invasion, metastasis, tumor development and tumor progression (26–30). To validate whether ZEB1 is a direct target of miR-342, a luciferase reporter assay was conducted. miR-342 mimics or NC, along with luciferase reporter vectors were transfected into 293T cells. The results demonstrated that there was a significant decrease in luciferase activities in psiCHECK-ZEB1-3′UTR WT but not in psiCHECK-ZEB1-3′UTR MUT following the transfection with miR-342 mimics (Fig. 3B; P<0.05). To confirm the regulatory effect of miR-342 on ZEB1 expression, RT-qPCR and western blotting were performed to determine ZEB1 expression in SUNE1 cells in response to the changes in miR-342 overexpression. The results indicated that restoration of miR-342 expression decreased ZEB1 expression at the mRNA and protein levels in SUNE1 cells (Fig. 3C and D; P<0.05).
Figure 3.
ZEB1 is a direct target of miR-342 in NPC. (A) Predicted human ZEB1 3′UTR binding site for miR-342. (B) A luciferase reporter assay was performed in 293T cells co-transfected with psiCHECK-ZEB1-3′UTR WT or psiCHECK-ZEB1-3′UTR MUT and miR-342 mimics or NC. *P<0.05 vs. NC. Quantification of ZEB1 (C) mRNA and (D) protein expression in SUNE1 cells following transfection with miR-342 mimics or NC. *P<0.05 vs. NC. (E) Reverse transcription quantitative polymerase chain reaction analysis of ZEB1 mRNA expression in NPC tissues compared with normal nasopharyngeal epithelium specimens. Data were presented as box plots. Top of the box indicates the upper quartile. Bottom of the box indicates the lower quartile. The central line in the box indicates the median. The whiskers indicate min to max. *P<0.05 vs. normal nasopharyngeal epithelium specimens. (F) An inverse correlation between miR-342 and ZEB1 mRNA expression was observed in NPC tissues. r=0.5127; P=0.0001. *P<0.05. ZEB1, zinc finger E-box-binding homeobox 1; 3′UTR, 3′ untranslated region; WT, wild type; MUT, mutated; NC, negative control; miR-342, microRNA-342; NPC, nasopharyngeal cancer.
ZEB1 expression was additionally detected in NPC tissues and normal nasopharyngeal epithelium specimens. As indicated in Fig. 3E, ZEB1 mRNA was significantly upregulated in NPC tissues in comparison with normal nasopharyngeal epithelium specimens (P<0.05). Additionally, the association between ZEB1 mRNA and miR-342 expression levels was evaluated in NPC tissues. Spearman correlation analysis indicated that there was a negative correlation between miR-342 and ZEB1 mRNA expression (Fig. 3F; r=0.5127; P=0.0001). Collectively, these results demonstrated that ZEB1 is a direct target of miR-342 in NPC.
ZEB1 is involved in the regulation of NPC cell proliferation and invasion by miR-342
ZEB1 was validated as a direct target of miR-342 in NPC in the present study; therefore, we hypothesized that ZEB1-knockdown may mimic the suppressive functions of miR-342 in NPC. To confirm this assumption, ZEB1 siRNA was introduced into SUNE1 cells to reduce its expression (Fig. 4A; P<0.05). Subsequently, the MTT and transwell invasion assays indicated that the downregulation of ZEB1 inhibited proliferation (Fig. 4B; P<0.05) and invasion (Fig. 4C; P<0.05) of SUNE1 cells. These results indicated that miR-342 inhibits cell proliferation and invasion of NPC by suppressing its target gene, ZEB1.
Figure 4.
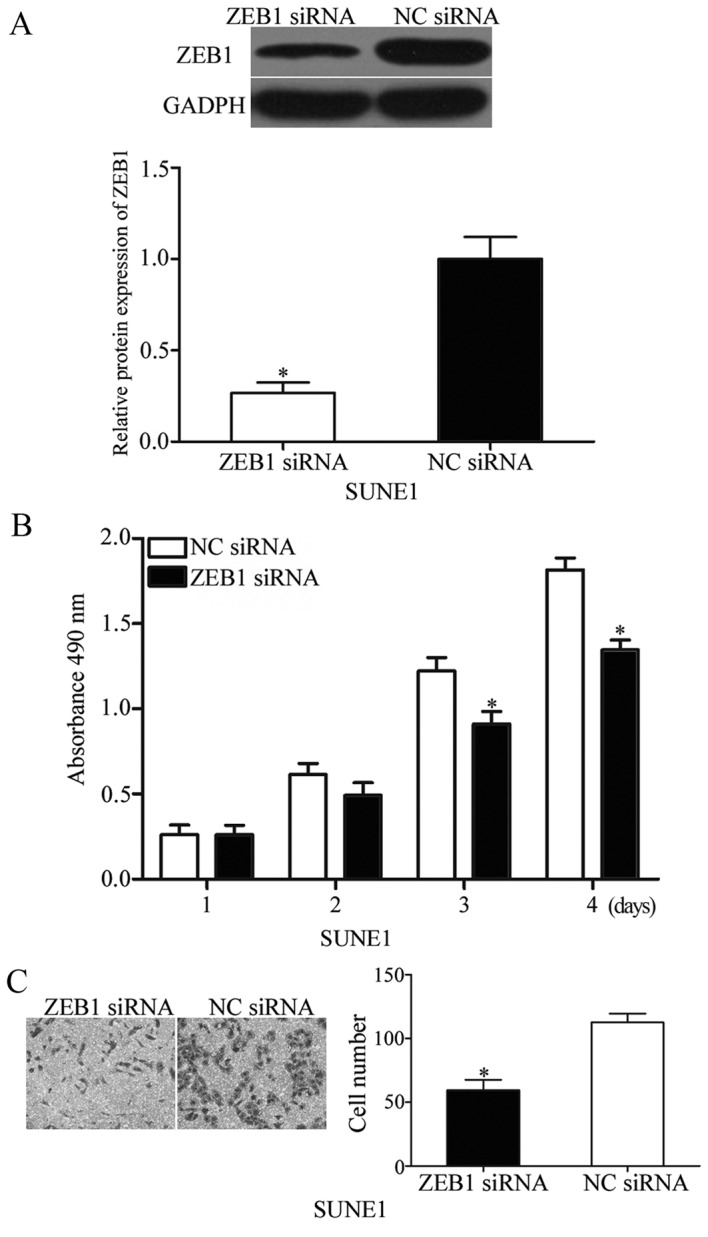
Inhibition of ZEB1 represses cell proliferation and invasion of nasopharyngeal cancer in vitro. (A) Western blot confirmed that ZEB1 protein expression was reduced by ZEB1 siRNA in SUNE1 cells. (B) The effect of ZEB1 knockdown on nasopharyngeal cancer cell proliferation was assessed by MTT assay. (C) Transwell invasion assay indicated a significant inhibition in cell invasion ability induced by ZEB1 knockdown in SUNE1 cells. *P<0.05 vs. NC siRNA. ZEB1, zinc finger E-box-binding homeobox 1; NC, negative control; siRNA, small interfering RNA.
Discussion
miR-342 has been previously demonstrated to be abnormally expressed in several types of tumors (18–23). In colorectal cancer, miR-342 was significantly downregulated in tumor tissues and cell lines (31). In breast cancer, the expression levels of miR-342 were demonstrated to be deregulated and markedly associated with estrogen receptor, human epidermal growth factor receptor 2 and vascular endothelial growth factor expression (18–20). In cervical cancer, miR-342 was expressed at low levels in tumor tissues (21). miR-342 was demonstrated to be downregulated in hepatocellular carcinoma (32) and lung cancer (22,23). However, miR-342 was identified to be highly expressed in multiple myeloma (33). In the present study, the results revealed that miR-342 expression was lower in NPC tissues and cell lines compared with that in normal nasopharyngeal epithelium specimens and the immortalized normal nasopharynx epithelial NP69 cell line. The expression levels of miR-342 were significantly associated with distant metastasis and TNM stage in patients with NPC. Therefore, miR-342 may contribute to tumorigenesis and tumor development, and may have a diverse expression pattern depending on the type of cancer.
mir-342 has previously been identified to generally serve important roles in several types of human cancer (21,31,32): For example, Wang et al (31) indicated that the ectopic expression of miR-342 induced the significant inhibition of colorectal cancer cell proliferation and invasion by the negative regulation of DNA (cytosine-5)-methyltransferase 1. A study by Li et al (21) indicated that the restoration of the expression of miR-342 targeted forkhead box protein M1 to inhibit cervical cancer cell growth and metastasis. Zhao et al (32) revealed that miR-342 overexpression repressed cell proliferation in hepatocellular carcinoma through targeting transforming growth factor-β activated kinase 1/mitogen-activated protein kinase kinase kinase 7 binding protein 2 (TAB2) and TAB3, consequently regulating the NF-κB pathway. Xie et al (22) demonstrated that, in non-small cell lung cancer, the upregulation of miR-342 markedly suppressed cell proliferation and invasion in vitro, and decreased tumor growth in vivo, via the blockage of Ras-related protein Rap-2b. Xu et al (23) revealed that miR-342 re-expression reduced cell proliferation of small cell lung cancer by directly targeting islet antigen-2. Together, these data indicated that miR-342 may provide a novel therapeutic target for the treatments of these types of cancer.
The mechanisms underlying how miR-342 affects cancer initiation and progression have been described previously in several types of human cancer (21,31,32). In the present study, ZEB1 was validated as a novel target of miR-342. First, bioinformatics analysis predicted that ZEB1 mRNA contained a miR-342 seed match at position 488–494 of the ZEB1 3′UTR. Second, the luciferase reporter assay demonstrated that miR-342 directly targeted the 3′UTR of ZEB1 mRNA. Third, RT-qPCR and western blotting indicated the negative regulation of miR-342 on ZEB1 mRNA and protein expression in NPC. Fourth, an inverse correlation was observed between ZEB1 mRNA and miR-342 expression in NPC tissues. Finally, ZEB1-knockdown attenuated cell proliferation and invasion of NPC, which was similar with the function induced by miR-342 overexpression. These results demonstrated that miR-342 may exhibit tumor-suppressive roles in NPC formation and progression by targeting ZEB1.
ZEB1, a member of the zinc finger family, is located on the short arm of human chromosome 10 (34). The expression level of ZEB1 has been demonstrated to be upregulated in various types of human cancer, including colorectal (35), thyroid (36), cervical (37), gastric (27) and bladder cancer (38). A number of studies have suggested that ZEB1 may be involved in a large number of cellular processes, including cell growth, apoptosis, migration, invasion, metastasis, tumor development and tumor progression (26–30). In the present study, it was identified that ZEB1 was significantly upregulated in NPC tissues. The downregulation of ZEB1 suppressed cell proliferation and invasion of NPC. Therefore, targeting ZEB1 may be an efficient strategy for NPC therapy. Previous studies have also identified that ZEB1 was regulated by various miRNAs in human cancer (39–41). In a previous study, miR-429 was reported to be downregulated in NPC and to suppress cell migration and invasion through targeting ZEB1 (42). Collectively, these results indicate that the miR-342/ZEB1 axis warrants investigation as a potential target of therapy to prevent the rapid growth and metastasis of NPC.
In summary, the present study demonstrated that miR-342 expression was reduced in NPC tissues and cells, and its tumor suppressive roles in NPC growth and invasion were mediated by direct targeting of its target ZEB1. miR-342 may be a useful potential therapeutic target in NPC.
References
- 1.Arango BA, Castrellon AB, Perez CA, Raez LE, Santos ES. Nasopharyngeal carcinoma: Alternative treatment options after disease progression. Expert Rev Anticancer Ther. 2010;10:377–386. doi: 10.1586/era.10.1. [DOI] [PubMed] [Google Scholar]
- 2.Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12:421–429. doi: 10.1016/S1044579X02000858. [DOI] [PubMed] [Google Scholar]
- 3.Chan AT, Teo PM, Johnson PJ. Nasopharyngeal carcinoma. Ann Oncol. 2002;13:1007–1015. doi: 10.1093/annonc/mdf179. [DOI] [PubMed] [Google Scholar]
- 4.Yip TT, Ngan RK, Fong AH, Law SC. Application of circulating plasma/serum EBV DNA in the clinical management of nasopharyngeal carcinoma. Oral Oncol. 2014;50:527–538. doi: 10.1016/j.oraloncology.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Chan AT, Grégoire V, Lefebvre JL, Licitra L, Hui EP, Leung SF, Felip E, EHNS–ESMO–ESTRO Guidelines Working Group Nasopharyngeal cancer: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl 7):vii83–vii85. doi: 10.1093/annonc/mds266. [DOI] [PubMed] [Google Scholar]
- 6.Lai SZ, Li WF, Chen L, Luo W, Chen YY, Liu LZ, Sun Y, Lin AH, Liu MZ, Ma J. How does intensity-modulated radiotherapy versus conventional two-dimensional radiotherapy influence the treatment results in nasopharyngeal carcinoma patients? Int J Radiat Oncol Biol Phys. 2011;80:661–668. doi: 10.1016/j.ijrobp.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 7.Zhang P, Hong H, Sun X, Jiang H, Ma S, Zhao S, Zhang M, Wang Z, Jiang C, Liu H. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/Notch1/E-cadherin in cisplatin-resistant nasopharyngeal carcinoma cells. Am J Cancer Res. 2016;6:141–156. [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Mack GS. MicroRNA gets down to business. Nat Biotechnol. 2007;25:631–638. doi: 10.1038/nbt0607-631. [DOI] [PubMed] [Google Scholar]
- 10.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 11.Kim NH, Cha YH, Kang SE, Lee Y, Lee I, Cha SY, Ryu JK, Na JM, Park C, Yoon HG, et al. p53 regulates nuclear GSK-3 levels through miR-34-mediated Axin2 suppression in colorectal cancer cells. Cell Cycle. 2013;12:1578–1587. doi: 10.4161/cc.24739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Ai Q, Cao H, Liu Q. miR-185-3p and miR-324-3p predict radiosensitivity of nasopharyngeal carcinoma and modulate cancer cell growth and apoptosis by targeting SMAD7. Med Sci Monit. 2015;21:2828–2836. doi: 10.12659/MSM.895660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan Y, Li X, You B, Shi S, Zhang Q, You Y. MicroRNA-338 inhibits migration and proliferation by targeting hypoxia-induced factor 1α in nasopharyngeal carcinoma. Oncol Rep. 2015;34:1943–1952. doi: 10.3892/or.2015.4195. [DOI] [PubMed] [Google Scholar]
- 15.Nie G, Duan H, Li X, Yu Z, Luo L, Lu R, Ji Z, Zhang W. MicroRNA205 promotes the tumorigenesis of nasopharyngeal carcinoma through targeting tumor protein p53-inducible nuclear protein 1. Mol Med Rep. 2015;12:5715–5722. doi: 10.3892/mmr.2015.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartels CL, Tsongalis GJ. MicroRNAs: Novel biomarkers for human cancer. Ann Biol Clin (Paris) 2010;68:263–272. doi: 10.1684/abc.2010.0429. (In French) [DOI] [PubMed] [Google Scholar]
- 17.Wu D, Niu X, Pan H, Zhou Y, Zhang Z, Qu P, Zhou J. Tumor-suppressing effects of microRNA-429 in human renal cell carcinoma via the downregulation of Sp1. Oncol Lett. 2016;12:2906–2911. doi: 10.3892/ol.2016.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savad S, Mehdipour P, Miryounesi M, Shirkoohi R, Fereidooni F, Mansouri F, Modarressi MH. Expression analysis of miR-21, miR-205, and miR-342 in breast cancer in Iran. Asian Pac J Cancer Prev. 2012;13:873–877. doi: 10.7314/APJCP.2012.13.3.873. [DOI] [PubMed] [Google Scholar]
- 19.He YJ, Wu JZ, Ji MH, Ma T, Qiao EQ, Ma R, Tang JH. miR-342 is associated with estrogen receptor-alpha expression and response to tamoxifen in breast cancer. Exp Ther Med. 2013;5:813–818. doi: 10.3892/etm.2013.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma T, Zhang J, Wu J, Tang J. Effect of miR-342-3p on chemotherapy sensitivity in triple-negative breast cancer. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2014;39:488–495. doi: 10.3969/j.issn.1672-7347.2014.05.009. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 21.Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588:3298–3307. doi: 10.1016/j.febslet.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 22.Xie X, Liu H, Wang M, Ding F, Xiao H, Hu F, Hu R, Mei J. miR-342-3p targets RAP2B to suppress proliferation and invasion of non-small cell lung cancer cells. Tumour Biol. 2015;36:5031–5038. doi: 10.1007/s13277-015-3154-3. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Cai T, Carmona GN, Abuhatzira L, Notkins AL. Small cell lung cancer growth is inhibited by miR-342 through its effect of the target gene IA-2. J Transl Med. 2016;14:278. doi: 10.1186/s12967-016-1036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.OuYang PY, Su Z, Ma XH, Mao YP, Liu MZ, Xie FY. Comparison of TNM staging systems for nasopharyngeal carcinoma, and proposal of a new staging system. Br J Cancer. 2013;109:2987–2997. doi: 10.1038/bjc.2013.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Kenney PA, Wszolek MF, Rieger-Christ KM, Neto BS, Gould JJ, Harty NJ, Mosquera JM, Zeheb R, Loda M, Darling DS, et al. Novel ZEB1 expression in bladder tumorigenesis. BJU Int. 2011;107:656–663. doi: 10.1111/j.1464-410X.2010.09489.x. [DOI] [PubMed] [Google Scholar]
- 27.Jia B, Liu H, Kong Q, Li B. Overexpression of ZEB1 associated with metastasis and invasion in patients with gastric carcinoma. Mol Cell Biochem. 2012;366:223–229. doi: 10.1007/s11010-012-1299-6. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Sun B, Qi L, Li H, Gao J, Leng X. Zinc finger E-box binding homeobox 1 promotes vasculogenic mimicry in colorectal cancer through induction of epithelial-to-mesenchymal transition. Cancer Sci. 2012;103:813–820. doi: 10.1111/j.1349-7006.2011.02199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quan Y, Jin R, Huang A, Zhao H, Feng B, Zang L, Zheng M. Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer Biol Ther. 2014;15:878–887. doi: 10.4161/cbt.28877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G, An H, Fang X. MicroRNA-144 regulates proliferation, invasion, and apoptosis of cells in malignant solitary pulmonary nodule via zinc finger E-box-binding homeobox 1. Int J Clin Exp Pathol. 2015;8:5960–5967. [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu R, Pan Z, Kang T, Huang W. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis. 2011;32:1033–1042. doi: 10.1093/carcin/bgr081. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, Zhang Y. miR-342-3p affects hepatocellular carcinoma cell proliferation via regulating NF-κB pathway. Biochem Biophys Res Commun. 2015;457:370–377. doi: 10.1016/j.bbrc.2014.12.119. [DOI] [PubMed] [Google Scholar]
- 33.Ronchetti D, Lionetti M, Mosca L, Agnelli L, Andronache A, Fabris S, Deliliers GL, Neri A. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-342, and miR-561 with deregulated host genes in multiple myeloma. BMC Med Genomics. 2008;1:37. doi: 10.1186/1755-8794-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen A, Zhang Y, Yang H, Xu R, Huang G. Overexpression of ZEB1 relates to metastasis and invasion in osteosarcoma. J Surg Oncol. 2012;105:830–834. doi: 10.1002/jso.23012. [DOI] [PubMed] [Google Scholar]
- 35.Zhang GJ, Zhou T, Tian HP, Liu ZL, Xia SS. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol Lett. 2013;5:564–568. doi: 10.3892/ol.2012.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Liu G, Wu S, Jiang F, Xie J, Wang Y. Zinc finger E-box-binding homeobox 1: Its clinical significance and functional role in human thyroid cancer. Onco Targets Ther. 2016;9:1303–1310. doi: 10.2147/OTT.S96723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y, Zheng X, Zhou J, Zhang Y, Chen K. ZEB1 promotes the progression and metastasis of cervical squamous cell carcinoma via the promotion of epithelial-mesenchymal transition. Int J Clin Exp Pathol. 2015;8:11258–11267. [PMC free article] [PubMed] [Google Scholar]
- 38.Ning Z, Wu K, Fan J, Wang B, Lv C, Zhu J, Wang X, Hsieh JT, He D. Aberrant expressions of β-catenin and ZEB1 in bladder cancer and their significance. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2014;30:1080–1083. (In Chinese) [PubMed] [Google Scholar]
- 39.Chen G, Huang P, Xie J, Li R. microRNA211 suppresses the growth and metastasis of cervical cancer by directly targeting ZEB1. Mol Med Rep. 2018;17:1275–1282. doi: 10.3892/mmr.2017.8006. [DOI] [PubMed] [Google Scholar]
- 40.Wu G, Wang J, Chen G, Zhao X. microRNA-204 modulates chemosensitivity and apoptosis of prostate cancer cells by targeting zinc-finger E-box-binding homeobox 1 (ZEB1) Am J Transl Res. 2017;9:3599–3610. [PMC free article] [PubMed] [Google Scholar]
- 41.Zou J, Liu L, Wang Q, Yin F, Yang Z, Zhang W, Li L. Downregulation of miR-429 contributes to the development of drug resistance in epithelial ovarian cancer by targeting ZEB1. Am J Transl Res. 2017;9:1357–1368. [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Jiang C, Sun Q, Yan F, Wang L, Fu Z, Liu T, Hu F. Downregulation of miR429 and inhibition of cell migration and invasion in nasopharyngeal carcinoma. Mol Med Rep. 2016;13:3236–3242. doi: 10.3892/mmr.2016.4940. [DOI] [PubMed] [Google Scholar]