Publisher's Note: There is a Blood Commentary on this article in this issue.
Key Points
DLBCL patients with concomitant HBV infection are characterized by distinct clinical features.
Genomic and transcriptomic analyses identified distinct mutation targets and tumorigenic pathways in HBV-associated DLBCLs.
Abstract
Hepatitis B virus (HBV) infection is endemic in some parts of Asia, Africa, and South America and remains to be a significant public health problem in these areas. It is known as a leading risk factor for the development of hepatocellular carcinoma, but epidemiological studies have also shown that the infection may increase the incidence of several types of B-cell lymphoma. Here, by characterizing altogether 275 Chinese diffuse large B-cell lymphoma (DLBCL) patients, we showed that patients with concomitant HBV infection (surface antigen positive [HBsAg+]) are characterized by a younger age, a more advanced disease stage at diagnosis, and reduced overall survival. Furthermore, by whole-genome/exome sequencing of 96 tumors and the respective peripheral blood samples and targeted sequencing of 179 tumors from these patients, we observed an enhanced rate of mutagenesis and a distinct set of mutation targets in HBsAg+ DLBCL genomes, which could be partially explained by the activities of APOBEC and activation-induced cytidine deaminase. By transcriptome analysis, we further showed that the HBV-associated gene expression signature is contributed by the enrichment of genes regulated by BCL6, FOXO1, and ZFP36L1. Finally, by analysis of immunoglobulin heavy chain gene sequences, we showed that an antigen-independent mechanism, rather than a chronic antigenic simulation model, is favored in HBV-related lymphomagenesis. Taken together, we present the first comprehensive genomic and transcriptomic study that suggests a link between HBV infection and B-cell malignancy. The genetic alterations identified in this study may also provide opportunities for development of novel therapeutic strategies.
Visual Abstract
Introduction
Diffuse large B-cell lymphoma (DLBCL) is one of the most common lymphoid malignancies, accounting for 30% to 40% of non-Hodgkin lymphomas (NHLs). Based on gene expression profiling, 2 major molecular subtypes with different chromosomal alterations and clinical outcomes have been identified: germinal center B-cell–like (GCB) and activated B-cell–like subtypes.1 Based on the latest 2016 World Health Organization classification, additional DLBCL associated subtypes have been defined, such as large B-cell lymphoma with IRF4 rearrangement, high-grade B-cell lymphoma with MYC, BCL2, and/or BCL6 translocations and Epstein-Barr virus (EBV)–positive DLBCL.2 These new entities highlight that specific chromosomal alterations or infectious agents are associated with different clinical manifestations. Further classification/subtyping of DLBCLs based on genetic characterizations is expected to have a significant impact on etiological studies, clinical management, and development of new therapeutic strategies for these patients.
Several studies, including the most recent one on 1001 patients, have explored the genome of DLBCL by using next-generation sequencing technologies.3-11 These studies have revealed recurrently occurring somatic mutations or structural variants that affect multiple cellular processes or signaling pathways, including apoptosis, B-cell receptor (BCR), and nuclear factor κB (NF-κB) signaling, epigenetic modifications, immune regulation/evasion and cell migration. The mutation spectra identified, however, differ somewhat in the published patient cohorts and the overlap of the mutated genes is still limited between these studies,7,8,12 which may reflect a high genetic heterogeneity in DLBCL. Of note, ethnic background seems to influence the mutational spectra, where some potential cancer-driver genes, such as CD70, CXCR4, DTX1, LYN, and TMSB4X, were mutated at a higher frequency in Chinese DLBCL patients, whereas GNA13 and EZH2 seemed to be more frequently mutated in patients from Western populations.8,13 The genomic changes in DLBCL may also be affected by the age of patients,14 alterations in DNA repair genes,15 exposure to different etiological agents,16 and/or autoimmune/inflammatory immune responses.17
Previous studies have shown that several types of virus, including HIV, EBV, human T-cell leukemia-lymphoma virus, and human herpesvirus-8, may contribute to the development of selected subtypes of lymphoma.18 A recent study has also shown that antiviral treatment can lead to a complete remission of hepatitis C virus (HCV)–associated, low-grade NHL, suggesting a causative role of HCV in these tumors.19 Meta-analyses have further shown that patients infected with hepatitis B virus (HBV) have a two- to threefold higher risk of developing NHL,20-23 including DLBCL.21,24 It also has been reported that DLBCL patients infected with HBV are diagnosed at a young age with a more advanced disease stage and show a significantly worse outcome.24 Thus, epidemiological and clinical studies have both suggested an association of HBV infection with DLBCL. However, the etiopathological role of HBV in lymphomagenesis remains largely unknown. Here, we performed a comprehensive genetic study on DLBCLs from patients with HBV infection and identified distinct molecular features of these tumors.
Methods
Patient information
Frozen tumor biopsy specimens from affected lymph nodes of 275 Chinese DLBCL patients were obtained from the Sun Yat-Sen University Cancer Center and Tianjin Medical University Cancer Institute and Hospital. DLBCL samples were classified as GCB or non-GCB based on the Hans algorithm.25 Of these, 266 were de novo and 9 might have resulted from the transformation of follicular lymphoma; 229 were taken at diagnosis, and 46 were taken at relapse (supplemental Table 1, available on the Blood Web site). Detection of HBV surface-antigen (HBsAg) and e-antigen and antibodies against HBV surface-antigen, e-antigen, and core-antigen was performed as routine blood tests in all patients, and the quantitation of viral DNA was performed in a subset of HBsAg+ patients (44 patients). The study was approved by the institutional review boards at the Sun Yat-Sen University Cancer Center, the Tianjin Medical University Cancer Institute, and Hospital and the Karolinska Institutet.
Whole-genome/exome sequencing (WGS/WES) analysis
DNA was extracted with the DNeasy Tissue and Blood Kit (Qiagen, Venlo, The Netherlands). WGS/WES was performed in BGI-Shenzhen using either HiSequation 2000 or HiSeq X10 platforms (Illumina, San Diego, CA) or Complete Genomics (BGI). 60 pairs of DLBCL/control samples were sequenced by WGS, and 47 pairs of samples were sequenced by WES. Among these, 11 pairs of samples have been sequenced by both WGS and WES. Analysis of significance of mutated genes was performed by 3 prediction methods,26-28 and the resulting top gene lists (q value < 0.1) were merged to identify significantly mutated genes in the discovery cohort. The mutational signature analysis was performed based on WGS data using a Bayesian version of the nonmatrix factorization method.29 More details are provided in supplemental Methods.
Targeted capture sequencing by lymphochip
Genes included in the targeted sequencing panel lymphochip were selected with the following criteria: (1) mutated in ≥3 cases within our discovery cohort and expressed in DLBCL samples, (2) recurrently mutated in ≥3 cases in other B-cell lymphoma cohorts, (3) important for DNA repair, and (4) important for targeted therapy. Together, this consists of the entire coding regions of 212 genes (supplemental Table 2), for a total size of 572 kb, and the lymphochip was synthesized at BGI. 100 ng DNA of each sample was used for the preparation of libraries. 2 × 50 bp pair-end sequencing was performed on BGISEQ500 platform.30
Transcriptome resequencing and gene set enrichment analysis (GSEA)
Transcriptome sequencing was performed on 108 DLBCL samples. Total RNA was extracted using Trizol (Invitrogen, Paisley, UK) and the libraries were prepared and sequenced at BGI (HiSequation 2000).31 Reads were aligned to the human reference genome and transcriptome hg19 by SOAP2.32 The number of transcripts per million was used to determine gene expression levels. Log2-transformed transcripts per million values were normalized by the R package Limma to remove the batch effect.33 Normalized expression levels were analyzed by Qlucore Omics Explorer (Qlucore AB, Lund, Sweden) and GSEA (Broad Institute).34
Functional characterization of TP73 mutants
The full length of the TAp73 sequence (tumor suppressor isoform of p73, a structural homology of p53) was cloned into a pcDNA 3.1 vector (Thermo Fisher Scientific, San Diego, CA). TAp73 mutants were generated by site-directed mutagenesis. TAp73 vectors and a reporter construct containing a p53-binding site in front of a GFP reporter (p53-responsive EGFP reporter) were cotransfected into H1299 cells (lacking p53 expression) with Lipofectamine 2000 (Thermo Fisher Scientific) in a 6-well plate. After 24 to 48 hours, cells were harvested and GFP level was measured by flow cytometry (Accuri C6; BD Biosciences).
Statistics
P values were calculated using appropriate statistical tests (χ2-test, Fisher’s exact test or Mann-Whitney U test [2 tailed]). Statistical tests were performed with GraphPad Prism (La Jolla, CA).
Results
Clinical features in HBsAg+ DLBCL patients
In our DLBCL cohort, 20% of patients were HBsAg+ (56/275), which is notably higher than the positive rate in the general population (7%).35 There was no significant difference between the HBsAg+ and HBsAg− groups in the percentage of GCB vs non-GCB patients or sex (Table 1). However, compared with HBsAg− cases, the HBsAg+ DLBCL patients displayed significantly younger age at diagnosis (median age, 42 vs 60 years; P < .0001), higher international prognostic index, more advanced disease at diagnosis (stage III/IV), and more frequent involvement of spleen (Table 1). Furthermore, they showed significantly reduced overall survival as compared with the HBsAg− patients (supplemental Figure 1A). When the 2 cell-of-origin subtypes or the treatment were considered separately, a significantly reduced overall survival was observed in HBsAg+ patients with the GCB subtype (supplemental Figure 1B-C). Patients with a relatively high HBV viral load (≥2000 IU/mL) had clinical characteristics similar to those of the remaining HBsAg+ patients, except that they appeared to be younger at diagnosis (median age, 32 vs 48 years; P = .0281; supplemental Figure 2A).
Table 1.
Clinical characteristics in HBsAg+ and HBsAg− DLBCL patients
| HBsAg+ DLBCL | HBsAg− DLBCL | P* | |
|---|---|---|---|
| No. of patients | 56 | 219 | — |
| Age, y | |||
| >60 | 10 (17.9) | 117 (53.4) | <.0001 |
| ≤60 | 46 (82.1) | 102 (46.6) | |
| Sex | |||
| Female | 16 (28.6) | 86 (39.3) | .1391 |
| Male | 40 (71.4) | 133 (60.1) | |
| Performance status† | |||
| 0-1 | 41 (73.2) | 176 (81.9) | .1491 |
| 2-4 | 15 (26.8) | 39 (18.1) | |
| Elevated LDH† | |||
| Yes | 32 (59.3) | 97 (46.4) | .0922 |
| No | 22 (40.7) | 112 (53.6) | |
| Subtype† | |||
| GCB | 21 (37.5) | 83 (38.1) | .9372 |
| Non-GCB | 35 (62.5) | 135 (61.9) | |
| Stage† | |||
| I-II | 14 (25.9) | 112 (54.6) | .0002 |
| III-IV | 40 (74.1) | 93 (45.4) | |
| IPI† | |||
| 0-2 | 28 (51.9) | 146 (71.2) | .0070 |
| 3-5 | 26 (48.1) | 59 (28.8) | |
| Spleen involvement† | |||
| Yes | 19 (38.0) | 34 (17.6) | .0019 |
| No | 31 (62.0) | 159 (82.4) | |
| Liver involvement† | |||
| Yes | 7 (14.0) | 14 (7.3) | .1303 |
| No | 43 (86.0) | 179 (92.7) |
Values are reported as n (%) of patients unless indicated otherwise.
IPI, international prognostic index; LDH, lactate dehydrogenase.
χ2 test was used for comparison. Significant values (P < .05) are highlighted in bold.
The calculation was based on 271, 263, 274, 259, or 243 samples with available data.
Genetic landscape of HBV-associated DLBCLs
Identification of significantly mutated genes in the discovery cohort
We next performed WGS/WES on DNAs prepared from tumor biopsy specimens and matched peripheral blood samples from 65 Chinese DLBCL patients. By including previously sequenced 31 Chinese DLBCL samples,8,36 altogether, 96 paired tumor/control samples were analyzed as a discovery cohort, including 20 HBsAg+ tumors. A median of 162 (11-460) or 10 835 (635-36 052) somatic mutations per tumor were observed in samples sequenced by WES or WGS, respectively. Among these, a median of 67 (5-160, WES) or 79 (9-159, WGS) nonsilent mutations in the coding region per tumor were observed.
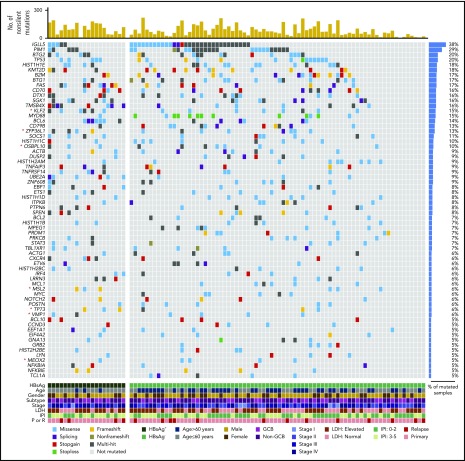
Three prediction methods were performed to define cancer-driver genes in our DLBCL discovery cohort.26-28 Altogether, 131 genes were affected by somatically occurring, nonsilent mutations in at least 5 samples (5%) and among these, 66 genes were considered to be statistically significantly mutated (Figure 1). The most frequently mutated genes (≥10%) were PIM1, BTG2, TP53, HIST1H1E, KMT2D/MLL2, B2M, BTG1, FAS, CD70, DTX1, SGK1, TMSB4X, KLF2, MYD88, BCL6, CD79B, ZFP36L1, SOCS1, HIST1H1C, and OSBPL10. Among the 66 potential cancer-driver genes, KLF2 (15%), ZFP36L1 (13%), OSBPL10 (10%), VMP1 (6%), TP73 (6%), MSL2 (6%), and MEOX2 (5%) have not been appreciated previously as being significant mutation targets in DLBCLs. Genes significantly mutated in our cohort were further assigned to the DAVID database for KEGG pathway analysis. Notably, consistent with the high HBV infection rate in our DLBCL cohort, one of the most significantly mutated pathways is the HBV-infection associated pathway, which has not been described previously. Additional significant pathways identified include BCR, JAK-STAT, and NF-κB signaling pathways (supplemental Table 3).
Figure 1.
List of the top somatic mutation targets in Chinese DLBCLs. Genes affected by nonsilent, somatically occurring mutations in ≥5 DLBCL samples in the discovery cohort (5% of 96 cases) and considered to be significantly mutated (q value < 0.1) are displayed. Novel significant mutation targets identified in our cohort are marked with an asterisk (red). IPI, international prognostic index; LDH, lactate dehydrogenase (level); P/R, primary/relapse.
Enhanced mutagenesis in HBV-associated DLBCLs
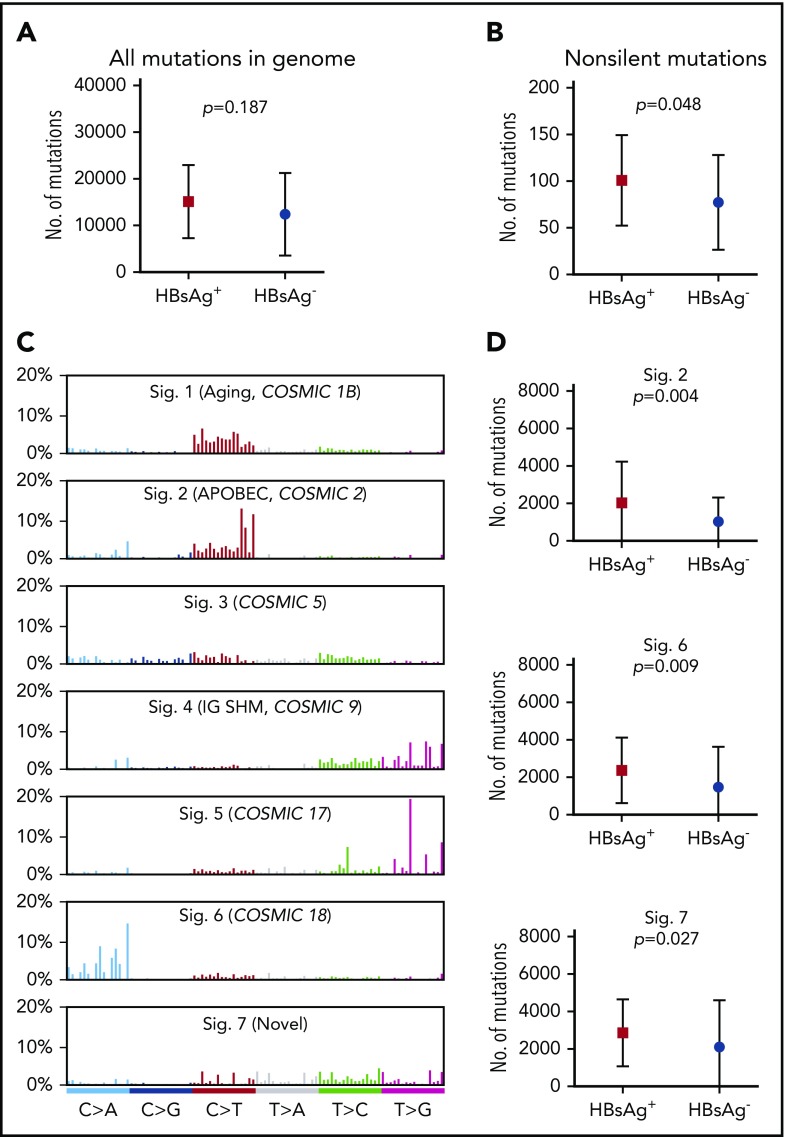
The general mutation profile in the genome of HBsAg+ and HBsAg− DLBCLs was subsequently compared. The total mutation load in the whole genome was higher in HBsAg+ DLBCLs, although this difference did not reach statistical significance (median, 15 036 vs 9902; Figure 2A). In the coding genome, significantly more nonsilent mutations were observed in HBsAg+ DLBCLs (median, 99 vs 66; Figure 2B).
Figure 2.
Enhanced mutagenesis in HBsAg+DLBCLs. (A-B) Comparison of the mutation load in the whole genome (A) or coding genome (B) between HBsAg+ and HBsAg− DLBCLs. (C) Major mutational signatures were identified according to the 96-substitution classifications from 60 Chinese DLBCLs characterized by WGS. (D) Signatures 2, 6, and 7 were enriched in the HBsAg+ samples. Error bars represent standard deviation. A Mann-Whitney U test was used to calculate the P value. Sig, signature.
The genome-wide mutational signatures of 60 cases with WGS data were subsequently characterized based on the 96 possible mutation types. Seven highly confident mutational signatures were extracted from our cohort (Figure 2C), and signatures 2, 6, and 7 were significantly enriched in HBsAg+ tumors (Figure 2D). Signatures 2 and 6 have previously been described,29 whereas signature 7, which was mainly characterized by T to V mutations (71%, V = A/C/G), was novel. Of these 3 mutation signatures, signature 2 has been linked to APOBEC enzymes, which belong to a family of proteins that usually function in the defense against viral infections, such as HIV and HBV.37,38 Taken together, our data suggest that HBV-associated DLBCLs display overall enhanced mutagenesis and are associated with selected mutational signatures, which may be partially resulted from APOBEC enzyme activity.
Distinct set of mutated genes in HBsAg+ DLBCLs
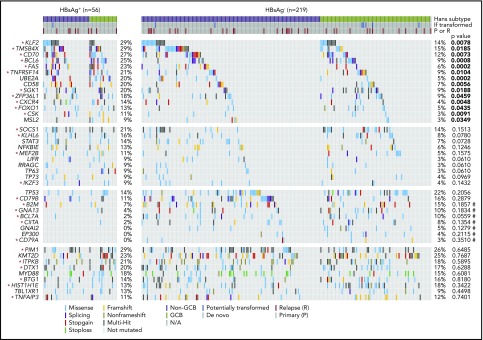
Based on the discovery cohort, we noted that some genes were significantly more mutated in the HBsAg+ group, such as TMSB4X, BCL6, FAS, UBE2A, DDX3X, CXCR4, KLF2, and SGK1 (supplemental Table 4). To further validate and define preferentially mutated genes in HBsAg+ DLBCLs, 198 DLBCL samples, including 19 samples that had been tested by WES and/or WGS, were analyzed by lymphochip, which targeted coding regions of 212 genes. The WES/WGS and lymphochip data were subsequently combined and analyzed in altogether 275 DLBCL samples (56 HBsAg+). 75 genes were affected by nonsilent mutations in at least 3 HBsAg+ DLBCLs (5% of cases), representing the most frequently mutated targets in these tumors (supplemental Table 5). The mutation profiles in HBsAg+ and HBsAg− DLBCLs were subsequently compared (Figure 3; supplemental Table 6). Firstly, 14 genes were confirmed to be preferentially mutated in the HBsAg+ group, including KLF2, TMSB4X, CD70, BCL6, FAS, TNFRSF14, UBE2A, CD58, SGK1, ZFP36L1, CXCR4, FOXO1, CSK, and MSL2. Of these, 11 genes are potentially off-targets of activation-induced cytidine deaminase (AID), a B-cell–specific factor that initiates somatic hypermutation (SHM) in the immunoglobulin genes (marked with stars in Figure 3; supplemental Table 7). Secondly, well-known mutation targets in DLBCL, such as B2M, GNA13, BCL7A, CIITA, and GNAI2, were less frequently mutated in HBsAg+ tumors. However, there are some recurrently mutated genes that are equally distributed in HBsAg+ and HBsAg− DLBCLs, such as PIM1, KMT2D, ITPKB, DTX1, MYD88, BTG1, HIST1H1E, TBL1XR1, and TNFAIP3. Among these, 6 genes are potentially AID off-targets (Figure 3; supplemental Table 7). Patients with a higher HBV viral load (supplemental Figure 2B-C) or occult HBV infection (supplemental Figure 3) might also be associated with certain frequently mutated genes. However, sample size for each specific group is notably limited. Taken together, these data showed a distinctive set of mutated genes in HBsAg+ DLBCLs, affecting multiple key pathways involving lymphomagenesis (Figure 4). Aberrant SHM mediated by AID activity may have contributed to the mutations observed in a subset of the highly mutated genes in HBsAg+ DLBCLs.
Figure 3.
Somatic mutation spectrum in HBsAg+and HBsAg−DLBCLs. The frequency of nonsilent mutations between HBsAg+ (n = 56) and HBsAg− (n = 219) DLBCLs that were sequenced by WES/WGS and/or lymphochip were compared by χ2 test or Fisher’s exact test. Significant values (P < .05) are highlighted in bold. Potential AID off-targets are marked by an asterisk, and P values from Fisher’s exact test are marked by a number sign.
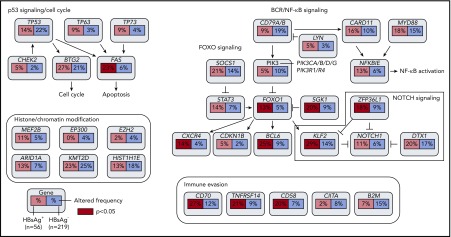
Figure 4.
Key signaling pathways that are affected by frequent somatic mutations in HBsAg+DLBCLs. The frequency of mutated genes in HBsAg+ and HBsAg− DLBCLs from Figure 3 are shown under the gene names.
Enriched BCL6 alterations in HBV-associated DLBCLs
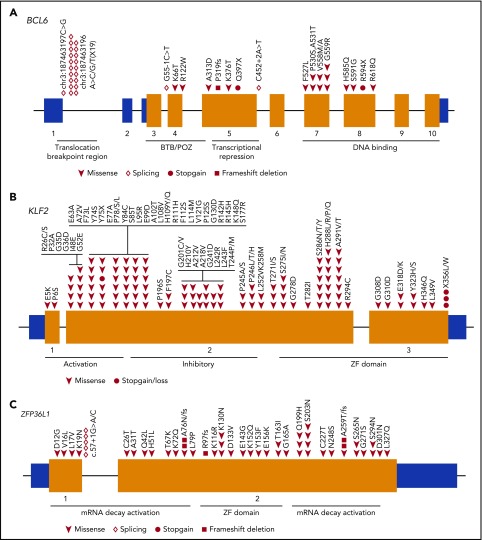
BCL6, a well-known proto-oncogene involved in the development of B-cell lymphoma, encodes a transcriptional repressor that is required for germinal center reactions. Close to half of the BCL6 mutations identified in our cohort were splicing mutations located at a hotspot position (chr3:187463196), which affects the splicing between the non-coding exons 1 and 2, resulting in retention of intron 1 (Figure 5A). The latter may affect RNA stability of the gene or lead to the disruption of the negative regulation loop.39,40 In samples characterized by WGS, the frequency of chromosomal translocation involving BCL6 was also significantly increased in HBsAg+ DLBCLs (57% vs 28%; P = .0472, χ2 test), with the BCL6 breakpoints occurring in a 3-kb region located at exon 1 and intron 1, the known major breakpoint region (Figure 5A). Taken together, the genetic alterations (nonsilent mutations and/or translocations) in BCL6 were significantly enriched in the HBsAg+ DLBCLs (79% vs 28%; P = .0013, Fisher’s exact test), suggesting a critical role of BCL6 dysregulation in the development of HBV-associated DLBCL.
Figure 5.
BCL6, KLF2, and ZFP36L1 are preferentially mutated in HBsAg+DLBCLs. (A) Diagram showing mutations identified in the BCL6 locus. Domains of BCL6 and a region containing translocation breakpoints identified in this report are indicated by horizontal lines. (B) Mutations identified in KLF2. (C) Mutations identified in ZFP36L1. BTB/POZ, broad-complex, Tramtrack, and bric a brac/poxvirus and zinc finger (ZF).
Enriched mutations in KLF2 or ZFP36L1 in HBV-associated DLBCLs
KLF2 encodes a transcription factor which is important for maintaining follicular B-cell identity, and its absence in mice leads to an expansion of marginal zone B cells.41 It is highly mutated (20% to 42%) in splenic marginal zone lymphoma42,43 but was found to be either not recurrently mutated3-9,42 or mutated at a relatively lower frequency in DLBCLs.43 In our cohort, KLF2 was affected by nonsilent mutations in 17% of all tumors analyzed, and it was one of the most significantly mutated genes in HBsAg+ DLBCLs (29%; Figures 3 and 5B).
It has recently been shown that KLF2 can be directly regulated by the messenger RNA decay activator protein ZFP36L1, which is required for the development and maintenance of marginal zone B-cell compartment.44 However, a significant frequency of mutation in ZFP36L1 has not been reported in splenic marginal zone lymphoma or DLBCL.12 In our DLBCL cohort, ZFP36L1 was mutated in 11% of all samples and preferentially mutated in HBsAg+ DLBCLs (Figures 3 and 5C). The KLF2 and ZFP36L1 mutations were mutually exclusive in most of the cases, and 41% of HBsAg+ DLBCL patients carried at least one nonsilent mutation in either gene, which was significantly higher than in HBsAg− DLBCL patients (41% vs 20%; P = .0011, χ2 test). This suggests that KLF2 and ZFP36L1 are functionally linked in lymphomagenesis and that mutations in these molecules may affect a common process or pathway.
Genetic alterations in the p53 signaling pathway
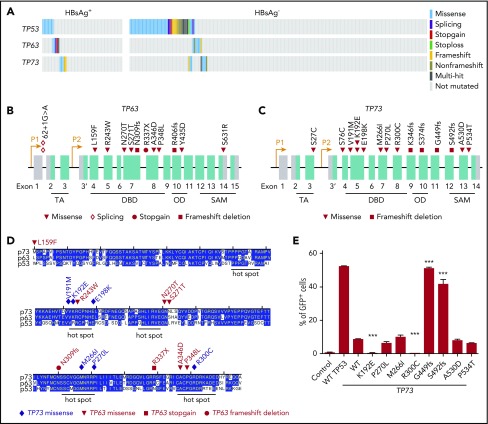
Approximately one-third of the HBsAg+ DLBCLs carried nonsilent, almost mutually exclusive mutations in TP53, TP63, or TP73 (Figure 6A-C). The DNA-binding domains (DBD) of p53, p63, and p73 show ∼63% identity in amino-acid sequence and are structurally highly conserved. Eight TP63 and 7 TP73 mutations were located in the DBD domain of the respective proteins, and most of these alterations are located at or close to TP53 mutation hotspots (Figure 6D). Furthermore, mutations in TP63 and TP73, but not in TP53, were enriched in HBsAg+ DLBCLs (18% vs 6%; P = .0066, χ2 test).
Figure 6.
Genetic alterations in TP53, TP63, and TP73 genes. (A) Mutations identified in TP53, TP63 and TP73 in HBsAg+ and HBsAg- DLBCLs. (B-C) Diagram showing mutations identified in TP63 and TP73, respectively. (D) Alignment of the DNA-binding domain (DBD) of TP53, TP63, and TP73. OD, oligomerization domain; SAM, sterile α motif; TA, transactivation. (E) Characterization of transcriptional activity of mutant TP73. Data shown are from 3 independent experiments. Error bars represent standard deviation. ***P < .0001.
Using a p53-responsive EGFP reporter assay,45 we further investigated whether TP73 mutations can affect the DNA-binding activity of p73. Cotransfection of a p53 binding site containing EGFP reporter and a wild-type (WT) TAp73-expressing construct into a p53-deficient cell line showed that WT TAp73 could bind to p53-responsive sites and transactivate GFP expression, although less efficient as compared with WT p53 (Figure 6E). Two mutations (K192E and R300C) in the DBD domain displayed a significantly impaired ability to transactivate GFP expression. Two truncating mutants (G449fs and S492fs) showed a significantly increased ability of transactivation in the current assay (Figure 6E). However, the loss of the SAM domain and the C terminus in these mutants may abolish the physical interaction between p73 and other proteins such as PTEN, resulting in an inability to induce apoptosis in response to DNA damage.46 The other four mutations (M266I and P270L in the DBD domain and A530D and P534T in the SAM domain) did not affect the transcriptional activity of TAp73. Taken together, these results suggest that a subset of the identified TP73 mutations may impact the DNA-binding activity of p73.
Gene expression analysis in HBsAg+ and HBsAg− DLBCLs
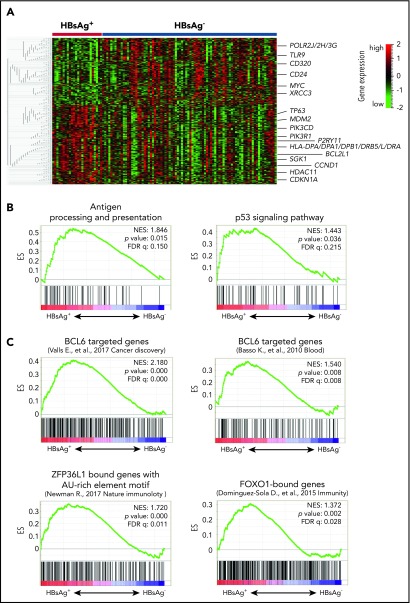
We next characterized the transcriptome of 108 Chinese DLBCLs, including 24 HBsAg+ samples. A distinctive gene expression profile was observed in HBsAg+ tumors compared with HBsAg− tumors, including a group of upregulated genes (n = 377), such as MDM2, PIK3CD, SGK1, BCL2L1, CCND1, TP63, and several major histocompatibility complex class II molecule–related genes and a group of downregulated genes (n = 324), such as TLR9, CD320, and MYC (Figure 7A; supplemental Table 8). Furthermore, GSEA showed that 2 gene sets were significantly upregulated in HBsAg+ DLBCLs, including the antigen processing and presentation and p53 signaling pathways (Figure 7B). The GSEAPreranked tool was further applied, and the BCL6-targeted,47,48 ZFP36L1-bound,44 and FOXO1-bound49 genes were significantly enriched in genes differentially expressed between HBsAg+ and HBsAg− DLBCLs (Figure 7C), which is consistent with the observation that these transcription factors were preferentially mutated in HBsAg+ DLBCLs.
Figure 7.
HBsAg+DLBCLs displays a distinctive gene expression pattern. (A) HBsAg+ DLBCLs showed a distinctive gene expression profiling. The normalized expression levels were analyzed using the Qlucore Omics Explorer software. Significantly and differentially expressed genes (P < .05) between HBsAg+ (n = 24) and HBsAg− (n = 84) DLBCLs were used to draw the heatmap. (B) GSEA revealed pathway alterations in HBsAg+ DLBCLs compared with that in HBsAg− DLBCLs. The GSEA was performed with 1000 sample permutations. Enrichments were considered significant at false discovery rate q < 0.25. (C) GSEAPreranked tool was used to analyze BCL6 core transcriptional signatures (120 genes,47 BCL6 targeted genes derived from DLBCL,48 ZFP36L1-bound genes,44 and FOXO1-targeted genes49 along the preranked genes differentially expressed in HBsAg+ vs HBsAg− DLBCLs. Genes were preranked based on the correlation between the gene expression by comparing HBsAg+ and HBsAg− DLBCLs. The enrichment was tested with 1000 gene set permutations. Enrichments were considered significant at false discovery rate q < 0.05. ES, enrichment score; NES, normalized enrichment score.
Characterization of V(D)J region of immunoglobulin heavy chain (IgH) in HBsAg+ and HBsAg− DLBCLs
By sequencing the V(D)J regions of IgH from 52 DLBCL samples using polymerase chain reaction cloning and/or high-throughput sequencing methods, we identified the major clone from 27 DLBCL samples, including 15 HBsAg+ samples (supplemental Table 9). The most frequent used heavy-chain variable (VH) region genes were VH4-34 (20%) and VH3-23 (20%) in HBsAg+ DLBCLs and VH4-34 in HBsAg− DLBCLs (30%). All VH region genes were mutated (6-86 mutations; 3% to 31%), and there was no significant difference in mutation frequency between the 2 groups. The amino acid sequences deduced from CDR3 sequences are shown in supplemental Table 9, and no sequences was qualified as stereotyped according to the criteria described previously.50 Furthermore, we did not find any significant sequence homology between the CDR3 region in HBsAg+ samples and the anti-HBsAg or HBsAg binding protein using the BLAST program.
Discussion
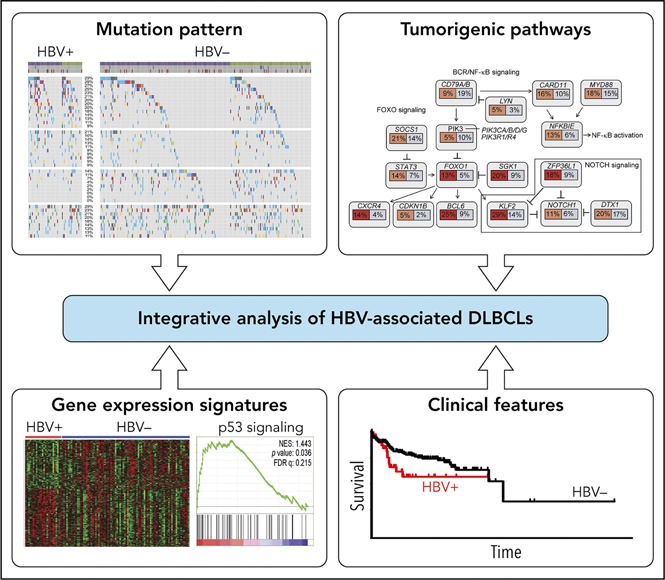
The genetics of DLBCL has been studied by various NGS approaches, and the landscape of somatic mutation of these tumors is emerging.3-9,11 However, most of the DLBCL patients previously studied belonged to Western populations. By analyzing a small cohort of patients, we have previously shown a different mutation spectrum in Chinese DLBCLs.8,13 Here, by integrating WGS, WES, targeted resequencing, and RNA sequencing, we performed a comprehensive genetic characterization of 275 DLBCL samples and identified 7 new potential cancer drivers for Chinese patients, including KLF2, ZFP36L1, OSBPL10, VMP1, TP73, MSL2, and MEOX2. Furthermore, 20% of our patients were HBsAg+, which provided an opportunity to explore the genetic alternations in HBV-associated DLBCLs. We observed overall enhanced mutagenesis in the HBsAg+ DLBCL genomes and identified a unique set of genetic alterations and a distinct gene expression profile in these tumors.
Together with the epidemiology and clinical evidence, our genomic data strongly suggest a causative role of HBV infection in the development of DLBCL. A previous study has suggested a chronic antigenic stimulation as a likely mechanism for the oncogenic role of HBV for lymphoma.24 This is supported by a history of chronic HBV infection, a frequent involvement of spleen and retroperitoneal lymph nodes, and biased usages of certain variable regions of IgH and κ light-chain genes (IGHV4-34 and IGKV4-1) in HBV-associated DLBCLs,24 which is reminiscent of HCV-associated NHL, where most lymphoma cells express IGVH1-69 and VK3-A27 genes that encode antibodies specific for HCV-E2 antigens.51,52 In our DLBCL cohort, the clinical presentation is very similar to that described previously,24 and we also observed that HBsAg+ patients had more frequent involvement of the spleen. However, by analyzing the IGVH of a subset of HBsAg+ DLBCL in our cohort, we did not find any evidence of a biased usage of IGVH genes or stereotyped CDR3 regions or any homology of CDR3 regions with anti-HBsAg antibodies. The latter was confirmed by reanalyzing the sequences published by Deng et al.24 Thus, unlike the classical antigen-driven, HCV-related lymphomas, the chronic antigenic stimulation model is less favored for HBV-associated DLBCLs. This is further supported by the clinical observations that HBV-associated DLBCLs do not respond to antiviral therapy.
An alternative mechanism could be that, like in EBV-driven lymphoma, HBV directly infects B cells, leading to the genetic alterations that contribute to the development of malignancies. HBV is by definition a hepatotropic virus, but it can also infect lymphocytes and the lymphoid system that has been shown to be an important reservoir for HBV.52,53 As HBV DNA has been previously found to be integrated into chromosomal DNA of cells from lymph nodes,54 one possible mechanism could be that like in HBV-induced hepatocellular carcinomas,55 HBV DNA may integrate into the B-cell genome and directly activate oncogenes or disrupt tumor suppressors. However, we were not able to detect HBV gene integration based on our WGS data. Another possibility, which is supported by our mutation signature analysis, is that HBV-induced, APOBEC-mediated mutagenesis contributes to the overall increased mutation burden in HBsAg+ DLBCLs. It is also of note that the majority of genes that were preferentially mutated in HBV-associated DLBCLs do not overlap with those in HBV-associate hepatocellular carcinomas56 or HBV-positive lung adenocarcinoma (supplemental Table 10),31 suggesting that the genetic alterations in HBV-associated DLBCL were most likely generated/selected in a B-cell–specific manner. Indeed, we noticed that majority of genes that are highly mutated in HBV-associated DLBCLs are potentially off-targets of the B-cell–specific factor AID. This may reflect a hyperactive status of B cells due to chronic infection, leading to an enhancement of mutational activity mediated by AID and further contributed to the mutation profiles we observed in HBV-associated DLBCLs. Finally, expression of HBV viral protein, in particular HBx, may be directly involved in regulation of the p53 and NF-κB signaling pathways, as well as modulation of transcriptional networks.57 Taken together, genetic lesions resulting from chronic HBV infection of B cells may be a plausible mechanism underlying the oncogenic role of HBV in lymphomagenesis.
The HBV-associated genetic changes in DLBCLs mainly affect several pathways, including p53 signaling, FOXO signaling, and immune evasion. It is notable that this is the first report showing that TP63 and TP73 are frequently mutated in any type of cancer, strongly suggesting a unique role of these 2 tumor suppressors in lymphomagenesis, especially in individuals infected with HBV. Four genes that frequently altered in HBsAg+ DLBCLs can be assigned to the FOXO signaling pathway (BCL6, CXCR4, KLF2, and SGK1), which regulates the dark zone program of germinal centers49 and is a hallmark of tonic BCR signaling in DLBCL.58 It is possible that the genetic changes in this pathway promote the growth and survival of tumor cells in HBsAg+ DLBCLs in an antigen-independent, tonic BCR-signaling–dependent manner. In addition, considering the exceptionally high frequency of BCL6 genetic alternations in HBsAg+ tumors, therapies targeting BCL6 may help to suppress the growth of the tumor cells in these patients.59 Finally, among the genes related to immune regulation/evasion, CD70, TNFRSF14, and CD58 were frequently mutated in HBsAg+ DLBCLs, whereas B2M and CIITA, both related to the function of major histocompatibility complex class II molecule, were less mutated. CD70 is critical for protection against EBV infection,60 and mutations in CD70 and TNFRSF14 in malignant B cells may also impair T- or natural killer cell–mediated antitumor responses, particularly in a microenvironment affected by HBV infection. Therapies that modulate the CD27-CD70, TNFRSF14-CD272, or CD58-CD2 pathways could thus be an interesting alternative to immunotherapy in HBsAg+ patients.61-63
In summary, using a comprehensive analysis of genetic alterations in HBV-infected DLBCLs, we present the first genetic evidence that suggest a direct link between HBV infection and B-cell lymphomagenesis. Based on the distinct clinical and molecular features, HBsAg+ DLBCL should be classified as a separate subtype of DLBCL. Further studies on larger cohorts of HBV-associated DLBCL, including those potentially with occult HBV infection,21 are required for identification and validation of candidate driver genes in these tumors and will shed light on the complex mechanism underlying HBV infection and B-cell lymphomagenesis.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank G. Melino for sharing the TP73 plasmid; L. Hammarström, R. Dalla-Favera, and L. Pasqualucci for helpful discussions and critically reading the manuscript; K. Basso for discussions on BCL6 gene signature; and the Science for Life Laboratory, the National Genomics Infrastructure, and Uppmax for providing assistance in sequencing and computational infrastructure.
This work was supported by the Swedish Cancer Society, the Swedish Research Council, the European Research Council (RNAEDIT-649019), the Swedish Childhood Cancer Fund, the Chinese Natural Science Foundation (81670184 and 81611130086), the Shenzhen Peacock Plan (KQTD20150330171505310), STINT (joint China-Sweden Mobility Program), Radiumhemmets, the Center for Innovative Medicine, and the KIDS Program at the Karolinska Institutet.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: W.R. collected, analyzed, and interpreted data and wrote the manuscript; X.Y., H.S., B.Z., M.P., and L.C. performed bioinformatics analysis; W.R., X.Y., Wei Li, M.P., Q.Z., M.N., and Y.L. performed experiments; B.M. reviewed the pathological data; Y.H., D.L., K.W., and S.Z. supervised bioinformatics analysis; X.W., Wenyu Li, H.Z., H.H., and R.P. collected samples and clinical information; K.G.W., Y.Z., W.J., and Z.L. were involved in supervision of the study; and Q.P.-H. designed and supervised the study and wrote the manuscript.
Conflict-of-interest disclosure: K.G.W. is cofounder and shareholder of Aprea Therapeutics AB, a company that develops p53-based cancer therapy including APR-246, and a member of its Clinical Advisory Board. Research in the K.G.W. laboratory has received financial support from Aprea Therapeutics AB. K.G.W. has received a salary from Aprea Therapeutics AB. The remaining authors declare no competing financial interests.
Correspondence: Qiang Pan-Hammarström, Department of Laboratory Medicine, Karolinska Institutet, 14186 Stockholm, Sweden; e-mail: qiang.pan-hammarstrom@ki.se.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, et al. . Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Pileri SA, et al. . The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morin RD, Mendez-Lago M, Mungall AJ, et al. . Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasqualucci L, Trifonov V, Fabbri G, et al. . Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohr JG, Stojanov P, Lawrence MS, et al. . Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109(10):3879-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin RD, Mungall K, Pleasance E, et al. . Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Grubor V, Love CL, et al. . Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2013;110(4):1398-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Miranda NF, Georgiou K, Chen L, et al. . Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 2014;124(16):2544-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novak AJ, Asmann YW, Maurer MJ, et al. . Whole-exome analysis reveals novel somatic genomic alterations associated with outcome in immunochemotherapy-treated diffuse large B-cell lymphoma. Blood Cancer J. 2015;5(8):e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgiou K, Chen L, Berglund M, et al. . Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. 2016;127(24):3026-3034. [DOI] [PubMed] [Google Scholar]
- 11.Reddy A, Zhang J, Davis NS, et al. . Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 2017;171(2):481-494.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenquist R, Beà S, Du MQ, Nadel B, Pan-Hammarström Q. Genetic landscape and deregulated pathways in B-cell lymphoid malignancies. J Intern Med. 2017;282(5):371-394. [DOI] [PubMed] [Google Scholar]
- 13.Ren W, Li W, Ye X, Liu H, Pan-Hammarström Q. Distinct subtype distribution and somatic mutation spectrum of lymphomas in East Asia. Curr Opin Hematol. 2017;24(4):367-376. [DOI] [PubMed] [Google Scholar]
- 14.Szczepanowski M, Lange J, Kohler CW, et al. . Cell-of-origin classification by gene expression and MYC-rearrangements in diffuse large B-cell lymphoma of children and adolescents. Br J Haematol. 2017;179(1):116-119. [DOI] [PubMed] [Google Scholar]
- 15.de Miranda NF, Peng R, Georgiou K, et al. . DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med. 2013;210(9):1729-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraan W, Horlings HM, van Keimpema M, et al. . High prevalence of oncogenic MYD88 and CD79B mutations in diffuse large B-cell lymphomas presenting at immune-privileged sites. Blood Cancer J. 2013;3(9):e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerhan JR, Ansell SM, Fredericksen ZS, et al. . Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110(13):4455-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos JC, Lossos IS. Newly emerging therapies targeting viral-related lymphomas. Curr Oncol Rep. 2011;13(5):416-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maciocia N, O’Brien A, Ardeshna K. Remission of follicular lymphoma after treatment for hepatitis C virus infection. N Engl J Med. 2016;375(17):1699-1701. [DOI] [PubMed] [Google Scholar]
- 20.Marcucci F, Mele A, Spada E, et al. . High prevalence of hepatitis B virus infection in B-cell non-Hodgkin’s lymphoma. Haematologica. 2006;91(4):554-557. [PubMed] [Google Scholar]
- 21.Chen MH, Hsiao LT, Chiou TJ, et al. . High prevalence of occult hepatitis B virus infection in patients with B cell non-Hodgkin’s lymphoma. Ann Hematol. 2008;87(6):475-480. [DOI] [PubMed] [Google Scholar]
- 22.Engels EA, Cho ER, Jee SH. Hepatitis B virus infection and risk of non-Hodgkin lymphoma in South Korea: a cohort study. Lancet Oncol. 2010;11(9):827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalia S, Chavez J, Castillo JJ, Sokol L. Hepatitis B infection increases the risk of non-Hodgkin lymphoma: a meta-analysis of observational studies. Leuk Res. 2013;37(9):1107-1115. [DOI] [PubMed] [Google Scholar]
- 24.Deng L, Song Y, Young KH, et al. . Hepatitis B virus-associated diffuse large B-cell lymphoma: unique clinical features, poor outcome, and hepatitis B surface antigen-driven origin. Oncotarget. 2015;6(28):25061-25073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hans CP, Weisenburger DD, Greiner TC, et al. . Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275-282. [DOI] [PubMed] [Google Scholar]
- 26.Kan Z, Jaiswal BS, Stinson J, et al. . Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869-873. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence MS, Stojanov P, Polak P, et al. . Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. ; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Liang X, Xuan Y, et al. . A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience. 2017;6(5):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu K, Zhang X, Li F, et al. . Frequent alterations in cytoskeleton remodelling genes in primary and metastatic lung adenocarcinomas. Nat Commun. 2015;6(1):10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia W, Qiu K, He M, et al. . SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 2013;14(2):R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritchie ME, Phipson B, Wu D, et al. . limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang X, Bi S, Yang W, et al. . Epidemiological serosurvey of hepatitis B in China--declining HBV prevalence due to hepatitis B vaccination. Vaccine. 2009;27(47):6550-6557. [DOI] [PubMed] [Google Scholar]
- 36.Qian J, Wang Q, Dose M, et al. . B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159(7):1524-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris RS, Bishop KN, Sheehy AM, et al. . DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803-809. [DOI] [PubMed] [Google Scholar]
- 38.Janahi EM, McGarvey MJ. The inhibition of hepatitis B virus by APOBEC cytidine deaminases. J Viral Hepat. 2013;20(12):821-828. [DOI] [PubMed] [Google Scholar]
- 39.Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101(8):2914-2923. [DOI] [PubMed] [Google Scholar]
- 40.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247(1):172-183. [DOI] [PubMed] [Google Scholar]
- 41.Hart GT, Wang X, Hogquist KA, Jameson SC. Krüppel-like factor 2 (KLF2) regulates B-cell reactivity, subset differentiation, and trafficking molecule expression. Proc Natl Acad Sci USA. 2011;108(2):716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clipson A, Wang M, de Leval L, et al. . KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia. 2015;29(5):1177-1185. [DOI] [PubMed] [Google Scholar]
- 43.Piva R, Deaglio S, Famà R, et al. . The Krüppel-like factor 2 transcription factor gene is recurrently mutated in splenic marginal zone lymphoma. Leukemia. 2015;29(2):503-507. [DOI] [PubMed] [Google Scholar]
- 44.Newman R, Ahlfors H, Saveliev A, et al. . Maintenance of the marginal-zone B cell compartment specifically requires the RNA-binding protein ZFP36L1. Nat Immunol. 2017;18(6):683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bykov VJ, Issaeva N, Shilov A, et al. . Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8(3):282-288. [DOI] [PubMed] [Google Scholar]
- 46.Lehman JA, Waning DL, Batuello CN, Cipriano R, Kadakia MP, Mayo LD. Induction of apoptotic genes by a p73-phosphatase and tensin homolog (p73-PTEN) protein complex in response to genotoxic stress. J Biol Chem. 2011;286(42):36631-36640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basso K, Saito M, Sumazin P, et al. . Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115(5):975-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valls E, Lobry C, Geng H, et al. . BCL6 Antagonizes NOTCH2 to Maintain Survival of Human Follicular Lymphoma Cells. Cancer Discov. 2017;7(5):506-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez-Sola D, Kung J, Holmes AB, et al. . The FOXO1 transcription factor instructs the germinal center dark zone program. Immunity. 2015;43(6):1064-1074. [DOI] [PubMed] [Google Scholar]
- 50.Murray F, Darzentas N, Hadzidimitriou A, et al. . Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 2008;111(3):1524-1533. [DOI] [PubMed] [Google Scholar]
- 51.Chan CH, Hadlock KG, Foung SKH, Levy S. V(H)1-69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood. 2001;97(4):1023-1026. [DOI] [PubMed] [Google Scholar]
- 52.Marcucci F, Mele A. Hepatitis viruses and non-Hodgkin lymphoma: epidemiology, mechanisms of tumorigenesis, and therapeutic opportunities. Blood. 2011;117(6):1792-1798. [DOI] [PubMed] [Google Scholar]
- 53.Pontisso P, Vidalino L, Quarta S, Gatta A. Biological and clinical implications of HBV infection in peripheral blood mononuclear cells. Autoimmun Rev. 2008;8(1):13-17. [DOI] [PubMed] [Google Scholar]
- 54.Umeda M, Marusawa H, Seno H, et al. . Hepatitis B virus infection in lymphatic tissues in inactive hepatitis B carriers. J Hepatol. 2005;42(6):806-812. [DOI] [PubMed] [Google Scholar]
- 55.Sung WK, Zheng H, Li S, et al. . Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44(7):765-769. [DOI] [PubMed] [Google Scholar]
- 56.Huang J, Deng Q, Wang Q, et al. . Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44(10):1117-1121. [DOI] [PubMed] [Google Scholar]
- 57.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52(4):594-604. [DOI] [PubMed] [Google Scholar]
- 58.Szydlowski M, Kiliszek P, Sewastianik T, et al. . FOXO1 activation is an effector of SYK and AKT inhibition in tonic BCR signal-dependent diffuse large B-cell lymphomas. Blood. 2016;127(6):739-748. [DOI] [PubMed] [Google Scholar]
- 59.Cardenas MG, Yu W, Beguelin W, et al. . Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma. J Clin Invest. 2016;126(9):3351-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abolhassani H, Edwards ES, Ikinciogullari A, et al. . Combined immunodeficiency and Epstein-Barr virus-induced B cell malignancy in humans with inherited CD70 deficiency. J Exp Med. 2017;214(1):91-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tiu RV, Visconte V, Elson P, et al. . LFA-3/CD2 pathway, potential target for immunosuppressive therapy in aplastic anemia: a phase I/II trial of alefacept in patients with relapsed/refractory aplastic anemia. Blood. 2013;122(21):3711-3712.24427808 [Google Scholar]
- 62.Jacobs J, Deschoolmeester V, Zwaenepoel K, et al. . CD70: an emerging target in cancer immunotherapy. Pharmacol Ther. 2015;155:1-10. [DOI] [PubMed] [Google Scholar]
- 63.Bullock TN. TNF-receptor superfamily agonists as molecular adjuvants for cancer vaccines. Curr Opin Immunol. 2017;47:70-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.