Abstract
The Universal Gene Set of Life (UGSL) is common to genomes of all extant organisms. The UGSL is small, consisting of <100 genes, and is dominated by genes encoding the translation system. Here we extend the search for biological universality to three dimensions. We characterize and quantitate the universality of structure of macromolecules that are common to all of life. We determine that around 90% of prokaryotic ribosomal RNA (rRNA) forms a common core, which is the structural and functional foundation of rRNAs of all cytoplasmic ribosomes. We have established a database, which we call the Sparse and Efficient Representation of the Extant Biology (the SEREB database). This database contains complete and cross-validated rRNA sequences of species chosen, as far as possible, to sparsely and efficiently sample all known phyla. Atomic-resolution structures of ribosomes provide data for structural comparison and validation of sequence-based models. We developed a similarity statistic called pairing adjusted sequence entropy, which characterizes paired nucleotides by their adherence to covariation and unpaired nucleotides by conventional conservation of identity. For canonically paired nucleotides the unit of structure is the nucleotide pair. For unpaired nucleotides, the unit of structure is the nucleotide. By quantitatively defining the common core of rRNA, we systematize the conservation and divergence of the translational system across the tree of life, and can begin to understand the unique evolutionary pressures that cause its universality. We explore the relationship between ribosomal size and diversity, geological time, and organismal complexity.
Keywords: last universal common ancestor, ribosome, tree of life, multiple sequence alignment, ribosomal RNA, structural bioinformatics
Introduction
The biological world is united by <100 genes. Orthologous genes shared by all living systems make up the Universal Gene Set of Life (UGSL) (Harris, et al. 2003; Koonin 2003; Charlebois and Doolittle 2004). Universal genes signal functions with special importance in evolution, the origin of life, medicine and chemical biology. The UGSL is dominated by genes encoding the translation system.
The translation system is characterized by:
Ubiquity: Genes for translation exist in every living system and dominate the UGSL (Harris et al. 2003; Koonin 2003; Charlebois and Doolittle 2004).
Similarity: Structure and function of the translation system are universally conserved (Hsiao et al. 2009; Melnikov et al. 2012). The genetic code is essentially universal.
Antiquity: Prebiological macromolecules are preserved in the translation system (Agmon 2009; Davidovich et al. 2009; Kovacs et al. 2017; Lupas and Alva 2017).
Centrality: The translation system is a nexus, dominating the interactome (Butland et al. 2005).
Abundance: Ribosomal RNAs (rRNAs) and ribosomal proteins (rProteins) are the most abundant biological macromolecules in the known universe (Ortiz et al. 2006; Scott et al. 2010).
Expenditure: Translation consumes the bulk of cellular resources (Li et al. 2014) and defines biological productivity.
Complexity: Ribosomal complexity is a proxy for organismal complexity (Petrov et al. 2014).
Here we quantitate universality of position and conformation of macromolecules encoded by the USGL. Sampling cytoplasmic ribosomes of all extant species, we explicitly define Common Core rRNA and begin to systematize conservation and divergence of the translational system in three dimensions across the tree of life. Conserved rRNA, along with universal rProteins, form the structural and functional basis of all cytoplasmic ribosomes and are essential foundations of life on earth. It is important to explicitly define conservation and to characterize its extent because conservation is commonly used as a proxy for significance; inaccurate or cherry-picked portrayals of conservation can lead to incorrect conclusions.
New Approaches
We develop methods to differentiate common core rRNA from rRNA that is domain or phyla or species specific. We have constructed and exhaustively aligned rRNA sequences from a database we call the Sparse and Efficient Representation of Extant Biology (the SEREB database). This database contains complete and cross-validated rRNA and rProtein sequences of species that are chosen as far as possible, to efficiently sample all phyla (Petrov et al. 2014). We believe the SEREB database will be useful for a variety of evolutionary studies including phylogenetic reconstructions and ancestral sequence reconstructions. This database documents an astounding degree of conservation of the translation system across the tree of life. With this database we can more successfully understand the unique evolutionary pressures that conserve translation and can explore biological variation and the acquisition of complexity.
We propose new statistical approaches for characterizing rRNA structure and thus determine which specific elements of rRNA are universally conserved in three-dimensions. We establish the pairing adjusted sequence entropy (PASE), which characterizes conservation of sequence simultaneously with conservation base pairing. Unpaired nucleotides are analyzed by standard measures of nucleotide similarity. Nucleotide pairs are analyzed by their adherence to rules of covariation (Holley et al. 1965; Shang et al. 2012). The resulting net measure of similarity controls for the differential restraints on the sequences of base paired nucleotides compared with sequences of unpaired nucleotides in rRNAs with conserved 3D structure. For canonically paired nucleotides the unit of structure is the nucleotide pair. For unpaired nucleotides the unit of structure is the nucleotide. PASE, combined with structural comparisons, was used here to define the common core of cytoplasmic rRNAs. The data suggest that since the Last Universal Common Ancestor (LUCA), rRNA has accreted onto the common core, primarily in eukaryotic lineages.
Results
Defining the Common Core
The rRNA of the common core is a collection of “elements”, described as helices, junctions, and loops, that are found in cytoplasmic ribosomes of all extant species. To characterize the extent of conservation of these elements in all extant species we have established and continue to refine the SEREB database. The statistics of the SEREB database represent information from across the tree of life.
The SEREB database is distinguished from conventional rRNA sequence databases that contain large numbers of entries, some of which are partial, or contain intervening sequences, errors and redundancies. The SEREB database contains only intact and accurate rRNA sequences; for some species multiple partial and fragmented rRNA sequences from disparate sources were assembled and cross-validated. Newly discovered phyla, such as Lokiarchaea (Da Cunha et al. 2017), are incorporated into the SEREB database as they become available. The current SEREB database contains 133 species. The list of SEREB organisms and the SEREB rRNA multiple sequence alignment (MSA) are provided in the Supplementary Material (supplementary table S1, supplementary dataset S1, Supplementary Material online).
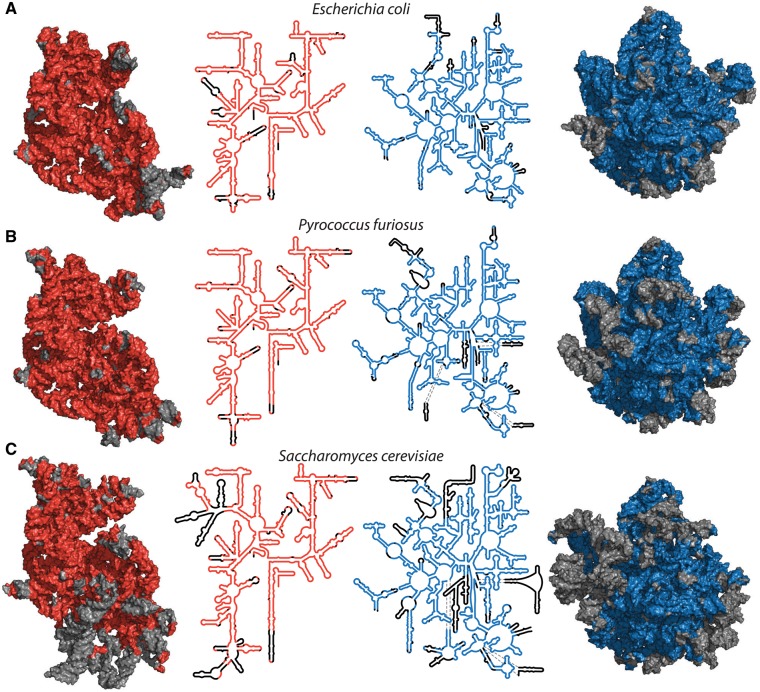
The rRNA common core is a 3D construct, and is an outcome of pioneering work of Ada Yonath, who showed that ribosomal particles from a variety of species can be crystallized and characterized by x-ray diffraction (Wittmann et al. 1982; Shevack et al. 1985). The common core is projected onto secondary and 3D structures of representative species in figure 1 and in supplementary figure S1, Supplementary Material online [bacteria: Escherichia coli, archaea: Pyrococcus furiosus, eukarya: Saccharomyces cerevisiae].
Fig. 1.
Common core of cytoplasmic rRNAs mapped onto 3D and secondary structures of a bacterium, an archaeon and a eukaryote. (A) RNAs of the bacterium E. coli, (B) the archaeon P. furiosus, (C) the eukaryote S. cerevisiae. Red (SSU, left) and blue (LSU, right) indicate common core rRNA. Black or gray indicates rRNA that is excepted from the common core and is variable in structure or absent from some species. Each subunit is viewed from the solvent exposed surface of the assembled ribosome, with the subunit interface directed into the page. A more detailed representation of these data, including nucleotide and helix numbers, is contained in supplementary figures S9 and S10, Supplementary Material online. E. coli: PDB ID 4V9D, P. furiosus: PDB ID 4V6U, and S. cerevisiae: PDB ID 4V88.
The rRNA Common Core
Around 90% of rRNA nucleotides of prokaryotes are incorporated into the common core. Most prokaryotic rRNA helices, 114 of 157 helices in the large subunit (LSU) and small subunit (SSU), are highly conserved in length and conformation in all species (supplementary table S2, Supplementary Material online), with few or no insertions and only subtle variation in nucleotide positions. The prokaryotic ribosome contains 11 helices with length polymorphism in one or more domains of life. We count 13 helices that undergo moderate divergence in structure and conformation. Ten helices vary significantly in structure and conformation, making their superimposition difficult. These helices generally contain sites of eukaryotic rRNA expansions. Additionally, there are nine helices that are completely absent from some members of the database.
rRNA that is excluded from the common core consists of 1) variable regions of helices associated with helical length polymorphism, 2) small variable bulges, 3) eukaryotic expansions and some regions immediately surrounding them, and 4) the 5’ and 3’ rRNA termini. As expected from previous work, (Ware et al. 1983; Bachellerie and Michot 1989; Gerbi 1996) divergent rRNA helices and helical extensions are restricted to the surface and nonfunctional regions of the ribosome. This observation is consistent with models of rRNA growth and evolution in which new rRNA is added onto old rRNA with minimal disturbance to the structure and function of existing rRNA (Petrov et al. 2014, 2015). Exceptions to this surface rule are the H16-H17-H18 group (supplementary fig. S2, Supplementary Material online) and the H54-H55 group. These helical elements appear to have expanded and/or contracted in their central regions, which are not on the surface of the ribosome.
Multiple Sequence Alignments
The process of characterization of the common core utilizes both structural and sequence information. The number of species whose rRNA sequences is known is very large (Cannone et al. 2002; Quast et al. 2012) compared with the number of species whose 3D ribosomal structures are known. Experimental structures of approximately 20 species have been determined (Ban et al. 2000; Yonath 2002a,b; Selmer et al. 2006). Accurate, structure-aware, sequence alignments were used to define the rRNA common core structure.
MSAs were performed by exhaustively iterating several recursive methods. Initial domain-specific MSAs were performed with MAFFT (Katoh and Standley 2016), then iterated with structure-based methods including PASE, local structural divergence (LSD), and global structural divergence (GSD) (see below) in analogy with previous structure-based alignments of proteins (Pei et al. 2008). Information from experimentally determined and thermodynamically predicted secondary structures was also incorporated into the MSA. Optimized domain-specific MSAs were combined and reoptimized in a universal MSA.
The final universal MSA has high completeness and accuracy. In the MSA, 76% of LSU E. coli rRNA and 84% of SSU rRNA are in universal columns. A universal column corresponds to a nucleotide with universal position, but possibly with polymorphic identity. When an allowance for 5% missing nucleotides is implemented, the numbers increase to 88% (LSU) and 90% (SSU) (table 1). The 5% tolerance reduces the impact of sequencing errors and rare idiosyncratic indels.
Table 1.
The rRNA Common Core.
Number of nucleotides in the common core. LSU includes the 5S rRNA.
Common core normalized to number of nucleotides of E. coli. (E. coli LSU = 3024 nts, includes 5S, SSU = 1542 nts).
Net number of nucleotide columns in the SEREB rRNA MSA including insertions.
Pairing Adjusted Sequence Entropy
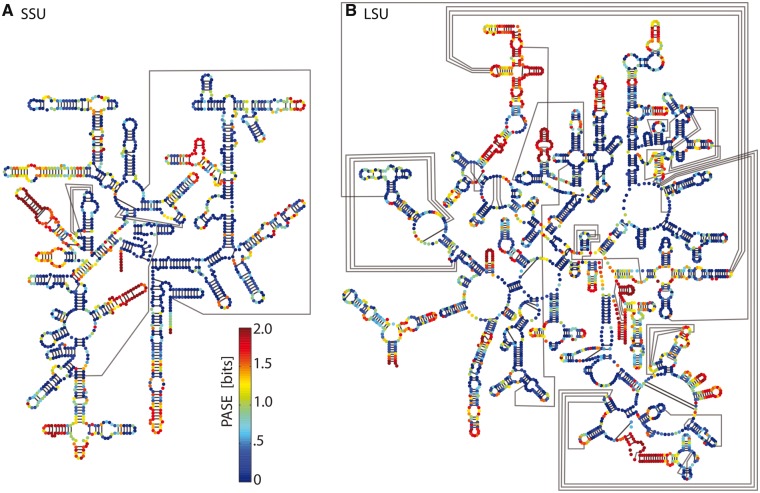
We developed a statistic called PASE (equation 4) that combines information on nucleotide identity with information on base pairing. A mapping of PASE onto the secondary structure of the E. coli rRNA is shown in figure 2. PASE is an extension of Shannon entropy (Shannon 1948; Gatlin 1966, 1972); PASE characterizes unpaired nucleotides by conventional conservation of identity. PASE characterizes paired nucleotides by their adherence to covariation. A canonically paired nucleotide is considered conserved if it is always canonically paired, even if its nucleotide identity is not conserved. The assumption is that if a nucleotide remains canonically paired, local rRNA conformation is likely to be conserved even if nucleotide identity varies. In contrast, when a base is unpaired or paired noncanonically, conformation is likely to be conserved only if the base identity is conserved. This assumption is validated by the close correspondence of PASE and local superimposition statistics.
Fig. 2.
Conservation of rRNA structure across the tree of life. PASE in the SEREB database mapped onto E. coli rRNA secondary structures. PASE represents nucleotide identity conservation for unpaired nucleotides and base pairing conservation for paired nucleotides. Blue indicates highly conserved, green moderately conserved, and red, not conserved. (A) SSU rRNA. (B) LSU rRNA. Watson-Crick pairing interactions are indicated by black lines.
Initially, the Shannon Entropy was calculated for each column of the MSA using the typical method (Gatlin 1966, 1972), to estimate conservation of nucleotide identity. Then, PASE was calculated to estimate the conservation of canonical Watson-Crick and GU wobble base pairing (cWW as defined by Leontis and Westhof 2001). For nucleotides treated as unpaired (supplementary figs. S4 and S5, Supplementary Material online), the extent of conservation is defined by a gap adjusted entropy score (GASE). For nucleotides treated as paired, conservation is defined by Shannon entropy taken over the probability of canonical base pairing (PASE). An example of an rRNA helix mapped with PASE data is shown in supplementary figure S3, Supplementary Material online.
Local Structural Divergence
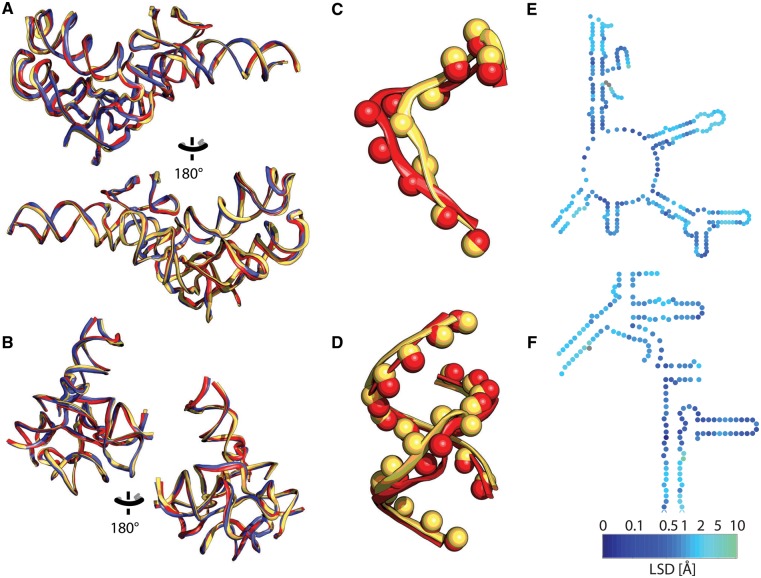
The MSA alone is not sufficient to define the common core because some rRNA is conserved in 3D space even though it lacks clear conservation of sequence or of canonical base pairing. We developed a structure-based statistic (LSD), to measure differences between positions of nucleotides in different ribosomes whose 3D structures are known. Each nucleotide is reduced to a pseudoatom (supplementary fig. S6, Supplementary Material online), as illustrated in figure 3C and D. A pseudoatom position is computed by the mass-weighted atomic positions of the phosphate-sugar linkage and the glycosidic nitrogen of the base. The pseudoatom definition smooths the structural distinction between pyrimidines and purines.
Fig. 3.
rRNAs from each domain of life, superimposed. The bacterium is red (E. coli), the archaeon is blue (P. furiosus), and the eukaryote is yellow (S. cerevisiae). rRNAs were superimposed based on (A) the PTC (LSU) and (B) the DCC (SSU). (C) Example pseudoatoms of E. coli and S. cerevisiae from the PTC. (D) Example pseudoatoms of E. coli and S. cerevisiae from the DCC. Mapping of LSD (difference in positions of pseudoatoms) onto relevant secondary structure of E. coli rRNA. (E) Secondary structures of the PTC and (F) DCC. Distances between pseudo atoms are indicated by color (dark blue lower divergence, to green higher divergence, gray indicates absence from S. cerevisiae).
LSD is calculated from pairwise differences in positions of corresponding pseudoatoms (of aligned nucleotides) of two superimposed rRNAs or rRNA elements (fig. 3). LSD is useful for assaying both MSA accuracy and actual structural divergence. During the process of MSA optimization, regions of rRNA with lower LSD were characterized by more accurate local MSA. Higher structural divergence suggested errors in the MSA, errors in rRNA sequence, or actual structural or conformational differences.
LSD is mapped onto primary, secondary and 3D levels of structure in supplementary fig. S7, Supplementary Material online). An example of a peak in LSD is h16 of the SSU, which bends one direction in prokaryotes and another direction in eukaryotes. An example of a more subtle conformational shift is in L12/P stalk of the LSU and the head of the SSU. An example of a loop with variable positions is seen at the termini of h6 which has variable length. Helix classifications are listed in supplementary table S2, Supplementary Material online.
GSD was obtained by taking the root mean square deviation of the LSD values over the LSU or SSU of two species (table 2). The GSD demonstrates that global differences within the common core are consistent with the major branching events in the phylogenetic tree. The GSDs indicate that the common core rRNAs of the bacterial and archaeal ribosomes are most similar, followed by those of the archaeal and eukaryotic ribosomes. The common cores for the bacterial and eukaryotic ribosomes are the most divergent. A full classification of helices is provided in supplementary table S2, Supplementary Material online. The LSD is elevated in loops that cap helices of variable length. Loops that are displaced by a great distance are excluded from the GSD calculation in figure 3.
Table 2.
rRNA Within the Common Core: GSD.
| GSDBAa | GSDAEb | GSDBEc | GSDBAd (Common Core) | GSDAEe (Common Core) | GSDBEf (Common Core) | |
|---|---|---|---|---|---|---|
| LSU | 5.26 | 4.37 | 6.36 | 3.52 | 3.94 | 4.82 |
| SSU | 5.17 | 4.78 | 6.23 | 4.84 | 4.33 | 5.87 |
GSD between E. coli (PDB ID 4V9D) and P. furiosus (PDB ID 4V6U).
GSD between P. furiosus and S. cerevisiae (PDB ID 4V88).
GSD between E. coli and S. cerevisiae.
GSD between bacteria and archaea (common core).
GSD between archaea and eukarya (common core).
GSD between bacteria and eukarya (common core).
Evaluation of the MSA
During optimization of the MSA, regions of poor MSA, PASE, or LSD statistics were inspected, adjusted, and resolved. Several rRNA elements, such as Helices 56, 57 and 88, do not align well in the MSA and are therefore red in the PASE mapping in figure 2. However, these regions show conserved structure by LSD and GSD, and therefore are included in the common core (fig. 1). These elements illustrate the truism that structure is more conserved than sequence (Illergard et al. 2009), even when sequence restrictions are relaxed by covariation as in PASE. In these regions, structure is conserved in the absence of any detectable signal for sequence conservation. Differences between PASE and GASE are mapped onto the E. coli rRNA secondary structure in supplementary figure S8, Supplementary Material online.
The final MSA was mapped onto the linear structure of the E. coli rRNA to compute alignment statistics on a per nucleotide basis. A gap-prorated version of the standard Shannon entropy (equation 2) was computed across the mapped alignment. The LSU has an average entropy of 1.04 and the SSU has an average entropy of 0.96 (table 3). For the common core only, the average entropy is 0.94 for the LSU and 0.84 for the SSU.
Table 3.
Conservation statistics.
| GASE Averagea | PASE Averagea | GASE Averageb | PASE Averageb | |
|---|---|---|---|---|
| LSU | 1.04 | 0.69 | 0.94 | 0.54 |
| SSU | 0.96 | 0.63 | 0.84 | 0.48 |
Computed over E. coli.
Computed over the common core.
PASE, computed over the E. coli mapped alignment, reveals a higher level of conservation than is evident in the standard Shannon Entropy. PASE shows that rRNA is more conserved than conventional sequence statistics indicate. The LSU has an average PASE of 0.69 and the SSU has an average PASE of 0.63 (table 3). When the common core alone is counted, the average PASE is 0.54 for the LSU and 0.48 for the SSU. These values indicate that the common core is highly conserved in cytoplasmic ribosomes across the tree of life.
Conservation within Common Core
Each nucleotide of a given species is classified as common core with conserved nucleotide identity, or with conserved cWW base pairing, or with conserved structure, or is classified as noncommon core. A detailed visualization of the relevant E. coli rRNA data is provided in supplementary figures S9 and S10, Supplementary Material online.
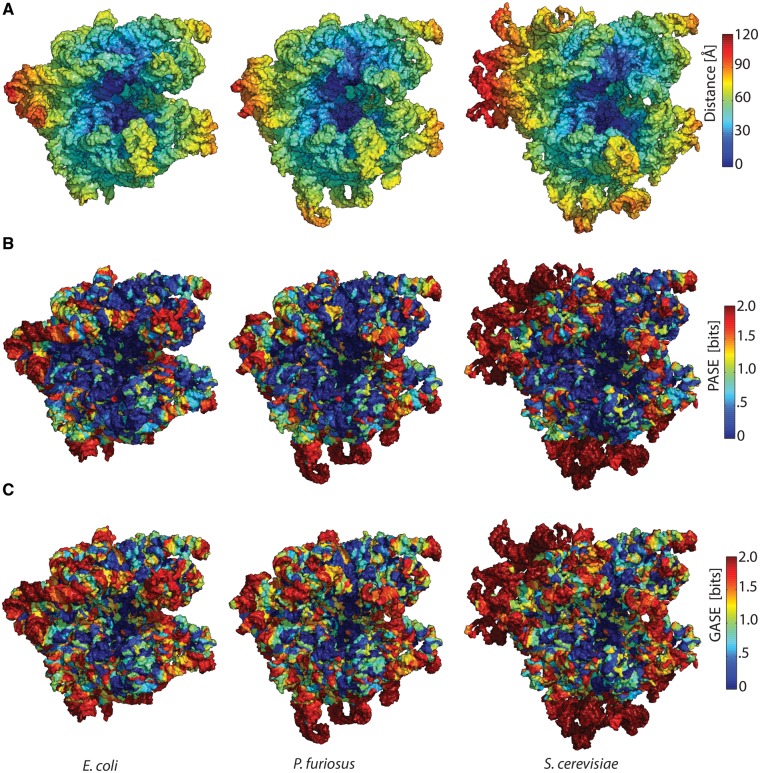
The degree of conservation of the common core shows clear and systematic variation in three dimensions. We have previously shown that it is useful to treat the ribosome as an onion (Hsiao et al. 2009). Various data are mapped and analyzed in the context of distance from functional foci (the PTC of the LSU and DCC or the SSU, fig. 4A). The majority of the common core is within 80 Å of the onion centers. A comparison of distance from functional foci and PASE reveals that extent of conservation is greatest near the onion centers and is most divergent on the onion periphery. PASE is mapped onto the ribosomal structures in figure 4B. The eukaryotic expansion segments (ESs) are clearly visible and are the most divergent regions of the ribosome, in dark red. PASE is an improved measure of conservation compared with nucleotide conservation entropy or GASE, which overstates the divergence of helical regions (fig. 4C).
Fig. 4.
Data mapping onto ribosomal structures of a bacterium (E. coli), an archaeon (P. furiosus) and a eukaryote (S. cerevisiae). (A) Distance: Assembled ribosomal subunits are represented as onions, using the PTC (LSU) or the DCC (SSU) as onion centers. rRNA is colored blue close to the centers of the onions, while red rRNA is remote. (B) PASE: rRNA is colored blue where PASE is low and red where PASE is high. (C) Standard nucleotide Shannon entropy: rRNA is colored blue where Shannon entropy is low and red where Shannon entropy is high. For the LSU, the center of the onion is the site of peptide bond formation. For the SSU, the center of the onion is the site of codon–anticodon interaction between mRNA and P-site tRNA.
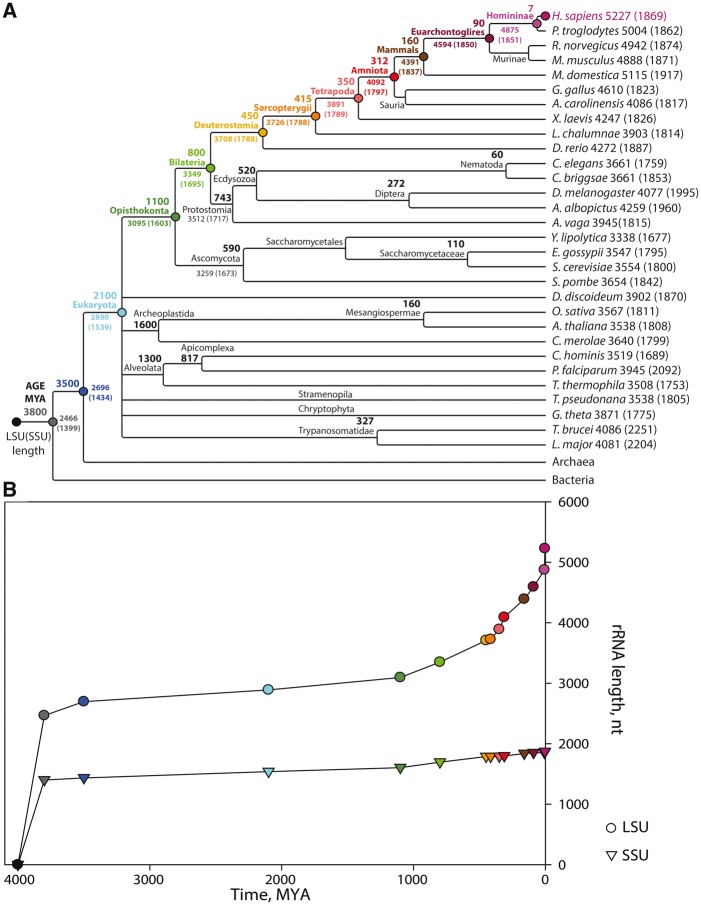
Ribosomal Size Evolution
The ribosomes of metazoans are larger than those of protists, which are larger than those of prokaryotes (Petrov et al. 2014). Birds and mammals (endothermic vertebrates) contain the largest ribosomes of all. Extant rRNA sizes along with estimates of sizes of various ancestral rRNAs are plotted in figure 5 in the context of evolutionary relationship and approximate time of evolutionary emergence (Hedges and Kumar 2009). For this representation, we have estimated sizes of ancestral rRNAs using the assumption that the most probable ancestral ribosomes contain rRNAs whose sequences align in both daughter species. Nonaligning nucleotides are assumed to be nonancestral. This sequence-based method will modestly underestimate sizes of ancestral rRNAs in part because sequence is less conserved than structure. The sequence-based method (fig. 5) gives LUCA rRNA sizes that are approximately 10% smaller than those of the structure-informed methods illustrated in figure 1.
Fig. 5.
rRNA size evolution. (A) A phylogenetic cladogram of eukaryotes contained in the SEREB database. Estimated dates of common ancestors (from Hedges et al. 2006) are indicated next to their names at the appropriate splits. The H. sapiens lineage is indicated by colored circles. (B) Estimated size evolution of ancestral LSU (circles) and SSU (triangles) rRNAs of the H. sapiens lineage. The colors in panel (B) point to data in panel (A). The timeline of the tree in panel (A) is not linear and does not to scale with panel (B). The origin of the ribosome is around 4.0 billion years ago.
The model timeline suggests that over evolution, rRNAs of prokaryotic lineages grew rapidly between the origin of life and LUCA and have remained essentially static in size for around 3.5 billion years. Within the limits of the model it appears that rRNAs within eukaryotic lineages have experienced distinct and well-separated phases of growth interleaved by a period of stasis. The rRNAs of endothermic vertebrate lineages began a more recent phase of rRNA growth that appears to be accelerating. We cannot exclude more complex models in which, for example, extinct ancestral prokaryotic ribosomes were larger or smaller than extant prokaryotic ribosomes. However, we consider those models less likely than the simpler model used here.
Eukaryotic ribosomes contain a shell that surrounds the common core (Melnikov et al. 2012). The eukaryotic shell is composed of both rProteins and rRNA (supplementary fig. S11, Supplementary Material online). Eukaryotic ESs emerge from common core rRNA at a few specific sites (Ware et al. 1983; Michot and Bachellerie 1987; Bachellerie and Michot 1989; Gerbi 1996). The ribosomes of endothermic vertebrae are further elaborated by rRNA tentacles that extend for hundreds of Å from the ribosomal surface.
In the Homo sapiens lineage, the period from the first multicellular organisms to vertebrates is characterized by a growth rate of about 0.65 nucleotides per million years. rRNA size increased more rapidly with emergence of endothermic vertebrates, with this effect being much more pronounced in the LSU than in the SSU. With the rise of endothermic vertebrates, the growth rate accelerated even more. The growth rate from the ancestor of vertebrates to the ancestor of mammals is about 2.5 nucleotides per million years. Human rRNA is about 370 nucleotides longer than rRNA of the last universal primate ancestor about 6 million years ago, which corresponds to a growth rate of 62 nucleotides per million years.
Common core rRNA is mapped onto H. sapiens rRNA in two and three dimensions in supplementary figure S12, Supplementary Material online. Entropy scores, radial distance from the functional centers of the LSU and SSU, and common core of the assembled human ribosome are visualized in supplementary figures S13 and S14, Supplementary Material online.
Discussion
The translation system provides our most extensive and complete view of universal biochemical processes and macromolecules. We define the common core as macromolecular assemblies with conserved 3D structure in essentially all living systems. The common core of the ribosome provides a window to the deepest roots of biology, which existed at and before LUCA. It has long been recognized that significant elements of rRNA secondary structure are conserved in all living systems (Clark et al. 1984; Hassouna et al. 1984; Gonzalez et al. 1985; Michot and Bachellerie 1987; Gerbi 1996; Mears et al. 2002). The concept of the common core has been discussed previously in Huang et al. (2005), Melnikov et al. (2012), Anger et al. (2013), and Doris et al. (2015). However, prior to this work, the common core of the ribosome has not been quantitatively defined or statistically described.
The rRNA Common Core
The rRNA common core is quantitatively characterized here at nucleotide resolution by comparisons of rRNA from ribosomes across the tree of life. The results incorporate information on sequence, secondary structure and 3D structure.
Around 90% of prokaryotic rRNA is contained in the common core. The common core contains the peptidyl transferase center, the polypeptide exit tunnel excluding the vestibule, the subunit interfaces, the A, P, and E sites and the tRNA rail, the bulk of the tRNA translocation machinery, the GTPase-associated region and essentially the entire SSU including the decoding center but excluding a few peripheral helices and the Shine-Dalgarno region. Antibiotic-binding sites (Auerbach et al. 2002; David-Eden et al. 2010) are contained within the common core, underscoring the subtlety required for species-specific inhibition. All rRNA pivoting positions identified by Fox and coworkers (Paci and Fox 2016) are contained within the common core.
Measures of Similarity
We have established a statistical measure of rRNA similarity (PASE) that simultaneously evaluates sequence and base pairing. PASE characterizes paired nucleotides by adherence to rules of canonical pairing and unpaired nucleotides by conventional conservation of identity. Helical regions are conserved in structure with low restraints on sequence, as long as base pairing is maintained (Smit et al. 2007). Therefore, efforts to assay rRNA conservation by sequence alone underestimate the extent of conservation. In helical regions, base pairs are the minimal units of selection (Parsch et al. 2000) consistent with the Kimura model of compensatory fitness interactions (Kimura 1985). In unpaired regions, nucleotides are the minimal units of selection, and their evolution tends to be driven by maximizing the fitness due to their functional or structural importance.
The rProtein Common Core
The rRNA common core is more extensive than the rProtein common core (Vishwanath et al. 2004). Bacteria, archaea, and eukarya contain domain-specific rProteins (Ban et al. 2014) that in some instances interact with common core rRNA. This observation suggests that some rProteins were remodeled after rRNA accreted and froze.
The SEREB Database
We have created a database of rRNA and rProtein sequences (the SEREB database) that samples extant biological species in a sparse, efficient and accurate manner. We have cross-validated the sequences and structurally refined a MSA of SEREB rRNA sequences (supplementary dataset S1, Supplementary Material online). The SEREB MSA, which is highly accurate, can be used as a seed for building MSAs from large databases.
Eukaryotic rRNA
Endothermic vertebrates appear to have entered a new and especially rapid phase of ribosomal growth (fig. 5). Ribosomes of these organisms contain rRNA “tentacles”, which are extended helical structures that attach to a protist-like base (Behrmann et al. 2015; Khatter et al. 2015) (fig. 1). These double helical rRNA tentacles are laden with defects such as bulges and mismatches. The tentacles are not tightly integrated with the ribosomal surface and appear to be dynamic and/or positionally disordered.
Why is rRNA larger in eukaryotes, especially in endothermic vertebrates? It is possible that rRNA expansions are directly adaptive in complex organisms, conferring immediate advantage in docking, trafficking, chaperoning, or biogenesis. Alternatively, in analogy with proposals for genome complexity (Lynch and Conery 2003), nonadaptive rRNA ESs and intervening spacer sequences may proliferate in the permissive environment of small populations; expansions would be eliminated by selection in large populations characteristic of protists or prokaryotes. This model does not explain the profound differences in the expansion of the LSU compared with the SSU. However, these differences might be attributable to gain function by ‘secondary deployment’ subsequent to nonadaptive rRNA expansion. Docking, trafficking and chaperoning functions generally involve the LSU rather than the SSU.
Summary
Our quantitative definition of common core rRNA can be used to help understand deep evolutionary events. Common core rRNA appears to be our best estimate of the rRNA contained of LUCA. Furthermore, the concept of the common core can be used to systematize and place in context the variability of the ribosome, especially in eukaryotes, which are characterized by numerous rRNA expansions. Each expansion of the common core can be analyzed for structural features and functional utility. One can address questions of where does rRNA grow, how does rRNA grow, and why does rRNA grow in some lineages but not others?
Materials and Methods
Sequences and Alignment
The SEREB database extends over all of major phyla, as far as available sequences allow. Appropriate rRNA sequences were compiled for both the LSU and the SSU. Sequences from representatives of major phyla were taken from SILVA (Silva et al. 2005), NCBI (2017), CRW (Cannone et al. 2002), and other databases and from the literature. Many of the required eukaryotic rRNA sequences were not accurately annotated or assembled. Accurate full-length sequences were in some instances were assembled from multiple independently sourced sequences. Contigs and chromosomal assemblies of DNA were examined to locate missing regions of rRNA. For species with fragmented rRNA, fragments were combined in the correct order. rRNA sequences were cross-validated by the MSA. Poorly aligning sequences were reinspected and adjusted.
Initial MSAs were performed independently for each of the three domains of life using MAFFT. To help reconcile the MSAs of the three domains, 3D superimpositions were used to establish correspondence of nucleotide sequences. The final three-domain MSA was optimized by an extended iterative process of 1) visual inspection of MSAs and superimposed structures, 2) minimizing PASE, LSD, and GSD while adjusting the MSA, 3) constraining experimentally determined and computational secondary structures and alignments of tetraloops, and 4) continuous reevaluation and refinement of rRNA sequences. This iterative method allowed unambiguous excision of eukaryotic ESs from the universal alignment and resulted in alignment of common core sequences with very few errors (supplementary dataset S1, Supplementary Material online).
Superimpositions
Three-dimensional structures of rRNAs were used to assist in defining the common cores of the LSU and SSU are: 3R8S and 4GD1 for E. coli, 3J2L and 3J20 for P. furiosus, 3U5D and 3U5B for S. cerevisiae, and 3J3F and 3J3D for H. sapiens. The structures were first placed into the same coordinate frame using the align function of PyMOL. Local superimpositions were performed independently for the LSU and SSU using a subset of the highly conserved nucleotides (in three dimensions) within each structure. The nucleotides used for superimpositions are localized near the PTC for the LSU (nucleotides 2061–2092, 2225–2245, and 2436–2501, for E. coli and their equivalents for the other structures) and the decoding center for the SSU (nucleotides 9–38, 548–569, 821–826, 872–927, 1390–1418, 1482–1530).
Pairing Adjusted Sequence Entropy
The standard sequence Shannon entropy (Gatlin 1966, 1972) recognizes conservation of nucleotide identity, but not conservation of paired nucleotides. We have developed PASE, which incorporates base pairing information. CG, GC, AU, UA, GU, UG pairs are equivalent in this scoring function. Unpaired bases are treated in the standard way (C, G, A, U are each different).
Entropy was calculated for each position, n, in the MSA. Since probabilities (P) are unknown, they are approximated by observed frequencies (f), which is the usual practice. Equation (1) is the standard Shannon entropy equation.
| (1) |
The variable c is the number of classes, which equals four, one for each nucleotide, A, C, U, and G. For each position, n, the frequencies of A, C, U, and G are calculated and used to compute the sequence Shannon entropy (HSE), which ranges from 0 to 2.
To account for gaps in the MSA, they were prorated, and were treated as a uniform distribution among all possible classes, such that a single gap character would count as 0.25 A, 0.25 C, 0.25, U, and 0.25 G. The gap adjusted Shannon entropy (HGASE) is calculated using equation (2). HGASE ranges between 0 and 2.
| (2) |
Each nucleotide is defined as paired (cWW) or unpaired at the level of the 3D structure by FR3D of E. coli (Sarver et al. 2008). For paired nucleotides the extent of conservation of base pairing, with no penalty for sequence variation, is determined. In addition to calculating the entropy of a single position in the alignment, we calculate the base pair shannon entropy (BPSE) accounting for the conservation of the canonical base pairs in each position per equation (3).
| (3) |
Each base pair is represented by two columns in an MSA. Within each column, an rRNA nucleotide can have one of five values, A, G, C, U, or gap. Therefore, there are 5 × 5 = 25 combinations of two characters. A base pair is defined as a dyad of CG, GC, AU, UA, GU, or UG. The fractions of dyads that fall into these classes are represented by fbp, therefore the fraction of dyads that do not fall into these classes is 1 – fbp. HBPSE ranges between 0 and 1. The structure of the E. coli ribosome was used to determine potential sites of base pairs, which are defined by FR3D as cWW (Sarver et al. 2008).
PASE, which represents the entropy of base pairs and single-stranded residues in a single statistic, is calculated as shown in equation (4). GASE is compared with twice the BPSE and the smaller value is retained. HPASE ranges between 0 and 2.
| (4) |
rRNA Size Evolution
A timeline of approximate ribosomal size as a function of evolutionary time was computed. LSU and SSU lengths are estimated from the SEREB database.
The accretion model implies that ribosomal rRNA has predominantly expanded in the species-specific lineages, although some reduction events are also possible. We use the assumption that the most probable ancestral macromolecules are similar to macromolecules common to daughter species; the most conservative changes are most likely. This subset of rRNA is, typically, most similar to the smaller rRNA (Petrov et al. 2014). This assumption, which is based on lack of our knowledge about the state of common ancestors at each node, overestimates the length of the ancestral rRNA if a particular accretion event occurred independently in two linages as a result of parallel evolution.
For size evolution we have used a sequence-based model of ancestral rRNAs that differs from our structure-based definition of the common core. Specifically, for each subset of sequences within a given group, we computed the number of columns that do not contain gaps at any position (i.e., describe a common state). This sequence-based method of estimating ancestral sizes is necessitated by the lack of 3D structures of ribosomes representing most phyla. This method will tend to slightly underrepresent the sizes of ancestral ribosome because structure is more conserved than sequence and because the MSA will always contain some level of error. The estimated time of a clade split was taken from TimeTree (Hedges et al. 2006; Hedges and Kumar 2009). Each time point represents a common ancestor.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Dr Emmanuel Tannenbaum. This work was supported by NASA Grant NNX16AJ29G. We thank Dr Vincent Daubin for helpful discussions.
Author Contributions
L.D.W. and A.S.P. conceived the study; A.S.P. and L.D.W. developed the theoretical PASE score; A.S.P. and C.R.B. and developed superimposition methodology; C.R.B. collected, analyzed, and visualized the data and generated most figures and tables; NAK generated 3D figures; P.I.P. and A.S.P. performed phylogenetic analysis; and L.D.W., C.R.B., A.S.P., P.I.P. and N.A.K. wrote the article.
References
- Agmon I. 2009. The dimeric proto-ribosome: structural details and possible implications on the origin of life. Int J Mol Sci. 107:2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R.. 2013. Structures of the human and Drosophila 80S ribosome. Nature 4977447:80–85. [DOI] [PubMed] [Google Scholar]
- Auerbach T, Bashan A, Harms J, Schluenzen F, Zarivach R, Bartels H, Agmon I, Kessler M, Pioletti M, Franceschi F, et al. 2002. Antibiotics Targeting Ribosomes: crystallographic Studies. Curr Drug Targ Infect Disord. 22:169–186. [DOI] [PubMed] [Google Scholar]
- Bachellerie JP, Michot B.. 1989. Evolution of large subunit rrna structure. the 3' terminal domain contains elements of secondary structure specific to major phylogenetic groups. Biochimie 716701–709. [DOI] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA.. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 2895481:905–920. [DOI] [PubMed] [Google Scholar]
- Behrmann E, Loerke J, Budkevich TV, Yamamoto K, Schmidt A, Penczek PA, Vos MR, Bürger J, Mielke T, Scheerer P, et al. 2015. Structural snapshots of actively translating human ribosomes. Cell 1614:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 4337025:531–537. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (Crw) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois RL, Doolittle WF.. 2004. computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 1412:2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CG, Tague BW, Ware VC, Gerbi SA.. 1984. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 1215:6197–6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cunha V, Gaia M, Gadelle D, Nasir A, Forterre P.. 2017. Lokiarchaea are close relatives of euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 136:e1006810.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Eden H, Mankin AS, Mandel-Gutfreund Y.. 2010. Structural signatures of antibiotic binding sites on the ribosome. Nucleic Acids Res. 3818:5982–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Belousoff M, Bashan A, Yonath A.. 2009. The evolving ribosome: from non-coded peptide bond formation to sophisticated translation machinery. Res Microbiol. 1607:487–492. [DOI] [PubMed] [Google Scholar]
- Doris SM, Smith DR, Beamesderfer JN, Raphael BJ, Nathanson JA, Gerbi SA.. 2015. Universal and domain-specific sequences in 23s–28s ribosomal rna identified by computational phylogenetics. Rna 2110:1719–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin L. 1966. The information content of DNA. J Theor Biol. 102:281–300. [DOI] [PubMed] [Google Scholar]
- Gatlin LL. 1972. Information theory and the living system. New York (NY: ): Columbia University Press. [Google Scholar]
- Gerbi SA. 1996. Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA—structure, evolution, processing, and function in protein synthesis. Boca Raton, FL: CRC Press; p. 71–87. [Google Scholar]
- Gonzalez IL, Gorski JL, Campen TJ, Dorney DJ, Erickson JM, Sylvester JE, Schmickel RD.. 1985. Variation among Human 28S Ribosomal RNA Genes. Proc Natl Acad Sci USA. 8222:7666–7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JK, Kelley ST, Spiegelman GB, Pace NR.. 2003. The genetic core of the universal ancestor. Genome Res. 133:407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouna N, Michot B, Bachellerie JP.. 1984. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 128:3563–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S.. 2006. Timetree: a public knowledge-base of divergence times among organisms. Bioinformatics 2223:2971–2972. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Kumar S.. 2009. The timetree of life. New York: Oxford University Press. [Google Scholar]
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A.. 1965. Structure of a ribonucleic acid. Science 1473664:1462–1465. [DOI] [PubMed] [Google Scholar]
- Hsiao C, Mohan S, Kalahar BK, Williams LD.. 2009. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol. 2611:2415–2425. [DOI] [PubMed] [Google Scholar]
- Huang HC, Nagaswamy U, Fox GE.. 2005. The application of cluster analysis in the intercomparison of loop structures in RNA. Rna 114:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illergard K, Ardell DH, Elofsson A.. 2009. Structure is three to ten times more conserved than sequence–a study of structural response in protein cores. Proteins 773:499–508. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 3213:1933–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP.. 2015. Structure of the human 80S ribosome. Nature 5207549:640–645. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1985. The role of compensatory neutral mutations in molecular evolution. J Genet. 641:7. [Google Scholar]
- Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nature Rev Microbiol. 12:127–136. [DOI] [PubMed] [Google Scholar]
- Kovacs NA, Petrov AS, Lanier KA, Williams LD.. 2017. Frozen in time: the history of proteins. Mol Biol Evol. 345:1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E.. 2001. geometric nomenclature and classification of RNA base pairs. Rna 74:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Burkhardt D, Gross C, Weissman JS.. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 1573:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Alva V.. 2017. Ribosomal proteins as documents of the transition from unstructured (poly) peptides to folded proteins. J Struct Biol 1982:74–81. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 3025649:1401–1404. [DOI] [PubMed] [Google Scholar]
- Mears JA, Cannone JJ, Stagg SM, Gutell RR, Agrawal RK, Harvey SC.. 2002. Modeling a minimal ribosome based on comparative sequence analysis. J Mol Biol. 3212:215–234. [DOI] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M.. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 196:560–567. [DOI] [PubMed] [Google Scholar]
- Michot B, Bachellerie JP.. 1987. Comparisons of large subunit rrnas reveal some eukaryote-specific elements of secondary structure. Biochimie 691:11–23. [DOI] [PubMed] [Google Scholar]
- NCBI. 2017. Database resources of the national center for biotechnology information. Nucleic Acids Res. 45(D1):D12–d17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz JO, Förster F, Kürner J, Linaroudis AA, Baumeister W.. 2006. Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J Struct Biol. 1562:334–341. [DOI] [PubMed] [Google Scholar]
- Paci M, Fox GE.. 2016. Centers of motion associated with EF-Tu binding to the ribosome. RNA Biol. 135:524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Braverman JM, Stephan W.. 2000. Comparative sequence analysis and patterns of covariation in RNA secondary structures. Genetics 1542:909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV.. 2008. Promals3d: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 367:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM et al. 2014. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci USA. 11128:10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, et al. 2015. History of the ribosome and the origin of translation. Proc Natl Acad Sci USA. 11250:15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO.. 2012. The silva ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(D1):D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB.. 2008. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J Math Biol. 56(1–2):215–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T.. 2010. Interdependence of cell growth and gene expression: origins and consequences. Science 3306007:1099–1102. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V.. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 3135795:1935–1942. [DOI] [PubMed] [Google Scholar]
- Shang L, Xu W, Ozer S, Gutell RR.. 2012. Structural constraints identified with covariation analysis in ribosomal RNA. PLoS One 76:e39383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. 1948. A mathematical theory of communication. At&T Tech J. 274:623–656. [Google Scholar]
- Shevack A, Gewitz HS, Hennemann B, Yonath A, Wittmann HG.. 1985. Characterization and crystallization of ribosomal particles from halobacterium-marismortui. FEBS Lett. 1841:68–71. [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. 2005. Second-generation shrna libraries covering the mouse and human genomes. Nat Gen. 3711:1281–1288. [DOI] [PubMed] [Google Scholar]
- Smit S, Widmann J, Knight R.. 2007. Evolutionary rates vary among rRNA structural elements. Nucleic Acids Res. 3510:3339–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanath P, Favaretto P, Hartman H, Mohr SC, Smith TF.. 2004. Ribosomal protein-sequence block structure suggests complex prokaryotic evolution with implications for the origin of eukaryotes. Mol Phylogenet Evol. 333:615–625. [DOI] [PubMed] [Google Scholar]
- Ware VC, Tague BW, Clark CG, Gourse RL, Brand RC, Gerbi SA.. 1983. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1122:7795–7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann HG, Mussig J, Piefke J, Gewitz HS, Rheinberger HJ, Yonath A.. 1982. Crystallization of Escherichia coli ribosomes. FEBS Lett. 1461:217–220. [DOI] [PubMed] [Google Scholar]
- Yonath A. 2002a. High-resolution structures of large ribosomal subunits from mesophilic eubacteria and halophilic archaea at various functional states. Curr Prot Pept Sci. 31:67–78. [DOI] [PubMed] [Google Scholar]
- Yonath A. 2002. The search and its outcome: high-resolution structures of ribosomal particles from mesophilic, thermophilic, and halophilic bacteria at various functional states. Annu Rev Biophys Biomol Struct. 311:257–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.