Abstract
Population genomic data can be used to infer historical effective population sizes (Ne), which help study the impact of past climate changes on biodiversity. Previous genome sequencing of one individual of the common bottlenose dolphin Tursiops truncatus revealed an unusual, sharp rise in Ne during the last glacial, raising questions about the reliability, generality, underlying cause, and biological implication of this finding. Here we first verify this result by additional sampling of T. truncatus. We then sequence and analyze the genomes of its close relative, the Indo-Pacific bottlenose dolphin T. aduncus. The two species exhibit contrasting demographic changes in the last glacial, likely through actual changes in population size and/or alterations in the level of gene flow among populations. Our findings suggest that even closely related species can have drastically different responses to climatic changes, making predicting the fate of individual species in the ongoing global warming a serious challenge.
Keywords: climate change, genome sequencing, population structure, Tursiops aduncus, Tursiops truncatus
Understanding the impact of a rapidly changing climate on biodiversity represents an urgent challenge due to the ongoing global warming (Thuiller 2007). Although the causes of past and present climatic changes may be different, the consequences of past climatic changes on biodiversity can provide useful insights. Hence, a promising approach is to use population genomic data to infer historical effective population sizes (Ne) and study how Ne has responded to past climatic change events such as Pleistocene glacial cycles. This approach has frequently revealed a reduction in Ne during the last glacial (110–12 kya) for temperate species, including marine cetaceans (Moura et al. 2014; Yim et al. 2014). Surprisingly, however, the genomic analysis of one individual of the common bottlenose dolphin Tursiops truncatus suggested a steep rise in Ne in the last glacial (Yim et al. 2014), raising questions about the reliability, generality, underlying cause, and biological implication of this finding. In the present study, we first verify the previous finding by additional sampling of T. truncatus. We then sequence and analyze the genomes of its closest congener, the Indo-Pacific bottlenose dolphin T. aduncus, for comparison. We report contrasting demographic changes of these two species in the last glacial, explore potential causes, and discuss implications.
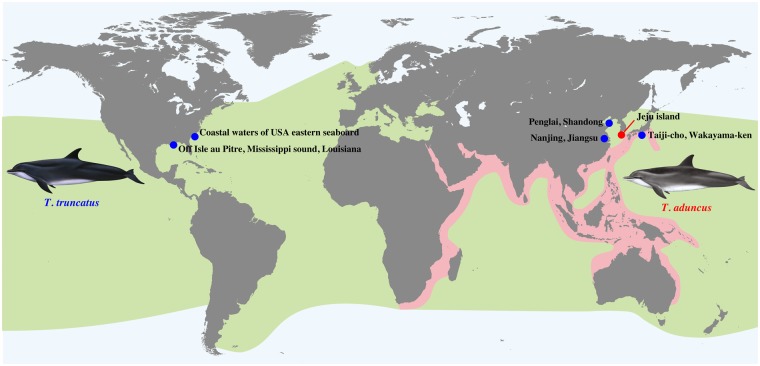
Bottlenose dolphins (genus Tursiops) include at least two taxonomically accepted, widespread species: T. truncatus and T. aduncus. According to a recent estimate based on both mitochondrial and nuclear markers, these two species diverged from each other ∼0.7 Ma (Gray et al. 2018). The use of more markers and widespread sampling makes this a more reliable estimate than the previously reported divergence time of ∼2.5 Ma (Vilstrup et al. 2011). The two dolphin species occupy distinct but overlapping habitats and regions (Hale et al. 2000): T. truncatus is distributed in the coastal, near-shore, and off-shore zones of most oceans worldwide, with the exception of polar areas, whereas T. aduncus is discontinuously distributed along the coastal areas of warm temperate and tropical Indian and Indo-Pacific oceans, from east Africa to the northwestern Pacific (fig. 1).
Fig. 1.
Map showing the geographic distributions of the common bottlenose dolphin Tursiops truncatus (green) and Indo-Pacific bottlenose dolphin T. aduncus (pink). The map is redrawn from the IUCN red list resource.shp file. See Materials and Methods for details.
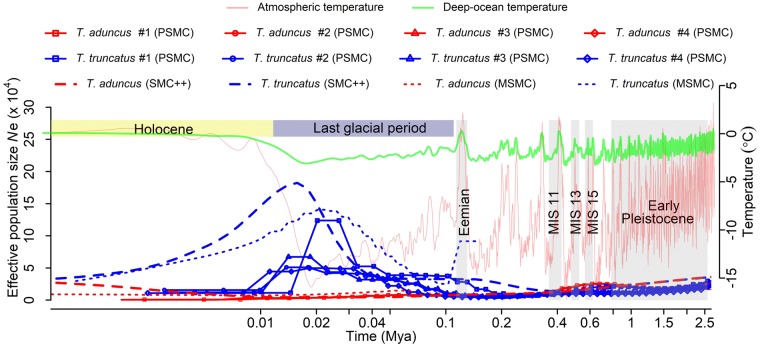
We used the Pairwise Sequentially Markovian Coalescent (PSMC) method (see Materials and Methods) to, respectively, infer the temporal changes in Ne from publically available genome sequences of four T. truncatus individuals, including the one previously analyzed (Yim et al. 2014; supplementary table S1, Supplementary Material online). For all four genomes, Ne rose sharply starting around the beginning of the last glacial, peaked when the atmosphere temperature reached the minimum, and then dropped as the temperature rebounded (fig. 2). Bootstrap analysis confirms the statistical robustness of these trends (supplementary fig. S1A, Supplementary Material online).
Fig. 2.
Contrasting demographic changes of the two bottlenose dolphin species in the last glacial, inferred from genome sequences using PSMC, MSMC, and SMC++. MIS, Marine Isotope Stage. All dolphins whose genomes are analyzed here are from Northwest Pacific. See supplementary table S1, Supplementary Material online for detailed information of individual dolphin genomes.
To probe whether this unusual demographic pattern is specific to T. truncatus, we generated a high-quality de novo genome assembly for T. aduncus, using 180× coverage of Illumina sequencing of an individual sampled from Korea, followed by sequencing of three additional individuals at lower coverages (22–32×;supplementary tables S1 and S2, Supplementary Material online; see Materials and Methods). Population genomic analyses were conducted using four T. truncatus and four T. aduncus individuals. Nucleotide diversity (π) is greater for T. truncatus (0.0015) than T. aduncus (0.00095), as expected from the wider geographic distribution of the former than the latter. The two species exhibit high levels of genome-wide differentiation (mean Fst = 0.61), consistent with their different geographic distributions and classification as distinct species (with the caveat that this conclusion is based on Northwest Pacific T. truncatus; see below). The PSMC analysis of each T. aduncus individual shows a decline in Ne during the last glacial (fig. 2 and supplementary fig. S1A, Supplementary Material online). This prolonged decline started >0.5 Ma, and no rebound since then is apparent (fig. 2). The PSMC-inferred, contrasting patterns of Ne between the two dolphin species are robust to different generation times and mutation rates assumed (supplementary fig. S1B, Supplementary Material online) and are confirmed by MSMC and SMC++ (fig. 2), which are improved methods that can simultaneously analyze multiple genome sequences (see Materials and Methods). The population size trajectories that have not been scaled by mutation rate and generation time also show a clear distinction between the two species (supplementary fig. S1C, Supplementary Material online).
All individuals of T. truncatus and T. aduncus analyzed above were sampled from Northwest Pacific (off the coasts of China, Korea, and Japan) and are thus more or less comparable. The public domain also houses the genome sequences from two individuals of T. truncatus from the Northwest Atlantic, sampled off Isle au Pitre in the Mississippi Sound, Louisiana and the coastal waters of the US eastern seaboard, respectively (supplementary table S1, Supplementary Material online). As expected, principal component analysis shows that all T. truncatus individuals are genetically distinct from T. aduncus (supplementary fig. S2, Supplementary Material online). The four T. aduncus individuals are genetically highly similar (supplementary fig. S2, Supplementary Material online). Within T. truncatus, however, the two Northwest Atlantic individuals are genetically quite distinct from each other and from the four Northwest Pacific individuals (supplementary fig. S2, Supplementary Material online), suggesting population differentiation of T. truncatus. Analyses of the 3rd and 4th principal components show population structure even within Northwest Pacific T. truncatus (supplementary fig. S2, Supplementary Material online). Strikingly, the Louisiana (Northwest Atlantic) individual has a temporal trend of Ne highly similar to those of T. aduncus individuals, with a decline in Ne in the last glacial (supplementary fig. S3, Supplementary Material online). The genome sequence coverage of the US eastern seaboard (Northwest Atlantic) individual (0.7×) is too low to allow a reliable PSMC analysis.
Because the tremendous temporal changes of Ne for (Northwest Pacific) T. truncatus coincided well with the drastic atmosphere and deep-ocean temperature changes of the last glacial (fig. 2), the demographic changes were potentially caused directly or indirectly by the climatic changes. Global cooling could directly affect dolphin habitats and distributions because of lowered sea levels and reduced ocean temperatures (Learmonth et al. 2006). Although these effects could explain the Ne decline for the coastal species T. aduncus, they cannot easily explain the Ne increase for the broadly distributed T. truncatus. Global climate change could have also affected dolphins through food availability due to alterations in currents, upwelling, and productivity, and through the altered ecology of pathogens, competitors, and predators (Hoegh-Guldberg and Bruno 2010). It is notable that the Ne’s of some dolphin predators declined during the last glacial, which could lead to a surge in dolphin Ne. For instance, dolphins are preyed upon by some large sharks, most of which are restricted to the warmer tropical climates. In general, sharks in warm zones show genetic signatures of reduced Ne in the last glacial (O’Brien et al. 2013). Furthermore, killer whales show a decline in Ne at the same period (Moura et al. 2014). Killer whales are more abundant in T. truncatus’ habitat than T. aduncus’ habitat at the present (Forney and Wade 2006). If these species had similar distributions during the last glacial, T. truncatus may have benefited more than T. aduncus from killer whale’s decline in the last glacial. Although dolphins are a relatively minor part of the diet of killer whales today, it is possible that climate changes indirectly caused the contrasting trends of Ne between T. truncatus and T. aduncus by differentially impacting their predators.
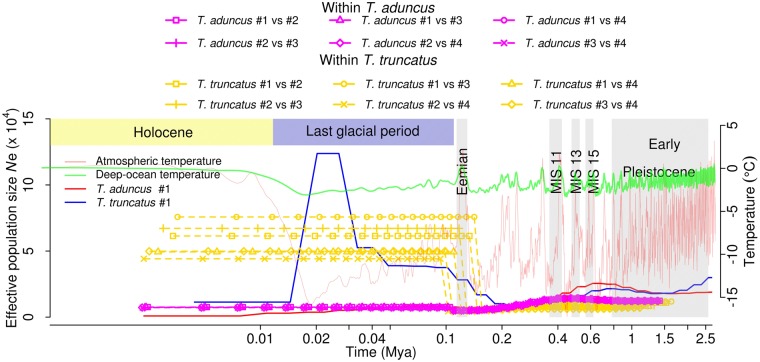
Temporal changes in Ne can also be caused by alterations in population structure or gene flow between populations (Mazet et al. 2016). Marine mammals with their complex patterns of ancestry can especially be influenced by such processes (Foote and Morin 2016). To examine this possibility in the dolphins, we conducted a PSMC analysis of pseudo-diploids artificially constructed from genomes of two different individuals (see Materials and Methods). Pseudo-diploids made from two different T. aduncus individuals and actual T. aduncus individuals show almost identical temporal trends of Ne (fig. 3), confirming a lack of population structure in this species. For (Northwest Pacific) T. truncatus, however, all pseudo-diploids show a much greater Ne from 0.1 to 0.2 Ma to the present (fig. 3), which is a sign of population structure, echoing a previous genetic-marker-based study that showed population structure of T. truncatus in Northeast Atlantic despite no obvious barrier to gene flow (Louis et al. 2014) and a report that coastal T. truncatus exhibits strong population structure even in geographically close populations (Martien et al. 2012). Our pseudo-diploid result, coupled with the hump in Ne from the real T. truncatus individuals during the last glacial (fig. 2), suggests a reduction in gene flow among T. truncatus populations in this period (Mazet et al. 2016), which was potentially caused directly or indirectly by the global cooling.
Fig. 3.
Pseudo-diploid analysis using PSMC suggests population structure in (Northwest Pacific) T. truncatus (yellow lines) but not T. aduncus (violet lines).
In line with the population structure inferred from the principal component analysis (supplementary fig. S2, Supplementary Material online), PSMC analysis shows an earlier substantial rise in Ne for pseudo-diploids constructed between Louisiana and Northwest Pacific T. truncatus individuals than for pseudo-diploids between Northwest Pacific individuals (supplementary fig. S4, Supplementary Material online). SMC++ split analysis using the joint frequency spectrum confirms the PSMC results of pseudo-diploids by showing much earlier split times between T. truncatus individuals than between T. aduncus individuals (supplementary fig. S5, Supplementary Material online). A previous study around the main Hawaiian Islands suggested hybridization between T. truncatus and T. aduncus (Martien et al. 2012). We thus also used PSMC to analyze pseudo-diploids constructed between T. truncatus and T. aduncus individuals. We found that the split times between the two species (≥0.3 Ma; supplementary fig. S6, Supplementary Material online) were much earlier than the split times found within T. truncatus (supplementary fig. S4, Supplementary Material online). The Ne’s of interspecific pseudo-diploids rose substantially in a step-wise manner, suggesting postdivergence gene flow (Song et al. 2014, 2017). The timing of the initial increase in Ne is thought to approximate the split time, and the presence of a series of split times instead of a single abrupt split time for all sampled individuals suggests a stepwise process of splitting up between the two species and/or ancient introgressions. However, more widespread sampling of individuals especially from populations that have been the focus of long-term observational studies will be required to acquire a better understanding of population structure. A recent study by Gray et al. (2018) supports this stepwise process of splitting of Tursiops lineages between 1 and 0.7 Ma.
In summary, our population genomic analysis revealed contrasting temporal trends of Ne in the last glacial between two closely related dolphin species and potentially even between populations within a single species, likely due to complex and often idiosyncratic ecological interactions that vary between species or populations, including for example changes in predator population sizes and migration rates among populations. Such variations make it possible for closely related species and populations to respond drastically differently to the same climate event. The pattern reported here is unlikely to be unique to bottlenose dolphins, because similar contrasts were previously inferred for sea snails (Albaina et al. 2012) and beltfish (He et al. 2015) on the basis of much smaller data and in minke whales on the basis of genomic data of fewer samples (Kishida 2017). Hence, predicting the impact of climate change on a particular species or population may be difficult without a much greater understanding of the specific ecological and biological factors involved.
Materials and Methods
Spatial Distribution Data
Spatial distribution data for >50 thousand species were collated by the International Union for Conservation of Nature (IUCN) as a resource to accompany the Red List of Threatened Species. This data set is available for download as .shp files at http://www.iucnredlist.org/technical-documents/spatial-data; last accessed May 25, 2018. We downloaded the data corresponding to Marine Mammals Group on December 29, 2016. Because this shapefile contains data for many marine mammals, we subsetted the data to the spatial distribution of T. truncatus and T. aduncus using the R package maptools. The spatial locations of the sampling points were then added to the figure. Dolphin images were added to the spatial distribution map.
Dolphin Samples
T. aduncus and T. truncatus were considered monospecific until the recognition of two species on the basis of genetic evidence and morphology (osteology and external morphology including beak morphology, dorsal fin shape, and the presence/absence of ventral spotting in adults; Wang et al. 1999, 2000a, 2000b). More recently, molecular evidence suggests that T. aduncus, long-beaked common dolphin Delphinus capensis, and striped dolphin Stenella coeruleoalba form a monophyletic clade that is sister to T. truncatus (Leduc et al. 1999; Vilstrup et al. 2011; Moura et al. 2013).
Tissue samples of four individuals of T. aduncus were obtained from stranded dead individuals on the coast of Jeju Island, Korea. Sex was recorded in the field, and confirmed by subsequent genetic analysis. We generated whole-genome sequencing data at ∼180× coverage (based on an estimated 2.29 Gb genome size; supplementary table S1, Supplementary Material online) for one individual and resequencing data of 22–32× coverage for the remaining three individuals (supplementary table S1, Supplementary Material online). These data were supplemented with publicly available sequences of six individuals of T. truncatus (four from Northwest Pacific and two from Northwest Atlantic) downloaded from the Short Read Archive (SRA). Sampling of T. truncatus from the Northwest Pacific is widespread, spanning over 1,000 km. Details of the coverage, sex, and SRA accession numbers of all samples are provided in supplementary table S1, Supplementary Material online.
Genomic DNA Library Construction, Sequencing, and Assembly
Genomic DNA was extracted from muscle tissues using the DNeasy Blood & Tissue kit (Qiagen, Germany). Extracted DNA was quantified by the Quant-iT BR assay kit (Invitrogen). For the reference genome of T. aduncus, we constructed short-insert (350 and 550 bp with 2 × 251 bp reads) and long-insert (5 and 10 kb with 2 × 101 bp reads) libraries using the standard protocol provided by Illumina (San Diego, USA). Three additional T. aduncus individuals were subjected to 2 × 150 bp paired-end sequencing of a 350 bp insert library. The raw reads were preprocessed using Trimmomatic v0.33 (Bolger et al. 2014) and Trim Galore (Martin 2011), in which reads containing adapter sequences, poly-N sequences, or low-quality bases (below a mean Phred score of 20) were removed. For the reference genome assembly of T. aduncus, we used all preprocessed reads from four (350 bp, 550 bp, 5 kb, and 10 kb) libraries and employed ALLPATHS-LG v52488 (Gnerre et al. 2011). Next, gaps (any nucleotide represented by “N” in scaffolds) were closed using GapCloser, a module of SOAPdenovo2 (Luo et al. 2012). For further analyses, any scaffold with >0.04% hits belonging to bacterial genomes from NCBI (release of Nov. 2016), downloaded from the RefSeq microbial genomes ftp site (Tatusova et al. 2014), was removed. A draft genome was assembled for T. aduncus using both paired-end and mate-pair libraries. The final assembled genome is 2.5 Gb in total length, close to the estimate by K-mer analysis (2.29 Gb), and was composed of 16,188 scaffolds (≥1 kb in their length) with an N50 value of 1,254 kb (supplementary table S2, Supplementary Material online). The assembly was found to be of good quality; 95.2% of the core eukaryotic genes (based on 248 core essential genes) from CEGMA v2.5 (Parra et al. 2007), 89.7% of the core vertebrate genes (based on 233 one-to-one orthologs in vertebrate genomes) previously identified (Hara et al. 2015), and 92.1% of the metazoan single-copy orthologs (based on 843 genes) from BUSCO v1.22 (Simão et al. 2015) could be identified in the genome assembly. Among these 92.1% of genes identified, 82.3%, 5.0%, and 4.7% are complete single-copy, complete duplicate, and partial genes, respectively.
Read Mapping and Variant Calling
The genome assembly and annotation of T. truncatus release-87 were downloaded from Ensembl. The sequencing reads from each dolphin species were mapped to the genome assembly using the Burrows–Wheeler aligner BWA-MEM (Li 2013), with default settings. PCR duplicate reads were collapsed using the rmdup option in SAMtools (v 0.1.19; Li et al. 2009). Regions of the genome annotated as repeats by RepeatMasker (http://www.repeatmasker.org/; last accessed May 25, 2018) or mitochondrial DNA transposed to the nuclear genome (numts) identified by blast hits to the mitochondrial sequence (at an E-value threshold of 10−3) were removed from further analysis. We identified variable sites across the genome using the “mpileup” command in samtools.
Identification of Sex Chromosome Markers and Sexing of Individuals
We searched for the previously identified Y chromosome marker from the SRY gene (Palsbøll et al. 1992) in sequencing reads to identify/validate the sex of each individual. The inferred sex always matched the previously recorded sex in the field. The scaffolds of the genome assembly from a female T. truncatus individual were grouped into autosomal and X chromosome scaffolds based on the ratio of sequencing coverage between female and male individuals, using a previously described approach (Bidon et al. 2015). Briefly, the mean coverage of all scaffolds longer than 200 kb was calculated using BEDTools (Quinlan and Hall 2010). Scaffolds that consistently showed a female/male coverage ratio near two were deemed to be from the X chromosome. As a negative control, we also examined the coverage ratio of male/male and female/female individuals.
Demographic History and Population Structure
Reads mapped to autosomal scaffolds longer than 100 kb were analyzed using the program PSMC (Li and Durbin 2011) to investigate temporal trends of Ne, under the assumption of a generation time of 21 years and a mutation rate of 1.21 × 10−9 per site per year (i.e., 2.5 × 10−8 per site per generation) for both dolphin species (Taylor et al. 2007; Yim et al. 2014). It is recognized that using ∼75% of the genome or more in the PSMC analysis results in reliable results (Nadachowska-Brzyska et al. 2016). Our PSMC analysis used 74% of the assembled genome after all the filtering. That is, regions of the genome annotated as repeats by RepeatMasker or mitochondrial DNA transposed to the nuclear genome (numts) identified by blast hits to the mitochondrial sequence (at an E-value threshold of 10−3) were removed from PSMC analysis. Only autosomal scaffolds longer than 100 kb were used for the analyses. Another common source of error in the PSMC analysis is the use of low-coverage genome sequences. By down-sampling a high-coverage genome sequence to various levels of coverage, Nadachowska-Brzyska et al. (2016) showed that results start to differ from the original result when the coverage is below 20×. In our study, we generated whole genome sequencing data from four individuals of T. aduncus, with 22–180× coverages (supplementary table S1, Supplementary Material online), and all four individuals show similar Ne plots from the PSMC analysis. We used publicly available data sets from six T. truncatus individuals in this study, including four from Northwest Pacific and two from Northwest Atlantic. Among the four Northwest Pacific samples, one has a 43× coverage and was previously analyzed using PSMC (Yim et al. 2014). Our reanalysis of this sample using PSMC yielded similar results. Three additional Northwest Pacific samples have 10–12× coverages and may not be ideal for the PSMC analysis. Nonetheless, we found the Ne plots from these three individuals highly similar to that of the 43× coverage individual. Hence, the relatively low coverage does not seem to affect the PSMC analysis in the present case. Of the two Northwest Atlantic samples, one has < 1× coverage and is excluded from PSMC, MSMC, and SMC++ analysis. The other Northwest Atlantic sample has 34× coverage, but interestingly its Ne plot is similar to that of T. aduncus instead of Northwest Pacific T. truncatus. This observation is likely genuine rather than artifactual, because even a variation in coverage from 10× to 43× among Northwest Pacific T. truncatus samples does not affect the Ne trend. We found that using the T. aduncus or T. truncatus genomes as the reference for mapping reads and performing the PSMC analysis did not show any difference. To ensure comparability of the results, the T. truncatus genome was used as the reference for read mapping in PSMC analysis of both species.
We parameterized the initial PSMC runs with 64 atomic time intervals and 28 free intervals with the –p parameter set to “4 + 25 × 2 + 4 + 6.” This parameter set was previously used for PSMC analysis of cetaceans (Moura et al. 2014). With these parameters, at least ten recombination events were inferred to occur in each of the time intervals within 20 iterations. However, these settings did not have sufficient resolution during the estimated split time of the species. Hence, we sought to optimize the –p and –t parameters that would provide a higher resolution of the data points. To choose an appropriate set of parameters allowing for increased resolutions, we tried a range of different values for –p (number of free atomic time intervals) and –t (upper limit of time to most recent common ancestor) parameters. For each set of parameters, we checked whether at least ten recombination events (fifth column of the PSMC output file) were inferred to occur in each of the time intervals within 20 iterations. Only those parameter combinations that satisfied these criteria were considered to avoid over-fitting. We noticed that, for higher values of the –t parameter, too few recombination events were inferred for the same values of the –p parameter. Hence, we used the PSMC default value of 5 for the parameter –t in our analysis. Increasing the –t parameter did not lead to any noticeable Ne increase in the PSMC plot into the very distant past. We used fewer time intervals and found that the parameter setting “20 + 13 × 2 + 4 + 6” was able to provide good resolution and still showed at least ten recombination events in each of the time intervals within 20 iterations.
To evaluate the robustness of the results to different parameters of mutation rate and generation time, we scaled the PSMC results for both species at generation times of 14, 21, and 28 years and mutation rates of 1.0 × 10−8, 1.5 × 10−8, 2.5 × 10−8, and 5 × 10−8 per site per year. The range of generation time was chosen based on realistic values across cetacean species (Taylor et al. 2007). Furthermore, we used 100 randomizations for each of the individuals in PSMC to obtain confidence intervals of Ne. Segments used for the 100 bootstraps were of 100 kb each. The contrasting patterns in Ne trends between the two species hold for a wide range of parameters. The parameters required to generate similar Ne trends in the two species would be highly unrealistic.
To examine the potential role of population structure in generating the observed temporal trends in Ne, we performed the pseudo-diploid analysis (Li and Durbin 2011; Prado-Martinez et al. 2013). Haploid sequences were generated from each individual using the seqtk program provided by Heng Li (https://github.com/lh3/seqtk; last accessed May 25, 2018). Sites with low quality were excluded using the flag –q 20, and one of the alleles was randomly chosen at heterozygous sites using the flag –r. The haploid genomes from two individuals were merged to create pseudo-diploid genomes, which were then subjected to the PSMC analysis.
Although the PSMC analysis is able to infer the temporal trends of Ne based on the whole genome sequence of a single individual, the MSMC (Multiple Sequentially Markovian Coalescent) method is able to integrate information from multiple individuals of the same species to make more robust inferences (Schiffels and Durbin 2014). We used the script bamCaller.py provided along with the MSMC program to separately identify SNPs from each dolphin species. Statistical phasing of the SNP calls by the program shapeit requires at least ten individuals from each population when a SNP reference panel is unavailable. However, we do not have a reference panel. Although a previous study (Zhou et al. 2017) phased SNP data with only four individuals, they had access to a recombination rate map, which is not available for the dolphins. Hence, we converted our data to MSMC input format without phasing, using the script generate_multihetsep.py. The MSMC program was run with the option of “fixedRecombination” for each species separately. We did not study population divergences using the relative gene flow analysis because using unphased data can bias this analysis (Schiffels and Durbin 2014).
The MSMC method is better suited for data in which the phases of the genotype calls are known. However, in our case, accurate phasing is not possible due to the lack of a large number of samples. In such cases, the SMC++ method is an excellent alternative to the MSMC analysis. In addition to the genomic distribution of variable sites, SMC++ also utilizes information from the site frequency spectrum across individuals and has been shown to perform more reliably than MSMC, especially when using unphased data (Terhorst et al. 2017). In addition to not requiring phased data, SMC++ is also able to infer split times in diverged populations. The VCF (Variant Call Format) file with genotype information for all sites, including nonvariant sites, was generated using samtools. This file was converted to SMC format using the vcf2smc option in SMC++ program for all individuals of each population separately. The repeat annotation and numt regions identified in the genome were passed as the mask file to vcf2smc command with the –m flag. Population size histories were estimated using the estimate option in SMC++ with a mutation rate of 1.21 × 10−9 per site per year for both dolphin species (Taylor et al. 2007; Yim et al. 2014). Because the ancestral state could not be reliably reconstructed for our focal species, we used only the folded frequency spectrum option while running SMC++. Documentation for SMC++ suggests various parameters that need to be experimented with to identify the settings best suited for each data set. With the default settings, our data set was not generating results for more recent times. Hence, we set the option –t1 to ten generations to extend the results to recent times. However, this resulted in overfitting of the curves as well as too much oscillation. To correct this issue, we decreased the regularization penalty to a value of 5 based on recommendations in the SMC++ documentation. The parameter for thinning was set at 3,000 and “ftol” was increased to 0.01. With these settings, we were able to extend the estimates of population size by SMC++ to recent times.
We next inferred the split times for the species pair using the split option of SMC++ after estimating the joint frequency spectrum for both species with the same parameter settings as described in the previous paragraph. The split option of SMC++ provides an independent validation of the results from the pseudo-diploid analysis from PSMC. The latest version of SMC++ (version 1.11.1) does not work with data from single individuals because the site frequency estimates from single individuals are unreliable. Hence, we grouped two randomly selected individuals of T. truncatus (from Northwest Pacific) into population 1 and two other individuals of T. truncatus (from Northwest Pacific) into population 2. These two populations were used to infer the split times between T. truncatus individuals. Similarly, T. aduncus individuals were randomly grouped into two populations and used to estimate the split times among T. aduncus individuals.
The population structure of all sampled individuals was investigated using Principal Component Analysis (PCA) with the software ngsTools (Fumagalli et al. 2014). First, the posterior probability of genotypes was calculated using ANGSD (Korneliussen et al. 2014). These posterior probability values were then used to estimate the covariance matrix between individuals using the program ngsCovar. Finally, this covariance matrix was decomposed into eigenvectors using R to obtain principal components. Pairs of principal components were compared to identify patterns of clustering of individuals.
Population Genetic Statistics
Single nucleotide variants at biallelic sites that had no missing data in any of the individuals were used to estimate population genetic statistics such as Fst (differentiation), dxy (divergence), and π (nucleotide diversity) using Heirfstat (Goudet 2005). We further used ANGSD (Korneliussen et al. 2014) and ngsTools (Fumagalli et al. 2014) to estimate population genetic summary statistics. The values estimated from the two methods were in agreement.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank researchers who provided public access to sequencing data through the Short Read Archive, Yun Song and Jonathon Terhorst for advice on SMC++ analysis, and Andy Foote for valuable comments on an early draft. Byung-Yeob Kim provided some T. aduncus tissue samples. This research was supported by a grant from the Collaborative Genome Program (20140428) of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries, Korea and a National Research Foundation of Korea (NRF) grant from the Korean government (MSIP) (NRF-2015R1A4A1041997) to J.K.P. J.Z. is supported by U.S. National Institutes of Health research grant R01GM120093.
References
- Albaina N, Olsen JL, Couceiro L, Ruiz JM, Barreiro R.. 2012. Recent history of the European Nassarius nitidus (Gastropoda): phylogeographic evidence of glacial refugia and colonization pathways. Mar Biol. 1599: 1871–1884. [Google Scholar]
- Bidon T, Schreck N, Hailer F, Nilsson MA, Janke A.. 2015. Genome-wide search identifies 1.9 Mb from the polar bear Y chromosome for evolutionary analyses. Genome Biol Evol. 77: 2010–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3015: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote A, Morin P.. 2016. Genome-wide SNP data suggest complex ancestry of sympatric North Pacific killer whale ecotypes. Heredity 1175: 316–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forney KA, Wade P.. 2006. Worldwide distribution and abundance of killer whales. In: Estes DA, Brownell RL Jr, DeMaster DP, Doak DF, Williams TM (eds) Whales, whaling and ocean ecosystems. Berkeley, CA: University of California Press, pp. 145–162. [Google Scholar]
- Fumagalli M, Vieira FG, Linderoth T, Nielsen R.. 2014. NgsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics 3010: 1486–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 1084: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. 2005. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 51: 184–186. [Google Scholar]
- Gray HWI, Nishida S, Welch AJ, Moura AE, Tanabe S, Kiani MS, Culloch R, Möller L, Natoli A, Ponnampalam LS, et al. 2018. Cryptic lineage differentiation among Indo-Pacific bottlenose dolphins (Tursiops aduncus) in the northwest Indian Ocean. Mol Phylogenet Evol 122:1–14. [DOI] [PubMed] [Google Scholar]
- Hale PT, Barreto AS, Ross GJB.. 2000. Comparative morphology and distribution of the aduncus and truncatus forms of bottlenose dolphin Tursiops in the Indian and Western Pacific Oceans. Aquat Mamm. 26:101–110. [Google Scholar]
- Hara Y, Tatsumi K, Yoshida M, Kajikawa E, Kiyonari H, Kuraku S.. 2015. Optimizing and benchmarking de novo transcriptome sequencing: from library preparation to assembly evaluation. BMC Genomics 16:977.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Zhang A, Weese D, Li S, Li J, Zhang J.. 2015. Demographic response of cutlassfish (Trichiurus japonicus and T. nanhaiensis) to fluctuating palaeo-climate and regional oceanographic conditions in the China seas. Sci Rep. 41: 6380.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg O, Bruno JF.. 2010. The impact of climate change on the world’s marine ecosystems. Science 3285985: 1523–1528. [DOI] [PubMed] [Google Scholar]
- Kishida T. 2017. Population history of Antarctic and common minke whales inferred from individual whole-genome sequences. Mar Mammal Sci. 332: 645–652. [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learmonth JA, MacLeod CD, Santos MB, Pierce GJ, Crick HQP, Robinson RA.. 2006. Potential effects of climate change on marine mammals. Oceanogr Mar Biol. 44:431–464. [Google Scholar]
- Leduc RG, Perrin WF, Dizon AE.. 1999. Phylogenetic relationships among the Delphinid cetaceans based on full cytochrome B sequences. Mar Mammal Sci. 153: 619–648. [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997.
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 4757357: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The sequence alignment/Map format and SAMtools. Bioinformatics 2516: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis M, Viricel A, Lucas T, Peltier H, Alfonsi E, Berrow S, Brownlow A, Covelo P, Dabin W, Deaville R, et al. 2014. Habitat-driven population structure of bottlenose dolphins, Tursiops truncatus, in the North-East Atlantic. Mol Ecol. 234: 857–874. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu YY, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 11: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martien KK, Baird RW, Hedrick NM, Gorgone AM, Thieleking JL, Mcsweeney DJ, Robertson KM, Webster DL.. 2012. Population structure of island-associated dolphins: evidence from mitochondrial and microsatellite markers for common bottlenose dolphins (Tursiops truncatus) around the main Hawaiian Islands. Mar Mammal Sci. 283: E208. [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 171: 10. [Google Scholar]
- Mazet O, Rodriguez W, Grusea S, Boitard S, Chikhi L.. 2016. On the importance of being structured: instantaneous coalescence rates and human evolution-lessons for ancestral population size inference? Heredity 1164: 362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura AE, Nielsen SCA, Vilstrup JT, Victor Moreno-Mayar J, Gilbert MTP, Gray HWI, Natoli A, Möller L, Rus Hoelzel A.. 2013. Recent diversification of a marine genus (Tursiops spp.) tracks habitat preference and environmental change. Syst Biol. 626: 865–877. [DOI] [PubMed] [Google Scholar]
- Moura AE, Van Rensburg CJ, Pilot M, Tehrani A, Best PB, Thornton M, Plön S, De Bruyn PJN, Worley KC, Gibbs RA, et al. 2014. Killer whale nuclear genome and mtdna reveal widespread population bottleneck during the last glacial maximum. Mol Biol Evol. 315: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadachowska-Brzyska K, Burri R, Smeds L, Ellegren H.. 2016. PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol Ecol. 255: 1058–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien SM, Gallucci VF, Hauser L.. 2013. Effects of species biology on the historical demography of sharks and their implications for likely consequences of contemporary climate change. Conserv Genet. 141: 125–144. [Google Scholar]
- Palsbøll PJ, Vader A, Bakke I, El-Gewely MR.. 1992. Determination of gender in cetaceans by the polymerase chain reaction. Can J Zool. 7011: 2166–2170. [Google Scholar]
- Parra G, Bradnam K, Korf I.. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 239: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, et al. 2013. Great ape genetic diversity and population history. Nature 4997459: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 266: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R.. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 468: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Song S, Sliwerska E, Emery S, Kidd JM.. 2017. Modeling human population separation history using physically phased genomes. Genetics 2051: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Sliwerska E, Kidd JM.. 2014. Population split time estimation and X to autosome effective population size differences inferred using physically phased genomes. BioRxiv. doi: 10.1101/008367. [DOI] [Google Scholar]
- Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I.. 2014. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 42(D1): D553–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Chivers SJ, Larese J, Perrin WF.. 2007. Generation length and percent mature estimates for IUCN assessments of cetaceans. Administrative report LJ-07-01. NOAA, La Jolla, USA; 2007. 24 pp.
- Terhorst J, Kamm JA, Song YS.. 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 492: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuiller W. 2007. Biodiversity: climate change and the ecologist. Nature 4487153: 550–552. [DOI] [PubMed] [Google Scholar]
- Vilstrup JT, Ho SY, Foote AD, Morin PA, Kreb D, Krützen M, Parra GJ, Robertson KM, De Stephanis R, Verborgh P, et al. 2011. Mitogenomic phylogenetic analyses of the Delphinidae with an emphasis on the Globicephalinae. BMC Evol Biol. 11:65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Chou LS, White BN.. 1999. Mitochondrial DNA analysis of sympatric morphotypes of bottlenose dolphins (genus: Tursiops) in Chinese waters. Mol Ecol. 810: 1603–1612. [DOI] [PubMed] [Google Scholar]
- Wang JY, Chou LS, White BN.. 2000. Differences in the external morphology of two sympatric species of bottlenose dolphins (genus Tursiops) in the waters of China. J. Mammal. 814: 1157–1165. [Google Scholar]
- Wang JY, Chou LS, White BN.. 2000. Osteological differences between two sympatric forms of bottlenose dolphins (genus Tursiops) in Chinese waters. J Zool. 2522: 147–162. [Google Scholar]
- Yim HS, Cho YS, Guang X, Kang SG, Jeong JY, Cha SS, Oh HM, Lee JH, Yang EC, Kwon KK, et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 46:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Massonnet M, Sanjak JS, Cantu D, Gaut BS.. 2017. Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proc Natl Acad Sci U S A. 144:11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.