Abstract
Aging is a complex process affecting different species and individuals in different ways. Comparing genetic variation across species with their aging phenotypes will help understanding the molecular basis of aging and longevity. Although most studies on aging have so far focused on short-lived model organisms, recent comparisons of genomic, transcriptomic, and metabolomic data across lineages with different lifespans are unveiling molecular signatures associated with longevity. Here, we examine the relationship between genomic variation and maximum lifespan across primate species. We used two different approaches. First, we searched for parallel amino-acid mutations that co-occur with increases in longevity across the primate linage. Twenty-five such amino-acid variants were identified, several of which have been previously reported by studies with different experimental setups and in different model organisms. The genes harboring these mutations are mainly enriched in functional categories such as wound healing, blood coagulation, and cardiovascular disorders. We demonstrate that these pathways are highly enriched for pleiotropic effects, as predicted by the antagonistic pleiotropy theory of aging. A second approach was focused on changes in rates of protein evolution across the primate phylogeny. Using the phylogenetic generalized least squares, we show that some genes exhibit strong correlations between their evolutionary rates and longevity-associated traits. These include genes in the Sphingosine 1-phosphate pathway, PI3K signaling, and the Thrombin/protease-activated receptor pathway, among other cardiovascular processes. Together, these results shed light into human senescence patterns and underscore the power of comparative genomics to identify pathways related to aging and longevity.
Keywords: evolution, longevity, primates, genotype-phenotype, aging
Introduction
Senescence, or biological aging, refers to the general deterioration of physiological function of an organism, leading to an increased susceptibility to diseases and, ultimately, death. One of the most intriguing and fundamental questions in biology is why and how such a dazzling array of aging rates exists in nature (Jones et al. 2014). Just within metazoans, aging rates vary by as much as 10,000-fold (Austad 2001), with, for instance, mammals differing >100-fold in maximum lifespan (or MLS, Foote et al. 2013). Many lines of evidence support the conclusion that longevity is influenced by genetic variation both between and within species (Christensen et al. 2006). First, estimates of the heritability of lifespan in human populations are ∼0.25 (Herskind et al. 1996; Hjelmborg et al. 2006). Second, numerous gene mutations have been documented to increase lifespan in a range of model organisms (Kenyon 2010; Barzilai et al. 2012), and third, different species of the same family, or even genus occupying similar environments can exhibit stark differences in their lifespans, reflecting both their unique ecology and genetics.
Over the years, numerous evolutionary aging theories and concepts have been postulated (see Trindade et al. 2013 for a review), although the two most established are the mutation accumulation (MA) theory (Medawar 1952) and the antagonistic pleiotropy (AP) theory of senescence (Williams 1957). Both hypotheses predict the existence of genetic variants with adverse effects expressed later in life (hence modulating lifespan), with the AP theory adding an adaptive aspect, since it poses that, damaging mutations could be favored by natural selection if they are advantageous early in life.
From an evolutionary perspective, aging is a polygenic and highly complex process whose expression evolves quite rapidly, with relatively close species displaying very different phenotypes. Of particular interest are current patterns of human aging, which have changed markedly since our divergence with other primates. Indeed, primates are an interesting order from the aging perspective, since they are evolutionary close to each other in terms of phylogenetic distances and have homogeneously slow life histories relative to other mammals, while displaying profound differences in lifespan. Among primates, New World monkeys comprise some of both, the shortest and longest-lived monkey species. Prosimians develop the most rapidly and are the shortest lived, and, in contrast, great apes have the slowest development, and live the longest among the primates (Finch and Austad 2012).
Contrary to average life expectancy, which may change depending on living conditions, MLS is a stable characteristic of a species and evolves rapidly (Oeppen and Vaupel 2002; de Magalhães et al. 2007; Foote et al. 2013). For instance, humans and macaques diverged only ∼30 Ma, yet over this time their MLS has diverged as much as 3-fold (Colman et al. 2014). Despite the remarkable evolutionary lability of MLS the existence and nature of the molecular mechanisms involved in such variation remains unclear.
Leveraging primate variation to identify molecular changes that contribute to increases in longevity among primate species may help shed light on the selection pressures that shaped, not only our distant past (Enard 2014) but also extant variation contributing to human lifespans. To identify such changes it is necessary to compare phenotypic diversity in aging patterns among primates to changes in DNA or protein sequences that occurred independently on different lineages of the primate phylogeny (O’Connor and Mundy 2009, 2013; Lartillot and Poujol 2011; Boddy et al. 2017). In fact, parallel routes of molecular evolution may be relatively common among protein-coding genes from different species (Scally et al. 2012), and comparative genomic approaches have already been applied to investigate numerous traits such as vocal learning (Wirthlin et al. 2014), echolocation (Parker et al. 2013), flight in bats (Zhang et al. 2013), adaptation to aquatic life (Yim et al. 2014), and evolution in horses (Orlando et al. 2013). A complete comparative approach of MLS needs to consider not only longevity but also other phenotypes, such as body size (Austad 2005; de Magalhães et al. 2007), and life-history traits that correlated with MLS and may thus confound the analysis and interpretation (Fushan et al. 2015; Ma and Gladyshev 2017). Surprisingly, this approach has rarely been applied to lifespan (Aledo et al. 2011; Li and de Magalhães 2013; Doherty and de Magalhães 2016), although a few studies have addressed the mechanisms of plasticity for lifespan within species, or established the contributions of known longevity-related pathways (Bonafè et al. 2003; Holzenberger et al. 2003; Kapahi et al. 2004; López-Otín et al. 2013). Perhaps due to their limited focus, many of the findings of these works are difficult to reconcile, and none of them seem to adequately explain all aspects of the aging process (Gladyshev 2013).
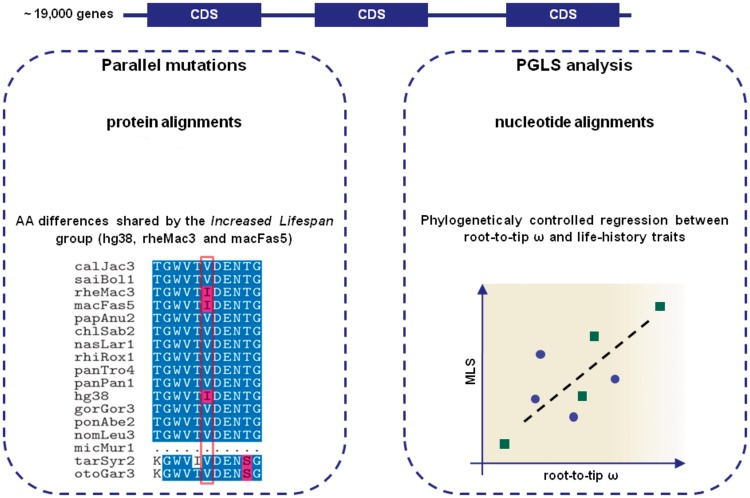
With such comparative phylogenetic strategy, we were able to identify molecular evolutionary correlates between genetic variation and MLS and other primate life-history traits. We deployed two approaches (fig. 1): one based on the detection of parallel mutations and the other one based on the study of gene–phenotype coevolution using phylogenetic generalized least squares (PGLS). The latter, has proven to be a powerful tool to detect gene–phenotype associations in combination with the root-to-tip dN/dS method (Montgomery et al. 2011; Montgomery and Mundy 2012a, 2012b; Lüke et al. 2014; Boddy et al. 2017). In summary, our study differs from, and extends, previous comparative studies in that we 1) consider several life-history variables and 2) perform analyses across the whole primate order. Both approaches allowed us to identify mutations, genes, and pathways that have putatively been fundamental to varying patterns of aging across primates.
Fig. 1.
Illustrated workflow of the two tests used in this study for measuring variation associated to MLS. Parallel amino-acid (AA) changes were investigated across all coding-regions for presence in the Increased Lifespan group of primate species compared with the rest, as shown in the example with a shared “I” (left panel). The second approach (right panel), was planned to test for coevolution between rates of protein evolution and life-history traits (or in short, gene–phenotype coevolution). First, root-to-tip dN/dS (ω) ratios were calculated for each species, for each gene. Using a phylogenetically controlled method, we tested for a correlation between the log10-transformed rates of protein evolution (i.e., root-to-tip ω, x axis) and each life-history trait (e.g., MLS, y axis). A significant linear relationship between gene and phenotype provides support for coevolution, as hypothetically illustrated in the figure.
Results
Relationship between Maximum Life Span and Other Life-History Traits
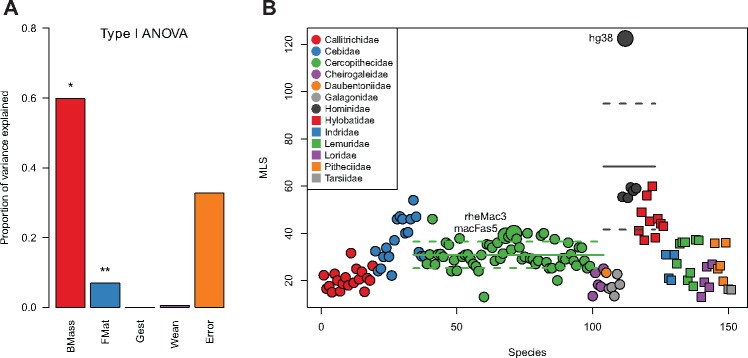
The life-history traits under study were largely obtained from the AnAge online database (de Magalhães and Costa 2009). They included MLS, body mass, female age at sexual maturity, gestation length, and weaning time. Longevity quotients (LQ) were calculated from the ratio of MLS to the predicted MLS based on the allometric equation for nonflying mammals (de Magalhães et al. 2007). These traits showed strong correlation among each other in primates (see supplementary fig. 1, Supplementary Material online). To test whether MLS covaries with other life-history traits, linear regression on the primate phenotype data was performed. Univariate linear regression showed that MLS covaries with body mass (adjR2=0.53, P < 2e-16), female age at maturity (adjR2=0.57, P < 2e-16), weaning time (adjR2=0.49, P < 2e-16), and gestation length (adjR2=0.32, P = 8.1e-12). Multiple linear regression followed by a type I analysis of variance (ANOVA) showed that, whereas female maturity presents the most significant nominal association with primate longevity (P = 0.002), body mass was the best predictor, accounting for ∼60% of the variance of MLS (P = 0.03). Age at female maturity accounted for almost 7% of the remaining variance, the remainder (∼30%) being residual error (fig. 2A).
Fig. 2.
(A) Bar plot showing the variance in MLS explained by each life-history trait as studied in a multivariate model. *P value < 0.05; **P value < 0.01. (B) Scatterplot showing variation in MLS (y axis) between primate species (x axis). Colors represent different primate families and species selected as Increased Lifespan in the parallel mutations analysis are displayed as bigger dots. Dark gray and green lines show, respectively, Hominidae and Cercopithecidae mean MLS (solid) and ±1 SD values (dashed). UCSC version names were used for species labeling. Correspondence to the species names can be found in supplementary table 5, Supplementary Material online. BMass, body mass; FMat, time to female maturity; Gest, gestation length; Wean, weaning time.
Pagel’s λ model was used to test for phylogenetic signals in primate traits. Consistent with previous findings, phylogeny explained a high proportion of the variance in primate MLS (λ = 0.87), as well as in the other life-history traits (female maturity = 0.9; weaning time = 0.71; gestation length = 0.93; body mass = 1). The only trait that showed a notably smaller λ was LQ, which had a value of λ = 0.69 (table 1).
Table 1.
Lambda (λ) Parameter Estimates for All Life-History Traits in Primates.
| Life-History Trait | λ | P (λ = 0)a |
|---|---|---|
| MLS | 0.87 | 7.9e-24 |
| LQ | 0.69 | 6.4e-11 |
| Adult body mass | 1 | 2.9e-49 |
| Female maturity | 0.90 | 2.5e-28 |
| Gestation length | 0.93 | 6.3e-42 |
| Weaning time | 0.71 | 8.2e-14 |
Significance of difference of the λ model from noise (λ = 0, LRT).
Parallel Mutations
To take a simple approximation to the study of any amino-acid changes shared by the species that increase their lifespan, we dichotomized MLS. Each primate species was classified in either of two categories according to the mean value of the primate family they belong to (see Materials and Methods). The species’ whose MLS is >1 SD from the mean of their family were classified within the Increased Lifespan group, whereas the rest were categorized as Control. The species in the Increased Lifespan group, which present the clearest increase in MLS since the ancestor of their family, included Homo sapiens, Macaca mulatta, and Macaca fascicularis (fig. 2B). To confirm the validity of considering human as an Increased Lifespan species, rather than discarding it due to culturally induced increase in lifespan, the same criteria was applied using 90 years as the MLS record for human, which could be more adequate for comparative purposes than 122, the commonly used value corresponding to the longest documented human lifespan (Lorenzini et al. 2005). Even in this case, humans are >1 SD from the mean of their family, and thus, we could safely consider humans as an Increased Lifespan species. Other thresholds besides 1SD MLS increase from the family mean were also considered to classify primate species. However, the results were exactly the same with any threshold between 0.6 and 1.4 SDs and poor information was obtained from more conservative or stringent cut-offs (supplementary fig. 2, Supplementary Material online). Consistently, discretizing LQ produced exactly the same classification.
Out of the ∼19,000 genes surveyed, 25 parallel amino-acid substitutions in 25 different genes were shared by all the species in the Increased Lifespan group while absent from the Control group (table 2 and supplementary fig. 3, Supplementary Material online). To ascertain whether this number of parallel changes could be expected by chance, we performed a series of four conservative resampling tests that produced distributions of parallel changes allowing us to assess the probability of our observation. In other words, we resampled as many species as within the Increased Lifespan group, three, from the phylogeny using four different criteria (see Materials and Methods). In the four sets of permutations, our observation of 25 parallel changes was situated in the empirical percentiles 83, 89, 92, and 93, respectively. Note that many of the sampled trios included more closely related species, for which a high number of parallel changes, or rather variants that are identical by descent, will naturally be observed. The mean number of parallel amino-acid changes found in the four resamplings was 71.6, 25.8, 8.6, and 5.7 and the median numbers of parallel changes were 3, 1, 1, and 0, respectively (supplementary fig. 4, Supplementary Material online). Thus, we conservatively concluded that our observation of 25 parallel changes in the three Increased Lifespan species and absent in the rest was not extreme in absolute terms. However, the observed value of parallel changes exceeded the median number of observations in the four random scenarios.
Table 2.
List of the 25 Genes Found Mutated in the Increased Lifespan Group, Together with a Description of Their Polymorphic State in Humans, the Reference and Alternative Alleles in Humans, Their Frequency, the Reported Change in 1 kG and the Amino-Acid Substitution in Primates.
| Gene | 1kG | ref | alt | Alternative Allele Frequency | 1kG_Change | Primates Change | SIFT | PolyPhen2 |
|---|---|---|---|---|---|---|---|---|
| AKAP9 | Not polymorphic | I3885V | ||||||
| ATG7a | Not polymorphic | T120A | ||||||
| BRD8 | rs412051 | T | C | AC=4894; AF=0.97 | p.Q1198R | Q1198R | 1, T | 0.0, B |
| C1QTNF2a | Not polymorphic | T9A | ||||||
| C9orf96/STKLD1 | Not polymorphic | L297V | ||||||
| DSC2a | rs561310777 | T | C | AC=1; AF=0.00019 | p.I520V | I520V | 0, D | 0.654, P |
| EFEMP2 | rs601314 | T | C | AC=4480; AF=0.89 | p.I259V | I259V | 1, T | 0.001, B |
| FCGBP | Not polymorphic | V499A | ||||||
| FGA | Not polymorphic | V244A | ||||||
| GP5a | Not polymorphic | Q68H | ||||||
| HEMK1a | rs192219149 | C | T | AC=2; AF=0.00039 | p.R98W | R98Q | 0.19, T | 0.001, B |
| IQCK | Not polymorphic | D21N | ||||||
| KIAA1614 | Not polymorphic | V29M | ||||||
| KLKB1 | Not polymorphic | A29T | ||||||
| MNTa | Not polymorphic | P392S | ||||||
| MYO16 | rs157024 | A | G | AC=665; AF=0.13 | p.I1171M | I1171M | 0.0, B | |
| MYOF | Not polymorphic | L135P | ||||||
| PLTPa | Not polymorphic | T435P | ||||||
| PRLa | Not polymorphic | M103I | ||||||
| RAD51AP1 | Not polymorphic | A161V | ||||||
| RXFP4 | Not polymorphic | A167V | ||||||
| STK31a | Not polymorphic | C933Y | ||||||
| SUPV3L1a | Not polymorphic | M330T | ||||||
| WDR87a | Not polymorphic | I36V | ||||||
| ZNF233a | Not polymorphic | Q556R |
Note.—When available, SIFT and PolyPhen2 scores are also reported for each change.
Discovered genes containing one gap in the control group.
Of course, the fact that 25 parallel changes are within random expectations does not exclude that the gene set is enriched with longevity-related genes. Indeed, the 25 detected genes were significantly enriched in genes previously related to aging, as determined from 10,000 bootstrap permutations of 25 genes from the genome (empirical P value = 6e-04) (supplementary fig. 5 and table 1, Supplementary Material online). Three genes: ATG7, MNT, and SUPV3L1 were categorized as aging-genes in model organisms from the GenAge database (Tacutu et al. 2013). Furthermore, we evaluated functional enrichment in the 25 geneset using GO and KEGG annotations. After false discovery rate (FDR) correction, several biological processes appeared as particularly enriched (fig. 3), including blood coagulation and intrinsic pathway and fibrin clot formation (adjP = 3.12e-02), negative regulation of hemostasis (adjP = 3.57e-02), wound healing (adjP = 3.57e-02), and regulation of body fluids levels (adjP = 3.57e-02). The KEGG pathway complement and coagulation cascades was also enriched (adjP = 7e-04), plus we found enrichment in genes related to advanced-age diseases, including blood coagulation disorders (adjP = 2e-04), coagulation protein disorders (adjP = 1.6e-3), heart diseases (adjP = 6.4e-03), cardiovascular diseases and thrombosis (both adjP = 7e-03), hemorrhagic disorders (adjP = 8.3e-03), arteriosclerosis, adhesion and arterial occlusive diseases (adjP = 0.0126), and coronary disease (adjP = 0.0133). These results were further validated using the expanded alignments generated in-house where new species (up-to 25) were included when available (fig. 3).
Fig. 3.
Heatmap of the enrichment analyses of genes disclosed in the Increased Lifespan group. Analyses are for gene ontology (A), pathway commons (B), KEGG pathways (C), and diseases (D). The first columns show pathway enrichments revealed when gaps were not allowed in the parallel mutation analysis (17sp and 25sp). The second columns show the results when one missing specie was allowed in the control group (17sp + gap, 25 sp + gap). Box diagrams roughly represent DNA Repair (dotted line) and Hemostasis (solid line) categories. As shown in the top right legend, green and yellow bars in top of the graphs show whether 17 or 25 primate species were used in the analyses. Color scale: in light yellow (nonsignificant, adjP ≥ 0.05), orange (marginally significant, 0.05 < adjP < 0.01), and red (significant, adjP ≤ 0.05).
Among the 25 nucleotide positions presenting parallel mutations, 20 were fixed for a novel variant in humans (1kGP Database), whereas the remaining five were segregating variants (table 2). Out of these latter five variants, only three had a frequency >1% in human populations (present in genes BRD8, EFEMP2, and MYO16) and four of them were defined as tolerated and benign by functional predictions of the SIFT and PolyPhen algorithms. The mutation in the DSC2 gene (p.Ile520Val) was predicted as highly deleterious and pathogenic by SIFT and PolyPhen, respectively. Another variant in the same amino-acid position (p.Ile520Thr) produces a dilated cardiomyopathy in humans according to the ClinVar database. In the ExAC Browser database (http://exac.broadinstitute.org/) which contains exome data on >60,000 individuals, only one individual was found to bear this allele (a frequency of 8.24e-06), thus it would seem that having an “I” in this position is fixed or almost fixed in humans.
The same amino-acid variants were evaluated in the 100-way alignment from UCSC (see Materials and Methods) in another set of three broadly studied long-lived mammals: the naked mole-rat and two long-lived bats. Out of the 25 amino-acid positions only 16 were present in the data set, among which 12 were different from the background primate state in at least one of these species; and two of them shared the very same change in these long-lived mammals: PRL and STK31 (supplementary table 2, Supplementary Material online).
Gene–Phenotype Evolution
To assess the relationship between rates of protein evolution and aging, we calculated root-to-tip ω values for each gene and species and evaluated their association with life-history traits using PGLS. For each gene, all the available primate sequences were used (at least n = 17). To correct for phylogenetic history, the median value for all the exome-wide, root-to-tip ωs for all species was included in the PGLS analyses as a covariate. We found no significant association between exome-wide values and either MLS (P = 0.89), body mass (P = 0.16), female age at maturity (P = 0.38), weaning time (P = 0.38), or LQ (P = 0.7) in individual PGLS regressions (supplementary fig. 6, Supplementary Material online). In contrast, gestation length and median root-to-tip ω values were nominally associated (P = 0.02).
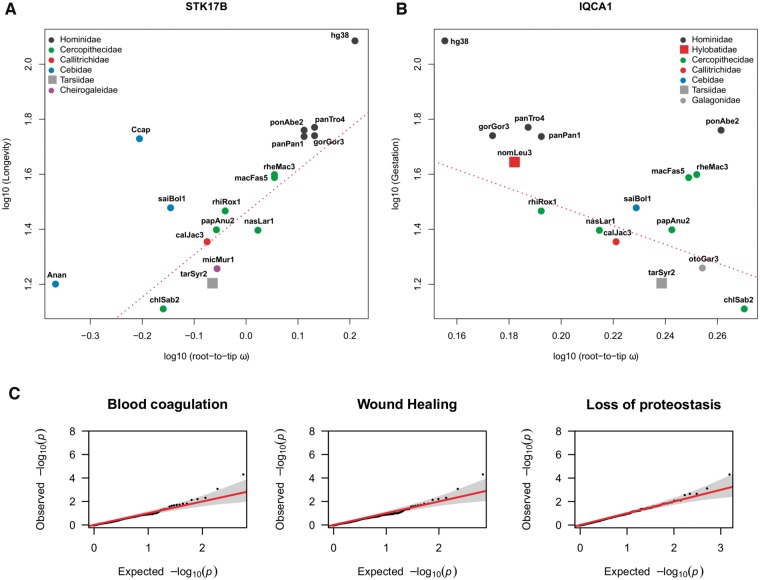
In the PGLS analyses of the life-history traits, only one gene exhibited marginally significant association to gestation length: IQCA1 (adjP = 0.049), whereas other genes showed a marginally nonsignificant association (FDR < 0.1) to the studied traits: STK17B (adjP = 0.060) in MLS, and CDC7 (adjP = 0.065), PER3 (adjP = 0.095), and SPRR2G (adjP = 0.095) in body mass (fig. 4A).
Fig. 4.
Phylogenetically controlled regression (PGLS) between log10 root-to-tip ω for STK17B (A) and IQCA1 (B) and log10 MLS and log10 Gestation length, respectively, across primate phylogeny. The dotted red line represents the regression intercept of the linear model (nonphylogenetically corrected). UCSC version names were used for species labeling. Correspondence to the species names can be found in supplementary table 5, Supplementary Material online. (C) QQ-plots illustrating the P values from each gene set of the root-to-tip ω in MLS. As depicted in the figure, outliers were above the null probability distribution, corresponding to ITPR1 and LBH genes, respectively.
We further assessed for deviation from the null straight line in a QQ-plot of MLS, using sets of genes related to senescence. These genesets were obtained from a review paper on the hallmarks of aging (López-Otín et al. 2013, see Materials and Methods for details), together with the major functional categories identified with our parallel mutations strategy (wound healing and blood coagulation). Most of the categories were not deviating from the null expectation, except Loss of Proteostasis, which revealed ITPR1 (adjP = 0.03). Moreover, wound healing and blood coagulation pathways presented the largest deviations from the null distribution, with two genes that were clearly above expectations, ITPR1 (adjP = 0.01) and LBH (adjP = 0.1), present in both pathways (fig. 4B).
The fact that almost no gene reaches significance is probably due to sample size and the drastic loss in statistical power induced by multiple-testing but, again, it does not exclude that the top associations are enriched with aging-related genes or pathways. The top genes (n = 26) with a nominal P value <1e-04 from all assessed traits are particularly enriched in aging genes (empirical P = 0.005) from the GenAge database (supplementary table 3, Supplementary Material online). This list of top genes was enriched in KEGG pathways such as cardiac muscle contraction (adjP = 0.015) and Alzheimer disease (adjP = 0.02); myometrial relaxation and contraction pathways (adjP = 0.015); and categories such as Sphingosine 1-phosphate pathway (adjP = 0.02), IFN-gamma pathway (adjP = 0.02), Class I PI3K signaling events (adjP = 0.02), and Thrombin/protease-activated receptor (PAR) pathway (adjP = 0.02) among many others; also diseases such as ventricular outflow obstruction (adjP = 0.005) were particularly enriched.
PGLS regressions were also performed for the 13 mitochondrial genes using a set of 93 primate mitochondrial genomes from MitoMap. Among all the evaluated life-history traits, only MLS and body mass showed significant associations with the root-to-tip ω of mitochondrial genes. Out of the 13 mitochondrial genes, ATP6, COX3, CYTB, and ND1 were correlated to MLS; and ATP6 and ND1 to body mass after mitochondrial-FDR correction. None of the genes associated to MLS survived when correction for mitochondrial-wide root-to-tip ω’s was applied in the model, but ATP6 remained associated to body mass after correction (supplementary table 4, Supplementary Material online).
Finally, to assess the soundness of our PGLS strategy, different primate phylogenetic trees were tested, resulting in consistent P values (supplementary fig. 7, Supplementary Material online).
Overlap between Life-History Traits
Nominally significant genes (P < 0.05) showing association between root-to-tip ω and each of the life-history traits under study (MLS, female maturity, gestation length, weaning time, and body mass) were retrieved and investigated for overlap. These five life-history traits shared 11 genes, which increases up to 19 genes when body mass is excluded. In both lists, the number of overlapping genes was significantly higher than expected by chance under the assumption of trait independence (empirical-P < 2.2e-16, supplementary fig. 8, Supplementary Material online). The 19 overlapping genes in MLS, weaning time, gestation, and female age at maturity (and body mass excluded) were: GP9, ADORA1, CCNJ, KCNQ5, ZNF300, NKRF, EMP1, RARS, GDF15, NMUR1, RP1, GAPDHS, GRAMD4, AKAP7, ACOT13, ZNF408, HSPB6, GABRG1, and BTBD9. This gene list showed enrichment in heart disorders and p73 transcription factor network, among other pathways (supplementary fig. 9, Supplementary Material online). Among them, GDF15 has been previously associated to aging in model organisms (GenAge database), and BTBD9 in humans (LongevityMap database).
Pathway Enrichment of Pleiotropies
As recently shown by Rodríguez et al. (2017), some variation in our genomes follows the predictions of the AP theory of senescence, with mutations providing selective advantage early in life at cost of having an adverse effect in old age. Using the list of pleiotropies provided by Rodríguez et al. (2017), we could confirm that aging pathways follow this expectation, with 5 out of the 9 curated hallmarks of aging (López-Otín et al. 2013) showing a significant enrichment of pleiotropic hits when compared with the genome-wide distribution of pleiotropies. This observation, validates our use of these sets as aging-related gene-sets. These categories were: altered intracellular communication, mitochondrial dysfunction, cellular senescence, genomic instability, and telomere attrition. The rest were not significant after FDR correction (table 3). Wound healing and blood coagulation, which are pathways identified here as having an evolutionary relationship to aging in primates, were also evaluated for enrichment in pleiotropies, and both turned out to be strongly enriched (wound healing, P = 2.6e-09; and blood coagulation, P = 2.33e-05) (table 3).
Table 3.
Pleiotropies Found in Each Category.
| Hallmarks of Aging | Genes | SNPs | n Pleios | Pleios/SNP | P value | SNPs/Genes |
|---|---|---|---|---|---|---|
| Cellular senescence | 51 | 44 | 29 | 0.65 | <2.2e-16* | 0.86 |
| Telomere attrition | 117 | 43 | 26 | 0.60 | 1.94e-15* | 0.36 |
| Mitochondrial dysfunction | 228 | 80 | 27 | 0.33 | 1.68e-08* | 0.35 |
| Genomic instability | 483 | 96 | 27 | 0.28 | 1.09e-06* | 0.19 |
| Altered intracellular communication | 624 | 354 | 65 | 0.18 | 4.68e-06* | 0.56 |
| Epigenetic alterations | 528 | 172 | 34 | 0.19 | 0.00018 | 0.32 |
| StemCell exhaustion | 105 | 75 | 13 | 0.17 | 0.0449 | 0.71 |
| Loss of proteostasis | 811 | 210 | 26 | 0.12 | 0.2 | 0.25 |
| Deregulated nutrient sensing | 185 | 58 | 8 | 0.13 | 0.25 | 0.31 |
| Wound healing | 561 | 181 | 47 | 0.26 | 2.66e-09* | 0.32 |
| Blood coagulation | 324 | 110 | 27 | 0.25 | 2.33e-05* | 0.34 |
Note.—*Asterisks represent significant categories after FDR correction.
Discussion
Biological mechanisms related to aging rates include, but are not limited to, the production of reactive oxygen species (ROS), control of oxidative damage, telomere shortening, various signaling pathways that produce antagonisms between development and aging, and inflammation responses that produce antagonisms between disease prevention and tissue damage (Ricklefs 2010; López-Otín et al. 2013). Other processes, particularly developmental mechanisms that influence life-history phenotypes, might be brought into play by evolution, including immune functions or inflammatory pathways (Finch 2010; Franceschi et al. 2017). How do these mechanisms account for variations in lifespan in the evolution of our species? From an evolutionary standpoint, it is surprising that, despite strong evidence that increases in lifespan have occurred in parallel in several independent primate species, such changes have been largely ignored in previous large-scale genomic comparisons. Here, we leveraged on the considerable variation in lifespans across the primate lineage to identify mutations and patterns of genic evolution that relate to aging. In particular, we targeted mutations that have occurred parallel to increases of lifespan and correlations between the rates of protein evolution and several life-history traits (fig. 1).
Mutations Parallel to Increased Lifespan
First, we tested for convergent changes occurring in long-lived primate species (to be precise, in primate species presenting relatively recent increases in lifespan). This is a reasonable hypothesis, especially since convergence at the genetic level seems to be more frequent than initially thought (Stern 2013). We identified a set of 25 genes sharing convergent nonsynonymous mutations in three Increased Lifespan primate species. About 3 of the 25 genes have been previously related to longevity in model organisms, as reported in the GenAge database: ATG7, MNT, and SUPV3L1. This is more than what would be expected by chance (P = 6e-04) and highlights the role of these genes in increasing aging via subtle amino-acid modifications. ATG7 encodes an essential enzyme for autophagy which also functions as a modulator of p53 in the regulation of survival during metabolic stress, among many other roles (Lee et al. 2012; Xiong 2015). Notably, the inactivation of this gene leads to reduced lifespan in model organisms (Jia and Levine n.d.; Juhász and Neufeld 2008; Eisenberg et al. 2009; Mizushima and Levine 2010; Pyo et al. 2013). More specifically, it results in loss of autophagy via deletion of ATG7-impaired DNA repair by homologous recombination through failed RAD51 recruitment (Liu et al. 2015). MNT is a tumor suppressor that has been also described to extend lifespan in Drosophila melanogaster and Caenorhabditis elegans (Demontis et al. 2014). Finally, mutants of the mitochondrial degradosome protein SUPV3L1 have been described to increase lifespan in a Saccharomyces cerevisiae model (Caballero et al. 2011). Although not directly associated to aging, a few other genes from the list of 25 hold promise for further experimental scrutiny, particularly considering their known functions or relation to aging phenotypes (supplementary table 1, Supplementary Material online). One example is DSC2, for which a predisposing mutation (c.631-2A > G) implicated in cardiopathy has been identified in one of the supercentenarians sequenced in a collection of the world’s oldest people (Gierman et al. 2014).
Although further functional data are necessary to demonstrate that these associations reflect causative relations with lifespan evolution, our results suggest that biological mechanisms related to wound healing, hemostasis, blood coagulation, clot formation, and many cardiovascular pathways are cornerstones of lifespan changes in the primate lineage. In agreement with our results, it has been largely documented that all phases of wound healing become delayed with age (Yanai et al. 2015; Keyes et al. 2016) and that imbalances in the hemostatic system are a common feature of the aging process (Franchini 2006; Mari et al. 2008). Additionally, gene expression signatures of blood coagulation are upregulated with age (de Magalhaes et al. 2009) and, interestingly, cardiac functioning improves in dogs after just weeks of an antiaging treatment with rapamycin (Urfer et al. 2017). Moreover, older age has long been associated with altered inflammation and hemostasis regulation since cardiovascular-related pathways have prominent roles influencing human longevity (Pilling et al. 2016). Thus, specific changes to wound healing and coagulation-related genes and pathways may underlie phenotypic changes in aged individuals and trigger lifespan increase, perhaps as a consequence of decreasing the risk of suffering cardiovascular diseases.
It is of note that most of the genetic variants that appear in parallel with increases in primate lifespan are fixed in humans, which highlights the importance of comparative genomics approaches, since they yield results that are complementary to those in within-species analysis that cannot access such variation. Further studies using intraspecific variation across nonhuman primates are needed to address whether these variants are or not also fixed in them.
Pleiotropies in the Aging Pathways
Genes affecting longevity might be crucial for organismic development, implying that these genes are under selective pressures so that mutations are subject to complex trade-offs. In fact, natural selection does not act on longevity per se, but rather on survival and, ultimately, on reproductive success. This makes interpretations very difficult, as the existence of pleiotropic effects in genes involved in aging could lead to a reduced capacity for their identification in the framework of an evolutionary approach, given that multiple functions can be under the control of each gene. In our analyses, we examined how genes previously described to be associated with aging were related to the pleiotropies described in Rodríguez et al. (2017) and we showed that 5 pathways out of 9 were highly enriched in such pleiotropies. Similarly, wound healing and blood coagulation categories, proposed by our evolutionary analysis in primates were also enriched in pleiotropic effects, highlighting that these genes are involved in multiple pathways and exhibiting the importance that these categories could have in the evolution of senescence, as predicted by the AP theory of aging. In agreement with our results, a recent study found that coronary artery disease loci are enriched in antagonistic-pleiotropic signals and suggests that natural selection may have maintained genetic variation contributing to cardiovascular disorders because of its beneficial effects in early life (Byars et al. 2017).
Gene–Phenotype Coevolution
PGLS treats both molecular and phenotypic data as continuous traits and can detect subtle variation between species as well as associations between genes and phenotypes that are consistent across a phylogeny. Since most of the variation in rate of actual senescence within species is caused by stochastic variation and measurement error (Ricklefs 2010; Ma and Gladyshev 2017), we also incorporated other life-history traits into the analyses. Our PGLS study identified key genes coevolving with interspecific variation in longevity-associated traits. IQCA1 (adjP = 0.04) was revealed to be significantly associated to gestation length. On the other hand, STK17B (adjP = 0.06) and CDC7, PER3 and SPRR2G were significantly associated to MLS and body mass (adjP <0.1), respectively. The 19 nominally significant genes (P < 0.05) overlapping in the four life-history traits evaluated (MLS, female maturity, weaning, and gestation length) disclosed pathways such as p73 transcription factor network and cardiovascular disorders. Interestingly, p73 has been previously linked to senescence and aging (Rufini et al. 2013) and the existence of cardiovascular phenotypes also found in the coevolution approach suggests that the same processes may have evolved along different routes to adapt to the long lifespans and slow development of primates. Moreover, the list of top genes (P < 1e-04) in all the studied phenotypes was particularly enriched in Sphingosine 1-phosphate pathway, IFN-gamma pathway, Class I PI3K signaling events, and Thrombin/protease-activated receptor (PAR) pathway, together with other cardiovascular pathways. Thrombin plays a critical role in hemostasis and coagulation, whereas Class I PI3K activation finally leads to mTORC1 activation triggering cellular growth (Dibble and Cantley 2015). Finally, Sphingosine-1-phosphate is primarily involved in supporting growth and survival, and it might contribute to aging by AP-like effects (Huang et al. 2014).
Furthermore, our analysis revealed associations between mitochondrial genes and MLS: ATP6, COX3, CYTB, and ND1. Among them, ATP6 has been previously associated with differences in metabolism and selection linked to energetics (da Fonseca et al. 2008), and CYTB has been found associated to longevity in mammals using a linear model analysis (Feng and Zhou 2017). Because molecular evolution rates correlate with generation times in mammals (Bromham et al. 1996), it is not surprising to find mitochondrial genes that correlate with life-history traits in our study. Even though subsequent analyses suggested that this association is due to covariation between generation time and longevity (Nabholz et al. 2008). In favor of the latter, when mean mitochondrial-wide ω values were included as covariate in our model, all signals associated to longevity disappeared.
Limitations of the Present Study
Our study did not examine genetic variation among individuals from the same species or over an individual lifespan but rather compared genetic variation among reference individuals of different species. At the time of analysis, only 17 full primate genomes were available, although we could gather up to 25 primate species for some of the genes. The addition of more species to the analysis has the potential to contribute independent variation in longevity, thus increasing power. On the other hand, and since more distant species may differ considerably in their developmental and/or genetic mechanisms, increasing the number of species may also result in adding more noise to the analysis. At any rate, despite the relatively small number of species in our analyses, we still find several genes with a phylogenetic association with MLS and other life-history traits, providing evidence for conserved molecular mechanisms on the evolution of senescence.
We have also identified several genes that coevolved with life-history traits in primates. However, our PGLS approach can only detect cases in which proteins and traits have evolved consistently (that is, when accelerated protein evolution has results in a systematic increment or decrement of a trait), so we cannot exclude neither the possibility that different species-specific mechanisms might have evolved in parallel, nor other, more complex, relationships between rates of protein evolution and the traits under study. All these possibilities remain to be assessed in future work.
Conclusions
To our knowledge, this is the first systematic report providing direct evidence of gene–phenotype evolution of aging-related traits in primates. Genes and biological processes reported in this study could be added to the list of genes that increase lifespan when overexpressed or mutated (gerontogenes) and represent a valuable resource for examination of new candidate interventions that mimic gene evolution associated with natural changes in lifespan.
Although our results may reflect local adaptive responses of species to their environment, we observed nonrandom association of gene evolution with pathways mainly related to wound healing, coagulation, and many cardiovascular processes. This would make sense from a biological perspective, since flexible and adjustable control of coagulation mechanisms is required for species that live longer. As far as we know for the first time, we report pathways putatively involved in maximum lifespan and correlated life-history traits. This can be explained in part by the fact that many studies in the field have been using models such as yeast or C. elegans. Although these model organisms have proven very useful in many aspects, they are quite different from humans and lack many of the molecular pathways that are fundamental in more complex animals living under natural conditions.
These findings provide direct insights into how nature reversibly adjusts lifespan and other traits during adaptive radiation of primates, and provide evidence that cardiovascular and hemostatic pathways evolve adaptively to allow for increases in longevity.
Materials and Methods
Phenotypic and Genomic Databases
The life-history traits evaluated in this study include MLS, adult body mass, female age at sexual maturity, gestation length, and weaning time. These variables were largely obtained from the AnAge online database (de Magalhães and Costa 2009). Longevity quotients (LQ) were calculated from the ratio of MLS to the predicted MLS based on the allometric equation for nonflying mammals (de Magalhães et al. 2007). When information was not available in AnAge, it was complemented by collecting data from additional sources such as the Animal Diversity Web (ADW) database (Myers et al. 2013) accessed at http://animaldiversity.org. Primate data from AnAge and ADW showed a strong correlation in life-history measures (supplementary fig. 10, Supplementary Material online). Correlation coefficients were 0.86 for MLS, 0.88 for gestation, 0.99 for adult weight, and 1 for birth mass. All data used in the analyses can be found in supplementary table 5, Supplementary Material online.
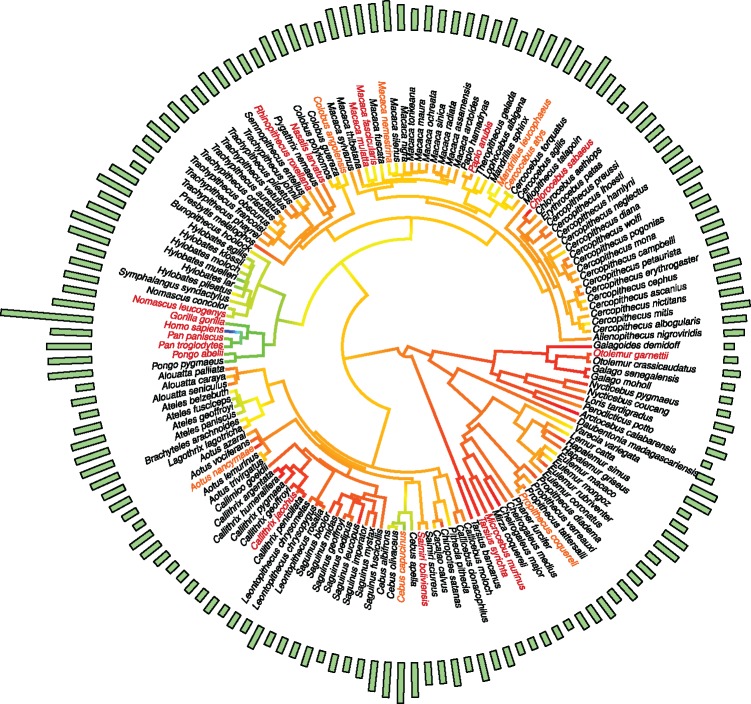
The consensus molecular chronogram tree for primates was downloaded from the 10kTrees project, version 3 (Arnold et al. 2010) at http://10ktrees.nunn-lab.org/. The 10kTrees data set is based on sequences of 11 mitochondrial and 6 autosomal genes sampled from GenBank. 10KTrees phylogenies contain 301 species of primates, out of which 174 were present in our phenotype data set (fig. 5). The database of life-history traits was adapted to the taxa from the phylogeny and the primate species tree was pruned to contain the species used in each analysis.
Fig. 5.
Circular tree created using all the primate species with longevity records from online databases (AnAge and Animal Diversity Web). In red, the 17 species from which genetic data were obtained from UCSC database are shown. New primate species added in this study to enrich the diversity of primates are shown in orange (n = 8). Species from which genetic data are not included in the study are shown in black. External green bars represent the MLS records obtained. Lower values for MLS are shown in darker red; higher values are shown in blue (n = 174 species).
Human 20-way multiple alignments were downloaded for 38,851 coding sequences from the UCSC browser at https://genome.ucsc.edu/ (accessed August 2016). These files included multiple alignments from 17 primate species plus Mus musculus, Tupaia belangeri, and Canis lupus, which were discarded from the analyses. Of all transcripts, only the longest one was kept for each gene. Gene alignments with an overall number of gaps > 50% and alignments where at least 2 of the species had >50% of the sequence in gaps were discarded. To avoid numerical problems with the log transformation and unrealistic substitution rates, the species for which root-to-tip ω had a value of 0 were discarded. After filtering, a total of 16,891 canonical genes were included in the study.
Additionally, protein multiple alignments of 179 primate species were downloaded from NCBI’s Organelle Genome Resources for the 13 mitochondrial genes. For 93 out of the 179 primate species, data on life-history traits were available, and were included in the consensus 10kTrees tree. The nucleotide alignment of the mitochondrial genes for the 93 primate species was carried out with MAFFT v7.271 (Katoh and Standley 2013).
Multiple Alignments of New Species
To enrich our set of primate alignments, we used the following pipeline. First, we retrieved transcript sequences in FASTA format from Aotus nancymaae, Cebus capucinus imitator, Cercocebus atys, Colobus angolensis, Macaca nemestrina, Mandrillus leucophaeus, Propithecus coquereli, Rhinopithecus bieti from NCBI via ftp (accessed in March 2017). To find putative orthologues, we ran BLASTN with an e-value cut-off of 1e-4 using the human sequences from the UCSC alignments as a query. Then we used an in-house python script to trim the untranslated regions from the retrieved sequences using the first and last nine nucleotides of the human sequences and allowing a maximum of two mismatches. Subsequently, we computed multiple alignments for each gene using MAFFT v7.271 (Katoh and Standley 2013). In a first filtering step, we calculated for each gene the average pairwise percentage of identity using trimAl v1.4.rev15 (Capella-Gutiérrez et al. 2009) and an in-house script, and we removed those sequences having <60% of average sequence identity. Then, we translated the nucleotide sequences into protein using an in-house script and we aligned the protein sequences using MAFFT v7.271. To remove incorrectly annotated sequences or those containing frame-shifts, we used trimAl as described above to discard sequences with <60% of average sequence identity at the protein level. Finally, we used pal2nal v14 (Suyama et al. 2006) to back translate the final protein alignments and obtain the final nucleotide alignments. After this process, 17,663 canonical genes were retained and 1,337 genes discarded due to poor similarity. In the final set, 8,554 alignments included 17 or more primate species (supplementary table 6, Supplementary Material online).
Relation between Life History and Phylogeny in Primates
Correlation between life-history traits was assessed using Spearman’s correlation. Nonphylogenetic regressions were used to test whether maximum lifespan covaries with other life-history traits (log-transformed). We used Pagel’s λ model to test for phylogenetic signals in primate life-history traits. This method estimates to which extent a correlation between given traits reflects the shared evolutionary history of the species (Pagel 1999). Pagel’s λ describes the proportion of variance that can be attributed to Brownian motion along a phylogeny (neutral drift). A value of λ equal or close to 1 suggests a character evolving stochastically, whereas λ < 1 indicates departure from neutral drift. Estimates of λ for all life-history traits were computed using the phylosig function in the phytools package in R (Revell 2012).
Parallel Amino-Acid Substitutions
To discretize continuous traits such as MLS and LQ, mean values of both traits were calculated for each of the 14 primate families, using all primate records (table 4). That allowed us to split the data set in two groups: Species that fall >1 SD from the mean of their family were considered as species having Increased Lifespan. The rest, were grouped together in the Nonincreased Lifespan or Control group (fig. 2B). Then, an in-house script was used to identify parallel amino-acid changes across all coding regions. Parallel mutations were identified as the same amino-acid change occurring in all the species labeled as Increased Lifespan and in none of the other species (the Control group). One gap at each amino-acid position was permitted in the Control group.
Table 4.
Mean MLS and LQ Computed Using All Records from the Primate Database.
| na | MLS Mean | MLS SD | MLS Limits | LQ Mean | LQ SD | LQ Limits | |
|---|---|---|---|---|---|---|---|
| Callitrichidae | 19 | 20.49 | 4.19 | 16.3–24.6 | 1.66 | 0.31 | 1.3–1.9 |
| Cebidae | 24 | 36.49 | 9.22 | 27.2–45.7 | 2.19 | 0.53 | 1.6–2.7 |
| Cercopithecidae | 66 | 30.86 | 5.59 | 25.2–36.4 | 1.63 | 0.26 | 1.3–1.8 |
| Cheirogaleidae | 6 | 19.44 | 4.67 | 14.7–24.1 | 1.7 | 0.4 | 1.3–2.1 |
| Daubentoniidae | 1 | 23.3 | – | – | 1.46 | – | – |
| Galagonidae | 8 | 17.62 | 3.37 | 14.2–20.9 | 1.48 | 0.14 | 1.3–1.6 |
| Hominidae | 6 | 68.22 | 26.66 | 41.5–94.8 | 2.66 | 1.12 | 1.5–3.7 |
| Hylobatidae | 10 | 45.92 | 7.37 | 38.5–53.2 | 2.45 | 0.41 | 2–2.8 |
| Indridae | 5 | 25.75 | 6.08 | 19.6–31.8 | 1.46 | 0.34 | 1.1–1.8 |
| Lemuridae | 12 | 30.01 | 8.04 | 21.9–38.0 | 1.87 | 0.45 | 1.4–2.3 |
| Loridae | 5 | 20.4 | 5.85 | 14.5–26.2 | 1.59 | 0.32 | 1.2–1.9 |
| Megaladapidae | 2 | – | – | – | – | – | – |
| Pitheciidae | 7 | 28.52 | 7.18 | 21.3–35.7 | 1.9 | 0.46 | 1.4–2.3 |
| Tarsiidae | 2 | 16.15 | 0.21 | 15.9–16.3 | 1.59 | 0.02 | 1.5–1.6 |
Number of species included in each primate family.
Parallel amino-acid substitutions were evaluated in the set of canonical genes across the 17 primate species phylogeny using multiple protein-coding alignments from UCSC. New primate species aligned in this study were used to further confirm the results whenever possible, given sequence availability. Finally, when available, sequences from other long-lived animals such as the naked-mole rat (Heterocephalus glaber) and two bats (Myotis davidii and Myotis lucifugus) which are part of the 100-way alignments from UCSC were evaluated for the observed amino-acid variants.
To assess whether the number of parallel changes found was higher than expected by chance, we used four series of conservative resampling tests. Three species were (1) randomly selected from the primate tree and the number of parallel changes evaluated for each combination and; in order to resemble as much as possible the Increased Lifespan set of species, three species were (2) randomly selected from the tree, but limiting the sampling to 2 species of Cercopithecidae (Old World catarrhine monkeys) and an outgrup species; (3) resampling was forced to contain 2 Cercopithecidae species and one Hominoidea; and (4) resampling was performed comparing humans to a combination of 2 Cercopithecidae (as shown in supplementary fig. 11, Supplementary Material online).
We assessed human polymorphism levels for all the genome positions corresponding to the list of identified parallel mutations. To do so, we used the 1000 Genomes Project panel (1kGP, 1000 Genomes Project Consortium 2015). The discovered positions in the human hg19 build were determined, and the frequencies of the reference and the alternative alleles were extracted from 1kGP for all human populations using Annovar (Wang et al. 2010). Functional prediction of variants from SIFT and PolyPhen2 scores were also obtained.
Evolutionary Analyses: Gene–Phenotype Coevolution across Primates
Estimates of the dN/dS during the evolution of each species as the divergence from their last common ancestor, termed “root-to-tip dN/dS”, were obtained for each gene using the free ratio model from PAML 4.9 (Yang 2007). The root-to-tip dN/dS (ω) is more inclusive of the evolutionary history of a locus and it is a property of the species tip rather than the terminal branch. Therefore, it is more suitable for regressions against phenotypic data from extant species (Montgomery and Mundy 2013). Briefly, for each gene and species, root-to-tip dNs were summed up (the number of nonsynonymous sites of a given gene in the corresponding species) and root-to-tip dSs were summed up (the number of synonymous sites). Finally, the ratio between the obtained dN and dS was used as a measure of root-to-tip ω for each species. The root-to-tip ωs were then subjected to regression analysis in the PGLS framework. As we were evaluating all the genes simultaneously (∼17,000) and the corresponding Bonferroni P value is ∼ 2e-06, which is overly stringent for this analysis, we instead adopted the 10% adjusted false discovery rate q value cutoff (Storey and Tibshirani 2003).
Association of gene and phenotypic changes were evaluated using the PGLS Brownian motion method from the R library nlme and P values for each gene root-to-tip ω were kept. Outliers can seriously affect the parameter estimates in any regression model thus, when necessary, species with studentized residuals > ±3 were removed, and PGLS was fitted again without the phylogenetic outliers (Jones and Purvis 1997).
Finally, associations between rates of protein evolution and life-history traits should also take into account that such traits are not independent from their phylogenetic history, thus genome-wide rates of protein evolution were also included in the PGLS regressions as an independent variable (see next section). In all analyses, life-history variables and root-to-tip ωs were log10 transformed to make variances closer, to linearize relationships between the variables and to make variation scale-independent (i.e., related to proportional rather than absolute differences between observations).
In total, 16,891 nuclear genes underwent PGLS analyses, we used in-house alignments for the 8554 multiple alignments that contained more than 17 species. For the rest, the alignments downloaded from UCSC were used to guarantee that the assessed gene alignments contained at least 17 primate species.
The 13 mitochondrial genes were assessed for gene–phenotype coevolution using the same PGLS regression pipeline described earlier. In that case, evaluation of Pagel’s λ was done simultaneously to fitting PGLS, using the method corPagel from R library nlme, which calculates the phylogenetic signal in the sample and accounts for it. This could be done in the mitochondrial analyses because of the larger sample size as compared with the whole-genome approach. In the latter, we did not have enough power to use corPagel method, and Brownian motion was chosen (supplementary fig. 12, Supplementary Material online).
Genome-Wide Rates of Evolution
Interspecific variation in effective population size could alter the rate of neutral substitution and the efficacy of selection to remove or fix nonsynonymous substitutions. This effect could bias our phylogenetic regression analyses if effective population size covaries with the studied traits. To address this, we obtained estimates of genome-wide root-to-tip ω by calculating the median root-to-tip values for each species and added them as covariates in the PGLS analyses. The same procedure was applied to mitochondrial gene sequences to obtain an estimate of mitochondrial-wide ω. We then used these values in a PGLS regression to test for an association with the studied traits and the median genome rate of evolution.
Pleiotropies and Aging-Related Genesets
To study genes that have been previously related to aging, a list of curated human genes associated with aging in different model systems was obtained from the GenAge data set (de Magalhães et al. 2005). We used gene ontology (GO) annotation, which describes how gene products behave in a cellular context, to select GO categories encompassing mechanisms or functions corresponding to processes considered hallmarks of aging in López-Otín et al. (2013) (supplementary table 7, Supplementary Material online). Additional GO categories such as wound healing (GO: 0042060), and blood coagulation (GO: 0007596) were also studied. Human GO gene sets were downloaded from AmiGO (Carbon et al. 2009), a tool for searching and browsing the Gene Ontology database (amigo.geneontology.org/amigo). To evaluate whether these gene sets were enriched in pleiotropic signals, we retrieved cases of putative pleiotropies from the GWAS Catalog (MacArthur et al. 2017), by selecting cases in which a given SNP or group of SNPs in LD have been associated with two or more pathologies by different GWAS studies, using the method described in Rodríguez et al. 2017. Binomial tests in each of the aging categories (López-Otín et al. 2013) were compared with genome-wide expectations (2,559 disease-associated alleles, and 266 pleiotropies, see Rodríguez et al. 2017).
Pathway Enrichment
Pathway analyses were performed to explore possible biological mechanisms that may underlie the associations between the identified genes and aging pathways. We used The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, GO ontology, Pathway commons, and disease-associated genes from WebGestalt for our analyses (Wang et al. 2013). For each pathway, the hypergeometric test was used to detect the overrepresentation of our set of genes among all genes in the pathway. Lastly, FDR was controlled using the Benjamini–Hochberg procedure. In all cases, the complete set of protein-coding genes was used as the background.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Ministerio de Ciencia e Innovación, Spain (BFU2012-38236 and BFU2015-68649-P (MINECO/FEDER, UE) to AN), by Direcció General de Recerca, Generalitat de Catalunya (2014SGR1311 and 2014SGR866 to A.N., and 2014BP-B00157 to G.M.), by the Spanish National Institute of Bioinfomatics of the Instituto de Salud Carlos III (PT13/0001/0026), by “Unidad de Excelencia María de Maeztu”, funded by the MINECO (ref: MDM-2014-0370), and by FEDER (Fondo Europeo de Desarrollo Regional)/FSE (Fondo Social Europeo). We would like to thank Greg Gibson for his helpful discussions and suggestions. This research was performed at Institut Biologia Evolutiva, Universitat Pompeu Fabra, Barcelona, Spain.
References
- 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aledo JC, Li Y, de Magalhães JP, Ruíz-Camacho M, Pérez-Claros JA.. 2011. Mitochondrially encoded methionine is inversely related to longevity in mammals. Aging Cell 102:198–207. [DOI] [PubMed] [Google Scholar]
- Arnold C, Matthews LJ, Nunn CL.. 2010. The 10kTrees website: a new online resource for primate phylogeny. Evol Anthropol Issues News Rev. 193:114–118. [Google Scholar]
- Austad SN. 2001. An experimental paradigm for the study of slowly aging organisms Exp Gerontol. 36:599–605. [DOI] [PubMed] [Google Scholar]
- Austad SN. 2005. Diverse aging rates in metazoans: targets for functional genomics. Mech Ageing Dev Funct Genomics Ageing II 1261:43–49. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Guarente L, Kirkwood TBL, Partridge L, Rando TA, Slagboom PE.. 2012. The place of genetics in ageing research. Nat Rev Genet. 138:589–594. [DOI] [PubMed] [Google Scholar]
- Boddy AM, Harrison PW, Montgomery SH, Caravas JA, Raghanti MA, Phillips KA, Mundy NI, Wildman DE.. 2017. Data from: Evidence of a conserved molecular response to selection for increased brain size in primates. Genome Biol Evol. 93:700–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafè M, Barbieri M, Marchegiani F, Olivieri F, Ragno E, Giampieri C, Mugianesi E, Centurelli M, Franceschi C, Paolisso G.. 2003. Polymorphic variants of insulin-like growth factor I (IGF-I) receptor and phosphoinositide 3-kinase genes affect IGF-I plasma levels and human longevity: cues for an evolutionarily conserved mechanism of life span control. J Clin Endocrinol Metab. 887:3299–3304. [DOI] [PubMed] [Google Scholar]
- Bromham L, Rambaut A, Harvey PH.. 1996. Determinants of rate variation in mammalian DNA sequence evolution. J Mol Evol. 436:610–621. [DOI] [PubMed] [Google Scholar]
- Byars SG, Huang QQ, Gray L-A, Bakshi A, Ripatti S, Abraham G, Stearns SC, Inouye M.. 2017. Genetic loci associated with coronary artery disease harbor evidence of selection and antagonistic pleiotropy. PLoS Genet. 136:e1006328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A, Ugidos A, Liu B, Öling D, Kvint K, Hao X, Mignat C, Nachin L, Molin M, Nyström T.. 2011. Absence of mitochondrial translation control proteins extends life span by activating sirtuin-dependent silencing. Mol Cell 423:390–400. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2515:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, Lomax J, Mungall C, Hitz B, Balakrishnan R, et al. . 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 252:288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW.. 2006. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 76:436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM.. 2014. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat Commun. 5:3557.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca RR, Johnson WE, O’Brien SJ, Ramos MJ, Antunes A.. 2008. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics 9:119.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP, Costa J.. 2009. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol. 228:1770–1774. [DOI] [PubMed] [Google Scholar]
- de Magalhães JP, Costa J, Church GM.. 2007. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol A Biol Sci Med Sci. 622:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP, Costa J, Toussaint O.. 2005. HAGR: the human ageing genomic resources. Nucleic Acids Res. 33(Database issue):D537–D543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Curado J, Church GM.. 2009. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 257:875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Patel VK, Swindell WR, Perrimon N.. 2014. Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell Rep. 75:1481–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Cantley LC.. 2015. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 259:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty A, de Magalhães JP.. 2016. Has gene duplication impacted the evolution of Eutherian longevity? Aging Cell 155:978–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. . 2009. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 1111:1305–1314. [DOI] [PubMed] [Google Scholar]
- Enard W. 2014. Comparative genomics of brain size evolution. Front Hum Neurosci. 8:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Zhou Q.. 2017. Absence of relationship between mitochondrial DNA evolutionary rate and longevity in mammals except for CYTB. J Mamm Evol. doi.org/10.1007/s10914-017-9399-4. [Google Scholar]
- Finch CE. 2010. Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci U S A. 107(suppl_1):1718–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE, Austad SN.. 2012. Primate aging in the mammalian scheme: the puzzle of extreme variation in brain aging. Age 345:1075–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, Kaschner K, Schultze SE, Garilao C, Ho SYW, Post K, Higham TFG, Stokowska C, van der Es H, Embling CB, et al. . 2013. Ancient DNA reveals that bowhead whale lineages survived Late Pleistocene climate change and habitat shifts. Nat Commun. 4:1677.. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Garagnani P, Vitale G, Capri M, Salvioli S.. 2017. Inflammaging and “Garb-aging.”. Trends Endocrinol Metab. 283:199–212. [DOI] [PubMed] [Google Scholar]
- Franchini M. 2006. Hemostasis and aging. Crit Rev Oncol Hematol. 602:144–51. [DOI] [PubMed] [Google Scholar]
- Fushan AA, Turanov AA, Lee S-G, Kim EB, Lobanov AV, Yim SH, Buffenstein R, Lee S-R, Chang K-T, Rhee H, et al. . 2015. Gene expression defines natural changes in mammalian lifespan. Aging Cell 143:352–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierman HJ, Fortney K, Roach JC, Coles NS, Li H, Glusman G, Markov GJ, Smith JD, Hood L, Coles LS, Kim SK.. 2014. Whole-genome sequencing of the world’s oldest people. PLoS One 911:e112430.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN. 2013. The origin of aging: imperfectness-driven non-random damage defines the aging process and control of lifespan. Trends Genet. 299:506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Holm NV, Sørensen TI, Harvald B, Vaupel JW.. 1996. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum Genet. 973:319–323. [DOI] [PubMed] [Google Scholar]
- Hjelmborg JB, Iachine I, Skytthe A, Vaupel JW, McGue M, Koskenvuo M, Kaprio J, Pedersen NL, Christensen K.. 2006. Genetic influence on human lifespan and longevity. Hum Genet. 1193:312–321. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Géloën A, Even PC, Cervera P, Le Bouc Y.. 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 4216919:182–187. [DOI] [PubMed] [Google Scholar]
- Huang X, Withers BR, Dickson RC.. 2014. Sphingolipids and lifespan regulation. Biochim Biophys Acta 18415:657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Levine B. n.d. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 36:597–599. [DOI] [PubMed] [Google Scholar]
- Jones KE, Purvis A.. 1997. An optimum body size for mammals? Comparative evidence from bats. Funct Ecol. 116:751–756. [Google Scholar]
- Jones OR, Scheuerlein A, Salguero-Gómez R, Camarda CG, Schaible R, Casper BB, Dahlgren JP, Ehrlén J, García MB, Menges ES.. 2014. Diversity of ageing across the tree of life. Nature 5057482:169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G, Neufeld TP.. 2008. Drosophila Atg7: required for stress resistance, longevity and neuronal homeostasis, but not for metamorphosis. Autophagy 43:357–358. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S.. 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 1410:885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 304:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. 2010. The genetics of ageing. Nature 4677315:622–622. [Google Scholar]
- Keyes BE, Liu S, Asare A, Naik S, Levorse J, Polak L, Lu CP, Nikolova M, Pasolli HA, Fuchs E, Mesirov JP.. 2016. Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell 1675:1323–1338.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Poujol R.. 2011. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Mol Biol Evol. 281:729–744. [DOI] [PubMed] [Google Scholar]
- Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJR, Motoyama N, Cao L, Finkel T.. 2012. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 3366078:225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, de Magalhães JP.. 2013. Accelerated protein evolution analysis reveals genes and pathways associated with the evolution of mammalian longevity. Age 352:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Xu N, O’Prey J, Lao LY, Joshi S, Long JS, O’Prey M, Croft DR, Beaumatin F, Baudot AD.. 2015. Loss of autophagy causes a synthetic lethal deficiency in DNA repair. Proc Natl Acad Sci U S A. 1123:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G.. 2013. The hallmarks of aging. Cell 1536:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini A, Tresini M, Austad SN, Cristofalo VJ.. 2005. Cellular replicative capacity correlates primarily with species body mass not longevity. Mech Ageing Dev. 12610:1130–1133. [DOI] [PubMed] [Google Scholar]
- Lüke L, Vicens A, Tourmente M, Roldan ERS.. 2014. Evolution of protamine genes and changes in sperm head phenotype in rodents. Biol Reprod. 903:67.. [DOI] [PubMed] [Google Scholar]
- Ma S, Gladyshev VN.. 2017. Molecular signatures of longevity: insights from cross-species comparative studies. Semin Cell Dev Biol. 70:190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et al. . 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45(D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari D, Coppola R, Provenzano R.. 2008. Hemostasis factors and aging. Exp Gerontol. 432:66–73. [DOI] [PubMed] [Google Scholar]
- Medawar P. 1952. An unsolved problem of biology an inaugural lecture delivered at University College, London, 6 December, 1951. London: H.K. Lewis and Co. [Google Scholar]
- Mizushima N, Levine B.. 2010. Autophagy in mammalian development and differentiation. Nat Cell Biol. 129:823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Venditti C, Barton RA, Mundy NI.. 2011. Adaptive evolution of four microcephaly genes and the evolution of brain size in anthropoid primates. Mol Biol Evol. 281:625–638. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI.. 2012a. Evolution of ASPM is associated with both increases and decreases in brain size in primates. Evolution 663:927–932. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI.. 2012b. Positive selection on NIN, a gene involved in neurogenesis, and primate brain evolution. Genes Brain Behav. 11:n/a. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI.. 2013. Microcephaly genes and the evolution of sexual dimorphism in primate brain size. J Evol Biol. 264:906–911. [DOI] [PubMed] [Google Scholar]
- Myers PR, Parr CS, Jones T, Hammond GS, Dewey TA.. 2013. The animal diversity web (online). Available from: http://animaldiversity.org.
- Nabholz B, Glémin S, Galtier N.. 2008. Strong variations of mitochondrial mutation rate across mammals – the longevity hypothesis. Mol Biol Evol. 251:120–130. [DOI] [PubMed] [Google Scholar]
- O’Connor TD, Mundy NI.. 2009. Genotype-phenotype associations: substitution models to detect evolutionary associations between phenotypic variables and genotypic evolutionary rate. Bioinformatics 2512:i94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TD, Mundy NI.. 2013. Evolutionary modeling of genotype-phenotype associations, and application to primate coding and non-coding mtDNA rate variation. Evol Bioinform Online 9:EBO.S11600–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeppen J, Vaupel JW.. 2002. Broken limits to life expectancy. Science 2965570:1029–1031. [DOI] [PubMed] [Google Scholar]
- Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, et al. . 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 4997456:74–78. [DOI] [PubMed] [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401:877–884. [DOI] [PubMed] [Google Scholar]
- Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, Stupka E, Rossiter SJ.. 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 5027470:228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling LC, Atkins JL, Bowman K, Jones SE, Tyrrell J, Beaumont RN, Ruth KS, Tuke MA, Yaghootkar H, Wood AR, et al. . 2016. Human longevity is influenced by many genetic variants: evidence from 75,000 UK Biobank participants. Aging (Albany NY) 83:547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo J-O, Yoo S-M, Ahn H-H, Nah J, Hong S-H, Kam T-I, Jung S, Jung Y-K.. 2013. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 4:307–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 32:217–223. [Google Scholar]
- Ricklefs RE. 2010. Life-history connections to rates of aging in terrestrial vertebrates. Proc Natl Acad Sci U S A. 10722:10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez JA, Marigorta UM, Hughes DA, Spataro N, Bosch E, Navarro A.. 2017. Antagonistic pleiotropy and mutation accumulation influence human senescence and disease. Nat Ecol Evol. 13:0055.. [DOI] [PubMed] [Google Scholar]
- Rufini A, Tucci P, Celardo I, Melino G.. 2013. Senescence and aging: the critical roles of p53. Oncogene 3243:5129–5143. [DOI] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LDeana. W, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, et al. . 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 4837388:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. 2013. The genetic causes of convergent evolution. Nat Rev Genet. 1411:751–764. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R.. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 10016:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, Costa J, Fraifeld VE, de Magalhães JP.. 2013. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 41(Database issue):D1027–D1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade LS, Aigaki T, Peixoto AA, Balduino A, Mânica da Cruz IB, Heddle JG.. 2013. A novel classification system for evolutionary aging theories. Front Genet. 4:25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urfer SR, Kaeberlein TL, Mailheau S, Bergman PJ, Creevy KE, Promislow DEL, Kaeberlein M.. 2017. A randomized controlled trial to establish effects of short-term rapamycin treatment in 24 middle-aged companion dogs. GeroScience 392:117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B.. 2013. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41(Web Server issue):W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 3816:e164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC. 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 114:398. [Google Scholar]
- Wirthlin M, Lovell PV, Jarvis ED, Mello CV.. 2014. Comparative genomics reveals molecular features unique to the songbird lineage. BMC Genomics 15:1082.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J. 2015. Atg7 in development and disease: panacea or Pandora’s Box? Protein Cell 610:722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Toren D, Vierlinger K, Hofner M, Nöhammer C, Chilosi M, Budovsky A, Fraifeld VE.. 2015. Wound healing and longevity: lessons from long-lived αMUPA mice. Aging 73:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yim H-S, Cho YS, Guang X, Kang SG, Jeong J-Y, Cha S-S, Oh H-M, Lee J-H, Yang EC, Kwon KK, et al. . 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 461:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, et al. . 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 3396118:456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.