Abstract
Exposures to diesel exhaust particles (DEP) from traffic and house dust mite (HDM) allergens significantly increase risks of airway diseases, including asthma. This negative impact of DEP and HDM may in part be mediated by epigenetic mechanisms. Beyond functioning as a mechanical barrier, airway epithelial cells provide the first line of immune defense towards DEP and HDM exposures. To understand the epigenetic responses of airway epithelial cells to these exposures, we exposed human bronchial epithelial cells to DEP and HDM and studied genome-wide 5-methyl-cytosine (5mC) and 5-hydroxy-methylcytosine (5hmC) at base resolution. We found that exposures to DEP and HDM result in elevated TET1 and DNMT1 expression, associated with 5mC and 5hmC changes. Interestingly, over 20% of CpG sites are responsive to both exposures and changes in 5mC at these sites negatively correlated with gene expression differences. These 5mC and 5hmC changes are located in genes and pathways related to oxidative stress responses, epithelial function and immune cell responses and are enriched for binding sites of transcription factors (TFs) involved in these pathways. Histone marks associated with promoters, enhancers and actively transcribed gene bodies were associated with exposure-induced DNA methylation changes. Collectively, our data suggest that exposures to DEP and HDM alter 5mC and 5hmC levels at regulatory regions bound by TFs, which coordinate with histone marks to regulate gene networks of oxidative stress responses, epithelial function and immune cell responses. These observations provide novel insights into the epigenetic mechanisms that mediate the epithelial responses to DEP and HDM in airways.
Keywords: 5-hydroxy-methylcytosine, 5-methyl-cytosine, TET1, traffic-related air pollution, allergen, airway epithelial cells
Introduction
Exposure to house dust mite (HDM) allergen is common, and as many as 50% of asthmatic patients are sensitized to HDM [1]. The causal role of HDM in allergic asthma is well established [2]. Importantly, allergic asthma phenotypes induced by HDM can be exacerbated by exposures to traffic-related air pollution [3–6]. Traffic-related air pollutants are a complex mixture of gaseous and particulate components with adverse effects on health supported by ample evidence [7–11]. Among traffic-related air pollution, diesel exhaust particles (DEP) are of particular concern with respect to health effects, due to their abundance and very small sizes. It is estimated that DEPs contribute up to 90% of the particulate matter (PM) derived from traffic sources. They are primarily ultrafine in size (<100 nm), and can be deposited in the nasal or peripheral airways. DEPs have been shown to induce oxidative stress and enhance allergic responses and airway inflammation induced by HDM [3–6]. Globally, a significant portion of all populations lives in regions with high traffic-related air pollution, including schools [7, 12–14]. Therefore, it is essential to understand how DEP exposure exacerbates HDM-induced allergic asthma.
Accumulating evidence supports that epigenetic mechanism(s) may explain the effects of DEP and HDM on airways and asthma. Both DEP and HDM have been shown to modify DNA methylation of the airways [15]. We recently demonstrated that TET1 (Ten Eleven Translocation 1) promoter methylation is associated with childhood asthma and exposure to traffic-related air pollution [16]. A member of the TET enzyme family (TET1/2/3), TET1 converts 5mC to oxi-mCs including 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), ultimately resulting in DNA demethylation. In contrast, the de novo addition of DNA methylation is achieved by DNA methyl-transferases 3 A and 3B (DNMT3A and 3B), while DNMT1 is responsible for 5mC addition during DNA replication. Among these cytosine modifications, 5hmC is often found at enhancers, with a direct role for 5hmC in demethylation of enhancers during development and gene activation [17–24]. In addition, similar to 5mC, 5hmC is also enriched in promoters and gene bodies and functions as a stable marker for setting up the epigenetic landscape through interactions with other proteins [17–19, 25–27] and functions in both transcriptional activation and repression [28]. TET1 has been shown to be regulated by DEP exposure in human bronchial epithelial cells [16]. TET1 expression is also regulated by HDM exposure in mouse lungs, while TET2 and TET3 remain unchanged [29]. In the same mouse model, the expression of DNMT3A was significantly decreased, while DNMT1 and DNMT3B remained unchanged. To date, it remains unclear how the dysregulation of TET1 by exposures alters 5mC and 5hmC in airway epithelial cells. This is likely due to the difficulty of measuring 5mC and 5hmC individually.
In this paper, we studied the genome-wide 5mC and 5hmC response to DEP and HDM by combining BiSulfite (BS) and oxidative BiSulfite (oxBS) conversion at base resolution. We examined the relationships among locations with differential 5mC and 5hmC in response to different exposures. In order to understand the functional relevance of identified changes, we performed transcriptomic analysis, gene ontology and pathway analysis, and functional genomics to examine TF binding and histone marks. To our knowledge, this is the first study to examine the impact of DEP and HDM exposures on 5mC and 5hmC at base-resolution on a genome-wide scale in airway epithelial cells.
Results
Exposure to Diesel Exhaust Particles and House Dust Mite Significantly Alters the Expression of TET1, DNMT1 and DNMT3A in Airway Epithelial Cells
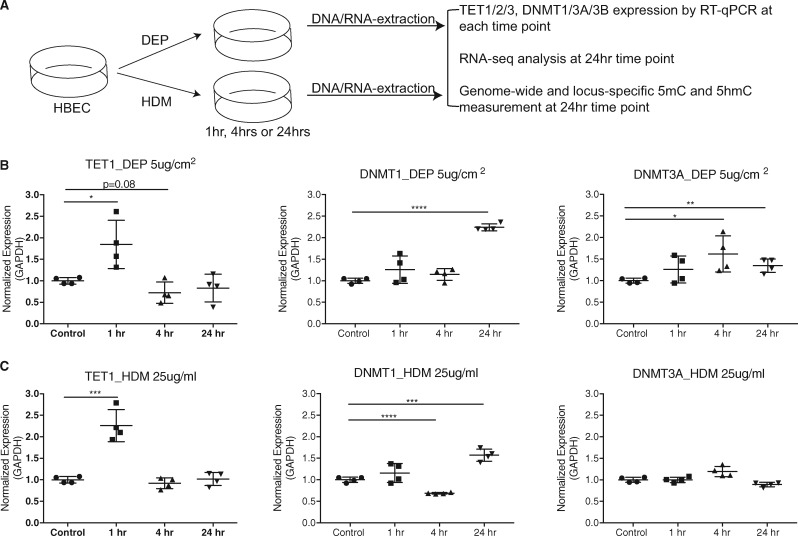
Previously we have shown in nasal epithelial cells that TET1 promoter methylation is associated with childhood asthma and traffic related air pollution [30]. To further understand the regulation of TET1/2/3 by DEP (major particulate matter in traffic-related air pollution) and HDM (an allergen known to increase asthma risk), we examined whether exposure to HDM or DEP alters the expression levels of TET1/2/3 (Fig. 1A). Consistent with our previous observations [16], in presence of 5μg/cm2 DEP, the expression of TET1 significantly increased at 1 h and returned to baseline at 4 h (Fig. 1B). We observed similar time-specific patterns when exposing HBECs with 25 μg/ml of HDM (Fig. 1C). No significant cell death was observed under these conditions [16]. As TET2 and TET3 are enzymes that have similar functions to TET1, we also assessed the expression of TET2 and TET3. In contrast to TET1, the expression of TET2 and TET3 did not significantly change following DEP and HDM challenge (Supplementary Fig. S1). DNMT1 showed significant upregulation at 24 hrs following either DEP or HDM exposure (Fig. 1B and C). Meanwhile, the expression of DNMT3A, a de novo DNA methylase, was significantly upregulated at 4 h by DEP (Fig. 1B and C), while the expression of DNMT3B trended higher at 4 h (Supplementary Fig. S1). In contrast HDM exposure significantly downregulated DNMT3B at 24 h (Supplementary Fig. S1). In conclusion, exposures to DEP and HDM regulate the expression of enzymes involved in DNA methylation maintenance in a sequential manner.
Fig. 1:
time-specific responses of TET1 and DNMT expression to DEP and HDM exposures in HBECs. (A) Experimental design. HBECs were exposed to Phosphate-buffered saline (PBS), DEP (5 μg/cm2) or HDM (25 μg/ml). Expression of TET1, DNMT1 and DNMT3A was measured by RT-qPCR. (B) and (C) Expression values (normalized to GADPH) were further normalized to the mean of controls at the indicated time points. Mean values from four biological replicates are represented as the mean ± SD. Treated group were compared to controls using a two-sided Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. represents not significant.
Exposures to DEP and HDM Altered 5mC and 5hmC in Airway Epithelial Cells
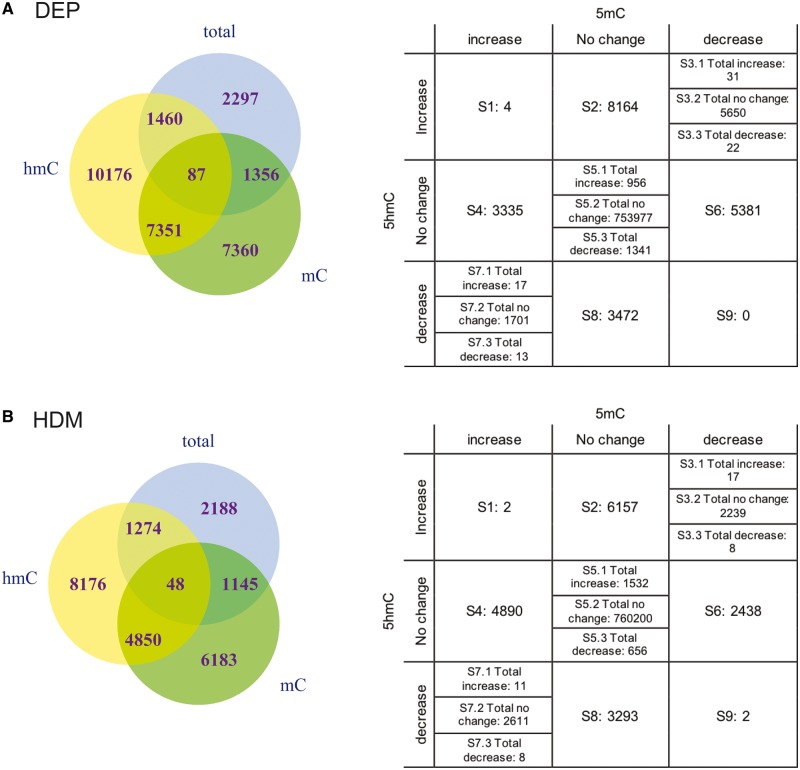
As TET1, DNMT3A and DNMT1 are involved in the DNA demethylation and methylation processes, we next assessed genome-wide 5mC and 5hmC levels by assaying DNA from bisulfite treatment (BS) and oxidative bisulfite treatment (oxBS) with Illumina MethylationEPIC chips [31, 32]. Of note, BS measures the sum of 5mC and 5hmC, while oxBS measures 5mC only. The levels of 5hmC can thus be derived as the difference between BS signal and oxBS signal. EPIC arrays interrogate 8 63 904 CpG sites, which covers CpG islands, non-CpG sites, cancer and reprogramming related differentially methylated CpG sites, FANTOM5 enhancers, ENCODE open chromatin, ENCODE transcription factor binding sites and miRNA promoter regions. Following 24 h exposures to 5 μg/cm2 DEP, 5200, 16 154 and 19 074 CpG sites underwent changes (Difference ≥ 0.10) in 5mC + 5hmC, 5mC and 5hmC, respectively (Fig. 2; Supplementary Table S3A, B and C). Among these, 87 sites had changes in 5mC + 5hmC, 5mC and 5hmC. Interestingly, there are 7351 sites with changes in both 5mC and 5hmC, but not in 5mC + 5hmC, due to opposite directions of change in 5mC and 5hmC (Fig. 2A). Overall, 68.6% of all sites with 5mC changes showed decreased levels and 72.7% sites of all sites with 5hmC changes showed increased levels of 5hmC following DEP exposure. In response to a 24 h stimulation with 25 μg/ml of HDM, similar patterns were observed for 5mC + 5hmC, 5mC and 5hmC (Fig. 2B and Supplementary Table S3D, E and F). These observations are consistent with the significant up-regulation of TET1, DNMT3A and DNMT1 following DEP exposure and similar changes of TET1 and DNMT1 following HDM exposure. Processes that possibly occur may include active conversion of 5mC to 5hmC by TET1, de novo methylation by DNMT3A and maintenance of only 5mC marks by DNMT1 through DNA replication that results in retention of 5mC marks and removal of 5hmC marks (passive demethylation).
Fig. 2:
exposures to DEP and HDM alter 5mC + 5hmC, 5mC and 5hmC in HBECs. (A) Venn diagram showing the degree of overlap between sites with 5mC + 5hmC, 5mC and 5hmC changes in response to 24 h treatment of 5 μg/cm2 DEP. The table lists number of CpG sites with different directions of changes in 5mC and 5hmC. (B) Venn diagram for 5mC + 5hmC, 5mC and 5hmC changes in response to 24 h treatment of 25 μg/ml HDM. The table lists number of CpG sites with different directions of changes in 5mC and 5hmC.
Exposures to DEP and HDM Induced Changes at Genomic Locations near Genes Involved in Oxidative Stress Responses, Epithelial Function and Immune Cell Responses
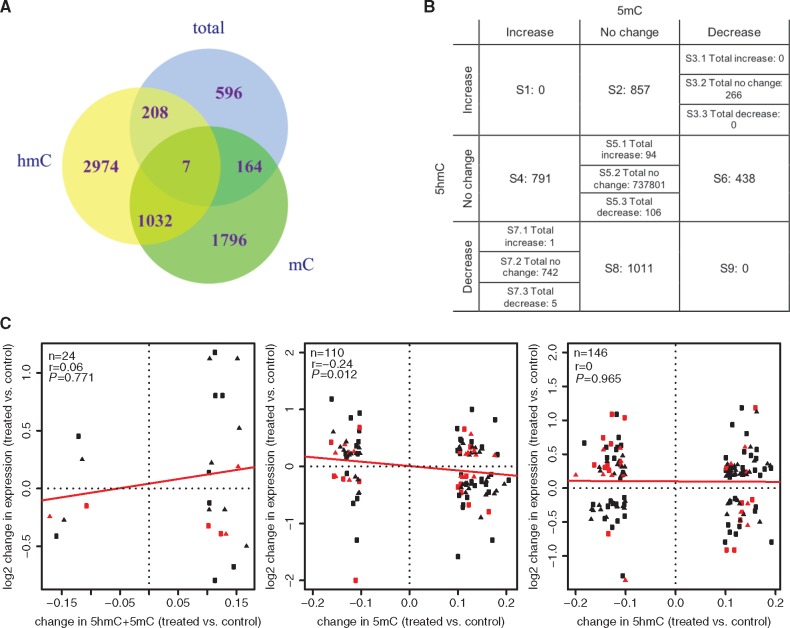
As co-exposure to DEP and HDM is associated with increased wheezing and asthma risk [5], we hypothesized that DEP and HDM exposures modify common genes and pathways through epigenetic mechanisms. For total DNA methylation levels (5mC + 5hmC), 5mC and 5hmC, respectively, 975, 2999 and 4221 sites were found that responded to both DEP and HDM (Fig. 3A;Supplementary Table S4A, B and C). Among 1960 CpG sites with changes in 5mC but not in 5hmC, 41% showed reduced 5mC level in both exposures; among the 3182 CpG sites with changes in 5hmC only, 45% of them showed increased 5hmC levels; among the 1039 CpG sites that showed changes in both 5mC and 5hmC, 1035 showed opposite directions of changes in 5mC and 5hmC and 26% of them showed reduced 5mC and increased 5hmC (Fig. 3A). Sites in S2 represent CpG sites with increase in 5hmC, while sites in S8 include those that have decreased 5hmC, both S2 and S8 have no changes in 5mC (Fig. 3B). Sites located in S4 are those with increased 5mC and sites in S6 represent CpG sites with decreased 5mC, both have no changes in 5hmC. Sites in S3 are those with decrease in 5mC and simultaneous increase in 5hmC (5mC loss/5hmC gain), while sites in S7 represent CpG sites with increase in 5mC and decrease in 5hmC (5mC gain/5hmC loss). These different combinations of changes in 5mC, 5hmC and 5mC + 5hmC suggest distinct underlying mechanisms resulting in such changes. Compared to the distribution of all CpG sites included on the EPIC arrays, fewer CpG sites whose 5hmC levels are responsive to DEP and HDM are located at CpG islands (11% vs. array 20%, p < 0.001) and promoters (defined as TSS200+TSS1500, 14% vs 20%, p < 0.001), more are located at open seas (64% vs. array 55%, p < 0.001) (Supplementary Fig. S2A), intergenic regions (29% vs. 32%, p < 0.001) and gene body (43% vs. 37%, p < 0.001) (Supplementary Fig. S2B). This is consistent with the notion that 5hmC is enriched at gene bodies and enhancers [33, 34]. Further, we examined the correlation between changes in 5mC + 5hmC, 5mC and 5hmC with changes in gene expression, and observed negative correlation between expression and changes in 5mC (Fig. 3C).
Fig. 3:
common CpG sites are responsive to DEP and HDM challenges, and 5mC changes at these sites are correlated with gene expression differences. (A) Venn Diagram showing the relationship of sites with 5mC + 5hmC, 5mC and 5hmC changes common to DEP and HDM. (B) Number of CpG sites with different directions of changes in 5mC and 5hmC. (C) Correlation between changes in gene expression and DNAm after DEP and HDM treatment. Only CpG sites responsive to both treatments were examined, with squares and triangles representing sites responsive to DEP and HDM, respectively. Sites in red were within the regulatory regions (TSS1500, TSS200, 5’UTR and 1stExon). Trend of correlation was indicated by the red lines. n: number of observations; r: Spearman’s correlation coefficient; P value testing if r is different from 0.
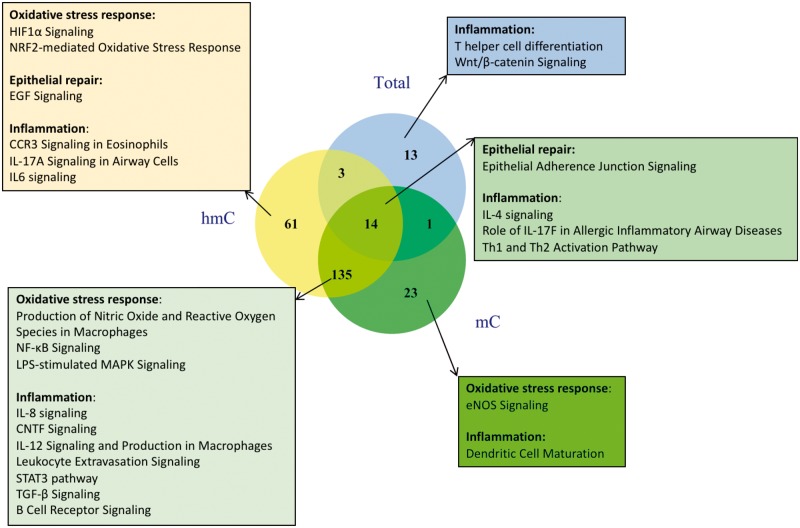
Among genes close to sites that were responsive to both DEP and HDM and had changes in 5mC + 5hmC, IPA analysis defined 31 enriched pathways (Supplementary Table S5A). There were 173 significantly enriched pathways among genes with changes in 5mC (out of 405 enriched pathways in total), and 213 significantly enriched pathways (400 enriched pathways in total) for genes with 5hmC changes (Supplementary Table S5B and C). The relationships between these pathways are shown in Fig. 4, with the most pathways identified from genes with opposite changes in 5mC and 5hmC. Several pathways known to be activated in response to DEP and HDM are highlighted, such as NRF2-mediated Oxidative Stress Response, Epithelial Adherence Junction Signaling, EGF signaling, NF-κB Signaling, and T helper cell differentiation and activation (Th1/Th2/Th17). In conclusion, our genome-wide analyses showed that DEP and HDM exposures epigenetically regulate the same pathways by altering the levels of 5mC and 5hmC and leading to changes in gene expression.
Fig. 4:
exposures to DEP and HDM modify 5mC and 5hmC levels of genes in relevant pathways. IPA analyses were performed for genes with 5mC + 5hmC, 5mC and 5hmC changes, respectively. The Venn diagram shows the relationship between significantly enriched pathways from the three datasets. Pathways known to be involved in DEP and HDM responses are indicated.
Genes with 5mC and 5hmC Levels Responsive to DEP and HDM Are Regulated by TET1
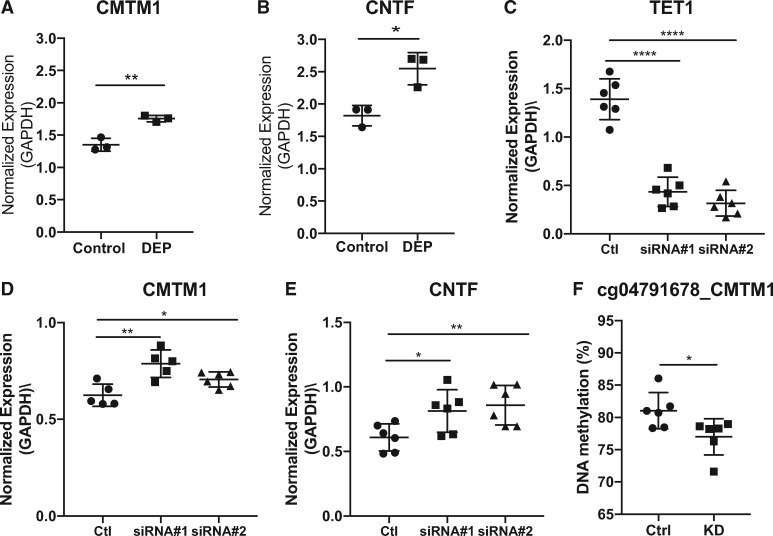
We chose 3 CpG sites located within the CMTM1, DNAH10 and CNTF genes with three different patterns of change and verified these changes using pyrosequencing in the same samples measured by arrays (Supplementary Fig. S3). These three sites were chosen based on their effect size in 5mC + 5hmC, 5mC or 5hmC (≥10%), representative patterns in 5mC and 5hmC changes, and biological relevance. CMTM1 belongs to the chemokine-like factor superfamily, links chemokines with the transmembrane 4 superfamily [35], and is widely expressed in lung tissues. DNAH10 (encoding dynein axonemal heavy chain) is expressed in cilia and flagella, and is the force-generating protein of respiratory cilia. CNTF (ciliary neurotrophic factor) is an IL6-type family cytokine released by airway smooth muscle cells in response to stimuli [36]. It enhances IgE production by B cells from atopic patients [37] and is upstream of STAT3 signaling to promote survival and/or differentiation in many cell types [38]. The CpG sites located within CMTM1 and DNAH10 showed a decrease in 5mC + 5hmC, no change in 5mC, and a decrease in 5hmC. In contrast, the CpG site located within CNTF showed no difference in 5mC + 5hmC, a decrease in 5mC and an increase in 5hmC.
We then tested whether different doses of DEP and HDM would cause similar changes in 5mC and 5hmC and found that higher doses of exposures (20 μg/cm2 for DEP and 100 μg/ml for HDM) did not further dysregulate 5mC + 5hmC, 5mC and 5hmC (Supplementary Fig. S4). Similar results were found for co-exposure to DEP and HDM for these two sites (data not shown). As predicted from previous studies showing that 5mC and 5hmC levels are associated with gene regulation [39–41], the expression levels of CMTM1 and CNTF were significantly increased 24 h after DEP exposure (Fig. 5A and B, did not measure DNAH10 due to technical difficulties). To further establish the direct involvement of TET1 in the regulation of these genes, TET1 was knocked down by over 70% using siRNAs (Fig. 5C), and the expression of CMTM1 and CNTF were significantly increased in HBECs (Fig. 5D and E). Along with the increase in gene expression, there was significant reduction of total DNAm (5mC + 5hmC) at CMTM1 promoter (Fig. 5F). Combined with data shown in Fig. 3C, our data support the regulation of gene expression by TET1 and DNAm changes following exposures.
Fig. 5:
regulation of CMTM1 and CNTF expression by exposures and TET1. HBECs were treated with 5 μg/cm2 DEP and expression levels of CMTM1 (A) and CNTF (B) were measured and normalized as described in Fig. 1. Mean values from three biological replicates are represented as the mean ± SD. The expression levels of TET1, CMTM1 and CNTF after TET1 siRNA knockdown are shown in C, D and E. F. Methylation of cg04791678 in CMTM1 after siRNA #1 knockdown of TET1. Mean values from six biological replicates are represented as the mean ± SD. Groups were compared using a two-sided Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. represents not significant.
5mC and 5hmC Changes Shared by DEP and HDM Challenges Are Located at TF Binding Sites and Co-Localize with Histone Marks for Promoters and Enhancers
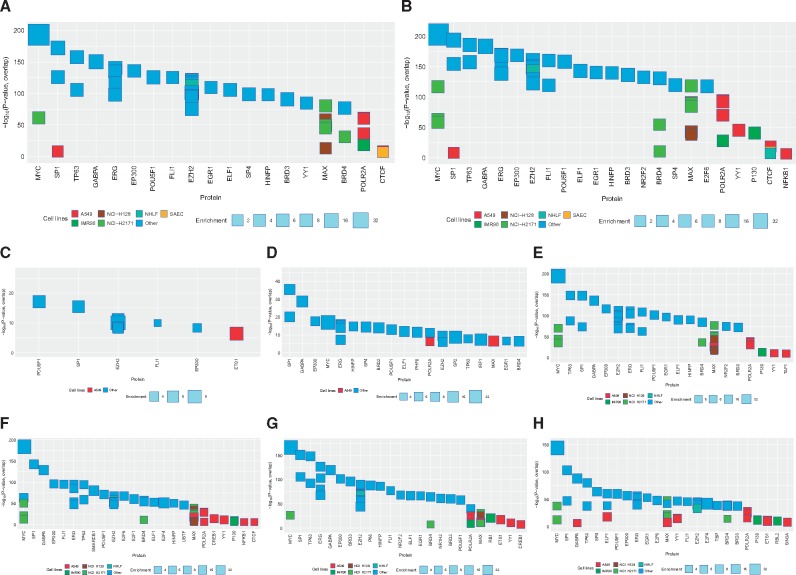
To understand the possible functions of the DNAm variations (5mC or 5hmC) shared by DEP and HDM, we performed comprehensive bioinformatic searches for possible TF binding around these sites. Two approaches were taken. First, we searched for enriched TF ChIP-seq peaks around these sites (see Methods) and found 273 and 409 datasets whose binding sites significantly overlap with exposure-induced 5hmC and 5mC changes, respectively (corrected P-value < 10−6) (Supplementary Table S6, top TFs shown in Fig. 6A and B). The large number of significant sites is perhaps not unexpected, given that 5hmC and 5mC-marked regions of the genome are often found in gene promoters. Additionally, different TFs seem to be enriched for regions with different directions of changes in 5mC, 5hmC and 5hmC + 5hmC (Fig. 6C–H). Among the top TFs associated with 5hmC changes (Fig. 6A), NRF2 (GABPA) is a known key player involved in oxidative stress response and detoxification processes [42] and asthma development [43, 44]. This TF was also associated with regions with exposure-induced 5mC changes (Fig. 6B and E–H), with the only exception of regions with increased 5hmC and no 5mC changes (6C). Besides NR2F2, there was an enrichment for NFKB1 binding in A549 cells around sites with overall 5mC changes (Fig. 6B and Supplementary Table S6B) and sites with 5mC loss/no 5hmC changes (Fig. 6F). NFKB1 is a subunit of NF-κB complex that is downstream of oxidative stress produced by exposures to HDM and DEP in airway epithelial cells [11, 45, 46]. In addition, there was enrichment for several TFs that either directly modify chromatin or interact with chromatin modifiers to regulate gene expression, such as EP300 [47], YY1 [48], MAX/MYC [49], HINFP [50, 51], CTCF [52–55], EHZ2 [56] and SIN3A [57]. Among these, EZH2 is part of the Polycomb repressive complexes 2/3 (PRC2/3, EZH2/SUZ12/EED) and is known to interact with Tet1 [58]. EZH2 was enriched in regions with 5hmC and/or 5mC changes (6B–6H) and SUZ12 was also found enriched in our dataset (Supplementary Table S6A). SIN3A, another known TET1 binding partner [59], was significantly enriched at regions with 5hmC loss and no 5mC changes (Fig. 6H). This supports a possible interaction between SIN3A, TET1 and local 5hmC marks. Importantly, EZH2 and SIN3A were found in lung-related cell lines including bronchial epithelial cells (A549) and lung fibroblasts (NHLF) (Fig. 6 and Supplementary Table S6A).
Fig. 6:
potential TF binding at regions with 5hmC and 5mC changes following DEP and HDM exposures. We estimated the significance of the degree of overlap between genomic regions with exposure induced 5hmC and/or 5mC changes and each member of a large library of transcription factor, co-factor, and other protein ChIP-seq datasets (see Methods). The top 30 significant results are shown regardless of cell type, along with results from lung related datasets passing a P < 10−6 significant threshold. The X-axis indicates the protein, in decreasing order of significance. The Y-axis indicates the significance (–log P-value) of the overlap of the given ChIP-seq dataset. The size of each box indicates the fold-enrichment relative to background (see Methods). The color of each box indicates the cell type: lung-related cell types are shown in multi-color, and non-lung cell types are shown in dark blue (see key below). (A) all locations with exposure-induced 5hmC changes; (B) all locations with exposure-induced 5mC changes; (C) 5hmC gain, 5mC no change; (D) 5mC loss, 5hmC gain; (E) 5mC gain, 5hmC no change; (F) 5mC loss, 5hmC no change; (G) 5mC gain, 5hmC loss; (H) 5hmC loss, no changes in 5mC. Detailed classification of 5mC and 5hmC changes are shown in Fig. 3B.
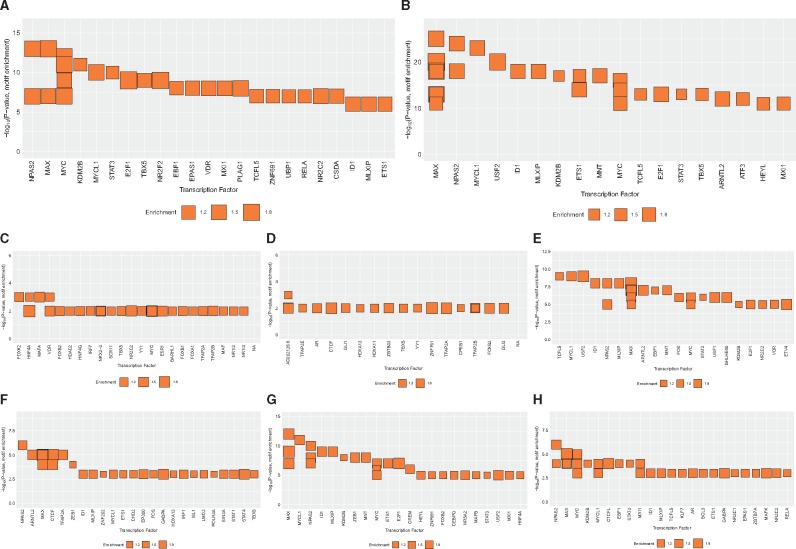
As a second approach to identify relevant TFs, we performed searches for enriched TF binding motifs within regions displaying 5mC and/or 5hmC changes in response to DEP and HDM in airway epithelial cells (top TFs shown in Fig. 7A and B). Similarly, different TFs seem to be enriched for regions with different directions of changes in 5mC, 5hmC and 5hmC + 5hmC (Fig. 7C–H). Consistent with the ChIP-seq analysis, motifs for MYC/MAX and NR2F2 are strongly enriched in regions with changes in 5hmC and 5mC status (Fig. 7A and B). Another top TF enriched at regions with 5hmC/5mC changes, STAT3, is known to be involved in epithelial responses to DEP [60] and LPS [61], activation of EGF signaling [62], T helper cell differentiation [63] and asthma [64]. It is specifically enriched at regions with 5mC gain/no 5hmC change (Fig. 7E) and regions with 5mC gain/5hmC loss (Fig. 7G). The roles of vitamin D and its receptor VDR in allergic disorders including asthma have also been widely studied [65–67]. Enrichment for VDR binding was found in regions with 5hmC gain/no 5mC changes (Fig. 7C) and regions with 5mC gain/no 5hmC changes (Fig. 7E).
Fig. 7:
transcription factor binding motif enrichment in regions with exposure-induced 5hmC and/or 5mC changes. Enrichment of transcription factor binding motifs in genomic regions with 5hmC and/or 5mC changes following exposures. Enrichment was calculated using the HOMER software package and the Cis-BP motif library (see Methods). The top 30 significantly enriched binding motifs are grouped by their corresponding transcription factors. The Y-axis indicates the significance (–log P-value) of the enrichment of the given motif. The X-axis indicates the corresponding TF, in decreasing order of significance. Box size indicates the fold-enrichment over background (see key below). (A) all locations with exposure-induced 5hmC changes; (B) all locations with exposure-induced 5mC changes; (C) 5hmC gain, 5mC no change; (D) 5mC loss, 5hmC gain; (E) 5mC gain, 5hmC no change; (F) 5mC loss, 5hmC no change; (G) 5mC gain, 5hmC loss; (H) 5hmC loss, no changes in 5mC.
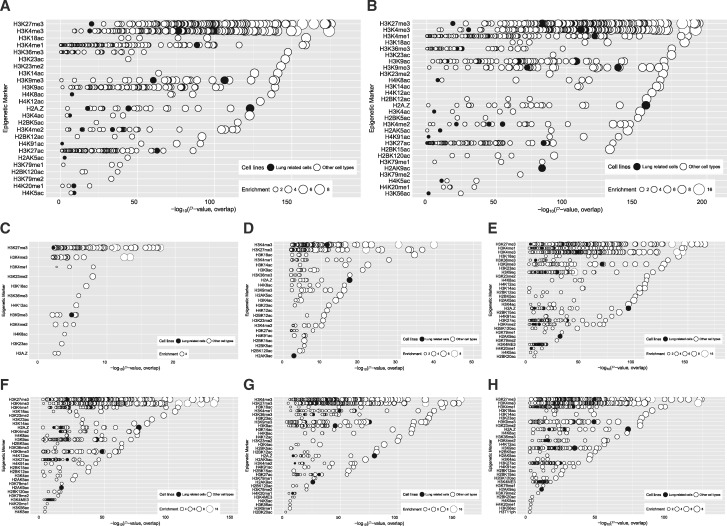
In addition to the study of TFs, our analysis also showed that genomic sites with 5hmC/5mC changes in response to HDM in HBEC have significant enrichment of both active histone marks (H3K4me3, H3K4me1, H3K36me3, H3K27ac, etc.) and repressive histone marks (H3K27me3, H3K9me3, H2AZ, etc.) that are associated with promoters and enhancer (Fig. 8A and B). Specifically, all of these active and repressive markers were found around CpG sites associated with 5hmC changes (Fig. 8C, D, G and H). This is consistent with previous observations that TET1 and 5hmC are associated with active “H3K4me3 only” promoters, bivalent poised “H3K4me3/H3K27me3” promoters, repressed “H3K27me3” promoters, “H3K4me1” enhancers, H3K36me3 marked gene bodies and CTCF marked insulators [27, 28, 68]. The same histone marks were also significantly enriched around sites with changes in both 5mC and 5hmC (Fig. 8D and G) and sites with changes in 5mC only (Fig. 8E and F). Collectively, our data suggest that 5mC and 5hmC differences induced by DEP and HDM may alter the binding of particular TFs and histone modifiers, which can in turn alter promoter/enhancer activity and regulate gene expression networks in airway epithelial cells.
Fig. 8:
overlap significance between 5mC or 5hmC-marked regions and histone marks. The significance of the degree of overlap between genomic regions with exposure induced 5mC/5hmC changes and each member of a large library of histone mark ChIP-seq datasets was estimated (see Methods). Histone marks with at least one significant result (P < 10−2) are shown. The Y-axis indicates the histone mark, in decreasing order of significance. The X-axis indicates the significance (–log P-value) of the overlap of the given dataset. The size of each circle indicates the fold-enrichment relative to background (see Methods). (A) all locations with exposure-induced 5hmC changes; (B) all locations with exposure-induced 5mC changes; (C) 5hmC gain, 5mC no change; (D) 5mC loss, 5hmC gain; (E) 5mC gain, 5hmC no change; (F) 5mC loss, 5hmC no change; (G) 5mC gain, 5hmC loss; (H) 5hmC loss, no changes in 5mC.
Discussion
In this paper, we studied the effects of DEP and HDM exposures on the expression of TET and DNA methylation on a genome-wide scale in airway epithelial cells (HBECs). We used a cutting-edge technology (oxidative bisulfite conversion) that allowed us to assess 5mC and 5hmC not only together (5mC + 5hmC) but also individually. We found that following DEP and HDM challenge in HBEC cells, there is a significant early increase in TET1 expression, and there are genome-wide changes in 5mC + 5hmC, 5mC and 5hmC modifications. Some of these changes are common to both exposures, and many of them occur at genes and pathways associated with oxidative stress responses, epithelial function, and immune cell responses. In general, changes in gene expression were correlated negatively with changes in 5mC, but not 5mC + 5hmC or 5hmC. For CMTM1 and CNTF, changes in 5mC and 5hmC are associated with gene expression differences and TET1 regulates their expression through modification of DNAm. Importantly, 5mC and 5hmC changes that are common to DEP and HDM exposures occur at binding sites for histone modifiers, as well as TFs involved in oxidative stress responses, epithelial function and immune cell responses. These DEP/HDM induced 5mC and 5hmC changes are associated with histone marks representative of promoters and enhancers. Taken together, our data suggest a novel regulation of gene networks in oxidative stress responses, epithelial function, and immune cell responses by TET1, 5mC and 5hmC following DEP and HDM challenges in airway epithelial cells.
We chose to treat airway epithelial cells using 5 μg/cm2 DEP and 25 μg/ml HDM because these doses upregulate the expression of TET1 without causing toxicity in airway epithelial cells. It has also been shown that these two treatments induced significant downstream aryl hydrocarbon receptor signaling, oxidative stress and proinflammatory responses and were used to examine the effects of real exposures [16, 69]. From candidates identified with the genome-wide array analysis, we verified three CpG sites using BS and oxBS pyrosequencing in the same DNA samples and then replicated our findings using a dose response design (Supplementary Figs S3 and S4). We correlated gene expression measured by RNA-seq with changes in 5mC + 5hmC, 5mC and 5hmC, and found a statistically significant negative correlation between changes in 5mC and gene expression (Fig. 3C). However, the two candidate CpG sites (cg26411814 and cg04791678) showed different patterns of associations between 5mC + 5hmC, 5mC and 5hmC with gene activation (CNTF and CMTM1, respectively) in response to DEP (Supplementary Figs S3 and S4, Fig. 5A and B) and TET1 knock down (Fig. 5D–F). This indicates that the regulation of gene expression by 5mC and 5hmC is both locus-specific and exposure-specific, thus highlights the importance of studying 5mC and 5hmC individually. Therefore, we recommend cautious interpretation of negative results in correlation studies between DNAm (5mC and 5hmC) and gene expression in samples collected at a certain time point.
Additionally, since TET2 and TET3 also regulate levels of 5hmC in terminally differentiated cells such as B and T cells [33, 70, 71], we measured their expression levels and observed no significant changes in TET2 and TET3 in response to DEP (Supplementary Fig. S1). Following the upregulation of TET1, we observed increased expression in DNMT1 and DNMT3A, consistent with the different patterns of 5mC and 5hmC changes we observed (Fig. 3B) and suggesting these 5mC and 5hmC changes may be a result of dysregulation in both TET1 and DNMTs. We speculate that regions with gain or loss in 5mC and no 5hmC changes (S4 and S6 in Fig. 3B table) are regulated by DNMTs, while regions with gain or loss in 5hmC and no 5mC changes (S2 and S8) as well as regions with 5mC loss and 5hmC gain (S3) are targeted by TET1. For regions with gain in 5mC and loss in 5hmC (S7), both enzymes are involved. Specifically, the changes in 5hmC in all scenarios may be either due to a redistribution of TET1 binding across genome, or result from a complex regulation of 5hmC by TET1 transcriptional upregulation and inhibition of TET1 enzymatic activity by oxidative stress produced by HDM/DEP [27]. The enrichment of different TFs and co-localization of both active and repressive histone marks is consistent with a dual role of TET1 and 5hmC in gene regulation [28, 68], which sometimes depends on the interacting partners of TET1 besides 5hmC levels [27]. For example, TET1 interacts with OGT and SET1/COMPASS complex to set up H3K4me3 and activate gene transcription; it can also interact with SIN3A/HDAC1, EZH2/SUZ12 and NURD complex to remove histone acetylation, set up H3K27me3 and silence gene expression. To support this, binding of SIN3A and EZH2/SUZ12 are enriched among the sites with 5mC/5hmC changes (Fig. 6 and Supplementary Table S6). However, not all sites interact with the same TFs. Further experiments are needed to determine whether TET1 directly bind to these loci in response to exposures and how they regulate local 5mC/5hmC levels, chromatin state, TF binding, and gene expression in a locus- and time-specific manner.
To assess the 5hmC levels, we used the oxidative bisulfite treatment, which involves harsh oxidation and reaction with sodium bisulfite. DNA methylation arrays also exhibited variability in measurement. All these introduce noises to the estimated beta value. One consequence is negative values when estimating 5hmC by subtracting oxBS signal from the BS signal. Though direct measurement of 5hmC by TAB-seq is possible [26], this technology is associated with incomplete oxidation by purified Tet1 protein (80–90%), making it harder to observe small differences. To solve the problem, we applied mathematical strategies with a recent methodology named OxyBS [72]. To account for the biological and experimental variability, we performed technical duplicate experiments using pooled biologic replicates as input and analysed the mean beta values. To enhance the potential of novel findings given the small sample size, instead of a strict P value cutoff, we used difference of beta values to indicate differential DNAm. We selected three biologically relevant CpG candidates. Out of 18 comparisons for these three sites, only one showed uncorrected P value ≤ 0.05 when array data were examined, while all of them were verified and replicated, suggesting the usefulness of this cost-efficient strategy.
In conclusion, using a cutting-edge technology and comprehensive bioinformatics analyses, our study identified novel 5mC and 5hmC changes following DEP and HDM challenges in human bronchial epithelial cells and presented plausible evidence supporting that these changes modify genes in oxidative stress responses, epithelial function and immune cell responses in coordination with specific transcription factors and histone modifications. As oxidative stress responses, epithelial function and immune cell responses are known important players in asthma pathogenesis, our results suggest novel epigenetic mechanisms contributing to asthma. Furthermore, a significant portion of 5mC and 5hmC changes are shared by DEP and HDM, which may mediate the interplay between these two exposures in asthma development and exacerbation [5, 6].
Methods
Cell Culture and Treatment
Human bronchial epithelial cells (HBECs) (a cell line obtained from Dr. John Minna’s lab at UT Southwestern Medical Center [73]) were grown to ∼70% confluence in Keratinocyte-SFM serum-free medium (10724-011, Life Technologies, Grand Island, NY) supplemented with human recombinant Epidermal Growth Factor (10450-013, Life Technologies, Grand Island, NY) and Bovine Pituitary Extract (13028-014, Life Technologies, Grand Island, NY). House dust mite extract (Dermatophagoides pteronyssinus) was purchased from Greer Laboratories (XPB70D3A2.5, Lenoir, NC). DEP was generated from a 4-cylinder Deutz diesel engine at the EPA (Research Triangle Park, NC); detailed characterization of this compressor DEP has been described previously and compared to other sources of DEP [74]. HBECs were cultured to 70% confluency in a 12 well plate and starved overnight (∼8 h), exposed to Saline, DEP (5 μg/cm2) or HDM (25 μg/ml) for 1–24 h in triplicate before harvest. For TET1 knockdown, cells were treated with two TET1 siRNA (#1: sc-90457 from Santa Cruz Biotechnology, Dallas, Texas; #2: AM16708 from Ambion, ThermoFisher Scientific, Waltham, MA). TET1 knockdown was performed according to the RNAiMAX protocol (ThermoFisher Scientific). 1ul Lipofectamine RNAiMAX transfection reagent (13778150, ThermoFisher) was added to 25ul Opti-MEM transfection medium (31985062, ThermoFisher). 1 μg TET1 siRNA from Santa Cruz Biotechnology or 30 pmole TET1 siRNA from Ambion, and their corresponding control scrambled siRNAs were added to 25ul Opti-MEM transfection medium. The DNA and lipid complex were then mixed together and incubated for 5 min. The DNA-Lipid complex was added to one well of a 12 well plate and cells were harvested 48 h later. DNA/RNA extraction from HBECs was performed using AllPrep DNA/RNA mini kit (80294, Qiagen, Hilden, Germany) according to the manufacturer’s protocols.
Sample Processing and methylationEPIC Beadchip Array
Genomic DNA from each treatment in triplicates was isolated and pooled. 1 μg DNA was then treated in duplicates with or without oxidation using the TrueMethyl® protocol for array analysis (NuGen) as follows: DNA was subjected to an initial buffer exchange step using a beads-based approach included in the kit and eluted in ultra-pure water. The full eluate (50 μl per sample) was carried forward. Samples were denatured using the provided denaturing solution for 30 min at 37°C, in a total reaction volume of 50 μl and immediately taken forward to oxidation. One μl of the provided oxidant solution was added to each sample undergoing the oxBS workflow only (1 μl of ultra-pure water was added to BS-only samples for mock oxidation). All samples were incubated for 30 min at 40°C. Oxidized samples were equilibrated to room temperature before proceeding immediately to bisulfite conversion. Bisulfite reagent solution was prepared as described in the TrueMethyl® protocol and 170 μl was added to each 25 μl oxidation reaction mixture. Finally, 5 μl of bisulfite additive was added to each reaction, bringing the total volume to 200 μl. All reactions were incubated using bisulfite-specific thermal cycling conditions. Converted DNAs were purified using the provided spin columns and 4 μl of each sample eluate was used as input into the Infinium EPIC array. All spike-in and controls in TrueMethyl®Array Kit were included to ensure the data quality and and the expected results were obtained.
Array Processing
Array quality was assessed using sample-independent and dependent internal control probes included on the array. All samples had >95% sites detected at P = 0.05 level; no quality issue was observed in staining, extension, hybridization, specificity or bisulfite conversion; therefore, samples were all included in the analysis. The signal intensities were then background-corrected using out-of-band probes (noob), and normalized using subset-quantile within array normalization (swan) with the R package “minfi”. CpG sites that were not detected in all samples at P = 0.01 level, and CpG sites on the X and Y chromosomes were excluded.
Methylation levels were measured as β values (), and used in analyses. For each of the samples, DNA without and with oxidation were assayed, which directly measured the sum of 5mC and 5hmC (βtotal), and the 5mC levels (βmC). The levels of 5hmC (βhmC) were estimated from BS and oxBS signals using the R package OxyBS [72]. Samples were assayed as technical duplicates; the average βtotal, βmC and βhmC were calculated and compared between control, DEP and HDM treated sample. An absolute difference of 0.1 or larger in β values was considered significant, which is consistent with previous publications [16, 30, 75].
RNA-Seq Library Preparation, Sequencing and Analysis
Cells were harvested and RNA were isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s instruction. The concentration and integrity of the isolated RNA was quantified using the Nanodrop 1000 (Thermo Fisher Scientific, Wilmington, DE) and 2100 Bioanalyzer (Agilent, Santa Clara, CA). A total of 1 μg RNA for each sample with RNA integrity number (RIN) values >8 was used for sequencing library construction. Briefly, polyA RNA was first purified via NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs, Ipswich, MA). Using the polyA RNA as input, the library for RNA-seq was prepared by using NEBNext Ultra Directional RNA Library Prep kit. The approach uses dUTP-based method to preserve strand information. After indexing via PCR (11 cycles) enrichment, the amplified libraries together with library preparation negative control were cleaned up by AMPure XP beads (Beckman Coulter, Brea, CA) for QC analysis using Bioanalyzer High Sensitivity chip (Agilent). To accurately quantify the library concentration for the cluster generation, the library was measured by qPCR using NEBNext Library Quant Kit (New England BioLabs) using QuantStudio 5 Real-Time PCR Systems (Thermo Fisher). Next, individually indexed and compatible libraries were proportionally pooled (∼25 million reads per sample in general) for clustering in cBot system (Illumina, San Diego, CA). Libraries at the final concentration of 15 pM were clustered onto a single read (SR) flow cell using Illumina TruSeq SR Cluster kit v3, and sequenced to 51 bp using TruSeq SBS kit on Illumina HiSeq system. All fastq files were adapter trimmed prior to alignment. Files were aligned against Human (GRch37/Ensembl). Sequence alignment was performed using Bowtie2 and RSEM [76]. Post-alignment, raw read counts were extracted and normalized to account for difference in read depth using DESeq [77]. Following normalization, gene read counts were filtered to remove low read values: maximum Reads Per Kilobase of transcript, per Million mapped reads (RPKM) across all samples must be ≥ 1. Differentially expressed (DE) genes were identified using DESeq, and genes with an FDR adjusted P-value (q-value) < 0.1 and fold of change ≥1.2, were considered significant.
Ingenuity Pathway Analysis
To better understand the biological meaning of the methylation changes, genes associated with identified differentially methylated CpG sites were extracted from the EPIC array annotation file and imported into Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA) for pathway mapping (canonical pathways). A cutoff of 0.05 was used for statistical significance in IPA analysis.
Bisulfite Pyrosequencing and Oxidative Bisulfite Pyrosequencing
A total of 200 ng genomic DNA was subjected to paired sodium bisulfite treatment and oxidative sodium bisulfite treatment. The TrueMethyl® protocol for array analysis (CEGX™) was used according to the manufacturer’s specifications. Standard PCR amplification reactions were performed to amplify targeted gene fragments at an annealing temperature of 50°C before being subjected to pyrosequencing. The generated pyrograms were automatically analysed using PyroMark analysis software (Qiagen, Valencia, CA, USA). Pyrosequencing assay design and genomic coordinates are documented in Supplementary Table S1.
Quantitative RT-qPCR
Total RNA was reverse-transcribed to cDNA using the VILO cDNA kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Real-time quantitative PCR was performed using the SYBR Green Master Kit and LightCycler® 480 instrument (Roche Diagnostics, Indianapolis, IN, USA). PCR was carried out in triplicate for each cDNA and the mean Ct value of the triplicate reaction was normalized against the mean Ct value of GAPDH. For the time course experiment, normalized values (to GAPDH) from treated samples at each time point were also normalized to their respective controls (values set to be 1) and resulting values were plotted. Primer sequences are provided in Supplementary Table S2.
Functional Genomics Analyses
To identify transcription factors (TFs) and other epigenetic marks that might be enriched at 5hmC and 5mC sites, we used a computational method that overlaps a set of genomic locations (e.g. regions with 5hmC marks) with a large collection of functional genomics datasets. To this end, we created a library of 1544 TF ChIP-seq datasets by compiling data from a variety of sources, including ENCODE [78], Cistrome [79], PAZAR [80] and Re-Map [81]. The library consists of ChIP-seq peaks monitoring the binding interactions of 390 different proteins in 487 cell lines and tissue types. We also created an analogous library consisting of 1207 epigenetic data sets (ChIP-seq for histone marks, DNase-seq, ATAC-seq, etc), from sources such as ENCODE, Roadmap Epigenomics [82], and a study performing ChIP-seq in many EBV-infected B cell lines [83]. In total, this library consists of ChIP-seq peaks monitoring the presence of 31 different histone marks in 204 cell lines and tissue types.
As input, the method takes a set of genomic loci (e.g. regions with differential 5hmC marks). The coordinates of each locus are padded by 100 bases in either direction (to account for experimental resolution). The resulting loci are then systematically intersected with the ChIP-seq and epigenetic data set libraries, and the number of input regions overlapping each dataset by at least one base is counted. Next, a P-value describing the significance of this overlap is estimated using a simulation-based procedure. To this end, a ‘negative set’ is created for comparison to the input set, which contain regions with 5mC and 5hmC marks (beta≥0.1) but no significant differences between control and treated cell lines (absolute paired difference < 0.1). A distribution of expected overlap values is then created from 1000 iterations of randomly sampling from the negative set, each time choosing a set of negative examples that match the input set in terms of the total number of genomic loci and the length of each locus. The distribution of the expected overlap values from the randomized data resembles a normal distribution, and can thus be used to generate a Z-score and corresponding P-value estimating the significance of the observed number of input regions that overlap each data set. Collectively, this procedure controls for the count and sizes of the input loci, and the count and sizes of each individual dataset in the library.
We also examined genomic loci with 5hmC and 5mC marks using standard TF motif enrichment analysis. To this end, we used the HOMER motif enrichment algorithm [84] and a large library of human position weight matrix (PWM) binding site models obtained from the CisBP database [85]. For each analysis, we used the aforementioned negative set to provide background sequences to HOMER.
Statistical Analysis
Expression in HBEC between control and DEP/HDM treated or TET1 knockdown groups were compared by two-sided t-tests in GraphPad Prism (CA, USA). A P value ≤ 0.05 was used to indicate statistical significance.
Supplementary Material
Acknowledgements
We thank the Genomics, Epigenomics and Sequencing Core at the University of Cincinnati for array processing and the Cincinnati Children’s Pyrosequencing Core Laboratory for Genomic and Epigenomic Research for assistance in pyrosequencing.
Funding
This work was supported by NIH/NIAID R21AI119236 (H.J.) and ALA/AAAAI Respiratory Diseases Research Award 515708 (H.J.). M.T.W. was supported by NIH R21 HG008186, NIH R01 NS099068-01A1, Cincinnati Children’s Hospital “Center for Pediatric Genomics” pilot study award, and a Cincinnati Children’s Hospital Research Fund “Endowed Scholar” award.
Availability of Data and Material
Array data have been deposited in GEO (GSE116402).
Competing Interests
Our authors do not have any conflicts of interest or financial disclosures related to this work, including declarations of financial interest, to report.
Supplementary Data
Supplementary data are available at EnvEpig online.
Conflict of interest statement. None declared.
References
- 1. Calderon MA, Linneberg A, Kleine-Tebbe J, De Blay F, Hernandez Fernandez de Rojas D, Virchow JC, Demoly P.. Respiratory allergy caused by house dust mites: what do we really know? J Allergy Clin Immunol 2015;136:38–48. [DOI] [PubMed] [Google Scholar]
- 2. Platts-Mills TA, Erwin EA, Heymann PW, Woodfolk JA.. Pro: the evidence for a causal role of dust mites in asthma. Am J Respir Crit Care Med 2009; 180:109–13. discussion 120-101. [DOI] [PubMed] [Google Scholar]
- 3. Takahashi G, Tanaka H, Wakahara K, Nasu R, Hashimoto M, Miyoshi K, Takano H, Yamashita H, Inagaki N, Nagai H.. Effect of diesel exhaust particles on house dust mite-induced airway eosinophilic inflammation and remodeling in mice. J Pharmacol Sci 2010; 112:192–202. [DOI] [PubMed] [Google Scholar]
- 4. Acciani TH, Brandt EB, Khurana Hershey GK, Le Cras TD.. Diesel exhaust particle exposure increases severity of allergic asthma in young mice. Clin Exp Allergy 2013; 43:1406–18. [DOI] [PubMed] [Google Scholar]
- 5. Brandt EB, Biagini Myers JM, Acciani TH, Ryan PH, Sivaprasad U, Ruff B, LeMasters GK, Bernstein DI, Lockey J, LeCras TD, Khurana Hershey GK.. Exposure to allergen and diesel exhaust particles potentiates secondary allergen-specific memory responses promoting asthma susceptibility. J Allergy Clin Immunol 2015; 136:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brandt EB, Kovacic MB, Lee GB, Gibson AM, Acciani TH, Le Cras TD, Ryan PH, Budelsky AL, Khurana Hershey GK.. Diesel exhaust particle induction of IL-17A contributes to severe asthma. J Allergy Clin Immunol 2013; 132:1194–204. e1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pollution. Traffic-related air pollution: a critical review of the literature on emissions, exposure, and health effects. Health Effects Institute, Special Report 17, January 2010.
- 8. Beelen R, Stafoggia M, Raaschou-Nielsen O, Andersen ZJ, Xun WW, Katsouyanni K, Dimakopoulou K, Brunekreef B, Weinmayr G, Hoffmann B.. Long-term exposure to air pollution and cardiovascular mortality: an analysis of 22 European cohorts. Epidemiology 2014; 25:368–78. [DOI] [PubMed] [Google Scholar]
- 9. Beelen R, Raaschou-Nielsen O, Stafoggia M, Andersen ZJ, Weinmayr G, Hoffmann B, Wolf K, Samoli E, Fischer P, Nieuwenhuijsen M, et al. Effects of long-term exposure to air pollution on natural-cause mortality: an analysis of 22 European cohorts within the multicentre ESCAPE project. Lancet 2014; 383:785–95. [DOI] [PubMed] [Google Scholar]
- 10. Molter A, Simpson A, Berdel D, Brunekreef B, Custovic A, Cyrys J, de Jongste J, de Vocht F, Fuertes E, Gehring U, et al. A multicentre study of air pollution exposure and childhood asthma prevalence: the ESCAPE project. Eur Respir J 2015; 45:610–24. [DOI] [PubMed] [Google Scholar]
- 11. Brandt EB, Myers JM, Ryan PH, Hershey GK.. Air pollution and allergic diseases. Curr Opin Pediatr 2015; 27:724–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Appatova AS, Ryan PH, LeMasters GK, Grinshpun SA.. Proximal exposure of public schools and students to major roadways: a nationwide US survey. J Environ Plann Manag 2008; 51:631–46. [Google Scholar]
- 13. Hystad P, Demers PA, Johnson KC, Brook J, van Donkelaar A, Lamsal L, Martin R, Brauer M.. Spatiotemporal air pollution exposure assessment for a Canadian population-based lung cancer case-control study. Environ Health 2012; 11:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mukherjee A, Agrawal M.. A global perspective of fine particulate matter pollution and its health effects. Rev Environ Contam Toxicol 2017; 244:5–51. [DOI] [PubMed] [Google Scholar]
- 15. Ji H, Biagini Myers JM, Brandt EB, Brokamp C, Ryan PH, Khurana Hershey GK.. Air pollution, epigenetics, and asthma. Allergy Asthma Clin Immunol 2016; 12:51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Somineni Hk, Zhang X, Biagini Myers JM, Kovacic MB, Ulm A, Jurcak N, Ryan PH, Khurana Hershey GK, Ji H.. Ten-eleven translocation 1 (TET1) methylation is associated with childhood asthma and traffic-related air pollution. J Allergy Clin Immunol 2016; 137:797–805. e795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE.. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 2011; 12:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Serandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Peron Cl, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res 2012; 40:8255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013; 341:1237905.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y.. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 2014; 28:2103–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 2014; 56:286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bogdanovic O, Smits AH, de la Calle Mustienes E, Tena JJ, Ford E, Williams R, Senanayake U, Schultz MD, Hontelez S, van Kruijsbergen I, et al. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat Genet 2016; 48:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kranzhofer DK, Gilsbach R, Gruning Ba, Backofen R, Nuhrenberg TG, Hein L.. 5'-Hydroxymethylcytosine precedes loss of CpG methylation in enhancers and genes undergoing activation in cardiomyocyte maturation. PLoS One 2016; 11:e0166575.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mahe EA, Madigou T, Serandour AA, Bizot M, Avner S, Chalmel F, Palierne G, Metivier R, Salbert G.. Cytosine modifications modulate the chromatin architecture of transcriptional enhancers. Genome Res 2017; 27:947–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011; 473:394–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012; 149:1368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delatte B, Deplus R, Fuks F.. Playing TETris with DNA modifications. EMBO J 2014; 33:1198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y.. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 2011; 25:679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheng RY, Shang Y, Limjunyawong N, Dao T, Das S, Rabold R, Sham JS, Mitzner W, Tang WY.. Alterations of the lung methylome in allergic airway hyper-responsiveness. Environ Mol Mutagen 2014; 55:244–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 2004; 64:9027–34. [DOI] [PubMed] [Google Scholar]
- 31. Stevens T, Cho SH, Linak WP, Gilmour MI.. Differential potentiation of allergic lung disease in mice exposed to chemically distinct diesel samples. Toxicol Sci 2009; 107:522–34. [DOI] [PubMed] [Google Scholar]
- 32. Houseman EA, Johnson KC, Christensen BC.. OxyBS: estimation of 5-methylcytosine and 5-hydroxymethylcytosine from tandem-treated oxidative bisulfite and bisulfite DNA. Bioinformatics 2016; 32:2505–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang X, Ulm A, Somineni HK, Oh S, Weirauch MT, Zhang HX, Chen X, Lehn MA, Janssen EM, Ji H.. DNA methylation dynamics during differentiation and maturation of human dendritic cells. Epigenet Chromatin 2014; 7:21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ji H, Zhang X, Oh S, Mayhew CN, Ulm A, Somineni HK, Ericksen M, Wells JM, Khurana Hershey GK.. Dynamic transcriptional and epigenomic reprogramming from pediatric nasal epithelial cells to induced pluripotent stem cells. J Allergy Clin Immunol 2015; 135:236–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anders S, Huber W.. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 2011; 12:R83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Portales-Casamar E, Arenillas D, Lim J, Swanson MI, Jiang S, McCallum A, Kirov S, Wasserman WW.. The PAZAR database of gene regulatory information coupled to the ORCA toolkit for the study of regulatory sequences. Nucleic Acids Res 2009; 37:D54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Griffon A, Barbier Q, Dalino J, van Helden J, Spicuglia S, Ballester B.. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res 2015; 43:e27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chadwick LH. The NIH roadmap epigenomics program data resource. Epigenomics 2012; 4:317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside P, Srivas R, Phanstiel DH, Pekowska A, et al. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell 2015; 162:1051–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014; 158:1431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nazor KL, Boland MJ, Bibikova M, Klotzle B, Yu M, Glenn-Pratola VL, Schell JP, Coleman RL, Cabral-da-Silva MC, Schmidt U, et al. Application of a low cost array-based technique—TAB-Array—for quantifying and mapping both 5mC and 5hmC at single base resolution in human pluripotent stem cells. Genomics 2014; 104:358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Booth MJ, Ost TW, Beraldi D, Bell NM, Branco MR, Reik W, Balasubramanian S.. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc 2013; 8:1841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsagaratou A, Aijo T, Lio CW, Yue X, Huang Y, Jacobsen SE, Lahdesmaki H, Rao A.. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc Natl Acad Sci USA 2014; 111:E3306–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu H, Zhang Y.. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014; 156:45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Han W, Ding P, Xu M, Wang L, Rui M, Shi S, Liu Y, Zheng Y, Chen Y, Yang T, Ma D.. Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics 2003; 81:609–17. [DOI] [PubMed] [Google Scholar]
- 50. Chiu HY, Chen CW, Lin HT, Hsieh CC, Lin SS, Cheng CM.. Study of gastric fluid induced cytokine and chemokine expression in airway smooth muscle cells and airway remodeling. Cytokine 2011; 56:726–31. [DOI] [PubMed] [Google Scholar]
- 51. Kimata H. Ciliary neurotrophic factor preferentially enhances spontaneous IgE production by B cells from atopic patients. Neuropeptides 2004; 38:92–7. [DOI] [PubMed] [Google Scholar]
- 52. Rezende LF, Vieira AS, Negro A, Langone F, Boschero AC.. Ciliary neurotrophic factor (CNTF) signals through STAT3-SOCS3 pathway and protects rat pancreatic islets from cytokine-induced apoptosis. Cytokine 2009; 46:65–71. [DOI] [PubMed] [Google Scholar]
- 53. Xu Y, Zhang M, Wang Y, Kadambi P, Dave V, Lu LJ, Whitsett JA.. A systems approach to mapping transcriptional networks controlling surfactant homeostasis. BMC Genomics 2010; 11:451.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Greco CM, Kunderfranco P, Rubino M, Larcher V, Carullo P, Anselmo A, Kurz K, Carell T, Angius A, Latronico MV, et al. DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy. Nat Commun 2016; 7:12418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin IH, Chen YF, Hsu MT.. Correlated 5-hydroxymethylcytosine (5hmC) and gene expression profiles underpin gene and organ-specific epigenetic regulation in adult mouse brain and liver. PLoS One 2017; 12:e0170779.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 2013; 53:401–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S.. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med 2005; 202:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sussan TE, Gajghate S, Chatterjee S, Mandke P, McCormick S, Sudini K, Kumar S, Breysse PN, Diette GB, Sidhaye VK, Biswal S.. Nrf2 reduces allergic asthma in mice through enhanced airway epithelial cytoprotective function. Am J Physiol Lung Cell Mol Physiol 2015; 309:L27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morgan MJ, Liu ZG.. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res 2011; 21:103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schuliga M. NF-kappaB signaling in chronic inflammatory airway disease. Biomolecules 2015; 5:1266–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang F, Marshall CB, Ikura M.. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci 2013; 70:3989–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E.. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev 2003; 17:1019–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Martinato F, Cesaroni M, Amati B, Guccione E.. Analysis of Myc-induced histone modifications on target chromatin. PLoS One 2008; 3:e3650.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sekimata M, Takahashi A, Murakami-Sekimata A, Homma Y.. Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG-binding protein, MBD2. J Biol Chem 2001; 276:42632–8. [DOI] [PubMed] [Google Scholar]
- 65. Sekimata M, Homma Y.. Sequence-specific transcriptional repression by an MBD2-interacting zinc finger protein MIZF. Nucleic Acids Res 2004; 32:590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bell AC, Felsenfeld G.. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000; 405:482–5. [DOI] [PubMed] [Google Scholar]
- 67. Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM.. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 2000; 405:486–9. [DOI] [PubMed] [Google Scholar]
- 68. Maurano MT, Wang H, John S, Shafer A, Canfield T, Lee K, Stamatoyannopoulos JA.. Role of DNA methylation in modulating transcription factor occupancy. Cell Rep 2015; 12:1184–95. [DOI] [PubMed] [Google Scholar]
- 69. Acemel RD, Maeso I, Gomez-Skarmeta JL.. Topologically associated domains: a successful scaffold for the evolution of gene regulation in animals. Wires Dev Biol 2017;6:e265.. [DOI] [PubMed] [Google Scholar]
- 70. Chittock EC, Latwiel S, Miller TC, Muller CW.. Molecular architecture of polycomb repressive complexes. Biochem Soc Trans 2017; 45:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kadamb R, Mittal S, Bansal N, Batra H, Saluja D.. Sin3: insight into its transcription regulatory functions. Eur J Cell Biol 2013; 92:237–46. [DOI] [PubMed] [Google Scholar]
- 72. Cartron PF, Nadaradjane A, Lepape F, Lalier L, Gardie B, Vallette FM.. Identification of TET1 partners that control its dna-demethylating function. Genes Cancer 2013; 4:235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K.. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 2011; 473:343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cao D, Tal TL, Graves LM, Gilmour I, Linak W, Reed W, Bromberg PA, Samet JM.. Diesel exhaust particulate-induced activation of Stat3 requires activities of EGFR and Src in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 2007; 292:L422–9. [DOI] [PubMed] [Google Scholar]
- 75. Severgnini M, Takahashi S, Rozo LM, Homer RJ, Kuhn C, Jhung JW, Perides G, Steer M, Hassoun PM, Fanburg BL, et al. Activation of the STAT pathway in acute lung injury. Am J Physiol Lung Cell Mol Physiol 2004; 286:L1282–92. [DOI] [PubMed] [Google Scholar]
- 76. Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, Bromberg JF.. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest 2007; 117:3846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Korn T, Bettelli E, Oukka M, Kuchroo VK.. IL-17 and Th17 Cells. Annu Rev Immunol 2009; 27:485–517. [DOI] [PubMed] [Google Scholar]
- 78. Simeone-Penney MC, Severgnini M, Tu P, Homer RJ, Mariani TJ, Cohn L, Simon AR.. Airway epithelial STAT3 is required for allergic inflammation in a murine model of asthma. J Immunol 2007; 178:6191–9. [DOI] [PubMed] [Google Scholar]
- 79. Mirzakhani H, Al-Garawi A, Weiss ST, Litonjua AA.. Vitamin D and the development of allergic disease: how important is it?. Clin Exp Allergy 2015;45:114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Han JC, Du J, Zhang YJ, Qi GB, Li HB, Zhang YJ, Yu XL.. Vitamin D receptor polymorphisms may contribute to asthma risk. J Asthma 2016; 53:790–800. [DOI] [PubMed] [Google Scholar]
- 81. Tizaoui K, Berraies A, Hamdi B, Kaabachi W, Hamzaoui K, Hamzaoui A.. Association of vitamin D receptor gene polymorphisms with asthma risk: systematic review and updated meta-analysis of case-control studies. Lung 2014; 192:955–65. [DOI] [PubMed] [Google Scholar]
- 82. Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y.. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 2011; 473:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McInnes N, Davidson M, Scaife A, Miller D, Spiteri D, Engelhardt T, Semple S, Devereux G, Walsh G, Turner S.. Primary paediatric bronchial airway epithelial cell in vitro responses to environmental exposures. Int J Environ Res Public Health 2016; 13:359.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ichiyama K, Chen T, Wang X, Yan X, Kim B-S, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 2015; 42:613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tsagaratou A, Lio CJ, Yue X, Rao A.. TET methylcytosine oxidases in T cell and B cell development and function. Front Immunol 2017; 8:220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.