Hsp70/Hsp90 co-chaperone Sti1/Hop bridges Hsp70 and Hsp90 to facilitate client transfer. Many Hsp90 mutations make Sti1 necessary for viability, implying the Hsp90-Hsp70 interaction is essential. These mutations cluster in regions important for...
Keywords: Sti1, Hop, Hsp90
Abstract
The Hsp90 chaperone is regulated by many cochaperones that tune its activities, but how they act to coordinate various steps in the reaction cycle is unclear. The primary role of Saccharomyces cerevisiae Hsp70/Hsp90 cochaperone Sti1 (Hop in mammals) is to bridge Hsp70 and Hsp90 to facilitate client transfer. Sti1 is not essential, so Hsp90 can interact with Hsp70 in vivo without Sti1. Nevertheless, many Hsp90 mutations make Sti1 necessary. We noted that Sti1-dependent mutations cluster in regions proximal to N-terminal domains (SdN) or C-terminal domains (SdC), which are known to be important for interaction with Hsp70 or clients, respectively. To uncover mechanistic details of Sti1–Hsp90 cooperation, we identified intramolecular suppressors of the Hsp90 mutants and assessed their physical, functional, and genetic interactions with Hsp70, Sti1, and other cochaperones. Our findings suggest Hsp90 SdN and SdC mutants depend on the same interaction with Sti1, but for different reasons. Sti1 promoted an essential Hsp70 interaction in the SdN region and supported SdC-region function by establishing an Hsp90 conformation crucial for capturing clients and progressing through the reaction cycle. We find the Hsp70 interaction and relationship with Sti1/Hop is conserved in the human Hsp90 system. Our work consolidates and clarifies much structural, biochemical, and computational data to define in vivo roles of Sti1/Hop in coordinating Hsp70 binding and client transfer with progression of the Hsp90 reaction cycle.
HSP90 is a highly abundant, essential protein chaperone that assists the maturation of folding of hundreds of “client” protein substrates (Karagöz and Rudiger 2015; Prodromou 2016). It functions as a C-terminally joined homodimer with a complex ATP-regulated cycle of binding and releasing substrates (Schopf et al. 2017). The cycle begins with Hsp90 in an “open” conformation with its N-terminal ATPase domains (NTDs) separated. Clients can bind Hsp90 in this open state and, upon binding ATP, the NTDs come together to form a “closed” client-bound state. The closed dimer then undergoes conformational changes during which Hsp90 forms a tighter secondary closed state. Subsequent ATP hydrolysis leads to release of the client and returns Hsp90 to the open conformation. The bacterial Hsp90 homolog HtpG can act without help from cofactors, but many cochaperones interact with eukaryotic Hsp90 to regulate various steps in this cycle. Much is known about their individual roles in regulating Hsp90, but how they cooperate to control the reaction cycle in cells remains uncertain.
Many clients of Hsp90 (Hsc82 and Hsp82 in yeast) interact first with Hsp70. The Hsp70–Hsp90 organizing protein (Hop in mammals, Sti1 in yeast) facilitates delivery of such clients to Hsp90 by bridging Hsp90 and client-bound Hsp70 (Chen and Smith 1998; Song and Masison 2005). Despite this apparently critical role of Sti1, cells lacking Sti1 grow normally under optimal conditions. Nevertheless, the importance of Sti1 for Hsp90 function is evident as sti1∆ yeast are exquisitely sensitive to Hsp90-inhibiting compounds and die or grow poorly under suboptimal conditions (Chang et al. 1997; Song and Masison 2005). Since Sti1 lacking ability to regulate Hsp70 can still restore growth under such conditions, Hsp90 likely depends on Sti1 for something more than recruiting Hsp70 (Song and Masison 2005). Sti1 also inhibits Hsp90 ATP hydrolysis by preventing dimerization of the NTDs (Prodromou et al. 1999; Richter et al. 2003), which is likely important for transfer of clients. Indeed, binding of Hop to Hsp90 alters Hsp90 conformation in a way that positions it not only to interact with Hsp70, but also to load clients (Southworth and Agard 2011).
A need for Sti1 to promote delivery of clients might be bypassed by the ability of Hsp90 to bind substrates directly (Lorenz et al. 2014), although experimental support that it occurs in vivo is lacking. Additionally, Hsp70 and Hsp90 can interact directly and cooperate to promote protein folding (Genest et al. 2015; Kravats et al. 2017, 2018), which must contribute enough to client transfer to support yeast growth when Sti1 is absent. Nonetheless, even if both of these mechanisms are active in cells, without Sti1 they cannot keep Hsp90 functioning effectively enough to support growth under adverse conditions, again suggesting that Sti1 does more than bring Hsp70-bound clients to Hsp90.
Underscoring the importance of Sti1 for Hsp90 function, several Hsp90 mutants depend on Sti1 for cell viability. As with sti1∆ cells, such mutants typically grow well under optimal conditions, but poorly under various stresses. The mechanistic basis of the Sti1 dependency, or whether different Sti1-dependent mutants require the same or different activities of Sti1 has not been investigated systematically. We observed that mutations that cause Sti1 dependency cluster in two regions of Hsp90, one important for interaction with Hsp70 and the other for interaction with client substrates (Genest et al. 2013; Kravats et al. 2017, 2018; Zuehlke et al. 2017). Suppressors of mutations in theses clusters point to separate roles for Sti1 in the two processes. The notion that Sti1 has dual roles agrees with structural data suggesting Hop binding promotes both Hsp70 interaction and a client-loading conformation of Hsp90 (Southworth and Agard 2011). Our findings consolidate much other data and clarify roles for Sti1/Hop in regulating the Hsp90 reaction cycle in eukaryotic cells.
Materials and Methods
Yeast strains, plasmids, and growth conditions
Yeast strains used are in Table 1. Standard techniques were used to generate and confirm gene knockouts (Sherman 2002).
Table 1. Yeast strains used in this study.
| Strain | Relevant genotype | Reference |
|---|---|---|
| MR318 | MATα hsc82::KanMX hsp82::KanMX/pMR62 (HSP82 CEN/URA3) | Genest et al. (2013) |
| 1670 | MR318, MATa sti1::HIS3/pMR62 | This study |
| MR908 | MR318, MATa sba1::ZeoR/pMR62 | This study |
| MR1020 | MR318, MATa aha1::ZeoR/pMR62 | This study |
| CLY7 | MR318, hch1::HIS3/pMR62 | Zuehlke et al. (2017) |
| MR1022 | MR318, sti1::HIS3 aha1::ZeoR/pMR62 | This study |
| MR1025 | MR318, sti1::HIS3 hch1::HIS3/pMR62 | This study |
| MR1048 | MR318, sti1::HIS3 sba1::ZeoR/pMR62 | This study |
All strains are isogenic to and derived from 779 to 6A: MATa kar1 SUQ5 ade2-1 his3∆202 leu2∆1 trp1∆63 ura3-52 (Jung and Masison 2001).
Plasmids used are listed in Table 2. Plasmid pMR349 (Zuehlke et al. 2017), which was used for most plasmid shuffle experiments, is a centromeric LEU2 plasmid with N-terminal His6-tagged Hsp82 under the control of the HSC82 (strong constitutive) promoter. Plasmid pMR325-K399C was used in the screen to identify suppressors of K399C Sti1 dependence (see below). It is p414-GPD (Mumberg et al. 1995) containing hsp82K399C on a SpeI–XhoI fragment. Plasmid pMR379 is pRS314 (Sikorski and Hieter 1989) with STI1 controlled by its own promoter cloned at the BamHI site. Plasmid pMR219W is pRS424 (Christianson et al. 1992) encoding AHA1 with its own promoter and terminator cloned between the NotI and SalI sites. Plasmid pMR365 is p414-GPD with human HSP90β on a SpeI–SalI fragment ligated into SpeI and XhoI. Plasmids p415-HSPA1A and p415-HSPA8 are p415-PSSA2 (Reidy et al. 2013) with HSPA1A (hHsp70) or HSPA8 (hHsc70) on a BamHI–NotI fragment regulated by the SSA2 promoter.
Table 2. Plasmids used in this study.
| Plasmid | Allele | Backbone | Reference |
|---|---|---|---|
| pRS314 | Empty vector | CEN/TRP1 | Sikorski and Hieter (1989) |
| pRS315 | Empty vector | CEN/LEU2 | Sikorski and Hieter (1989) |
| pMR62 | PHSC82::HSP82 | pRS316 | Genest et al. (2011) |
| pMR349 | PHSC82::His6-HSP82 | pRS315 | Zuehlke et al. (2017) |
| pMR325-K399C | PGPD::hsp82K399C | p414-GPD (Mumberg et al. 1995) | This study |
| pMR379 | STI1 | pRS314 | This study |
| pMR365 | PGPD::HSP90β | p414-GPD (Mumberg et al. 1995) | This study |
| pMR219W | AHA1 | pRS424 | This study |
| p415-HSPA1A | PSSA2::HSPA1A | p415-PSSA2 (Reidy et al. 2013) | This study |
| p415-HSPA8 | PSSA2::HSPA8 | p415-PSSA2 | This study |
| pSK59 | HSP82 | pET28b | This study |
| pSK92 | AHA1 | pET28b | This study |
| pMR299 | SSA2 | pET24a | This study |
| pMR373 | HSP90β | pET28b | This study |
| pET24-HSPA1A | HSPA1A | pET24b | Genest et al. (2013) |
| pMR372 | HSPA8 | pET41b | This study |
The following plasmids were used for expressing proteins in Escherichia coli: Plasmid pSK59 is pET28b with the HSP82 open reading frame on a SpeI–XhoI fragment between NheI and XhoI sites. Plasmid pMR373 is similar, but contains HSP90β. Plasmid pSK92 has AHA1 ligated into pET28b digested with NheI and XhoI. Plasmid pMR299 is pET24a with SSA2 on a NdeI–XhoI fragment. pMR372 is pET41b with nontagged HSPA8, which encodes human Hsc70, on a NdeI–XhoI fragment. pET24-HSPA1A encodes human Hsp70. Mutations on plasmid-borne genes were introduced using the QuickChange Lightning or QuickChange Lightning Multi Site-Directed Mutagenesis Kits (Agilent).
YPAD (rich medium) contains 1% yeast extract, 2% peptone, 0.04% adenine, and 2% dextrose. SC media contain 2% dextrose, 0.7% yeast nitrogen base, and dropout mix lacking appropriate amino acids (Sunrise Scientific). Solid media contained 2% agar. Where used, radicicol (AG Scientific) was included in plates at a concentration of 25 mg/L. Plates were incubated for 2–3 days at 30° unless indicated otherwise.
Plasmid shuffling
Strains isogenic to MR318 were transformed with plasmids encoding the indicated HSP90 alleles expressed from LEU2 (HSP82) or TRP1 (HSP90β) centromeric plasmids and selected on plates lacking leucine or tryptophan, respectively, and containing uracil to allow loss of resident plasmid pMR62. Individual transformants were grown as patches on a master plate of the same selective medium and then replica plated onto the same medium lacking or containing 1 g/L 5-fluoroorotic acid (FOA), which kills cells that failed to lose the parental URA3 plasmid. Failure to grow on plates containing FOA indicates inability of the Hsp90 allele expressed from the LEU2 or TRP1 plasmid to support growth as the only source of Hsp90. Each experiment was repeated at least three times with three to four individual transformants. For most images presented, pools of transformants were grown as 0.5-cm patches and treated similarly.
Genetic screens
To identify intragenic suppressors of the Hsp90-lethal, Sti1-dependent C-terminal domain proximal (SdC) mutant E507R and cold sensitive W585T (Genest et al. 2013); plasmids pMR62L-E507R and pMR62L-W585T were mutagenized with hydroxylamine (Schatz et al. 1988) and used to transform strain 1670. Among ∼15,000 transformants of each plasmid, 8 that conferred resistance to FOA were isolated. Sequencing the Hsp82 alleles on these plasmids showed second-site mutations that restored function of E507R. These mutations included A107P (twice), A107T (twice), A107G, and A577V. Those suppressing W585T included D113N and T159I. Testing for cross-complementation showed A577V suppressed W585T. Additionally, when introduced by site-directed mutagenesis, A107N suppressed both E507R and W585T. Individual substitutions A107N and A577V did not cause significant growth defects.
To identify intragenic suppressors of K399C Sti1 dependence, plasmid pMR325-K399C was mutagenized with hydroxylamine. This mutagenized pool was then used to transform strain 1670. Primary transformation plates were replica plated to media lacking or containing FOA. The plasmid from FOA-resistant isolates was recovered and sequenced. From ∼10,000 primary transformants, the suppressing mutation E402K was isolated twice. No other suppressors were isolated. The substitution E402R requires two nucleotide base changes, which explains why we did not isolate it in the screen (based on frequency of recovery of E402K, changes producing E402R would be expected to occur in 1 of 25 million transformants).
To identify novel mutations in Hsp82 that conferred dependence on Sti1, strain 1670 was transformed with pMR349 mutagenized by propagation in E. coli strain XL-1 Red (Agilent) and plated on SC lacking Leu. After 3 days of incubation at 30°, primary transformation plates were replica plated to the same medium with and without FOA. Colonies that failed to grow on FOA were recovered from the plates without FOA and transformed with plasmid pMR379 (STI1) and plated on SC lacking Leu and Trp. Transformants were replica plated to the same medium with and without FOA. The mutated pMR349 (hsp82) plasmids in isolates that became FOA resistant upon transformation with plasmid pMR379 (STI1) were recovered and sequenced. Six Sti1-dependent mutations in Hsp82 (E199K, Y344C, I388N, S485T, M593T, and G655D) were found among ∼3000 transformants.
Growth assays
To assess relative growth rates, overnight cultures of cells recovered from FOA plates were grown in nonselective medium, diluted in water to OD600 = 0.25, serially diluted fivefold, and 5-μl drops were plated and incubated as noted.
In vivo pull downs and Western blotting
Hsp90 complexes from yeast cells were isolated as described (Zuehlke et al. 2017). Briefly, 50 OD600 units of cells expressing His6-Hsp82 were washed, suspended in 0.5 ml of lysis buffer (25 mM HEPES, pH 7.4, 100 mM NaCl, 2 mM MgCl2, 0.1% Tween 20, protease and phosphatase inhibitors), and broken by agitation with glass beads. Lysates were cleared by centrifugation and diluted 1:2.5 with lysis buffer. Talon resin (50 μl slurry) was added to 400 μl of diluted lysate and the mixture was incubated end over end at 4° for 30 min. Resin was washed with lysis buffer containing 10 mM imidazole and Hsp90 complexes were eluted with buffer containing 250 mM imidazole.
Antibody ADI-SPA-840 (Enzo) was used for detection of Hsp82 on Western blots. Hsp70 was detected using ADI-SPA-757 (Enzo). Antibodies to Sti1 were described previously (Song and Masison 2005). Antisera specific to Sba1 and Aha1 were gifts from J. Johnson and P. LaPointe, respectively.
Protein purification
Proteins were expressed in Rosetta 2 (DE3) E. coli grown in 2× YT medium. Log-phase cultures (OD600 = 0.4–0.8) were induced with 1 mM IPTG for either 3–4 hr at 30° or overnight at 20°. Cells were washed and stored at −80°. All protein purifications were performed at 4°.
Hsp90 proteins were purified from 800-ml cultures. Cells were suspended in 50 ml of lysis buffer Q/W1 (25 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 2 mM imidazole, 10% glycerol) supplemented with 0.1% Triton X-100, protease inhibitors, lysozyme (0.5 mg/ml), DNase I (20 μg/ml), and RNase A (20 μg/ml) and lysed by sonication (Branson Sonifier 250) on ice for 4 min at power setting 5 with a 50% duty cycle. Lysates were cleared by centrifugation at 12,500 relative centrifugal force (rcf) for 20 min, passed through a 0.22 μm filter, and applied to 10 ml of Talon resin packed into a XK 16 Column (GE) at a flow rate of 2 ml/min. The resin was washed with 50 ml of buffer Q/W1 and then with 20 ml of buffer W2 (buffer Q/W1 with 10 mM imidazole). Hsp90 was eluted using buffer E (buffer Q/W1 with 200 mM imidazole) in five 5-ml fractions. Peak fractions were pooled and concentrated to ∼0.5 ml using Amicon ultracentrifugation filters (Millipore, Bedford, MA) before being applied to a Superdex 200 10/300 GL gel filtration column equilibrated with buffer GF (25 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol) at a constant flow rate of 0.4 ml/min. Purified Hsp90 (at a concentration of 1–1.5 mg/ml) was eluted from the column at 10.6–13.6 ml. Yields of Hsp90 typically ranged from 3 to 5 mg of purified protein per liter of initial culture, with a purity of >95%.
Nontagged yeast and human Hsp70 proteins were purified similarly. Cell pellets from 800-ml cultures were suspended in 40 ml DEAE Q/L/W buffer (20 mM Tris–HCl, pH 7.5, 20 mM NaCl, 2 mM MgCl2, 1 mM DTT) supplemented with protease inhibitors, DNase I, RNase A, and antifoam agent (0.8 ml). Cells were lysed via a French press, clarified by centrifugation (20 min at 12,500 rcf) and filtration (0.22 μm), and loaded onto 20-ml DEAE sepharose packed into a XK16 column at a flow rate of 2 ml/min. The resin was then washed with 100 ml of DEAE Q/L/W buffer. A total of 10 ml of DEAE elution buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT) was then applied and the flow through was discarded. Proteins were eluted with 50 ml DEAE elution buffer, which was then mixed immediately with 5 ml ATP-agarose (Sigma Chemical, St. Louis, MO) that had been equilibrated with DEAE elution buffer and incubated end over end at 4° for 30–60 min. The mixture was then packed into an empty XK 16 column and washed with 25 ml DEAE elution buffer at a flow rate of 1 ml/min. The resin was further washed with 25 ml buffer ATP-W2 (20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 2 mM MgCl2, 1 mM DTT), followed by 10 ml of DEAE elution buffer. Purified Hsp70 was eluted from the column using buffer ATP-E (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, 5 mM ATP) in 10 2.5 ml fractions. Peak fractions were pooled and yields ranged from 4 to 10 mg Hsp70 per liter of culture, with >95% purity.
Aha1 protein was purified similarly to Hsp90, except the gel filtration step was omitted. After Talon affinity chromatography, the His6 tag was removed using the Thrombin Cleavage Kit (Millipore) according to the manufacturer’s instructions. Cleaved Aha1 was then reapplied over Talon resin to remove any residual uncleaved His6-Aha1 protein.
In vitro pull downs
In vitro pull-down assays were performed by first mixing 0.33 μM purified His6-tagged Hsp90 with 1 μM Hsp70 in 25 mM HEPES, pH 7.3, 100 mM NaCl, 2 mM MgCl2, 0.1% Tween 20, with 0.1 mg/ml BSA in a final volume of 250 μl. BSA was added to reduce nonspecific binding of Hsp70 to the Talon resin. After incubation at room temperature for 10 min, 30 μl Talon resin slurry was added and the mixture was incubated end over end for 10 min at room temperature. The resin was collected in a spin column and quickly washed with 75 μl of 25 mM HEPES, pH 7.3, 100 mM NaCl, 2 mM MgCl2, 0.1% Tween 20, 10 mM imidazole. Bound proteins were eluted with 30 μl of the same buffer containing 250 mM imidazole but lacking Tween 20. Eluted proteins were separated on 4–20% Criterion TGX SDS-PAGE gels (Bio-Rad, Hercules, CA) and stained with PageBlue (Thermo Scientific). Hsp90:Hsp70 ratios were determined by quantifying band intensities using ImageJ. Each experiment was repeated at least three times from at least two independent preparations of each protein. Values are reported as proportion of bound Hsp70 relative to wild-type Hsp90.
ATPase assays
A 1 μM concentration of purified Hsp82 was mixed with 50 μM γ-32P labeled ATP in 250 μl of 25 mM Tris–HCl, pH 7.4, 100 mM KCl, 5 mM MgCl2, 1 mM DTT. Aliquots were removed at certain time points and the rate of ATP hydrolysis was determined as described (Chock and Eisenberg 1979). ATPase stimulation by Aha1 was performed similarly except purified Aha1 was included at a final concentration of 1.25 μM.
Cochaperone profiles
His6-Hsp82 complexes (wild type and K399C) were isolated as described above and stored at −80°. Cells expressing nontagged Hsp82 were treated identically to serve as a control for nonspecific Talon resin binders. Subsamples were subjected to reductive di-methylation essentially as described (Boersema et al. 2009) using protocol C on four-layered large bore Stage Tips. Peptides derived from HIS-tagged wild type Hsp82, HIS-tagged hsp82K399C and non-tagged Hsp82 (negative control) pull-downs were labeled with light (L), medium (M) or heavy (H) isotopes, respectively. Subsamples were then combined and fractionated using strong cation-exchange chromatography as described (Kulak et al. 2014), which yields six subfractions. In hopes of obtaining the best quantitative data (as well as to support potential secondary goals of the experiment), each fraction was separated using a very long 300-min gradient (2% level for 5 min, 2–20% in 180 min, 20–40% in 40 min, 40–80% in 10 min) using a Waters nanoACQUITY using a vented-column configuration (analytical: 1.7 μM BEH130, 75 μM by 25 cm; trap:180 μM by 2-cm Symmetry C18) at 50°. The outflow of the analytical column was connected to a pre-Thermo Proxeon nano flow source using a conductive union (Upchurch) and distally coated silica emitter (New Objective) mounted on a Q-Exactive which collected a single MS1 spectrum (70K resolution) followed by up to 10 MS2 fragmentation spectra (17.5K). Raw data were analyzed March 2016 using MaxQuant 1.5317 (Cox and Mann 2008). To provide for the best comparisons, (L-H)/(M-H) values for any observed peptides of each protein were used and the median values were considered representative.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://figshare.com/s/5f48fd1e1880a3acbbc2.
Results
Hsp90 mutations causing Sti1 dependence cluster in two regions
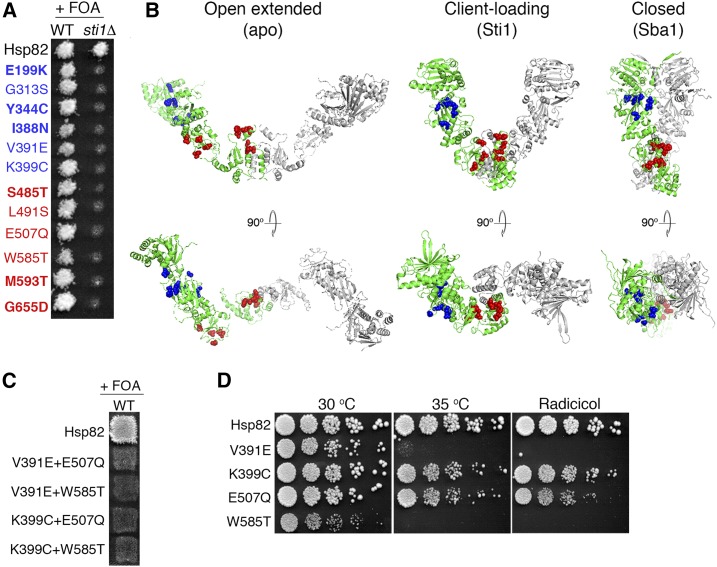
We mapped previously described mutations in Hsp82 that caused dependence on Sti1 for cell viability (see Figure 1A and Table 3) (Flom et al. 2007; Genest et al. 2013; Kravats et al. 2017) and noted that they clustered into two separate regions on the tertiary structure of the client-loading and closed conformations of Hsp82 (Figure 1B, middle and right panels). This clustering suggests these two sites are critically important for activities of Hsp90 that are regulated by Sti1. We designated these sites Sti1-dependent N-terminal domain proximal (SdN) and SdC.
Figure 1.
Hsp90 mutations that cause Sti1 dependence cluster in two regions. (A) Plasmid shuffle of indicated Hsp82 mutants in strains MR318 (STI1+, WT) and 1670 (sti1∆). All of the mutants support growth of the STI1+ strain, but none support growth of the sti1∆ strain on FOA, indicating dependence on Sti1. Those indicated in bold were identified in this study. (B) Mutations in (A) are represented as spheres on the various tertiary conformations of Hsp90. For clarity, only one monomer of the Hsp90 homodimer is colored. Left, the open-extended, apo form of Hsc82 (Krukenberg et al. 2009); middle, the Hop-bound conformation of Hsp90 (Southworth and Agard 2011); right, the closed, Sba1-bound form of Hsp82 (Ali et al. 2006). (C) A similar plasmid experiment as in (A) with the indicated double mutants in strain MR318 (WT). Combining SdN and SdC mutations was lethal even in these STI1+ cells. (D) Log-phase cultures of cells recovered from STI1+ cells on FOA plates in (A) that express the indicated Hsp82 mutants as the only source of Hsp90 were serially diluted and plated on rich medium without (left and middle) or with radicicol (right). All Hsp82 mutants have growth defects with V391E and W585T exhibiting the most severe phenotypes. WT, wild type.
Table 3. Identification and characterization of Sti1-dependent mutations of Hsp90.
| Mutation | How identified | Sti1 dependence demonstrated | |
|---|---|---|---|
| Hsc82 | Hsp82 | ||
| E199K | Sti1-dependent screen (this study) | This study | |
| G309S | G313S | Temperature-sensitive screen (Flom et al. 2007) | Flom et al. (2007) |
| Y344C | Sti1-dependent screen (this study) | This study | |
| I388N | Sti1-dependent screen (this study) | This study | |
| V391E | Defect in Aha1 interaction (Retzlaff et al. 2010) | This study | |
| K399C | Defect in Hsp70 interaction (Kravats et al. 2018) | Kravats et al. (2018) | |
| S481Y | Temperature-sensitive screen (Flom et al. 2007) | Flom et al. (2007) | |
| S485T | Sti1-dependent screen (this study) | This study | |
| L487S | L491S | Temperature-sensitive screen (Flom et al. 2007) | Flom et al. (2007) |
| E507Q | Defect in client interaction (Genest et al. 2013) | This study | |
| W585T | Defect in client interaction (Genest et al. 2013) | This study | |
| M589A | Temperature-sensitive screen (Flom et al. 2007) | Flom et al. (2007) | |
| M593T | Sti1-dependent screen (this study) | This study | |
| G655D | Sti1-dependent screen (this study) | This study | |
As no Sti1-dependent mutations were found outside these regions, we hypothesized that Sti1 regulation of Hsp90 was limited to two activities. To test this idea, we performed a random, unbiased genetic screen to identify additional mutations of Hsp82 that cause dependency on Sti1 to support growth. All of the six new mutations that we isolated were in these two regions. Three (E199K, Y344C, and I388N) were located in the SdN region and three (S485T, M593T, and G655D) were in the SdC region (Figure 1A). Although our screen was not saturated, these results are precisely what would be expected if the roles of Sti1 in the Hsp90 cycle were related only to the functions determined by these two sites of Hsp82.
Two of these mutations are in residues (S485 and M593) homologous to those identified previously in a screen for similar mutations using the Hsc82 isoform of yeast Hsp90 (Flom et al. 2007), implying conservation of the specific need for Sti1 by both Hsp82 and Hsc82. This notion is supported by the observation that G313S and L491S in Hsp82, homologous to G309S and L487S in Hsc82, are also Sti1 dependent (Figure 1A). For further study, we chose two from each class that had been characterized previously: V391E and K399C from the SdN region and E507Q and W585T from the SdC region.
The SdN mutations K399C and V391E are involved in the physical and functional interaction of Hsp90 with Hsp70 and Aha1 (see below), respectively (Meyer et al. 2004; Retzlaff et al. 2010; Wolmarans et al. 2016; Kravats et al. 2017, 2018). Although a relationship between V391E and Sti1 was not established previously, we included it in our study because we found that it was lethal in sti1∆ cells (discussed below, see Figure 1A). The SdC mutations E507Q and W585T are analogous to those of E. coli Hsp90 (HtpG) that weaken client interaction (Genest et al. 2013; Zuehlke et al. 2017). Thus, SdN and SdC regions correspond to locations on Hsp90 important for interactions with Hsp70 and clients, respectively.
To characterize how these four Hsp82 mutants function in vivo, we expressed them from plasmids in place of wild-type Hsp90. Strain MR318 (STI1+) and 1670 (sti1∆) lack chromosomal HSC82 and HSP82 genes and express wild-type Hsp82 from the URA3-marked plasmid pMR62. Cells cannot lose this plasmid because it encodes the only source of Hsp90. We transformed these strains with versions of LEU2 plasmid pMR349 encoding wild-type or mutant Hsp82. Transformants of either strain that express wild-type Hsp82 from pMR349 can lose pMR62 and grow on plates containing FOA, which kills cells expressing Ura3 (Figure 1A, top row). Each of the mutant versions of Hsp82 also allowed growth of the STI1+ strain on FOA (Figure 1A, left column), indicating they supported growth as the only source of Hsp90 when Sti1 was present; but none of them supported growth of sti1∆ cells (Figure 1A, right column), illustrating their dependence on Sti1. Combining SdN and SdC mutations in the same Hsp82 protein was lethal even in STI1+ cells (Figure 1C), suggesting these sites have nonoverlapping functions that are independently dispensable, but together are essential for Hsp82 to support viability.
From the FOA plate we recovered the STI1+ cells expressing the mutant versions of Hsp82 as the only source of Hsp90 to study further. All of the Hsp82 mutants exhibited growth defects compared with cells expressing wild-type Hsp82, in particular regarding sensitivity to both elevated temperature and radicicol, a competitive inhibitor of Hsp90 ATPase (Figure 1D). Differences did not correlate with location as V391E (SdN) and W585T (SdC) caused greater sensitivity than K399C (SdN) or E507Q (SdC). The temperature sensitivity was suppressed by adding the osmotic stabilizer sorbitol to the growth medium (Supplemental Material, Figure S1A), indicating that the sensitivity is not due to inactivation of the Hsp82 mutants at elevated temperature, but rather to impairment of an Hsp82 function in cell wall integrity, such as Slt2 kinase signaling (Millson et al. 2005; Truman et al. 2007). Sorbitol did not rescue Sti1 dependency (Figure S1B), however, which is consistent with a direct role of Sti1 for Hsp82 function. Thus, all the mutations reduce overall Hsp90 activity and do so to varying degrees.
To determine if other cochaperones are involved in functions of the SdN or SdC sites, we assessed effects of deleting the nonessential cochaperones Aha1, Hch1, or Sba1. Aha1 stimulates Hsp90 ATPase activity and promotes release of clients (Mayer et al. 2002; Panaretou et al. 2002; Hessling et al. 2009; Retzlaff et al. 2010), Hch1 reduces client binding, and exacerbates effects of SdC mutant W585T (Zuehlke et al. 2017), and Sba1 prolongs client interactions by binding an NTD-closed state of Hsp90 (McLaughlin et al. 2006).
Deleting AHA1 failed to overcome Sti1 dependence of any of the mutants (Figure S2A), but it slightly improved the ability of all Hsp82 mutants to support growth of STI1+ cells at optimal temperature. Deleting AHA1 also modestly improved the growth of cells expressing wild-type Hsp82 or the Hsp82K399C and Hsp82E507Q mutants at 35°, but did not restore growth of cells expressing the Hsp82W585T or Hsp82V391E mutants (Figure S2B). Thus, Aha1 did not have a strong or specific influence on Sti1-related phenotypes caused by the different mutations.
Deleting HCH1 weakly suppressed Sti1 dependence of Hsp82W585T, but did not affect Sti1 dependence of the other mutants (Figure S2C). Deleting HCH1 was shown to improve growth of cells with Hsp82W585T as the only Hsp90 at 30° (Zuehlke et al. 2017), and here we find it modestly and nonspecifically improved growth of the Sti1-dependent mutants in general (Figure S2D). Thus, like Aha1, the presence of Hch1 generally exacerbates the effects of the SdN and SdC mutants.
We found Hsp82W585T required Sba1 to support growth, even when Sti1 was present (Figure S2E, left). This result indicates that this mutant is defective beyond its need for Sti1, which explains its reduced growth even at optimal temperature. Deleting SBA1 did not affect the Sti1 dependence of any of the other mutants tested (Figure S2E, right) and it exacerbated the thermosensitivity of all the mutants that supported growth (Figure S2F). Unlike Aha1 and Hch1, both of which promote substrate release, the Sti1-dependent mutants functioned better when Sba1 was present. Together these results indicate that Sti1-dependent mutants are generally defective in binding clients.
Sti1 dependency of SdN is relieved by increasing Hsp70 interaction
Some SdN mutations reduce interaction with Hsp70 (Flom et al. 2007; Genest et al. 2015; Kravats et al. 2017, 2018), which is in line with the view that they cause Hsp82 to rely more on Sti1 to interact with Hsp70. To test this idea, we used pull downs with purified proteins to assess effects of the Hsp82 mutations on direct physical interactions with Ssa2, the major yeast cytosolic Hsp70. In agreement with the SdN region being important for Hsp70 binding, V391E and K399C mutations measurably reduced this binding (Figure 2A). In contrast, neither E507Q nor W585T had an appreciable effect on Ssa2 binding, which suggests that the SdC mutants depend on Sti1 for something other than recruiting Hsp70.
Figure 2.
Sti1 dependency caused by K399C is relieved by increasing Hsp70 interaction. (A) A portion of a Coomassie-stained gel of a representative in vitro pull-down experiment of purified His6-tagged wild-type and mutant Hsp82 with Ssa2 is shown above a graph displaying quantification of three replicate experiments (see also Figure S3). SdN mutants reduce interaction with Ssa2 to 50% of wild type. Values are averages relative to wild type and normalized to amount of Hsp82. Error bars are the SD. (B) Relative abundance of cochaperones copurifying in early and late complexes with His6-Hsp82K399C were derived from quantitative mass-spec. Left, early complexes; right, late complexes enriched by addition of AMP-PNP. (C) Cultures of sti1∆ strain 1670 expressing the indicated Hsp82 mutants as the sole source of Hsp90 were diluted and plated as in Figure 1D. Removal of negative charge at position 402 (E402A, F or S) partially rescues K399C Sti1 dependence, while positive charge (E402K or R) restored wild-type growth. (D) Western analysis of indicated proteins from lysates and His6-Hsp82 complexes (Pull-down) isolated from hsc82∆,hsp82∆ strain MR318 expressing the indicated Hsp82 mutants as the sole source of Hsp90. Aha1 and Sba1 samples were treated with AMP-PNP. E402R, alone or in combination with K399C (C + R), increased the amount of Hsp70 recovered. (E) Quantification of the amounts of Hsp70 (Ssa2) recovered by in vitro pull down with indicated Hsp82 mutants. E402R, alone or with K399C (C + R), increased the amount of Hsp70 recovered. Values are averages of three replicate experiments and error bars are the SD. (F) Intrinsic (−Aha1) or Aha1-stimulated (+Aha1) ATPase activity of Hsp82. Neither were affected by K399C or E402R, alone or in combination (C + R). Values are averages of three replicates and error bars are the SD. nt, nontagged wild-type Hsp82; WT, wild type.
As a separate approach to quantify the extent of this effect and determine if K399C affected interactions with other cochaperones, we developed a highly quantitative mass spectrometry (mass-spec) method for analyzing interaction of Hsp90 with cochaperones in vivo. In agreement with earlier work (Kravats et al. 2017, 2018) and our other approaches (e.g., Figure 2, A, D, and E), we found Hsp82K399C bound only 50% as much Hsp70 (Ssa1 and Ssa2) as wild-type Hsp82 (Figure 2B, left). K399C had little effect on binding of Hsp82 to any of the other cochaperones present in the samples (Sti1, Cpr6, or Ppt1).
We created similar mass-spec interaction profiles for late-cycle Hsp90 complexes by treating lysates with the nonhydrolyzable ATP analog AMP-PNP (Johnson et al. 2007). Sba1 binds an N-terminally dimerized form of Hsp90 which is considered to be late in the reaction cycle, and this binding is widely used as a sensor for this closed conformational state (Zuehlke and Johnson 2012). The K399C mutation reduced the amount of Sba1-containing complexes to only 30% of wild type (Figure 2B, right). We suspected K399C reduced Sba1 binding by causing a stall in the cycle before the NTDs dimerize so fewer complexes have reached the later conformation that binds Sba1, although it is possible it was due to a conformational alteration.
A stall would be consistent both with Sba1 binding Hsp82 after Sti1 (Richter et al. 2004; Li et al. 2011) and with these same late K399C complexes containing more Sti1, which keeps the NTDs apart (Figure 2B, right). Relative to wild-type Hsp82, late Hsp82K399C complexes also contained more Hsp70 (75%) than early Hsp82K399C complexes (50%). Since Hsp70 binds both Sti1 and Hsp82, the increased Hsp70 in late Hsp82K399C complexes could be due to its binding to the additional Sti1 in the complexes. These results could all be explained easily by Sti1 persisting to bind the mutant Hsp90 with Hsp70 until the client is transferred to Hsp90. Only when Sti1 and Hsp70 then leave the complex would the NTDs dimerize and allow binding of Sba1.
To gain more insight into the mechanism of the effects of K399C on Hsp82 function, we mutagenized Hsp82K399C and looked for second mutations that relieved the dependence on Sti1. Among ∼10,000 transformants, we twice isolated E402K, which is very close to K399C and on the same surface-exposed face of an α-helix. We presumed that E402K restored affinity for Hsp70 by restoring positive charge to this region. Tellingly, reversing the charge of E402 by substituting with arginine (E402R) restored function even better than lysine (Figure 2C), while the conservative acidic substitution E402D did not relieve the Sti1 dependence (Figure S3B). Eliminating the negative charge by making noncharged substitutions at E402 restored function, but much less effectively. Thus, net positive charge in this region is clearly important for Sti1-independent function of Hsp82, presumably by promoting interaction with Hsp70. We selected the strongest suppressor, E402R, for continued work.
We tested if E402R affected interaction of Hsp82 with Hsp70 or cochaperones in vivo by isolating Hsp82 complexes from cells and comparing the abundance of copurifying proteins. Although K399C had no effect on the amount of Sti1 copurifying in Hsp82 complexes from cell lysates, it clearly reduced the amount of Hsp70 that copurified (Figure 2D), as seen previously (Kravats et al. 2018). E402R also did not affect interaction of Hsp82 with Sti1, but it considerably improved binding of Hsp70, whether it was alone or combined with K399C. The amount of Hsp70 bound to wild-type Hsp82 and the Hsp82K399C,E402R double mutant was similar. Overall, these results suggest that E402R caused a general increase in affinity of Hsp82 for Hsp70, which can explain how it overcomes the need for Sti1 caused by K399C.
The similar recovery of Sti1 by wild type, Hsp82K399C, and Hsp82E402R suggested the effects of E402R on binding to Hsp70 were direct. Confirming this conclusion, we found using purified proteins that E402R increased the amount of direct Hsp70 interaction and that combining E402R with K399C restored Hsp70 binding to wild-type levels (Figure 2E). The magnitude of these effects of the Hsp82 mutations on Hsp70 binding were the same as those from cell lysates and are consistent with our in vivo data, suggesting the mutations have a direct effect on Hsp70–Hsp90 interaction in vivo. These in vitro and in vivo results are consistent with the conclusion that K399C impairs Hsp90 function by reducing a direct interaction with Hsp70 and that E402R overcomes this defect by restoring affinity of Hsp82K399C for Hsp70.
In AMP-PNP-treated cell lysates, the amounts of Aha1 and Sba1 copurifying with Hsp82K399C was less than that copurifying with wild-type Hsp82; while the amounts copurifying with Hsp82E402R and Hsp82K399C,E402R were greater (Figure 2D). Since Aha1 and Sba1 act in the Hsp90 cycle after Hsp70, more efficient client recruitment to Hsp82 caused by E402R could lead to a subsequent increase in binding of Aha1 and Sba1. Aha1 binds Hsp82 in our defined SdN region, however, so E402R might affect Aha1 interaction directly. We therefore tested if K399C or E402R affected a functional interaction of Hsp82 with Aha1 by measuring the ability of Aha1 to stimulate ATPase of purified Hsp82 mutant proteins (Figure 2F). Neither intrinsic nor Aha1-stimulated ATPase activity of Hsp82 was affected by K399C, E402R, or the combination of these mutations; indicating that E402R does not suppress K399C by affecting regulation of Hsp82 ATPase by Aha1.
Since E402R, either alone or with K399C, increased the amount of copurifying Sba1 (Figure 2D), we tested whether the ability of E402R to suppress K399C Sti1 dependence relied on Sba1. By using a plasmid shuffle in the sti1∆, sba1∆ strain MR1048, we found that lethality caused by K399C was rescued by E402R, indicating that E402R relieves Sti1 dependence of K399C independently of Sba1 (Figure S3C).
Taken together, these results strengthen the conclusion that the SdN region of Hsp82 is the site of Hsp70 interaction and that reduced affinity for Hsp70 caused by the SdN mutations imposes a need for Sti1 to improve interaction of Hsp70 with Hsp82.
E402R suppresses Sti1 dependence of SdN mutants and exacerbates SdC phenotypes
We expected E402R would also relieve the Sti1 dependence of other SdN mutants (e.g., V391E) by similarly restoring Hsp70 interaction, but would not help SdC mutants because they did not show reduced ability to bind Hsp70 in vitro. As anticipated, E402R suppressed the Sti1 dependence caused by V391E, but not by E507Q (Figure 3A). E402R displayed a very weak suppressive effect on Hsp82W585T Sti1 dependence, as seen by the slight growth on FOA plates (Figure 3A), but we were unable to work with these cells because they grew extremely slowly. Thus, ability of E402R to effectively overcome Sti1 dependence was restricted to SdN mutants.
Figure 3.
E402R suppresses Sti1 dependence of SdN mutants and exacerbates SdC phenotypes. (A) Plasmid shuffle experiment was done as in Figure 1A using strains expressing Hsp82 mutants with the additional E402R substitution. E402R suppressed the Sti1 dependence of the SdN mutants (V391E and K399C) but not the SdC mutants (E507Q and W585T). (B) STI1+ cells expressing the indicated Hsp82 mutants as the only source of Hsp90 were diluted and plated as in Figure 1D. E402R suppressed the sensitivity to high temperature and radicicol of the SdN mutants, but exacerbated SdC growth defects. (C) E402R did not affect intrinsic (−Aha1) or Aha1-stimulated (+Aha1) ATPase activity of purified Hsp82 when present alone or combined with V391E (E + R). Values are averages of three replicates and error bars are the SD. (D) Combining E402R with V391E (E + R) restored physical interaction with Ssa2 as determined by in vitro pull down. Values are averages of three replicates and error bars are the SD. WT, wild type.
In STI1+ cells, E402R completely suppressed the temperature sensitivity of SdN mutants V391E and K399C, but exacerbated the weak high temperature growth caused by E507Q (Figure 3B). Additionally, combining E402R with W585T exacerbated the growth defect at optimal temperature caused by W585T (Figure 3B, left panel), although it weakly suppressed the temperature sensitivity (Figure 3B, middle panel). Similarly, E402R suppressed the radicicol sensitivity of the SdN mutant Hsp82V391E but made the SdC mutant Hsp82E507Q more sensitive to radicicol (Figure 3B, right panel). These results show that improving Hsp70 interaction is not enough to overcome loss of all Sti1 functions, again implying that Sti1 does more than bridge Hsp70 and Hsp90.
Knowing V391E reduces interaction of Hsp82 with Aha1, we tested if E402R might restore the loss of Aha1 stimulation of Hsp82V391E ATPase activity by restoring this interaction. We found that combining E402R with V391E did not affect Aha1 stimulation of Hsp82 ATPase (Figure 3C), but did restore direct Hsp70 interaction (Figure 3D). These results suggest Sti1 dependency caused by V391E is due to reduced ability of Hsp82V391E to interact with Hsp70 rather than with Aha1.
Mutations promoting NTD dimerization suppress Sti1 dependence of SdC mutants
The Hsp82 charge-reversal substitution E507R is more detrimental than E507Q, causing lethality even in STI1+ cells (Genest et al. 2013). We used this phenotype to screen for second-site suppressors and identified A107P, A107G, A107T, and A577V. The A107N mutation was shown earlier to stabilize ATP binding and promote dimerization of the NTDs (Prodromou et al. 2000; Vaughan et al. 2009), so we suspected the other substitutions at this residue had a similar effect on Hsp90 function that mediated their suppressive effect. Indeed, A107N also suppressed both E507R lethality and W585T cold sensitivity (Zuehlke et al. 2017).
Closing of the NTDs is needed for progression through the Hsp90 cycle and, by promoting this step, A107N consequently increases intrinsic Hsp82 ATPase. Since Sba1 binds the NTD-dimerized conformation of Hsp82, A107N also leads to an increase in Sba1 binding, which prolongs the client-bound state. Accordingly, a defect in binding of Sba1 by Hsp82W585T is overcome by adding A107N, pointing to an indirect effect of W585T on client binding (Zuehlke et al. 2017). Thus, A107N likely suppresses the growth defects caused by W585T by promoting allosteric changes that aid client binding, which implies W585T causes defective client binding because it is impaired in adopting an NTD-dimerized conformation.
We hypothesized that such a defect could also underlie a common SdC-related need for Sti1 and that A107N would overcome the Sti1 dependency caused by W585T and the other SdC mutations. We found A107N did suppress the Sti1 dependence of both Hsp82W585T and Hsp82E507Q, but it did not relieve Sti1 dependence of Hsp82V391E or Hsp82K399C (Figure 4A). We further tested if the temperature sensitivity of Hsp82 mutants could be suppressed by A107N in STI1+ cells. A107N confers resistance to radicicol (Millson et al. 2010), precluding use of this compound for these growth assays. A107N fully rescued growth of the W585T at elevated temperature, but did not suppress V391E (Figure 4B). Thus, A107N specifically suppressed Sti1 dependence of SdC mutations, which is consistent with these mutants needing Sti1 to overcome a defect in NTD dimerization.
Figure 4.
Mutations promoting NTD dimerization suppress Sti1 dependence of SdC mutants. (A) Plasmid shuffle as in Figure 1A with Hsp82 mutants combined with A107N. A107N relieved Sti1 dependence caused by SdC, but not SdN mutations. (B) STI1+ cells expressing the indicated Hsp82 mutants as the only source of Hsp90 were diluted and plated as in Figure 1D. A107N relieved the temperature sensitivity of SdC mutants, but not the V391E SdN mutant. (C) In vitro pull down using purified Hsp82 and Ssa2. A107N did not affect interaction with Hsp70. (D) As in (A) using Hsp82 mutants combined with A577V. A577V behaved like A107N. (E) As in (B) using A577V in place of A107N. A577V behaved like A107N. (F) A107N and A577V increased intrinsic ATPase of Hsp82. (G) In vivo, A107N and A577V increased the amount of Sba1 recovered from AMP-PNP-treated lysates. (H) As in (A) using indicated Hsp82 mutants. The nitrosylation mimetic A577N is an SdC mutant that is suppressed by combining it with A107N. Combining both SdC suppressors A107N and A577V in the same allele was lethal even in STI1+ cells. Values in (C and F) are averages of three replicates and error bars are the SD. nt, nontagged wild-type Hsp82; WT, wild-type.
These results also suggest that the effects of A107N were unrelated to binding of Hsp70. To test this prediction, we performed pull downs with purified proteins. We found A107N had no effect on direct Hsp70 interaction (Figure 4C). These results, together with our genetic data indicating that increasing binding to Hsp70 makes effects of SdC mutations worse, provide additional evidence of dual roles for Sti1 in Hsp90 regulation.
The identification of A577V with A107N as a suppressor of E507R lethality points to A577 as sharing an overlapping function with A107. In support of this deduction, we found A577V similarly suppressed the growth defects and Sti1 dependence caused by the SdC mutations E507Q and W585T, but it did not suppress SdN mutations (Figure 4D). Additionally, in STI1+ cells, A577V rescued the thermosensitivity and radicicol hypersensitivity caused by W585T, but not V391E (Figure 4E). Thus, despite residing on the opposite end of the molecule from A107, A577V altered Hsp82 function in vivo like A107N.
In humans, the amino acid homologous to A577 is cysteine-597, which can be modified by nitrosylation to reduce Hsp90 activity (Martinez-Ruiz et al. 2005). Correspondingly, changing yeast A577 to cysteine (or isoleucine) enhances NTD dimerization and ATPase, while changing it to asparagine, a nitrosylation mimetic (Sieracki and Komarova 2013), diminishes NTD dimerization and therefore ATPase (Retzlaff et al. 2009). Because A577V suppressed SdC mutants like A107N, we expected it would act like the structurally related A577I and increase NTD dimerization. Consistent with this prediction, A577V increased intrinsic Hsp82 ATPase activity compared to wild type, although to a lesser extent than A107N (Figure 4F). In support of A577V promoting closure of Hsp90 NTDs, it also increased the amount of Sba1 recovered from AMP-PNP-treated lysates, but again less than A107N (Figure 4G). These results are consistent with the conclusion that the SdC mutations cause a defect in client capture by reducing efficiency of NTD dimerization.
If this inference regarding the molecular defects of SdC mutants is correct, then the nitrosylation mimetic mutation A577N, which impairs NTD dimerization, should cause an SdC-like defect. As expected, we found Hsp82A577N was indeed dependent on Sti1 and this dependence was suppressed by A107N (Figure 4H). Notably, combining the SdC-suppressing mutation of the same residue (A577V) with A107N was lethal in STI1+ cells, underscoring the importance of residue 577 in regulating the Hsp90 cycle and suggesting unrestrained NTD dimerization is deleterious. This ability of mutations in residue A577 to either cause or suppress SdC phenotypes is consistent with its identification as an allosteric switch point (Retzlaff et al. 2009) and links it to an indirect role in client binding.
These results suggest A577V suppresses the SdC mutants like A107N by priming the closure of the NTDs and that increased ATPase activity is a consequential effect. In support of the idea that NTD closure rather than acceleration of ATPase activity per se is the basis for suppression of SdC mutants, we found that overexpressing Aha1, which stimulates Hsp90 ATPase, did not relieve Sti1 dependency of SdC mutants (Figure S4).
Combining SdN and SdC suppressor mutations obviates the need for Sti1
Our conclusion that Sti1 is needed for two different functions in the Hsp90 cycle is in line with the fact that all known mutations in Hsp82 that cause Sti1 dependency reside only in the SdN and SdC regions, as described above. As another way to confirm Sti1 has two functions in the Hsp90 cycle, we tested the effects of combining the suppressing mutations. If the SdN and SdC mutants rely on Sti1 for different reasons, then a suppressing mutation of either class (e.g., E402R or A107N) in an otherwise wild-type Hsp82 might partially overcome the need for Sti1 at elevated temperature and, if so, combining them should be additive. Indeed, E402R and A107N each partially suppressed the thermosensitivity phenotype of sti1∆ cells, and combining them obviated the need for Sti1 at elevated temperature (Figure 5). This essentially complete independence of Sti1 for the Hsp82A107N,E402R double mutant is consistent with Sti1 being needed for two, and likely only two, Hsp90 activities.
Figure 5.
Combining SdN and SdC suppressors overcomes need for Sti1. Wild-type (STI1+) or sti1∆ cells expressing wild-type Hsp82, A107N, E402R, or the A107N,E402R double mutant (N + R) as the sole source of Hsp90 were plated on rich medium as in Figure 1D and incubated at the indicated temperatures. A107N and E402R each partially suppressed the high temperature growth defect of sti1∆ cells, while combining them (N + R) fully restored growth. WT, wild type.
Sti1 dependence of Hsp90β resembles SdN mutants and is relieved by human Hsc70
Yeast Hsc82 and Hsp82 are 97% identical, while the two human Hsp90 isoforms, Hsp90α and Hsp90β, are 85% identical to each other and 60% identical to the yeast Hsp90s. Hsp90β can support growth of yeast as the sole source of Hsp90, but this ability depends on Sti1 (Piper et al. 2003), which we confirmed in our strains (Figure 6A). In light of our findings, we presumed this need for Sti1 reflected a similar limitation of Hsp90 activities demanding one or both specific functions of Hop/Sti1.
Figure 6.
In yeast, human Hsp90β is an SdN mutant that is suppressed by human Hsc70. (A) A plasmid shuffle experiment was done as in Figure 1A with human Hsp90β using strains MR318 (wild type, WT) and 1670 (sti1∆). E414R in Hsp90β (homologous to E402R) suppressed Sti1 dependency of Hsp90β. All images are from the same plate. (B) As in (A), but coexpressing the human constitutive Hsp70 (Hsc70, HspA8) or the inducible isoform (Hsp70, HspA1A). Hsc70, but not Hsp70, overcame the Sti1 dependence of Hsp90β. (C) By in vitro pull down, Hsp90β (WT) bound approximately half as much Ssa2 as Hsc70. The E414R substitution in Hsp90β (E412R) increased direct interaction with both Ssa2 and Hsc70. Values are averages of three replicates and error bars are the SD.
Informed by our earlier studies of interspecies chaperone cooperation in yeast (Tutar et al. 2006; Miot et al. 2011; Reidy et al. 2012, 2013), we hypothesized that the Hsp90β dependence on Sti1 was due to coevolved alterations in Hsp90β and Hsp70 that reduced interaction of Hsp90β with yeast Hsp70s like an SdN mutant. We tested this idea by introducing the E414R mutation, which is homologous to E402R of Hsp82, into Hsp90β. Hsp90βE414R no longer required Sti1 to support growth (Figure 6A), suggesting that Hsp90β needs Sti1 because it cannot interact productively enough with yeast Hsp70s without it and that E414R overcomes this need by increasing affinity of Hsp90β for yeast Hsp70.
This conclusion implies that human Hsp70 has a higher affinity for Hsp90β than yeast Hsp70 and suggests it could alleviate dependence of Hsp90β on Sti1. We tested this prediction by coexpressing the constitutive (hHsc70; HspA8) or stress-inducible (hHsp70; HspA1A) human Hsp70 in cells expressing Hsp90β as the only source of Hsp90. We found hHsc70, but not hHsp70, allowed Hsp90β to support growth of sti1∆ cells (Figure 6B). This difference is probably related to the reported differences in ability of constitutive and inducible mammalian Hsp70 isoforms to support growth of yeast in place of yeast Hsp70s (Tutar et al. 2006). These results suggest that human Hsc70 interacts with Hsp90β better than yeast Hsp70s and the reduced affinity of yeast Hsp70s causes a need for Sti1.
In agreement with these in vivo results, we found purified Hsp90β bound roughly half as much Ssa2 as hHsc70 (Figure 6C) and, as predicted, Hsp90βE414R bound roughly 2.5-fold more of both hHsc70 and Ssa2 than wild-type Hsp90β bound to hHsc70 (Figure 6C). These results suggest that the Hsp90–Hsp70 interaction site in the Hsp90 SdN region is evolutionarily conserved.
Unexpectedly, wild-type Hsp90β bound more hHsp70 than hHsc70 (Figure 6C). Thus, the failure of hHsp70 to overcome Sti1 dependence of Hsp90β was not simply due to reduced Hsp70 interaction and must reflect differences in the ability of hHsc70 and hHsp70 to cooperate with Hsp90β, at least in yeast. Together our results uncover a clear functional difference between hHsc70 and hHsp70, which is probably related more to how they cooperate with yeast Hsp70 system components than with Hsp90β. This conclusion implies J-proteins, nucleotide exchange factors, and other Hsp70 cochaperones are crucial for Hsp90 function. Overall, our findings strongly support the idea that SdN mutants cause Sti1 dependency because of a disruption in a conserved physical contact between Hsp90 and Hsp70, and that this Hsp90–Hsp70 interaction is essential, but not sufficient, for Hsp90 functions required for viability.
Discussion
Our findings substantiate a postulated direct interaction of Hsp70 with Hsp90 in eukaryotic cells and clarify a role for Sti1 beyond recruiting Hsp70 that provides new insight into the importance of Sti1 for the Hsp90 reaction cycle (Southworth and Agard 2011; Kravats et al. 2018).
We define two sites on Hsp82 (SdN and SdC) important for different aspects of the Hsp90 reaction cycle that are regulated by Sti1. One of these functions is to promote a physical interaction of Hsp90 with substrate-bound Hsp70, which is well characterized (Chen and Smith 1998; Song and Masison 2005; Flom et al. 2007; Schmid et al. 2012; Alvira et al. 2014; Genest et al. 2015; Röhl et al. 2015; Kravats et al. 2017, 2018). The E402R alteration in Hsp82 essentially replaces this bridging function of Sti1/Hop by increasing the intrinsic ability of Hsp90 to interact directly with Hsp70, which increases the efficiency of client delivery. The lethality of combining SdN mutations of Hsp82 with depletion of Sti1 suggests that a direct or a Sti1/Hop-facilitated Hsp90–Hsp70 interaction is essential. The findings that dependence of human Hsp90β on Sti1 in yeast is suppressed by E414R or coexpression of human Hsc70 further suggest that this essential Hsp70–Hsp90 interaction is conserved in humans.
Another proposed role of Sti1 is to facilitate loading of the delivered substrates (Southworth and Agard 2011). This idea is based on structural data that show binding of Hop alters Hsp90 conformation in a manner that brings the dimer arms closer, while aligning hydrophobic residues on the inner face of the dimer. At the same time, Hop binding causes a rotation of the NTDs in a way that poises them for dimerization, yet keeps them apart (see Figure 1B). Hop therefore drives conformational rearrangements in Hsp90 that orient it to bind Hsp70 for client transfer while simultaneously poising the NTDs for subsequent dimerization and capture of the clients. The authors propose that stabilization of this poised Hsp90 conformation constitutes a distinct function of Hop/Sti1 (Southworth and Agard 2011).
Our data agree with this hypothesis of a role for Hop/Sti1 in client loading as an explanation of what is occurring in cells. Both Hsp70 recruitment and client-loading functions presumably occur during the same interaction of Sti1 with Hsp82. However, the general dispensability of the Hsp70-binding region of Sti1 (TPR1), but not the Hsp90-binding region (TPR2), for Hsp82 function suggests these allosteric changes in Hsp82 are driven adequately by the TPR2 region, which is consistent with structural data (Song and Masison 2005; Flom et al. 2007; Southworth and Agard 2011; Alvira et al. 2014). Thus, Sti1 lacking TPR1 could still be expected to enhance a direct interaction of Hsp82 with Hsp70 by inducing or prolonging a client-loading conformation of Hsp82 that is more accessible to Hsp70. TPR1 might have a separate role mediated by its ability to regulate Hsp70 independently of Hsp90 (Song and Masison 2005).
Our data suggest that Sti1 regulates Hsp90 to coordinate transfer of clients from Hsp70 with advancement of the Hsp90 cycle. Binding of Sti1 to Hsp90 both prevents commitment to subsequent steps until a productive interaction with Hsp70-delivered clients has occurred and positions Hsp90 to advance in its cycle promptly after client delivery. As release of Sti1 is associated with binding of Sba1 to Hsp82 (Richter et al. 2004; Li et al. 2011), this action of Sti1 would couple client binding with subsequent binding of Hsp90 to Sba1, a conclusion supported by our data showing prolonged binding of Sti1 and reduced binding of Sba1 to the late Hsp82K399C complexes. Rather than Sba1 “expelling” Sti1 (Li et al. 2011), Sti1 prevents Sba1 from binding by impeding NTD dimerization of Hsp90 until the client is properly positioned. When Sti1 exits the complex, the NTDs would readily dimerize, leading to binding of Sba1.
In contrast to the SdN mutants needing Sti1 to help Hsp82 stay open, the SdC mutants need Sti1 to help Hsp82 close. This need of SdC mutants is bypassed by A107N or A577V. The NTD mutation A107N strengthens the closed position of a lid structure over the ATP binding site that stabilizes the ATP-bound state and promotes NTD dimerization (Prodromou et al. 2000). The C-terminal residue A577 influences Hsp90 function similarly and alterations in A577 can promote NTD closure (Retzlaff et al. 2009). Computer modeling suggests the related effects of A107N and A577V can be explained by long-range interdomain communication between the N- and C-terminal regions that could be associated with allosteric regulation of Hsp90 activity (Morra et al. 2009). Our data showing that A577V increases both Sba1 interaction in vivo and Hsp82 ATPase in vitro are consistent with it promoting NTD closure and with earlier data showing that similarly altered A577I does the same (Retzlaff et al. 2009). The ability of the distantly separated A107N and A577V to counteract SdC mutations by promoting NTD dimerization implies that the SdC mutations impair NTD dimerization.
This conclusion is consistent with our earlier work suggesting that mutations in the SdC region have possible indirect or long-range effects on Hsp90 that influence client binding (Genest et al. 2013; Zuehlke et al. 2017) and with data showing that mutations in S485, defined here as an SdC residue, impair NTD dimerization and the binding of Sba1 upon addition of AMP-PNP (Soroka et al. 2012). Taking our data in the context of these and much other biochemical, structural, and molecular modeling work, we suppose the SdC mutations weaken client binding by disturbing long-range allosteric changes induced upon client interactions that are needed for appropriate NTD function. This allosteric communication is apparently essential as SdC mutant cells cannot survive unless Sti1 is present to compensate for such a defect by its ability to position Hsp90 in a way that primes the NTDs for dimerization.
In agreement with these ideas, recent computational analysis showed potential allosteric coupling between residue S485 and the NTD region that could be critical for interdomain communication (Stetz et al. 2018). Our findings emphasize the importance of the SdC region in Hsp90 cycle regulation in vivo and suggest S485 and other SdC residues are involved in such signaling that results in promoting NTD dimerization.
The NTD-proximal residue W300 of Hsp82 was recently identified as a switch point linked to transmitting information between domains in response to client interaction (Rutz et al. 2018). W300 is on the opposite side of Hsp90 from our defined SdN site, however, suggesting its role might not be related to Sti1 function. Accordingly, unlike loss of Sti1, the W300A mutation causes pronounced growth defects under optimal conditions and we found that, rather than being suppressed by E402R or A107N, it was lethal when combined with either of them (Figure S4C). Thus, W300A likely disrupts Hsp90 functions differently than our SdN or SdC mutations.
Current concepts for how Sti1 positions Hsp70 and Hsp90 for efficient client transfer and how Hsp70 and Hsp90 interact in a Sti1-independent manner are based primarily on biochemical studies focusing on the structure and function of Hsp90. Our findings here confirm and consolidate these proposed mechanisms by demonstrating conserved physiological roles for Sti1/Hop and for the Hsp70 (SdN) and client (SdC) interaction sites on Hsp90, which together cooperate in the transfer and capture of Hsp70-bound clients. By clarifying details of how of Sti1 regulates Hsp90 activities, our work provides insight into how cochaperones cooperate to coordinate progression of the Hsp90 reaction cycle. Important questions raised by our work that remain for future studies are how and to what extent Hsp70 cofactors might be involved in Hsp90 substrate capture and cycle regulation and the degree to which post-translational modifications of Sti1 or Hsp82 influence these interactions and functions.
Acknowledgments
We thank our National Institutes of Health colleagues for insightful discussions and help with the manuscript. This work was supported by the Intramural Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney diseases.
Footnotes
Supplemental material available at Figshare: https://figshare.com/s/5f48fd1e1880a3acbbc2.
Communicating editor: A. Mitchell
Literature Cited
- Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., et al. , 2006. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440: 1013–1017. 10.1038/nature04716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvira S., Cuellar J., Rohl A., Yamamoto S., Itoh H., et al. , 2014. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 5: 5484 10.1038/ncomms6484 [DOI] [PubMed] [Google Scholar]
- Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J., 2009. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4: 484–494. 10.1038/nprot.2009.21 [DOI] [PubMed] [Google Scholar]
- Chang H. C., Nathan D. F., Lindquist S., 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17: 318–325. 10.1128/MCB.17.1.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Smith D. F., 1998. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 273: 35194–35200. 10.1074/jbc.273.52.35194 [DOI] [PubMed] [Google Scholar]
- Chock S. P., Eisenberg E., 1979. The mechanism of the skeletal muscle myosin ATPase. I. Identity of the myosin active sites. J. Biol. Chem. 254: 3229–3235. [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M., 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Flom G., Behal R. H., Rosen L., Cole D. G., Johnson J. L., 2007. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem. J. 404: 159–167. 10.1042/BJ20070084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O., Hoskins J. R., Camberg J. L., Doyle S. M., Wickner S., 2011. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc. Natl. Acad. Sci. USA 108: 8206–8211. 10.1073/pnas.1104703108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O., Reidy M., Street T. O., Hoskins J. R., Camberg J. L., et al. , 2013. Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol. Cell 49: 464–473. 10.1016/j.molcel.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O., Hoskins J. R., Kravats A. N., Doyle S. M., Wickner S., 2015. Hsp70 and Hsp90 of E. coli directly interact for collaboration in protein remodeling. J. Mol. Biol. 427: 3877–3889. 10.1016/j.jmb.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessling M., Richter K., Buchner J., 2009. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 16: 287–293. 10.1038/nsmb.1565 [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Halas A., Flom G., 2007. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol. Cell. Biol. 27: 768–776. 10.1128/MCB.01034-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Masison D. C., 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43: 7–10. 10.1007/s002840010251 [DOI] [PubMed] [Google Scholar]
- Karagöz G. E., Rudiger S. G., 2015. Hsp90 interaction with clients. Trends Biochem. Sci. 40: 117–125. 10.1016/j.tibs.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Kravats A. N., Doyle S. M., Hoskins J. R., Genest O., Doody E., et al. , 2017. Interaction of E. coli Hsp90 with DnaK involves the DnaJ binding region of DnaK. J. Mol. Biol. 429: 858–872. 10.1016/j.jmb.2016.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravats A. N., Hoskins J. R., Reidy M., Johnson J. L., Doyle S. M., et al. , 2018. Functional and physical interaction between yeast Hsp90 and Hsp70. Proc. Natl. Acad. Sci. USA 115: E2210–E2219. 10.1073/pnas.1719969115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg K. A., Bottcher U. M., Southworth D. R., Agard D. A., 2009. Grp94, the endoplasmic reticulum Hsp90, has a similar solution conformation to cytosolic Hsp90 in the absence of nucleotide. Protein Sci. 18: 1815–1827. 10.1002/pro.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M., 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11: 319–324. 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- Li J., Richter K., Buchner J., 2011. Mixed Hsp90-cochaperone complexes are important for the progression of the reaction cycle. Nat. Struct. Mol. Biol. 18: 61–66. 10.1038/nsmb.1965 [DOI] [PubMed] [Google Scholar]
- Lorenz O. R., Freiburger L., Rutz D. A., Krause M., Zierer B. K., et al. , 2014. Modulation of the Hsp90 chaperone cycle by a stringent client protein. Mol. Cell 53: 941–953. 10.1016/j.molcel.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Martinez-Ruiz A., Villanueva L., Gonzalez de Orduna C., Lopez-Ferrer D., Higueras M. A., et al. , 2005. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc. Natl. Acad. Sci. USA 102: 8525–8530. 10.1073/pnas.0407294102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. P., Nikolay R., Bukau B., 2002. Aha, another regulator for hsp90 chaperones. Mol. Cell 10: 1255–1256. 10.1016/S1097-2765(02)00793-1 [DOI] [PubMed] [Google Scholar]
- McLaughlin S. H., Sobott F., Yao Z. P., Zhang W., Nielsen P. R., et al. , 2006. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 356: 746–758. 10.1016/j.jmb.2005.11.085 [DOI] [PubMed] [Google Scholar]
- Meyer P., Prodromou C., Liao C., Hu B., Roe S. M., et al. , 2004. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 23: 1402–1410. 10.1038/sj.emboj.7600141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millson S. H., Truman A. W., King V., Prodromou C., Pearl L. H., et al. , 2005. A two-hybrid screen of the yeast Proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot. Cell 4: 849–860. 10.1128/EC.4.5.849-860.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millson S. H., Prodromou C., Piper P. W., 2010. A simple yeast-based system for analyzing inhibitor resistance in the human cancer drug targets Hsp90alpha/beta. Biochem. Pharmacol. 79: 1581–1588. 10.1016/j.bcp.2010.01.031 [DOI] [PubMed] [Google Scholar]
- Miot M., Reidy M., Doyle S. M., Hoskins J. R., Johnston D. M., et al. , 2011. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. USA 108: 6915–6920. 10.1073/pnas.1102828108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra G., Verkhivker G., Colombo G., 2009. Modeling signal propagation mechanisms and ligand-based conformational dynamics of the Hsp90 molecular chaperone full-length dimer. PLoS Comput. Biol. 5: e1000323 10.1371/journal.pcbi.1000323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M., 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. 10.1016/0378-1119(95)00037-7 [DOI] [PubMed] [Google Scholar]
- Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J. K., et al. , 2002. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell 10: 1307–1318. 10.1016/S1097-2765(02)00785-2 [DOI] [PubMed] [Google Scholar]
- Piper P. W., Panaretou B., Millson S. H., Truman A., Mollapour M., et al. , 2003. Yeast is selectively hypersensitised to heat shock protein 90 (Hsp90)-targetting drugs with heterologous expression of the human Hsp90beta, a property that can be exploited in screens for new Hsp90 chaperone inhibitors. Gene 302: 165–170. 10.1016/S0378-1119(02)01102-2 [DOI] [PubMed] [Google Scholar]
- Prodromou C., 2016. Mechanisms of Hsp90 regulation. Biochem. J. 473: 2439–2452. 10.1042/BCJ20160005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C., Siligardi G., O’Brien R., Woolfson D. N., Regan L., et al. , 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18: 754–762. 10.1093/emboj/18.3.754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C., Panaretou B., Chohan S., Siligardi G., O’Brien R., et al. , 2000. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 19: 4383–4392. 10.1093/emboj/19.16.4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M., Miot M., Masison D. C., 2012. Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions. Genetics 192: 185–193. 10.1534/genetics.112.142307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M., Sharma R., Masison D. C., 2013. Schizosaccharomyces pombe disaggregation machinery chaperones support Saccharomyces cerevisiae growth and prion propagation. Eukaryot. Cell 12: 739–745. 10.1128/EC.00301-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzlaff M., Stahl M., Eberl H. C., Lagleder S., Beck J., et al. , 2009. Hsp90 is regulated by a switch point in the C-terminal domain. EMBO Rep. 10: 1147–1153. 10.1038/embor.2009.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzlaff M., Hagn F., Mitschke L., Hessling M., Gugel F., et al. , 2010. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol. Cell 37: 344–354. 10.1016/j.molcel.2010.01.006 [DOI] [PubMed] [Google Scholar]
- Richter K., Muschler P., Hainzl O., Reinstein J., Buchner J., 2003. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the atpase cycle. J. Biol. Chem. 278: 10328–10333. 10.1074/jbc.M213094200 [DOI] [PubMed] [Google Scholar]
- Richter K., Walter S., Buchner J., 2004. The Co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J. Mol. Biol. 342: 1403–1413. 10.1016/j.jmb.2004.07.064 [DOI] [PubMed] [Google Scholar]
- Röhl A., Wengler D., Madl T., Lagleder S., Tippel F., et al. , 2015. Hsp90 regulates the dynamics of its cochaperone Sti1 and the transfer of Hsp70 between modules. Nat. Commun. 6: 6655 10.1038/ncomms7655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz D. A., Luo Q., Freiburger L., Madl T., Kaila V. R. I., et al. , 2018. A switch point in the molecular chaperone Hsp90 responding to client interaction. Nat. Commun. 9: 1472 10.1038/s41467-018-03946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Solomon F., Botstein D., 1988. Isolation and characterization of conditional-lethal mutations in the TUB1 alpha-tubulin gene of the yeast Saccharomyces cerevisiae. Genetics 120: 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A. B., Lagleder S., Grawert M. A., Rohl A., Hagn F., et al. , 2012. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 31: 1506–1517. 10.1038/emboj.2011.472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopf F. H., Biebl M. M., Buchner J., 2017. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18: 345–360. 10.1038/nrm.2017.20 [DOI] [PubMed] [Google Scholar]
- Sherman F., 2002. Getting started with yeast. Methods Enzymol. 350: 3–41. 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Sieracki N. A., Komarova Y. A., 2013. Studying cell signal transduction with biomimetic point mutations, pp. 381–392 in Genetic Manipulation of DNA and Protein-Examples from Current Research, edited by Figurski D. InTech, Rijeka, Croatia. [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Masison D. C., 2005. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1). J. Biol. Chem. 280: 34178–34185. 10.1074/jbc.M505420200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroka J., Wandinger S. K., Mausbacher N., Schreiber T., Richter K., et al. , 2012. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol. Cell 45: 517–528. 10.1016/j.molcel.2011.12.031 [DOI] [PubMed] [Google Scholar]
- Southworth D. R., Agard D. A., 2011. Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-EM structure of the human Hsp90:Hop complex. Mol. Cell 42: 771–781. 10.1016/j.molcel.2011.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetz G., Tse A., Verkhivker G. M., 2018. Dissecting structure-encoded determinants of allosteric cross-talk between post-translational modification sites in the Hsp90 chaperones. Sci. Rep. 8: 6899 10.1038/s41598-018-25329-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman A. W., Millson S. H., Nuttall J. M., Mollapour M., Prodromou C., et al. , 2007. In the yeast heat shock response, Hsf1-directed induction of Hsp90 facilitates the activation of the Slt2 (Mpk1) mitogen-activated protein kinase required for cell integrity. Eukaryot. Cell 6: 744–752. 10.1128/EC.00009-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutar Y., Song Y., Masison D. C., 2006. Primate chaperones Hsc70 (constitutive) and Hsp70 (induced) differ functionally in supporting growth and prion propagation in Saccharomyces cerevisiae. Genetics 172: 851–861. 10.1534/genetics.105.048926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan C. K., Piper P. W., Pearl L. H., Prodromou C., 2009. A common conformationally coupled ATPase mechanism for yeast and human cytoplasmic HSP90s. FEBS J. 276: 199–209. 10.1111/j.1742-4658.2008.06773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolmarans A., Lee B., Spyracopoulos L., LaPointe P., 2016. The mechanism of Hsp90 ATPase stimulation by Aha1. Sci. Rep. 6: 33179 10.1038/srep33179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuehlke A. D., Johnson J. L., 2012. Chaperoning the chaperone: a role for the co-chaperone Cpr7 in modulating Hsp90 function in Saccharomyces cerevisiae. Genetics 191: 805–814. 10.1534/genetics.112.140319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuehlke A. D., Reidy M., Lin C., LaPointe P., Alsomairy S., et al. , 2017. An Hsp90 co-chaperone protein in yeast is functionally replaced by site-specific posttranslational modification in humans. Nat. Commun. 8: 15328 10.1038/ncomms15328 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://figshare.com/s/5f48fd1e1880a3acbbc2.