Proofreading during DNA replication and post-replication mismatch repair are two major defenses against mutations. Foster et al. and Niccum et al. used mutation accumulation and whole genome sequencing to assemble a database of thousands...
Keywords: DNA proofreading, mutation accumulation, mutation hotspots, DNA replication fidelity, mismatch repair
Abstract
When the DNA polymerase that replicates the Escherichia coli chromosome, DNA polymerase III, makes an error, there are two primary defenses against mutation: proofreading by the ϵ subunit of the holoenzyme and mismatch repair. In proofreading-deficient strains, mismatch repair is partially saturated and the cell’s response to DNA damage, the SOS response, may be partially induced. To investigate the nature of replication errors, we used mutation accumulation experiments and whole-genome sequencing to determine mutation rates and mutational spectra across the entire chromosome of strains deficient in proofreading, mismatch repair, and the SOS response. We report that a proofreading-deficient strain has a mutation rate 4000-fold greater than wild-type strains. While the SOS response may be induced in these cells, it does not contribute to the mutational load. Inactivating mismatch repair in a proofreading-deficient strain increases the mutation rate another 1.5-fold. DNA polymerase has a bias for converting G:C to A:T base pairs, but proofreading reduces the impact of these mutations, helping to maintain the genomic G:C content. These findings give an unprecedented view of how polymerase and error-correction pathways work together to maintain E. coli’s low mutation rate of 1 per 1000 generations.
ACCURATE mutation rates have recently been determined for a variety of wild-type and mutant strains of Escherichia coli using mutation accumulation (MA) experiments coupled with whole-genome sequencing (WGS). Such experiments revealed that, at least in a laboratory setting, few DNA repair pathways are essential for maintaining E. coli’s low mutation rate of 1 mutation per 103 generations (Lee et al. 2012; Foster et al. 2015). Of 11 E. coli strains each defective in a major DNA repair pathway, only those unable to repair oxidative damage showed a substantial increase in spontaneous mutation rates (Foster et al. 2015). Thus, the major determinants of replication accuracy are the intrinsic fidelity of DNA replication, replication proofreading, and postreplication mismatch repair (MMR).
E. coli’s replicative DNA polymerase, polymerase III (Pol III), is a multiprotein machine. As measured in vitro, the polymerase subunit, α (encoded by the dnaE gene), has an intrinsic error rate of one per 104–105 nucleotides incorporated (Bloom et al. 1997). The major determinant of this accuracy is a restrictive active site that sterically prevents most mismatches (Johnson 2010). The 3′ to 5′ exonuclease of the proofreading subunit of Pol III, ϵ (encoded by the dnaQ gene), improves accuracy by removing mismatched nucleotides, allowing polymerase to resynthesize the DNA. In vitro, proofreading improves the accuracy of DNA synthesis 10- to 100-fold (Bloom et al. 1997). Based on the mutation rates of reporter genes, estimates of proofreader’s contribution to replication accuracy in vivo have ranged from 102- to 105-fold (Fowler et al. 1974; Cox and Horner 1982; Schaaper 1988, 1993; Nowosielska et al. 2004). Using an MA protocol, Tsuru et al. (2015) reported that proofreading improved accuracy only 25-fold (Tsuru et al. 2015). However, the E. coli strain used in that study carried a deletion of the dnaQ gene, and such strains rapidly accumulate suppressor mutations in the dnaE gene, some of which lower the mutation rate (Lancy et al. 1989; Fijalkowska and Schaaper 1995).
To obtain an accurate estimate of the intrinsic error rate of DNA Pol III in vivo, proofreading must be eliminated. However, in addition to its proofreading functions, ϵ is an important structural component of the core polymerase and its loss causes severe growth defects. Partial function alleles of dnaQ can overcome this problem and allow the contribution of proofreading to the overall mutation rate to be evaluated (Cox and Horner 1982; Taft-Benz and Schaaper 1998). For the study reported here, we used the mutD5 allele of dnaQ, which reduces the exonuclease activity by 98% while maintaining the core polymerase structure (Fijalkowska and Schaaper 1996; Taft-Benz and Schaaper 1998; Perrino et al. 1999). The mutational phenotypes of the mutD5 allele have been extensively investigated using reporter gene assays (Fowler et al. 1974; Cox and Horner 1982; Schaaper 1988, 1993). Here, we extend this work to the entire chromosome by using an MA protocol followed by WGS.
Several factors complicate the mutational analysis of mutD5 mutant strains. Strains carrying certain mutant dnaQ alleles are induced to various degrees for the SOS response (Slater et al. 1994; Gautam et al. 2012; Whatley and Kreuzer 2015), which could alter the mutational profile. In addition, the mutator phenotype of mutD5 mutant strains is medium-dependent; mutation rates are 10- to 1000-fold higher when mutD5 mutant strains are grown on rich medium rather than on minimal medium (Cox and Horner 1982; Schaaper 1988). Finally, as mentioned above, suppressor mutations may arise that could alter the mutational profile.
To obtain a better estimate of the intrinsic error rate of DNA Pol III and a more complete understanding of the role of ϵ in replication fidelity, we used the MA/WGS approach to analyze the mutation rates and mutational spectra of a mutD5 mutant strain and a mutD5 mutant strain also defective in MMR. We evaluated the impact of growth on rich vs. minimal media. In addition, we show that the SOS-induced error-prone polymerases do not contribute to the mutation rate or spectra of E. coli strains carrying the mutD5 allele.
Materials and Methods
Bacterial strains and media
All strains used in this study, the methods of their construction, and the media used are given in Supplemental Material, Table S1. Genetic constructions were confirmed by PCR analyses using the oligonucleotides listed in Table S2. Further details are in the supplemental materials and methods.
Estimation of mutation rate from fluctuation assays
Mutation rates were determined as described (Foster 2006; Hall et al. 2009), using mutation to nalidixic acid resistance (NalR) as the reporter.
MA experiments
The MA procedure has been described previously (Lee et al. 2012; Foster et al. 2015). Generations were estimated from the colony diameters as previously described (Lee et al. 2012). More details are given in the supplemental materials and methods.
With these highly mutating strains, several precautions were taken to minimize the occurrence of mutations before or during the MA procedure that might modify the mutation rates or spectra. MA lines were initiated from at least two founders so that lines derived from founders that, after sequencing, proved to carry known mutator or antimutator mutations could be eliminated. The MA procedure was restricted to three to six passages to minimize selection. After sequencing, any MA lines that had known mutators or antimutators, or had mutation rates > 2 SD above or below the mean, were eliminated.
Genomic DNA preparation, library construction, sequencing, and SNP and insertion/deletion calling
Genomic DNA (gDNA) was isolated from an aliquot of an overnight culture (in rich or minimal medium as appropriate) inoculated from freezer stocks made after the last passage of each MA line. That the constructed deletions were present in each MA line was confirmed using diagnostic PCR of the gDNA before library construction; the oligonucleotides used are listed in Table S2. Library construction, sequencing, SNP and insertion/deletion (indel) calling, and mutation annotation are described in the supplemental materials and methods.
Some MA lines were eliminated because of poor sequence coverage. Identical mutations in two or more lines arose if mutations occurred in the founder colony or if cross contamination occurred during streaking. If lines shared > 50% of their mutations, then only one line was retained for analysis. If lines shared < 50%, each mutation was randomly assigned to only one of the lines.
Estimation of mutation rates from MA experiments
For each experiment, the mutation rate was estimated by dividing the total number of mutations accumulated by all the MA lines by the total number of generations that were undergone. This value for mutations per generation was then divided by the appropriate number of sites (A:T sites, G:C sites, etc.) to give the conditional mutation rate. The individual mutation rates for each line were used to compute confidence limits (CLs) (see the supplemental materials and methods for further details).
Statistical analysis
Standard statistical analysis was used (Zar 1984). Means and CLs were calculated from the MA lines for each experiment as described (Foster et al. 2018). Values and 95% CLs for ratios between variables were calculated as in Rice (1995). The expected values for χ2 tests were calculated from the numbers of the relevant feature in the genome or from the results of 1000 Monte Carlo simulations for each strain, as described (Lee et al. 2012).
Data availability
Strains are available upon request. File S1 contains the supplemental materials and methods. File S2 contains supplemental tables, which include strain genotypes, methods of strain construction, oligonucleotide sequences, and detailed data from each experiment. File S3 contains supplemental figures that are referenced in the text. The sequences, SNPs, and indels reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive [https://trace.ncbi.nlm.nih.gov/Traces/sra/ (accession no. SRP013707)] and in the IUScholarWorks Repository (hdl.handle.net/2022/20340). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6513035.
Results
Mutational profile of mutD5 and mutD5 mutL mutant strains growing on rich medium
Base pair substitution rates:
The base pair substitution (BPS) rate of the mutD5 mutant strain growing on rich (LB) medium was 84 × 10−8 BPS/generation/nt, 4000-fold greater than that of the wild-type strain and 35-fold greater than that of the MMR-defective strains (Table 1). This increase relative to wild-type is in the middle of the 102–104-fold range reported in previous studies (Fowler et al. 1974; Schaaper 1988, 1993; Fijalkowska and Schaaper 1996; Nowosielska et al. 2004). To estimate the intrinsic error rate of DNA Pol III, we deleted the mutL gene in the mutD5 mutant strain, creating a strain deficient in the two most important pathways for correcting replication errors: MMR and proofreading. The BPS rate of the mutD5 mutL mutant strain was 125 × 10−8 BPS/generation/nt, 1.5-fold greater than that of the mutD5 mutant strain (Table 1), an increase slightly smaller than the 1.6–3.4 range previously reported (Schaaper 1993).
Table 1. Comparisons of BPS rates among strains.
| Strain | Description | BPS per generation ± 95% CL | BPS per generation per nt (×108) ± 95% CL | Rate compared to | ||
|---|---|---|---|---|---|---|
| WT ± 95% CL | MMR ± 95% CL | mutD5 ± 95% CL | ||||
| MA experiments on LB | ||||||
| WT | WT | (9.6 ± 0.1) × 10−4 | 0.021 ± 0.003 | 1 | (8.5 ± 0.2) × 10−3 | (2.5 ± 0.04) × 10−4 |
| MMR | mmr− | 0.11 ± 0.002 | 2.4 ± 0.04 | 118 ± 3 | 1 | (4.0 ± 0.6) × 10−3 |
| PFM163 | mutD5 | 3.9 ± 0.03 | 84 ± 0.6 | 4081 ± 66 | 35 ± 1 | 1 |
| PFM165/397/399a | mutD5 mutL | 5.8 ± 0.5 | 125 ± 10 | 6049 ± 483 | 51 ± 4 | 1.5 ± 0.1 |
| PFM479 | mutD5 dinB | 3.3 ± 0.5 | 72 ± 12 | 3478 ± 555 | 29 ± 5 | 0.85 ± 0.14 |
| PFM515/517b | mutD5 dinB umuDC | 4.2 ± 0.5 | 90 ± 11 | 4358 ± 521 | 37 ± 4 | 1.1 ± 0.1 |
| PFM686 | mutD5 lexA3 | 5.4 ± 0.6 | 116 ± 12 | 5619 ± 598 | 48 ± 5 | 1.4 ± 0.1 |
| MA experiments on minimal medium | ||||||
| PFM163 | mutD5 | 0.68 ± 0.10 | 15 ± 2.2 | 713 ± 102 | 6.0 ± 0.9 | 0.17 ± 0.03 |
| PFM165 | mutD5 mutL | 3.9 ± 0.9 | 84 ± 20 | 4058 ± 901 | 34 ± 7 | 1.0 ± 0.2 |
95% CLs are the SDs of the BPSs of the MA lines for each strain multiplied by the critical value of the t distribution (see Materials and Methods). All comparisons are with strains grown on LB medium. Values labeled WT are the combined data from eight MA experiments previously reported (Lee et al. 2012; Foster et al. 2015); the strains used are PFM2, wild-type (two data sets, 3K and 6K generations); PFM35, uvrA; PFM40, alkA tagA; PFM88, ada ogt; PFM91, nfi; PFM101, umuDC dinB; and PFM133, umuDC dinB polB, all of which had nearly identical BPS rates and spectra. Values labeled MMR are the combined data from 10 experiments with mmr− mutant strains reported in Lee et al. (2012) and Foster et al. (2018); the strains used are PFM5, mutL; PFM144, mutL; PFM288, mutL; PFM304, mutLS; PFM342, mutS; PFM343, mutS; PFM555/556, mutS; PFM197, mutH; and PFM567/568, mutLSH, all of which had nearly identical BPS rates and spectra. BPS, base pair substitution; CL, confidence limit; WT, wild-type; MMR, Mismatch repair; MA, mutation accumulation; nt, nucleotide.
Two separate transductions of the ΔmutLSc allele into the mutD5 mutant strain, PFM163; PFM397 and PFM399 are two isolates from the same transduction.
Two isolates from the same transduction.
Selective pressure during the MA experiment:
Selective pressure is usually evaluated by the ratio of nonsynonymous to synonymous (NS/S) BPSs. Based on the codon usage in E. coli MG1655, the expected NS/S ratio is 3.25 (Lee et al. 2012), and this was significantly greater than the ratios for the mutD5 and mutD5 mutL mutant strains (Table 2). One thousand Monte Carlo simulations using the BPS spectra of the mutant strains yielded NS/S ratios of ∼2, slightly (6%), but statistically significantly, greater than the observed ratios (Table 2). Thus, the mutD5 and the mutD5 mutL mutant strains appear to be under mild selective pressure, likely because they have poor viability, as previously observed (Fijalkowska and Schaaper 1996).
Table 2. Evaluation of selective pressure during the MA experiments: type of BPS.
| Nonsynonymous/synonymous BPSs | |||||||
|---|---|---|---|---|---|---|---|
| Strain | Observed | Expected from genomea | χ2a | P | Expected from simulationsb | χ2b | P |
| MA experiments on LB | |||||||
| mutD5 | 2.02 | 3.25 | 260 | < 0.001 | 2.14 | 4.0 | 0.04 |
| mutD5 mutL | 1.81 | 3.25 | 1235 | < 0.001 | 1.93 | 16 | < 0.001 |
| mutD5 dinB | 1.95 | 3.25 | 275 | < 0.001 | 2.09 | 5.4 | 0.02 |
| mutD5 dinB umuDC | 1.93 | 3.25 | 862 | < 0.001 | 2.09 | 7.83 | 0.005 |
| mutD5 lexA3 | 1.92 | 3.25 | 507 | < 0.001 | 2.05 | 8.4 | 0.004 |
| MA experiments on minimal medium | |||||||
| mutD5 | 1.97 | 3.25 | 93 | < 0.001 | 2.17 | 3.8 | 0.05 |
| mutD5 mutL | 1.75 | 3.25 | 291 | < 0.001 | 1.90 | 5.4 | 0.02 |
BPS, base pair substitution; MA, mutation accumulation.
χ2 values were calculated comparing the observed value to the expected value calculated from the ratio of all possible nonsynonymous vs. synonymous changes in the MG1655 genome, which is 3.25 (Lee et al. 2012).
χ2 values were calculated comparing the observed value to the value expected from 1000 Monte Carlo simulations using the BPS spectra of each strain.
If mutations accumulate in a neutral manner, the number of BPSs in coding and in noncoding (C/NC) DNA should reflect the numbers of base pairs in each (Lee et al. 2012). We previously observed that the C/NC ratio was significantly less than expected in wild-type strains, but slightly greater than expected in MMR-defective strains, suggesting that MMR preferentially repairs coding DNA (Lee et al. 2012). The C/NC ratio of the mutD5 strain was 5.47, not significantly different from the 5.74 ratio based on the genome or the 5.51 ratio obtained from Monte Carlo simulations using the BPS spectrum of the mutD5 mutant strain (Table 3). However, the C/NC ratio in the mutD5 mutL mutant strain, 6.96, was a significant 20% greater than the expected ratios both from the genome and from simulations, and 30% greater than the ratio of the mutD5 strain (χ2 = 74.5, P < 0.001) (Table 3). However, it was close to the 6.63 reported for a mutL mutant strain (Lee et al. 2012) (χ2 = 0.4, P = 0.5), suggesting that the apparent preference to repair coding DNA in wild-type strains is solely due to MMR, and that proofreader does not have this preference.
Table 3. Evaluation of selective pressure during the MA experiments: position of BPS.
| BPSs in coding/noncoding DNA | |||||||
|---|---|---|---|---|---|---|---|
| Strain | Observed | Expected from genomea | χ2a | P | Expected from simulationsb | χ2b | P |
| MA experiments on LB | |||||||
| mutD5 | 5.47 | 5.74 | 2.0 | 0.15 | 5.51 | 0.05 | 0.83 |
| mutD5 mutL | 6.96 | 5.74 | 89 | < 0.001 | 5.87 | 70 | < 0.001 |
| mutD5 dinB | 5.50 | 5.74 | 1.35 | 0.24 | 5.54 | 0.04 | 0.85 |
| mutD5 dinB umuDC | 5.84 | 5.74 | 0.80 | 0.37 | 5.59 | 1.6 | 0.21 |
| mutD5 lexA3 | 5.73 | 5.74 | 0.01 | 0.93 | 5.59 | 0.55 | 0.5 |
| MA experiments on minimal medium | |||||||
| mutD5 | 4.03 | 5.74 | 41 | < 0.001 | 5.68 | 38 | < 0.001 |
| mutD5 mutL | 6.88 | 5.74 | 17 | < 0.001 | 5.81 | 14 | < 0.001 |
BPS, base pair substitution; MA, mutation accumulation.
χ2 values were calculated comparing the observed value to the expected value calculated from the ratio of coding vs. noncoding nucleotides in the MG1655 genome, which is 5.74 (Lee et al. 2012).
χ2 values were calculated comparing the observed value to the value expected from 1000 Monte Carlo simulations using the BPS spectra of each strain.
The BPS spectra:
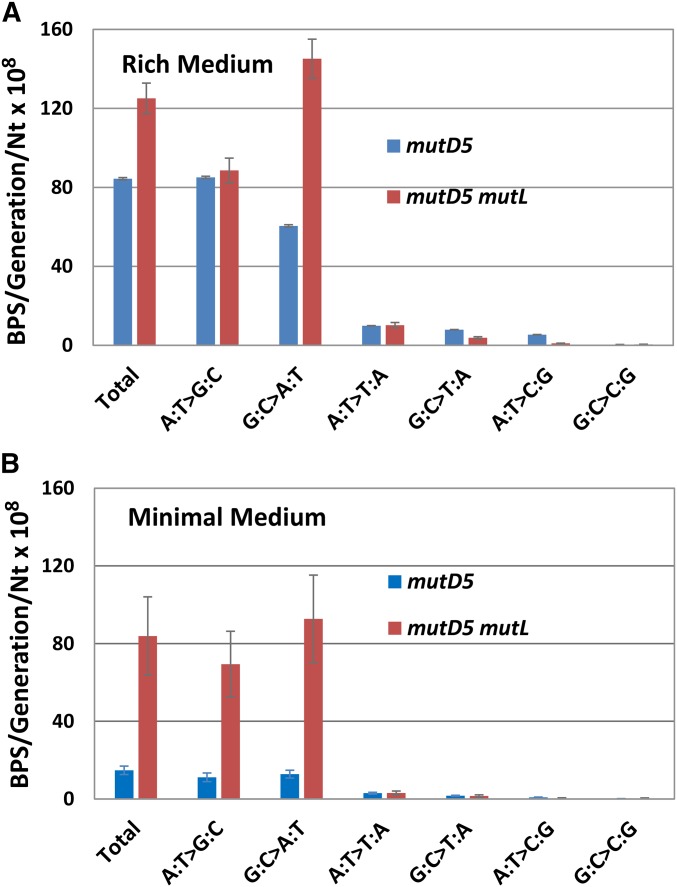
The spectrum of BPS accumulated by the mutD5 mutant strain growing on rich medium is shown in Figure 1A and detailed in Table 4 (the numbers of BPSs are given in Table S3). As has been previously reported (Schaaper 1988, 1993; Fijalkowska and Schaaper 1996; Nowosielska et al. 2004), transitions occurred sixfold more frequently than transversions. The A:T transition rate was only 1.4-fold greater than the G:C transition rate (χ2 = 138, P < 0.001), less than the threefold observed with MMR-defective strains (Lee et al. 2012; Foster et al. 2018). The rates of the various transversions also varied significantly (χ2 = 501, P < 0.001) in the order A:T to T:A > G:C to T:A > A:T to C:G >> G:C to C:G, a pattern similar to that observed for MMR-mutant strains (Lee et al. 2012; Foster et al. 2018).
Figure 1.
The conditional BPS rates and spectra accumulated by the mutD5 and mutD5 mutL-mutant strains. The bars represent the BPSs per generation per number of A:T or G:C base pairs in the genome; the error bars are 95% CLs. (A) Cells were grown on LB medium; mutD5, PFM163; mutD5 mutL, PFM165/397/399. (B) Cells were grown on minimal glucose medium; mutD5, PFM163; mutD5 mutL, PFM165. The data in (B) for the mutD5 strain on minimal medium is presented with an expanded axis in Figure S4A. BPS, base pair substitution; CL, confidence limit.
Table 4. Conditional BPS rates during the MA experiments.
| Conditional BPS rates × 108 ± 95% CL | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MA experiments on LB medium | MA experiments on minimal medium | ||||||||
| Types of BPS | WT × 102 | MMR | mutD5 | mutD5 mutL | mutD5 dinB | mutD5 dinB umuDC | mutD5 lexA3 | mutD5 | mutD5 mutL |
| Total | 2.1 ± 0.03 | 2.45 ± 0.05 | 84 ± 0.6 | 125 ± 10 | 72 ± 12 | 90 ± 11 | 116 ± 13 | 15 ± 2.2 | 84 ± 20 |
| Transitions | 1.1 ± 0.01 | 2.38 ± 0.05 | 73 ± 0.6 | 117 ± 10 | 63 ± 11 | 79 ± 10 | 104 ± 12 | 12 ± 2.1 | 81 ± 19 |
| A:T > G:C | 0.8 ± 0.02 | 3.65 ± 0.10 | 85 ± 0.6 | 89 ± 8.8 | 71 ± 11 | 86 ± 10 | 110 ± 12 | 11 ± 2.2 | 69 ± 17 |
| G:C > A:T | 1.4 ± 0.01 | 1.15 ± 0.03 | 60 ± 0.6 | 145 ± 14 | 56 ± 11 | 72 ± 12 | 99 ± 12 | 13 ± 2.0 | 93 ± 23 |
| Transversions | 0.9 ± 0.02 | 0.07 ± 0.002 | 12 ± 0.1 | 7.7 ± 0.7 | 8.6 ± 1.7 | 11 ± 1.3 | 12 ± 1.0 | 2.8 ± 0.3 | 2.7 ± 0.8 |
| A:T > T:A | 0.4 ± 0.01 | 0.05 ± 0.002 | 10 ± 0.09 | 10 ± 1.5 | 8.1 ± 1.4 | 10 ± 1.2 | 12 ± 1.0 | 3.0 ± 0.4 | 3.1 ± 1.0 |
| G:C > T:A | 0.6 ± 0.02 | 0.03 ± 0.001 | 8.0 ± 0.08 | 3.8 ± 0.7 | 6.2 ± 1.5 | 8 ± 1.0 | 8.7 ± 0.9 | 1.7 ± 0.2 | 1.5 ± 0.6 |
| A:T > C:G | 0.7 ± 0.02 | 0.04 ± 0.002 | 5.4 ± 0.1 | 1.0 ± 0.2 | 2.8 ± 0.8 | 4 ± 1.0 | 2.8 ± 0.5 | 0.8 ± 0.2 | 0.4 ± 0.2 |
| G:C > C:G | 0.2 ± 0.01 | 0.02 ± 0.001 | 0.4 ± 0.02 | 0.5 ± 0.1 | 0.2 ± 0.1 | 0.5 ± 0.2 | 0.7 ± 0.2 | 0.1 ± 0.06 | 0.33 ± 0.2 |
| A:T sites | 1.9 ± 0.04 | 3.74 ± 0.10 | 100 ± 1 | 100 ± 8 | 82 ± 13 | 100 ± 11 | 124 ± 13 | 15 ± 2.4 | 73 ± 18 |
| G:C sites | 2.3 ± 0.03 | 1.19 ± 0.03 | 69 ± 0.7 | 150 ± 14 | 62 ± 12 | 80 ± 12 | 108 ± 13 | 15 ± 2.1 | 95 ± 23 |
| Synonymous | 0.7 ± 0.02 | 1.18 ± 0.02 | 39 ± 0.3 | 65 ± 5.5 | 34 ± 6.0 | 44 ± 5.9 | 56 ± 6.6 | 6.6 ± 1.2 | 44 ± 11 |
| Nonsynonymous | 0.6 ± 0.01 | 0.72 ± 0.01 | 24 ± 0.2 | 36 ± 2.8 | 21 ± 3.3 | 26 ± 3.1 | 33 ± 3.6 | 4.0 ± 0.6 | 24 ± 6 |
| Position | |||||||||
| Noncoding DNA | 3.2 ± 0.07 | 2.18 ± 0.05 | 88 ± 0.7 | 106 ± 9 | 74 ± 13 | 89 ± 11 | 116 ± 11 | 20 ± 2.7 | 72 ± 18 |
| Coding DNA | 1.9 ± 0.03 | 2.49 ± 0.05 | 84 ± 0.6 | 128 ± 10 | 71 ± 12 | 90 ± 11 | 116 ± 13 | 14 ± 2.1 | 86 ± 21 |
| Amino acid changes | |||||||||
| Conservative | 0.6 ± 0.01 | 1.00 ± 0.04 | 30 ± 0.2 | 46 ± 3.5 | 25 ± 4.0 | 31 ± 3.7 | 41 ± 4.6 | 4.6 ± 0.8 | 31 ± 8 |
| Nonconservative | 0.6 ± 0.01 | 0.48 ± 0.04 | 20 ± 0.1 | 27 ± 2.2 | 17 ± 2.8 | 21 ± 2.7 | 27 ± 2.9 | 3.5 ± 0.4 | 17 ± 4 |
Data are the conditional mutation rates, which are the numbers of mutations/numbers of generations/relevant numbers of nucleotides or possible amino acid changes; for example, the conditional mutation rate of A:T > G:C transitions is the number of A:T > G:C transitions/generations/A:T base pairs in the genome. The 95% CLs are the SDs of the BPSs of the MA lines for each strain multiplied by the critical value of the t distribution (see Materials and Methods). Data for WT and MMR are from Foster et al. (2018). BPS, base pair substitution; CL, confidence limit; MA, mutation accumulation; WT, wild-type; MMR, Mismatch repair.
Deleting MMR repair in the mutD5 mutL mutant strain resulted in a 2.4-fold increase in the G:C transition rate, which in this strain exceeded the A:T transition rate by 1.6-fold (χ2 = 1229, P < < 0.001) (Figure 1A, Table 4, and Table S3). This increase in G:C transitions entirely accounted for the difference in mutation rates between the mutD5 and mutD5 mutL mutant strains, and resulted in a spectrum of BPSs closely resembling that of the wild-type strain (Table 4). In contrast, other studies have found that the rate of A:T transitions exceeds that of G:C transitions in MMR-defective mutD5 mutant strains (Schaaper 1993) (see Discussion). The rates of the various transversions occurred in the same pattern in the mutD5 mutL mutant strain as in the mutD5 mutant strain.
The DNA strand bias of BPSs:
In MMR-defective strains, A:T transitions are 2.4-fold more frequent when A is on the lagging strand template (LGST) and T is on the leading strand template (LDST) than in the opposite orientation. Likewise, G:C transitions are 2.3-fold more frequent when C is on the LGST and G is on the LDST than in the opposite orientation (Lee et al. 2012; Bhagwat et al. 2016; Foster et al. 2018). Neither the mutD5 nor the mutD5 mutL mutant strain exhibited these strong strand biases (Table 5). In the mutD5 mutant strain, A:T transitions were only 1.17-fold more frequent when A was on the LGST than on the LDST, and there was no strand bias for G:C transitions. In the mutD5 mutL mutant strain, A:T and G:C transitions occurred 1.19- and 1.03-fold more frequently with A and C on the LGST. While statistically significant, these 10–20% strand biases are much less prominent than the twofold biases exhibited by MMR-defective proofreading-proficient strains, suggesting that nucleotide misincorporation during DNA replication is not strand biased but proofreading is (see Discussion).
Table 5. DNA strand biases of the BPSs accumulated in the MA experiments.
| LGST | LDST | LGST/LDST | LGST | LDST | LGST/LDST | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Aobs | Aexp | Aobs | Aexp | Aobs | Aexp | Pa | Cobs | Cexp | Cobs | Cexp | Cobs | Cexp | Pa |
| MA experiments on LB | ||||||||||||||
| A:T transitions | G:C transitions | |||||||||||||
| mutD5 | 3649 | 3365 | 3108 | 3392 | 1.17 | 0.99 | < 0.001 | 2440 | 2402 | 2522 | 2560 | 0.97 | 0.94 | 0.4 |
| mutD5 mutL | 7702 | 7046 | 6450 | 7105 | 1.19 | 0.99 | < 0.001 | 12194 | 11628 | 11824 | 12390 | 1.03 | 0.94 | < 0.001 |
| mutD5 dinB | 3403 | 3038 | 2698 | 3063 | 1.26 | 0.99 | < 0.001 | 2498 | 2374 | 2406 | 2530 | 1.04 | 0.94 | 0.5 |
| mutD5 dinB umuDC | 3314 | 2967 | 2644 | 2991 | 1.25 | 0.99 | < 0.001 | 2599 | 2491 | 2547 | 2655 | 1.02 | 0.94 | 0.03 |
| mutD5 lexA3 | 5523 | 5001 | 4520 | 5042 | 1.22 | 0.99 | < 0.001 | 4694 | 4525 | 4652 | 4821 | 1.01 | 0.94 | 0.01 |
| A:T transversions | G:C transversions | |||||||||||||
| mutD5 | 613 | 608 | 608 | 613 | 1.01 | 0.99 | 0.8 | 311 | 332 | 374 | 353 | 0.83 | 0.94 | 0.3 |
| mutD5 mutL | 881 | 899 | 924 | 906 | 0.95 | 0.99 | 0.6 | 299 | 344 | 412 | 367 | 0.73 | 0.94 | 0.02 |
| mutD5 dinB | 447 | 464 | 485 | 468 | 0.92 | 0.99 | 0.4 | 226 | 275 | 342 | 293 | 0.66 | 0.94 | 0.003 |
| mutD5 dinB umuDC | 447 | 489 | 536 | 494 | 0.83 | 0.99 | 0.06 | 282 | 295 | 327 | 314 | 0.86 | 0.94 | 0.5 |
| mutD5 lexA3 | 643 | 654 | 670 | 659 | 0.96 | 0.99 | 0.67 | 408 | 432 | 484 | 460 | 0.84 | 0.94 | 0.26 |
| MA experiments on minimal medium | ||||||||||||||
| A:T transitions | G:C transitions | |||||||||||||
| mutD5 | 967 | 855 | 750 | 862 | 1.29 | 0.99 | < 0.001 | 1073 | 976 | 943 | 1039 | 1.14 | 0.94 | 0.002 |
| mutD5 mutL | 1964 | 1722 | 1496 | 1737 | 1.31 | 0.99 | < 0.001 | 2555 | 2342 | 2284 | 2496 | 1.12 | 0.94 | < 0.001 |
| A:T transversions | G:C transversions | |||||||||||||
| mutD5 | 278 | 289 | 302 | 291 | 0.92 | 0.99 | 0.5 | 133 | 137 | 149 | 145 | 0.89 | 0.94 | 0.8 |
| mutD5 mutL | 82 | 85 | 89 | 86 | 0.92 | 0.99 | 0.7 | 42 | 48 | 58 | 51 | 0.72 | 0.94 | 0.4 |
LGST, lagging strand template; LDST, leading strand template; obs, observed; exp, expected; MA, mutation accumulation experiments.
P is the probability of the χ2 value calculated comparing the observed to the expected values. The expected values were calculated from the ratios of nucleotides of each type on the LGST vs. the LDST in the MG1655 genome, which are 0.99 for adenines and 0.94 for cytosines.
The local sequence context of BPS:
The sequence context in which a base pair appears affects its mutability (Lee et al. 2012; Sung et al. 2015). In both the mutD5 and the mutD5 mutL mutant strains, the adjacent bases are the most important determinants (Figures S1 and S2). Therefore, we analyzed the influence of only the bases immediately 5′ and 3′ to the mutated base. While there are 64 possible triplets, in double-stranded DNA only 32 are nonredundant. A triplet and its reverse complement (each read 5′ to 3′) are equivalent since each pairs with the other on the opposite DNA strand.
As shown in Figure 2, the mutation rate of A:T base pairs in the triplets 5′NAC3′+5′GTN3′ was ≈twofold greater than the average rate of A:T base pairs in the other triplets (throughout this report, a triplet and its complement are both presented 5′ to 3′ with the mutated base in the middle). This pattern is similar to that observed for both the wild-type and MMR-mutant strains, except that the dominance of 5′NAC3′+5′GTN3′ sites (10- to 16-fold) was more dramatic in the MMR-defective strains (Lee et al. 2012; Foster et al. 2018). In the mutD5 mutant strain, the mutation rate of G:C base pairs in the triplets 5′NGC3′+5′GCN3′ was also about twofold greater than the average mutation rate of G:C base pairs in the other triplets, but in the mutD5 mutL mutant strain, this ratio dropped to 1.2- to 1.6-fold. Thus, these sites are not as prominent in the mutD5 mutL spectrum as 5′NAC3′+5′GTN3′ sites. Based on the results from all the strains examined to date, mutations are potentiated by a C 3′ to the purine or a G 5′ to the pyrimidine at A:T base pairs and, to a lesser extent, at G:C base pairs. Interestingly, the context bias of BPSs in the mutD5 mutL mutant strain is similar in pattern and relative magnitude to that in the wild-type strain (Figure S3), which will be further discussed below (see Discussion).
Figure 2.
The context bias of the base pair substitutions accumulated by the mutD5 and mutD5 mutL strains. The x-axis labels are the 32 nonredundant triplets oriented 5′NMN3′ with the mutated base in the center. The bars represent the BPS per generation per triplet in the genome; the error bars are 95% CLs. (A) Cells were grown on LB medium; mutD5, PFM163; mutD5 mutL, PFM165/397/399. (B) cells were grown on minimal glucose medium; mutD5, PFM163; mutD5 mutL, PFM165. The data in (B) for the mutD5 strain on minimal medium is presented with an expanded axis in Figure S4B. BPS, base pair substitution; CL, confidence limit.
Conclusions based on the phenotype of mutD5 mutant strains are complicated by the possibility that MMR becomes saturated when proofreader is deficient (Schaaper 1988). Comparison of the mutation rates of the mutD5 and mutD5 mutL mutant strains (Table 4) indicates that the ability of MMR to prevent A:T mutations was saturated, but not its ability to prevent G:C mutations; the G:C mutation rate of the mutD5 mutant strain increased an additional 2.17 ± 0.05-fold with the loss of MMR (mean ± 95% CL) (see Discussion).
Spontaneous indel rates and spectra:
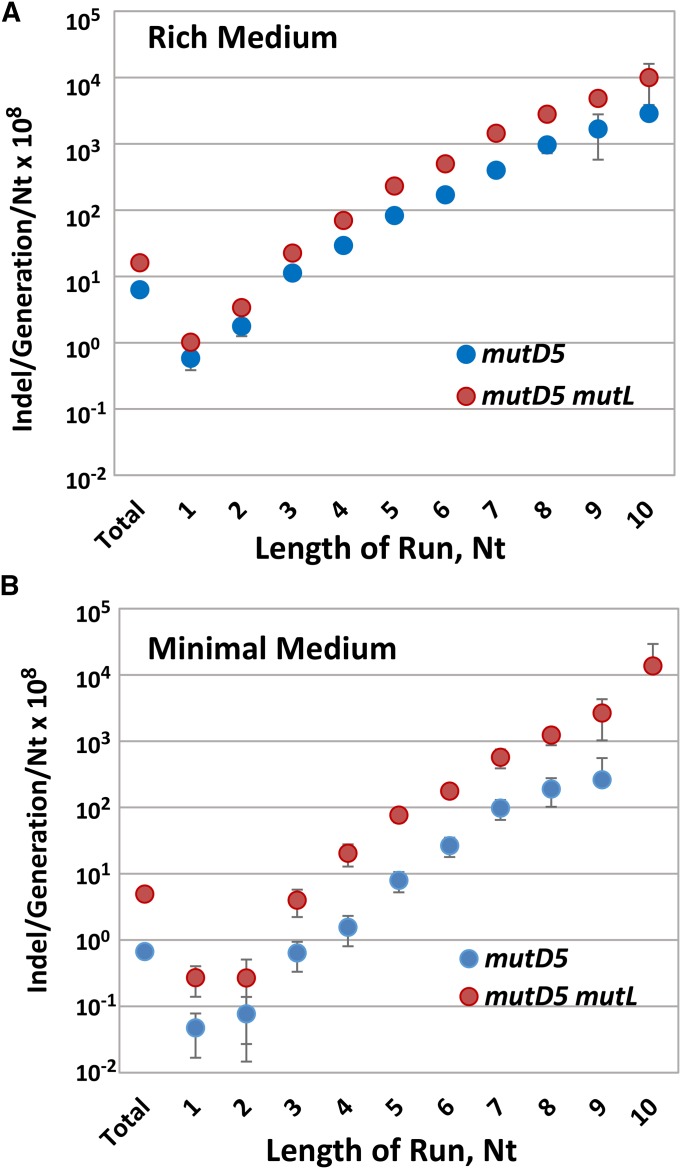
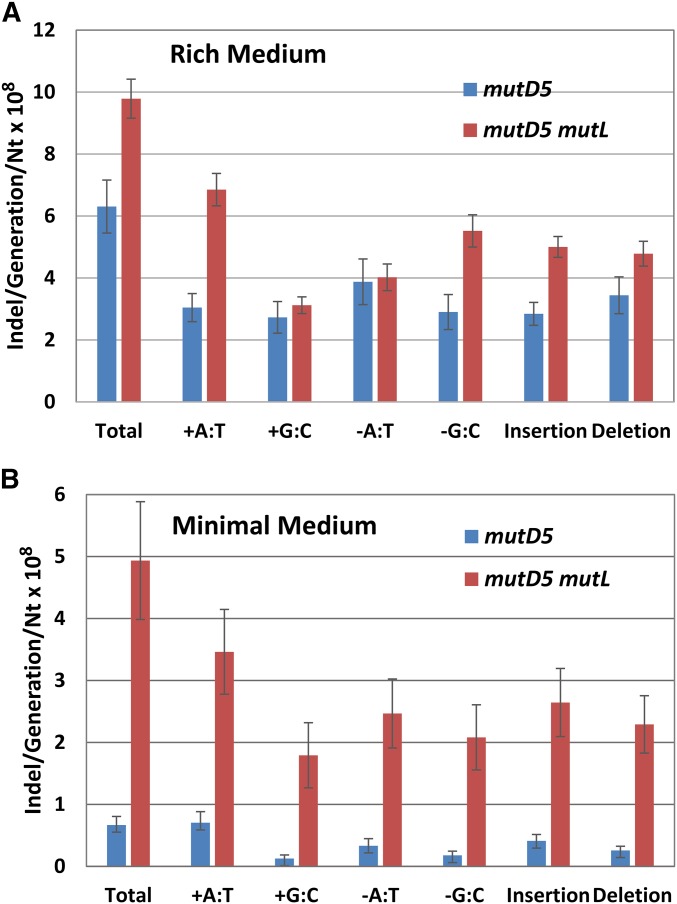
As previously observed for wild-type and MMR-defective strains (Lee et al. 2012), in both the mutD5 and mutD5 mutL mutant strains, the rates of small (≤ 4 bp) indels were one-tenth the BPS rates (Table 6). Also as expected from previous studies (Streisinger et al. 1966; Lee et al. 2012), in both the mutD5 and the mutD5 mutL mutant strains, homopolymeric runs were hotspots for indels and the indel rate increased exponentially with the length of a run (Figure 3A). In the mutD5 mutant strain, all types of indels occurred at nearly the same rates. However, in the mutD5 mutL mutant strain, A:T insertions dominated, occurring 1.7-fold more often as A:T deletions (χ2 = 60, P < 0.001) and 2.1-fold more often as G:C insertions (χ2 = 108, P < 0.001) (Figure 4A and Table 6). G:C deletions were also prominent, occurring 1.8-fold more frequently than G:C insertions (χ2 = 56, P < 0.001) and 1.4-fold more frequently than A:T deletions (χ2 = 23, P < 0.001).
Table 6. Conditional rates of indels accumulated in the MA experiments.
| Conditional indel rates ×108 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MA experiments on LB medium | MA experiments on minimal medium | ||||||||
| Type of indel | Wild-type ×103a | mutLb | mutD5 | mutD5mutL | mutD5dinB | mutD5dinBumuDC | mutD5lexA3 | mutD5 | mutD5mutL |
| Total | 2.06 ± 0.05 | 0.48 ± 0.01 | 6.3 ± 0.9 | 9.8 ± 0.5 | 4.9 ± 0.6 | 6.5 ± 0.8 | 7.0 ± 0.8 | 0.67 ± 0.14 | 4.9 ± 1.0 |
| +1 bp | 0.53 ± 0.02 | 0.26 ± 0.01 | 2.8 ± 0.4 | 5.0 ± 0.3 | 2.7 ± 0.2 | 3.5 ± 0.5 | 3.5 ± 0.4 | 0.41 ± 0.10 | 2.6 ± 0.5 |
| −1 bp | 1.40 ± 0.05 | 0.20 ± 0001 | 3.4 ± 0.6 | 5.0 ± 0.4 | 2.2 ± 0.4 | 3.0 ± 0.4 | 3.5 ± 0.4 | 0.25 ± 0.07 | 2.3 ± 0.5 |
| +>1 bp | 0.02 ± 0.004 | 0.01 ± 0.001 | 0.01 ± 0.02 | 0.04 ± 0.02 | 0.006 ± 0.0003 | 0.02 ± 0.02 | 0.02 ± 0.02 | < 0.003 | 0.04 ± 0.06 |
| −>1 bp | 0.11 ± 0.01 | 0.01 ± 0.0004 | 0.006 ± 0.02 | 0.003 ± 0.007 | < 0.006 | 0.01 ± 0.01 | 0.01 ± 0.02 | 0.003 ± 0.007 | 0.02 ± 0.04 |
| +1 A:T | 0.74 ± 0.04 | 0.10 ± 0.002 | 3.0 ± 0.5 | 6.9 ± 0.5 | 3.1 ± 0.2 | 4.1 ± 0.7 | 4.4 ± 0.6 | 0.70 ± 0.18 | 3.5 ± 0.7 |
| +1 G:C | 0.34 ± 0.01 | 0.41 ± 0.01 | 2.6 ± 0.5 | 3.1 ± 0.3 | 2.3 ± 0.3 | 2.8 ± 0.5 | 2.6 ± 0.5 | 0.13 ± 0.06 | 1.8 ± 0.5 |
| −1 A:T | 1.91 ± 0.08 | 0.24 ± 0.004 | 4.0 ± 0.7 | 4.0 ± 0.4 | 2.6 ± 0.2 | 3.5 ± 0.6 | 3.5 ± 0.5 | 0.33 ± 0.12 | 2.5 ± 0.6 |
| −1 G:C | 0.91 ± 0.09 | 0.16 ± 0.002 | 2.9 ± 0.6 | 5.5 ± 0.5 | 1.9 ± 0.6 | 2.6 ± 0.5 | 3.5 ± 0.5 | 0.18 ± 0.07 | 2.1 ± 0.5 |
| In run | 10 ± 0.3 | 2.9 ± 0.04 | 33 ± 4 | 55 ± 3 | 24 ± 3 | 34 ± 4 | 37 ± 4 | 3.7 ± 0.8 | 29 ± 5 |
| Not in run | 0.47 ± 0.03 | 0.01 ± 0.001 | 1.0 ± 0.2 | 0.9 ± 0.2 | 1.1 ± 0.1 | 1.1 ± 0.3 | 1.0 ± 0.2 | 0.07 ± 0.03 | 0.27 ± 0.15 |
| Noncoding | 7.0 ± 0.2 | 1.3 ± 0.02 | 11 ± 2 | 19 ± 1 | 9.7 ± 0.6 | 11 ± 2 | 13 ± 2 | 1.7 ± 0.5 | 11 ± 2 |
| Coding | 1.22 ± 0.04 | 0.34 ± 0.004 | 5.4 ± 0.7 | 8.3 ± 0.5 | 4.1 ± 0.6 | 5.7 ± 0.7 | 6.0 ± 0.7 | 0.48 ± 0.12 | 3.9 ± 0.9 |
A run is three or more of the same base pairs in sequence. Data are the conditional mutation rates, which is the number of mutations/number of generations/relevant number of nucleotides. When few indels were recovered, the 95% CL can include 0. MA, mutation accumulation experiments.
Data from Foster et al. (2015) for strains PFM2, PFM35, PFM40, PFM88, PFM91, PFM101, and PFM122.
Combined results from the mutL mutant strains PFM5, PFM144, and PFM288.
Figure 3.
The rates of the indels in homopolymeric runs accumulated by the mutD5 and mutD5 mutL mutant strains. The bars represent the indels per generation per number of base pairs in each run of nt length in the genome. The error bars are 95% CLs, some of which are smaller than the symbols. (A) Cells were grown on LB medium; mutD5, PFM163; mutD5 mutL, PFM165/397/399. (B) Cells were grown on minimal glucose medium; mutD5, PFM163; mutD5 mutL, PFM165. CL, confidence limit; indel, insertion/deletion.
Figure 4.
The conditional rates and spectra of the indels accumulated by the mutD5 and mutD5 mutL− mutant strains. The bars represent the indels per generation per number of relevant base pairs in the genome; the error bars are 95% CLs. (A) Cells were grown on LB medium; mutD5, PFM163; mutD5 mutL, PFM165/397/399. (B) Cells were grown on minimal glucose medium; mutD5, PFM163; mutD5 mutL, PFM165. The data in (B) for the mutD5 strain on minimal medium is presented with an expanded axis in Figure S5. CL, confidence limit; indel, insertion/deletion.
Mutational profile of mutD5 and mutD5 mutL mutant strains growing on minimal medium
The BPS profile:
Growing strains carrying the mutD5 allele on minimal rather than on rich medium lowers the mutation rate (Fowler et al. 1974; Cox and Horner 1982; Schaaper 1988). To evaluate the resulting mutational profile, we performed MA/WGS experiments with the mutD5 and mutD5 mutL mutant strains growing on glucose minimal medium. Relative to growth on rich medium, the BPS rate of the mutD5 mutant strain declined sixfold, to 15 × 10−8 BPS/generation/nt, whereas the BPS rate of the mutD5 mutL mutant strain declined only 1.5-fold, to 84 × 10−8 BPS/generation/nt (Table 4).
The ratios of NS/S BPSs of the mutD5 and the mutD5 mutL mutant strains growing on minimal medium, 1.97 and 1.75, were significantly less than expected from the genome or from simulations (Table 2), but not significantly different from that observed when the cells were grown on LB medium (χ2 = 0.4, P = 0.53 and χ2 = 1.5, P = 0.23, respectively). Thus, the mutD5 and mutD5 mutL mutant strains appear to be under some selective pressure whether they are growing on rich or on minimal medium.
The ratio of BPSs in C/NC DNA for the mutD5 mutant strain grown on minimal medium was significantly less than expected based on the genome or simulations (Table 3), and also significantly less than the ratio obtained when it was grown on rich medium (χ2 = 31, P < 0.001). Thus, the lower mutation rate of the mutD5 strain on minimal medium results in a slight bias for BPSs to occur in noncoding DNA. In contrast, the mutD5 mutL mutant strain grown on minimal medium showed the same bias for BPS to occur in coding DNA as it did when it was grown on rich medium (χ2 = 0.1, P = 0.7) (Table 3). Thus, on both types of media, MMR appears to preferentially prevent mutations in coding DNA, as previously observed (Lee et al. 2012).
The BPS spectra for the mutD5 and mutD5 mutL strains grown on minimal medium are shown in Figure 1B and given in Table 4 (also see Figure S4A and Table S3). Overall, the differences in the BPS spectra between the two growth media were modest. When the mutD5 mutant strain was grown on minimal medium, A:T transitions declined disproportionally relative to G:C transitions (7.6-fold vs. 4.7-fold; χ2 = 160, P < 0.001). When the mutD5 mutL mutant strain was grown on minimal medium, the ratio of transitions to transversions was double the ratio seen when the strain was grown on rich medium, largely due to a threefold decline in the relative rate of transversions. G:C transitions also declined slightly; their rate was 1.3-fold higher than that of A:T transitions (χ2 = 87 P < 0.001), compared to 1.6-fold higher when the mutD5 mutL mutant strain was grown on LB.
DNA strand bias:
Overall, growth on minimal medium did not change the strand biases from those observed when the strains were grown on LB. The one exception was a 1.2-fold increase in the frequency at which G:C transitions occurred with C on the LGST in the mutD5 mutant strain, which was significantly greater than expected (Table 5).
The local sequence context of BPSs:
Growing the mutD5 mutL strain on minimal medium resulted in nearly the same pattern of local sequence biases as growth on rich medium (Figure 2B). In particular, mutations at A:T base pairs were two- to threefold more frequent in the context 5′NAC3′+5′GTN3′, just as they were when the cells were grown on LB, indicating that DNA polymerase makes these errors frequently when cells are growing on either medium. However, in the mutD5 mutant strain, the influence of the 3′C was nearly gone, suggesting that MMR is better able to correct these errors when the cells are growing on minimal medium, probably because of the lower error rate (Figure 2B and Figure S4B).
Spontaneous indel rates and spectra:
As observed when strains were grown on rich medium, when the mutant strains were grown on minimal medium, indel rates were 10-fold lower than BPS rates (Table 6), homopolymeric runs were hotspots for indels, and the indel rate increased exponentially with the length of the run (Figure 3B). The spectra of indels in the two media were also similar (Figure 4B, Figure S5, Table 6, and Table S4). The only striking difference was the dominance of A:T insertions in the mutD5 mutant strain, which occurred at a twofold higher rate than A:T deletions (χ2 = 11, P = 0.001) and a fivefold higher rate than G:C insertions (χ2 = 34, P = < 0.001.
The SOS response does not contribute to the mutational load of the mutD5 mutant strain
Previous studies have reported that the SOS response is induced to various degrees in cells carrying mutant alleles of dnaQ (Slater et al. 1994; Gautam et al. 2012; Whatley and Kreuzer 2015). However, we have not investigated the extent to which SOS may be induced in our strains under the conditions of the MA experiments. The SOS response controls the expression of the two error-prone DNA polymerases, DNA Pol IV (encoded by the dinB gene) and Pol V (encoded by the umuDC genes) (Kenyon and Walker 1980; Fernández de Henestrosa et al. 2000), which could contribute to the mutational load of the mutD5 mutant strain. To test this hypothesis, we performed MA/WGS experiments with mutD5 mutant strains in which dinB, or both dinB and umuDC, were deleted, or which carried an allele of the SOS repressor, lexA3, that constitutively represses the SOS response (Mount et al. 1972).
As shown in Tables 1 to 5, deletion of the dinB gene, or both the dinB and the umuDC genes, in the mutD5 mutant strain made no significant difference in the BPS rates, spectra, or the other mutational parameters tested. Likewise, the rates and spectra of indels were unaffected by the deletions (Table 6). Surprisingly, the BPS rate of the mutD5 lexA3 strain was 1.4-fold higher than that of the mutD5 strain (t = 13, d.f. = 2, P = 0.005) (Table 4), suggesting that some other LexA-repressed gene may act to prevent some BPSs. Otherwise, the lexA3 allele did not affect the mutational profile of the mutD5 mutant strain. All of these results indicate that neither the error-prone polymerases nor the SOS response overall contributes to the mutational load of the mutD5 mutant strain in our MA experiments.
Discussion
The results of our studies of E. coli with a deficiency in proofreading can be summarized as follows.
The mutation rate of strains carrying the mutD5 mutant allele is ≈4000-fold higher than the mutation rate of the wild-type strain. This factor falls in the middle of previous estimates of 102–105. Loss of MMR increases this factor 1.5-fold.
As revealed in a strain defective for both proofreading and MMR, the replicative polymerase, Pol III, has a bias for making the errors that produce transitions, especially A:T transitions at 5′NAC3′+5′GTN3′ sites and, to a lesser degree, G:C transitions at 5′NGC3′+5′GCN3′ sites. However, overall, the spectrum of replication errors is dominated by G:C transitions.
Pol III has little strand bias for making errors. However, proofreading is strand-biased, resulting in the 2× bias observed for G:C transitions in wild-type strains, and for both G:C and A:T transitions in MMR-deficient strains.
Both proofreading and MMR have a bias for correcting the errors that produce A:T transitions, thus these transitions become prominent when either one is defective. Proofreader is also efficient at correcting the mismatches leading to G:C transitions, but, since these are the more prominent replication errors, G:C transitions dominate the wild-type spectrum.
When the mutD5 mutant strain is grown on minimal medium, its mutation rate is sixfold lower than when it is grown on LB, but this factor is only 1.5-fold if MMR is defective.
Neither the activities of the error-prone polymerases nor the SOS response overall contributes to the mutation load of the mutD5 mutant strain.
Our results differ in certain respects to those of previous studies of mutD5 and mutD5 mutL mutant strains. Using mutation to LacI−d as the reporter, Schaaper (1988, 1993) found that the BPS spectrum of the mutD5 mutant strain was dominated by G:C transitions, whereas that of the mutD5 mutL mutant strain was dominated by A:T transitions. In contrast, our results showed that the BPS spectrum of the mutD5 mutant strain was slightly biased toward A:T transitions, whereas the BPS spectrum of the mutD5 mutL mutant strain was biased toward G:C transitions. There are a number of possible reasons for these differences. First, while the LacI−d phenotype can result from a number of mutational events (Schaaper and Dunn 1991), the target is only 210 bp and does not include every possible sequence context in the genome. Second, the mutD5 alleles may differ. The mutD5 allele used in early studies by Schaaper and others had a long history of passages and genetic manipulations. Indeed, we sequenced the dnaQ gene of a strain derived from the original mutD5 mutant isolate (Degnen and Cox 1974) and discovered that it was actually the gene from E. coli B, not E. coli K12. Finally, as mentioned above, these highly mutating strains accumulate mutational enhancers and suppressors that can change the mutational profile.
Because of these considerations, we took precautions to ensure that our results were due only to loss of proofreading. First, we used recombineering to transfer only the E. coli K12 dnaQ gene carrying the mutD5 mutation, a C to T mutation at position 44 of the coding sequence (Fijalkowska and Schaaper 1996), to our parental strain. Before being used in a MA experiment, we sequenced the dnaQ and dnaE genes of each derived strain to verify that the dnaQ gene carried only the mutD5 mutation and that the dnaE gene was wild-type. Also, before use, we performed fluctuation tests to ensure that strains had the expected mutation rates. After sequencing the MA lines, we eliminated any that had known mutators or antimutators, or that had mutation rates > 2 SD above or below the mean, which would indicate that unknown mutation rate modifiers had appeared during the experiment.
Previous studies have found that MMR is saturated, at least partially, in strains that carry the mutD5 allele (Schaaper 1988). Here, we show that when growing on LB, the mutD5 mutL− mutant strain has a BPS rate 1.5-fold higher and an indel rate 1.6-fold higher than the mutD5 mutant strain, indicating that MMR is able to correct errors in the mutD5 strain. While this difference is much less than the ≈120-fold increase in the BPS rate observed when MMR is inactive in a proofreading-proficient strain (Table 1), the number of BPSs that MMR prevents in the mutD5 mutant strain, ≈2 per generation, is greater than the number that MMR prevents in the wild-type strain, ≈0.1 per generation. A similar conclusion can be made for the effect of MMR on indel formation; ≈0.2 indels per generation are prevented by MMR in the mutD5 mutant background, but only 0.02 in the wild-type background (Table 6 and Lee et al. 2012). However, although MMR may be working at high efficiency in the mutD5 mutant strain, it clearly cannot drive the mutation rate down to wild-type levels. In addition, when proofreader is defective, MMR appears to be nearly saturated for BPSs at A:T sites but not at G:C sites. Although in the absence of proofreading 5′NAC3′+5′GTN3′ are hotspots, A:T BPSs also arise at high rates at the other A:T sites, and these are relatively poor substrates for MMR [see the accompanying article in this issue by Foster et al. (2018)].
Most, if not all, of the increase in mutation rate of mutD5 mutant strains when growing in LB rather than minimal medium is due to the thymidine in LB (Degnen and Cox 1974; Erlich and Cox 1980). The most likely mechanism is a direct interaction between dTTP and ɛ that partially inactivates proofreading (Biswas and Kornberg 1984). Previous work has shown that because of the lower error rate, MMR is not saturated when mutD5 strains are grown on minimal medium (Schaaper 1988). From our data, MMR was able to prevent ≈3 BPSs per generation when the mutD5 mutant strain was growing on minimal medium, less than a twofold increase in efficiency over when the mutD5 mutant strain was growing on LB. Thus, in confirmation of previous results, the sixfold increase in mutation rate when the mutD5 strain is growing on LB medium must be due to some factors in addition to further saturation of MMR.
Our data show that neither the SOS response overall, nor the error-prone polymerases specifically, contribute to the mutation rate of strains carrying the mutD5 allele. The error-prone polymerases also did not add to the mutation rate during MA experiments with wild-type E. coli (Foster et al. 2015). We hypothesize that, in our strains and under our conditions, the SOS response may not be induced to sufficient levels to produce mutations. In support of this hypothesis, overproduction of the mutD5 allele, which is dominant, did not induce the SOS-response as measured by prophage induction (Gautam et al. 2012). Whatley and Kreuzer (2015) found that even in highly mutating dnaQ mutant strains the level of SOS induction, as measured by a lacZ fusion to the SOS-induced gene dinD, was only twofold higher than in wild-type strains; these authors concluded that mutation rate and SOS induction were not coupled in dnaQ mutant strains.
In the absence of MMR and proofreading, mutations were biased toward conversion of G:C to A:T base pairs and for creation of +1 A:T indels (Table 4 and Table 6). We assume that these biases are intrinsic to DNA Pol III. A long-standing a hypothesis, called the “A-rule,” postulates that some (but not all) DNA polymerases are biased for binding and inserting As when replicating past abasic sites and certain other DNA lesions [reviewed in Strauss (2002)]. However, this process creates mainly transversions, whereas the spectrum in our MA experiments is dominated by transitions, and also would be unlikely to produce +1 A:T indels. In addition, the estimated rate of spontaneous depurination during replication fails by two orders of magnitude to account for the mutation rates observed in the mutD5 mutant strains (Lee et al. 2012). Thus, our results suggest that DNA Pol III has a preference for inserting A’s even when replicating undamaged DNA.
The spectrum and context bias of BPSs in the mutD5 mutL mutant strain is similar in pattern and relative magnitude to that in the wild-type strain (Figure S3 and Table 4), suggesting that the effects of MMR and proofreading are synergistic, but nonetheless leave the signature of replication errors to appear in wild-type cells, albeit at a 6000-fold lower rate. Both MMR and proofreading are more efficient at preventing BPSs at A:Ts than at G:Cs, but this factor for MMR is about fourfold whereas for proofreader it is only twofold. Given that replication errors are biased toward G:C transitions, and that proofreader is 40-fold more powerful than MMR but only slightly biased against preventing G:C mutations, the result is that G:C transitions dominate the wild-type spectrum. But within that context, A:T BPSs at 5′NAC3′+5′GTN3′ sites and, to a lesser degree, G:C BPSs at 5′NGC3′+5′GCN3′ sites, are hotspots in every genetic background. These mutations, particularly the A:T mutations, are well corrected by MMR but not preferentially corrected by proofreader, and so also appear in the wild-type spectrum.
The G:C content of the E. coli genome is ∼50%. In the absence of error correction, the mutational bias of replication would tend to increase the A:T content unless selection reversed the trend. The results presented here indicate that proofreading is the major error-correcting activity maintaining the G:C content of the genome, reducing the nearly twofold bias for replacing G:C with A:T base pairs to the 1.4-fold bias seen in wild-type cells.
Acknowledgments
We thank the following past members of the P.L.F. laboratory for technical assistance: H. Bedwell-Ivers, C. P. Coplen, J. Eagan, N. Gruenhagen, N. Ivers, E. Popodi, I. Rameses, D. Simon, K. Smith, K. Storvik, J. P. Townes, and L. Whitson.; Roel Schaaper for the strain provided; and the anonymous reviewers of this paper for helpful suggestions. The National BioResource Project at the (Japanese) National Institute of Genetics provided bacterial strains and plasmids. This work was supported by the National Institutes of Health (T32 GM-007757 to B.A.N.) and the US Army Research Office Multidisciplinary University Research Initiative Award (W911NF-09-1-0444 to P.L.F. and H.T.).
Note added in proof: See Foster et al. 2018 (pp. 1029–1042) in this issue for a related work.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6513035.
Communicating editor: J. Nickoloff
Literature Cited
- Bhagwat A. S., Hao W., Townes J. P., Lee H., Tang H., et al. , 2016. Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc. Natl. Acad. Sci. USA 113: 2176–2181. 10.1073/pnas.1522325113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S. B., Kornberg A., 1984. Nucleoside triphosphate binding to DNA polymerase III holoenzyme of Escherichia coli. A direct photoaffinity labeling study. J. Biol. Chem. 259: 7990–7993. [PubMed] [Google Scholar]
- Bloom L. B., Chen X., Fygenson D. K., Turner J., O’Donnell M., et al. , 1997. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J. Biol. Chem. 272: 27919–27930. 10.1074/jbc.272.44.27919 [DOI] [PubMed] [Google Scholar]
- Cox E. C., Horner D. L., 1982. Dominant mutators in Escherichia coli. Genetics 100: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen G. E., Cox E. C., 1974. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J. Bacteriol. 117: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich H. A., Cox E. C., 1980. Interaction of an Escherichia coli mutator gene with a deoxyribonucleotide effector. Mol. Gen. Genet. 178: 703–708. 10.1007/BF00337881 [DOI] [PubMed] [Google Scholar]
- Fernández de Henestrosa A. R., Ogi T., Aoyagi S., Chafin D., Hayes J. J., et al. , 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35: 1560–1572. 10.1046/j.1365-2958.2000.01826.x [DOI] [PubMed] [Google Scholar]
- Fijalkowska I. J., Schaaper R. M., 1995. Effects of Escherichia coli dnaE antimutator alleles in a proofreading-deficient mutD5 strain. J. Bacteriol. 177: 5979–5986. 10.1128/jb.177.20.5979-5986.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska I. J., Schaaper R. M., 1996. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc. Natl. Acad. Sci. USA 93: 2856–2861. 10.1073/pnas.93.7.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2006. Methods for determining spontaneous mutation rates. Methods Enzymol. 409: 195–213. 10.1016/S0076-6879(05)09012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Lee H., Popodi E., Townes J. P., Tang H., 2015. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 112: E5990–E5999. 10.1073/pnas.1512136112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, P. L., B. A. Niccum, E. Popodi, J. P. Townes, H. Leeet al., 2018 Determinants of base-pair substitution patterns revealed by whole-genome sequencing of DNA mismatch repair defective Escherichia coli. Genetics 209: 1029–1042. [DOI] [PMC free article] [PubMed]
- Fowler R. G., Degnen G. E., Cox E. C., 1974. Mutational specificity of a conditional Escherichia coli mutator, mutD5. Mol. Gen. Genet. 133: 179–191. 10.1007/BF00267667 [DOI] [PubMed] [Google Scholar]
- Gautam S., Kalidindi R., Humayun M. Z., 2012. SOS induction and mutagenesis by dnaQ missense alleles in wild type cells. Mutat. Res. 735: 46–50. 10.1016/j.mrfmmm.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. M., Ma C. X., Liang P., Singh K. K., 2009. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25: 1564–1565. 10.1093/bioinformatics/btp253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. A., 2010. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim. Biophys. Acta 1804: 1041–1048. 10.1016/j.bbapap.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C., 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. USA 77: 2819–2823. 10.1073/pnas.77.5.2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancy E. D., Lifsics M. R., Kehres D. G., Maurer R., 1989. Isolation and characterization of mutants with deletions in dnaQ, the gene for the editing subunit of DNA polymerase III in Salmonella typhimurium. J. Bacteriol. 171: 5572–5580. 10.1128/jb.171.10.5572-5580.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Popodi E., Tang H., Foster P. L., 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 109: E2774–E2783. 10.1073/pnas.1210309109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D. W., Low K. B., Edmiston S. J., 1972. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet light-induced mutations. J. Bacteriol. 112: 886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowosielska A., Janion C., Grzesiuk E., 2004. Effect of deletion of SOS-induced polymerases, pol II, IV, and V, on spontaneous mutagenesis in Escherichia coli mutD5. Environ. Mol. Mutagen. 43: 226–234. 10.1002/em.20019 [DOI] [PubMed] [Google Scholar]
- Perrino F. W., Harvey S., McNeill S. M., 1999. Two functional domains of the epsilon subunit of DNA polymerase III. Biochemistry 38: 16001–16009. 10.1021/bi991429+ [DOI] [PubMed] [Google Scholar]
- Rice J. A., 1995. Mathematical Statistics and Data Analysis. Wadsworth Publishing Company, Belmont, CA. [Google Scholar]
- Schaaper R. M., 1988. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc. Natl. Acad. Sci. USA 85: 8126–8130. 10.1073/pnas.85.21.8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., 1993. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 268: 23762–23765. [PubMed] [Google Scholar]
- Schaaper R. M., Dunn R. L., 1991. Spontaneous mutation in the Escherichia coli lacI gene. Genetics 129: 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S. C., Lifsics M. R., O’Donnell M., Maurer R., 1994. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J. Bacteriol. 176: 815–821. 10.1128/jb.176.3.815-821.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss B. S., 2002. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst.) 1: 125–135. 10.1016/S1568-7864(01)00014-3 [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., et al. , 1966. Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 31: 77–84. 10.1101/SQB.1966.031.01.014 [DOI] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Gout J. F., Miller S. F., Williams E., et al. , 2015. Asymmetric context-dependent mutation patterns revealed through mutation-accumulation experiments. Mol. Biol. Evol. 32: 1672–1683. 10.1093/molbev/msv055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft-Benz S. A., Schaaper R. M., 1998. Mutational analysis of the 3′→5′ proofreading exonuclease of Escherichia coli DNA polymerase III. Nucleic Acids Res. 26: 4005–4011. 10.1093/nar/26.17.4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuru S., Ishizawa Y., Shibai A., Takahashi Y., Motooka D., et al. , 2015. Genomic confirmation of nutrient-dependent mutability of mutators in Escherichia coli. Genes Cells 20: 972–981. 10.1111/gtc.12300 [DOI] [PubMed] [Google Scholar]
- Whatley Z., Kreuzer K. N., 2015. Mutations that separate the functions of the proofreading subunit of the Escherichia coli replicase. G3 (Bethesda) 5: 1301–1311. 10.1534/g3.115.017285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar J. H., 1984. Biostatistical Analysis. Prentice Hall, Englewood Cliffs, NJ. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. File S1 contains the supplemental materials and methods. File S2 contains supplemental tables, which include strain genotypes, methods of strain construction, oligonucleotide sequences, and detailed data from each experiment. File S3 contains supplemental figures that are referenced in the text. The sequences, SNPs, and indels reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive [https://trace.ncbi.nlm.nih.gov/Traces/sra/ (accession no. SRP013707)] and in the IUScholarWorks Repository (hdl.handle.net/2022/20340). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6513035.