Abstract
The treatment of acute promyelocytic leukemia (APL) with all-trans retinoic acid (ATRA) induces granulocytic differentiation. This process renders APL cells resistant to cytotoxic chemotherapies. Epigenetic regulators of the histone deacetylases (HDACs) family, which comprise four classes (I–IV), critically control the development and progression of APL. We set out to clarify the parameters that determine the interaction between ATRA and histone deacetylase inhibitors (HDACi). Our assays included drugs against class I HDACs (MS-275, VPA, and FK228), pan-HDACi (LBH589, SAHA), and the novel HDAC6-selective compound Marbostat-100. We demonstrate that ATRA protects APL cells from cytotoxic effects of SAHA, MS-275, and Marbostat-100. However, LBH589 and FK228, which have a superior substrate–inhibitor dissociation constant (Ki) for the class I deacetylases HDAC1, 2, 3, are resistant against ATRA-dependent cytoprotective effects. We further show that HDACi evoke DNA damage, measured as induction of phosphorylated histone H2AX and by the comet assay. The ability of ATRA to protect APL cells from the induction of p-H2AX by HDACi is a readout for the cytoprotective effects of ATRA. Moreover, ATRA increases the fraction of cells in the G1 phase, together with an accumulation of the cyclin-dependent kinase inhibitor p21 and a reduced expression of thymidylate synthase (TdS). In contrast, the ATRA-dependent activation of the transcription factors STAT1, NF-κB, and C/EBP hardly influences the responses of APL cells to HDACi. We conclude that the affinity of HDACi for class I HDACs determines whether such drugs can kill naïve and maturated APL cells.
Keywords: APL, ATRA, DNA damage, HDAC, HDACi, Histone acetylation
Introduction
Under leukemic conditions, hematopoietic stem or precursor cells fail to differentiate. Instead, they proliferate in an uncontrolled manner (Coombs et al. 2015). Acute myeloid leukemia (AML) results from the unregulated proliferation and survival of myeloid cells (Hu and Zuckerman 2014; di Masi et al. 2015; Krämer et al. 2013). Acute promyelocytic leukemia (APL) is a subtype of AML. Nearly all APL cases carry the chromosomal translocation t(15;17) which fuses the promyelocytic leukemia (PML) gene with the retinoic acid receptor-α (RARα) gene. This balanced translocation gives rise to the PML–RARα protein (Hu and Zuckerman 2014; di Masi et al. 2015; Krämer et al. 2013). Its remaining RARα part binds retinoic acid (RA)-dependent promoters of differentiation genes and suppresses them via the recruitment of corepressors. This inhibitory effect involves HDACs (Altucci and Gronemeyer 2001; Breitman et al. 1981). Moreover, PML–RARα disrupts the nuclear PML body organization. These aberrations concertedly promote the survival and accumulation of immature, nonfunctional promyelocytes (Grignani et al. 1998; de Thé et al. 2012).
A new era of treatment began with the introduction of the vitamin A derivative all-trans retinoic acid (ATRA) for the treatment of APL in the 1980s (Altucci and Gronemeyer 2001; Breitman et al. 1981). Pharmacological doses of ATRA overcome the PML–RARα-induced maturation block by the recruitment of co-activators, such as histone acetyltransferases, to retinoic acid-dependent genes. ATRA also induces the degradation of PML–RARα, and both effects propel the functional differentiation of leukemic cells (Altucci and Gronemeyer 2001; Breitman et al. 1981). APL is additionally treated with arsenic trioxide (ATO/As2O3) (Fang et al. 2002). ATO directly binds and destroys the PML–RARα protein and often leads to an eradication of the leukemic clone(s) in combinatory schedules (Zhang et al. 2010). While ATO effectively cures APL in about 70% of all cases (de Thé et al. 2012), the remaining patients relapse and prognosis is poor if stem cell transplantation is not feasible (Sanford et al. 2015; Iriyama et al. 2014; Thirugnanam et al. 2009). Hence, additional treatment options are necessary.
HDACs are a group of 11 proteins that remove acetyl groups from lysine residues in histone tails and in other proteins (Spange et al. 2009). The differentiation, proliferation, and survival of transformed cells often depend on HDACs, which makes these enzymes valid pharmacological targets (Dokmanovic et al. 2007; Göttlicher et al. 2001; Iriyama et al. 2014). HDACi are epigenetic drugs that are relatively nontoxic to normal cells (Dokmanovic and Marks 2005; Müller and Krämer 2010), and the HDACi suberoylanilide hydroxamic acid (SAHA, vorinostat) and depsipeptide (FK228, romidepsin) are used for the treatment of cutaneous T cell lymphoma (Ellis and Pili 2010). Moreover, the FDA has recently approved LBH589 (panobinostat, Farydak) for the treatment of multiple myeloma (Lee et al. 2016a, b).
Since HDACs contribute to the differentiation block imposed by PML–RARα, a combination therapy of ATRA and HDACi appears as a logical combination. Furthermore, as well as ATRA and ATO, HDACi accelerate the degradation of PML–RARα (Hennig et al. 2015; Krämer et al. 2008b). Experiments in mice and clinical pilot trials suggested that combinations of HDACi with ATRA show promising effects against leukemic cells (Leiva et al. 2012; Cimino et al. 2006). Nevertheless, the clinical outcome was disappointing in most AML patients who received ATRA and HDACi in combination (Kuendgen et al. 2006; Tassara et al. 2014; Quintás-Cardama et al. 2011; Fredly et al. 2013).
We recently provided a possible explanation for the poor efficacy of ATRA/HDACi combinations in the clinic. We showed that application schedules as well as the choice of a particular HDACi determined the effectiveness of a treatment involving ATRA and HDACi (Hennig et al. 2015). If APL cells have differentiated toward the granulocytic lineage with ATRA for 24 h, HDACi that are specific for the class I HDACs HDAC1, 2, 3, and 8 (VPA, MS-275) do not induce apoptosis (programed cell death). In contrast, the pan-specific HDACi LBH589 still induces apoptosis of NB4 cells, irrespective of a previous exposure to ATRA ((Hennig et al. 2015) and Supplemental Fig. S1 and Table 1). We collected these results in NB4 cells, which are a prototypical human APL cell line with PML–RARα (Lanotte et al. 1991).
Table 1.
Inhibitory characteristics of selected HDACi
| HDAC1 | HDAC2 | HDAC3 | HDAC4 | HDAC5 | HDAC6 | HDAC7 | HDAC8 | HDAC9 | |
| Bradner et al. (2010a)a | |||||||||
| FK228 | 0.00005 | 0.00005 | 0.00005 | 0.012 | 0.84 | 0.01 | 1.2 | 0.003 | 0.86 |
| LBH589 | 0.0005 | 0.002 | 0.004 | 0.62 | 0.2 | 0.04 | 5.1 | 0.3 | 2.8 |
| MS-275 | 0.04 | 0.15 | 0.8 | >10 | >10 | >10 | >10 | >10 | >10 |
| SAHA | 0.003 | 0.004 | 0.01 | >10 | 7.8 | 0.02 | >10 | 1.1 | >10 |
| Tubacin | 0.07 | 0.09 | 0.6 | >10 | 2.2 | 0.02 | >10 | 0.4 | >10 |
| VPA | 51 | 75 | 131 | >1000 | >1000 | >1000 | >1000 | 850 | >1000 |
| Bradner et al. (2010b)b | |||||||||
| FK228 | 0.0000015 | 0.000038 | 0.00015 | 0.0205 | 0.55 | 0.0095 | 0.0095 | 0.00015 | 1.1 |
| LBH589 | 0.001 | 0.00065 | 0.0011 | 0.55 | 0.08 | 0.0015 | 4.55 | 0.105 | 3.2 |
| MS-275 | 0.022 | 0.065 | 0.36 | – | – | – | – | – | |
| SAHA | 0.0013 | 0.0016 | 0.005 | – | 3.6 | 0.0016 | – | 0.48 | – |
| HDAC1 | HDAC2 | HDAC4 | HDAC6 | ||||||
| Furumai et al. (2002)c | |||||||||
| FK228 | 0.036 | 0.048 | 0.51 | 0.014 | |||||
| HDAC1 | HDAC6 | ||||||||
| Yurek-George et al. (2007)d | |||||||||
| FK228 | 0.004 | 0.787 | |||||||
| SAHA | 0.775 | 0.196 | |||||||
IC50 of selected histone deacetylase inhibitors (µM) (adapted from Bradner et al. 2010a)
Ki of several histone deacetylase inhibitors (µM) (adapted from Bradner et al. 2010b)
IC50 of selected histone deacetylase inhibitors (µM) (adapted from Furumai et al. 2002)
IC50 of selected histone deacetylase inhibitors (µM) (adapted from Yurek-George et al. 2007)
When we collected the above-mentioned results, we considered effects of ATRA and HDACi on factors that control apoptosis and differentiation. Caspases are at the heart of the initiation and execution of apoptosis (Droin et al. 2013; Samudio et al. 2010). The intrinsic apoptosis pathway is triggered by the efflux of mitochondrial cytochrome C, APAF1, and caspase-9, and the extrinsic apoptosis pathway relies on caspase-8. The activation of these initiator caspases culminates in the activation of the apoptosis executioner caspase-3 (Droin et al. 2013; Samudio et al. 2010). Mitochondrial proteins belonging to the BCL family critically control mitochondrial integrity and the induction of apoptosis at the level of mitochondria (Droin et al. 2013; Samudio et al. 2010). Pro-survival molecules, like the NF-κB target gene BCL-XL which stabilizes mitochondria, as well as the myeloid differentiation marker C/EBPε, are downregulated in NB4 cells after the treatment with LBH589, but not after an exposure to MS-275 or VPA. Therefore, we speculated that altered gene expression profiles and cellular differentiation protected NB4 cells from HDACi-induced cytotoxic effects (Hennig et al. 2015). This hypothesis is supported by the literature. For example, ATRA is able to induce NF-κB-dependent prosurvival genes in NB4 and U937 AML cells (Mathieu et al. 2005; Mathieu and Besançon 2006; Altucci et al. 2001) and in other leukemic cells (Nagel et al. 2015; Demchenko and Kuehl 2010). NF-κB also contributes to the myeloid maturation process. NF-κB family members directly interact with C/EBPβ (Xia et al. 1997; Weber et al. 2003; Zwergal et al. 2006), and NF-κB binds synergistically with other C/EBP family members to target genes (Stein et al. 1993). For example, NF-κB p50 binds together with C/EBPα, C/EBPδ, and C/EBPε at the 5-lipoxygenase-activating protein promotor in LPS-treated THP-1 AML cells (Serio et al. 2005). Moreover, C/EBPε binds more avidly to its target sequences upon an interaction with NF-κB p65 (Chumakov et al. 2007). ATRA additionally activates the inducible transcription factor STAT1 and its target genes via an enhanced phosphorylation of STAT1 at Ser727 (Shang et al. 1999; Dimberg et al. 2003). This phosphorylation allows the full induction of some target genes of STAT1 (Wen et al. 1995) as well as the maturation of leukemic cells (Ying et al. 2013; Fang et al. 2013).
It is presently unclear whether such mechanisms promote the relative resistance of APL cells against HDACi. In addition, ATRA might protect cells from the recently reported HDACi-dependent DNA damage. For example, MS-275 promotes the accumulation of p-H2AX in multiple myeloma cells (Cai et al. 2013) and FK228 causes DNA damage that is linked to an augmented production of reactive oxygen species (ROS) and the activation of p53 in human lung and colon cancer cells (Wang et al. 2012). Furthermore, HDACi promote the phosphorylation of H2AX and its upstream kinase ataxia telangiectasia mutated (ATM) in various cell types (Quintás-Cardama et al. 2011). HL60 AML cells phosphorylate ATM and H2AX in response to the HDACi trichostatin-A and apicidin, and K562 chronic myeloid leukemia cells show p-H2AX-positive nuclei after a treatment with the naturally occurring HDACi butyrate (Gaymes et al. 2006). Further studies report that lymphoid Molt4 cells and HL60 cells carry increased p-H2AX in the presence of SAHA (Sanchez-Gonzalez et al. 2006). As different HDACi induce a variable amount of DNA damage (Lee et al. 2016b), such differences may well determine whether cells undergo apoptosis in response to a treatment with HDACi and ATRA.
We set out to identify the parameters and pathways that determine the interplay between ATRA and HDACi-induced cell death. Our results show that HDACi induce the DNA damage marker p-H2AX in leukemic cells. The data further suggest that the extent of HDACi-induced DNA damage and cell cycle arrest determine whether NB4 cells undergo apoptosis. Moreover, we demonstrate that the HDAC/HDACi dissociation constant Ki and the resulting ability to block class I HDACs indicates the robustness of HDACi treatment schedules against ATRA-induced survival mechanisms.
Materials and methods
Chemicals
LBH589, MS-275, ABT-737, Obatoclax, and FK228 were from Selleck Chemicals (Houston, TX, USA) or Sigma-Aldrich (Munich, Germany), or they were gifts from Prof. W.-G. Zhu, Beijing, or Dr. C. Wichmann, Munich. Valproic acid (VPA), doxycycline, ATRA, and propidium iodide (PI) were from Sigma-Aldrich (Munich, Germany). IFN-α was from Roche (Germany), TNF-α from ReliaTech (Wolfenbüttel, Germany), carboxy-H2DFFDA from Thermo Fischer (Erlangen, Germany), and SAHA from Cayman Chemical (Ann Arbor, MI, USA). For Marbostat-100, see patent Mahboobi et al. (2016).
Constructs
The cDNA of rat C/EBPα p42 (Grebien et al. 2015) was cloned into pSIN-TREtight-Gateway cassette-StrepII-HA-IRES-GFP-PGK-BlastR by Gateway cloning. Primers and vector information are available upon request.
Cell culture
The promyelocytic cell line NB4 grows in Roswell Park Memorial Institute (RPMI) medium containing 10% fetal calf serum (FCS) and 0.5% gentamicin in a humidified incubator at 37 °C and 5% CO2. NB4 cells harbor a PML/RARA fusion gene and stem from a relapsed APL patient (Lanotte et al. 1991). Correct cell identity was verified via DNA fingerprint at the Leibniz Institute, DSMZ GmbH, Braunschweig. 5 × 105 NB4 cells/ml were seeded and immediately stimulated with ATRA or HDACi for the indicated times and concentrations. FDCP-1 cells were cultivated in RPMI medium with 10% FCS, glutamate, penicillin/streptomycin, and 2 ng/ml IL-3 (Immunotools, Friesoythe, Germany). Tet-On competent FDCP-1 cells were generated by retroviral introduction of pMSCV-rtTA3-PGK-PuroR (Zuber et al. 2011) as described in Bigenzahn et al. (2016). FDCP-1 rtTA3 cells were subsequently transduced with pSIN-TREtight-C/EBPα p42-StrepII-HA-IRES-GFP-PGK-BlastR and selected with blasticidin (10 μg/ml). FDCP-1 rtTA3 cells were seeded at 5 × 105 cells/ml and treated with doxycycline (Sigma) at 1 µg/ml for 24 h to induce gene expression. MS-275 treatment of FDCP-1 rtTA3 C/EBPα cells was initiated 24 h after doxycycline treatment for 48 h using the indicated concentrations.
Flow cytometry
Propidium iodide (PI) staining for fragmented DNA: 5× 105 cells were washed twice with phosphate-buffered saline buffer (PBS), fixed with 1 ml of 70% ethanol in PBS, and stored over night at −20 °C. Cells were then washed with PBS, resuspended in PBS, supplemented with 125 µg/ml RNase-A (Roche, Germany) and 31 µg/ml PI (Biotium, CA, USA), and incubated for 20 min at 37 °C. DNA content of the cells was measured by flow cytometry with a fluorescence-activated cell sorter (FACS). Dead cells were identified by gating on the fraction of cells with a subG1 (<2 N) DNA content. FDCP-1 rtTA3 and FDCP-1 rtTA3 C/EBPα-expressing cells were washed with PBS supplied with 0.1% FBS. Viable cells were identified as FSChigh SSClow cells, by flow cytometry, while dead/apoptotic cells were FSClow SSChigh.
Intracellular FACS staining
After washing with PBS, cells were fixed with 4% paraformaldehyde/PBS for 20 min at room temperature (RT). Subsequently, cells were permeabilized in 0.1% Triton X-100/PBS for five min at RT. Afterward, cells were blocked in 10% FCS/PBS for 15 min at RT and then incubated with 1:50 anti-pY701 STAT1-PE antibody (BD Biosciences, #612564) or IgG2a-PE Isotype control (BD Biosciences, #553457) in 10% FCS/PBS for 1 h at 4 °C. After three washing steps, the cells were analyzed on a FACS-Canto cytometer (BD Biosciences). Histogram overlays were created using the Weasel.jar software.
Annexin V/propidium iodide staining
Annexin V/propidium iodide staining allows the simultaneous detection of early apoptotic, late apoptotic, and necrotic cells. FITC-labeled Annexin V (MACS Miltenyi Biotec) binds to the phospholipid phosphatidylserine, which translocates from the inner leaflet of the plasma membrane to the outer leaflet in early and late apoptotic cells. Propidium iodide (PI, Sigma) is a standard flow cytometric dye used to distinguish viable from nonviable cells. The membranes of dead and damaged, necrotic cells are permeable to PI, but usually negative for Annexin V (viable cells exclude PI). Late apoptotic cells are PI positive, but additionally stain positive for Annexin V. NB4 cells were seeded into 12-well plates at a density of 2 × 105 cells/ml; 24 h later, cells were treated with 2 µM MS-275, 30 nM LBH589, and 5 nM FK228 for 24–48 h. Subsequently, cells were harvested in FACS tubes, washed once with cold PBS, and centrifuged at 188×g for 5 min. Then, the cell pellet was gently resuspended in a mixture of 50 µl 1 × Annexin V binding buffer (10 × Annexin V binding buffer: 100 mM HEPES 1.4 M NaCl 25 mM CaCl2, 1% BSA in 50 ml H2Obidest., pH 7,4) and 2.5 µL FITC Annexin V. Cells were incubated in the dark on ice for 15 min. The DNA of the cells was stained with 430 µl 1 × Annexin V binding buffer and 10 µl PI (stock solution: 50 µg/µl). Samples were directly measured on a FACSCanto II flow cytometer (BD Biosciences). Data were evaluated with the FACSDiva software 7.0 (BD Biosciences).
Immunoblot and antibodies
Cells were collected, washed with PBS, and then lysed with NETN buffer for 10 min. NETN buffer: 150 mM NaCl, 50 mM Tris/HCl pH 8.0, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, supplemented with protease inhibitor cocktail PIC (2 µg/ml leupeptin, 4 µg/ml antipain, 0.2 µg/ml benzamidine, 20 U/ml aprotinin), 1 mM sodium vanadate, 1 mM sodium fluoride, phenylmethylsulfonyl fluoride (1:500 of a saturated solution), 40 mM β-glycerophosphate. After a sonification step, cell debris was removed via centrifugation at 20,000×g. For detection of C/EBPα in FDCP-1 rTA3 cells, lysates were prepared in RIPA buffer. The protein concentration was determined by Bradford assay (Roth, Karlsruhe, Germany) according to the manufacturer’s instructions. Equal amounts of proteins were loaded on SDS gels for immunoblot. Antibodies were from Santa-Cruz (TX, USA): anti-acetylated tubulin SC-23950, anti-C/EBPβ SC-150, anti-C/EBPε SC-158, anti-HSP70 SC-24, anti-NF-κB SC-8008 and SC-372, anti-phosphorylated STAT1 (Y701) SC-7988, anti-STAT1 SC-346, anti-β-actin sc-47778, and anti-p21 SC-6246; from Cell Signaling Technology (MA, USA): anti-caspase-3 cleaved #9664, anti-phosphorylated NF-κB (Ser 536) #3036, anti-phosphorylated p38 (Y180/T182) #9215, anti-p38 #9212, and anti-thymidylate synthase #9045; from Millipore/Upstate (Darmstadt, Germany): anti-acetylated histone H3 (H3) #06-599 and anti-p-H2AX #05-636; from BD Biosciences (Heidelberg, Germany): anti-PARP cleaved #552596; and from Sigma-Aldrich (München, Germany): anti-β-actin #A5316; for anti-acetylated histone 4 antibody (ac-H4), see (Göttlicher et al. 2001).
ROS measurement
Cells were seeded at 1 × 106 cells/ml. After stimulation, they were washed once with warm RPMI without FCS and then twice with warm PBS. Cells were resuspended in PBS and rotated with 20 µM carboxy-H2DFFDA for 30 min at room temperature in the dark. Then, cells were washed twice with warm PBS and resuspended in 500 µl PBS and mean fluorescence intensities were measured with flow cytometry.
qRT-PCR
RNA quantification was done as described in (Pietschmann et al. 2012). Data obtained were analyzed with the delta-Cq quantification model (Hellemans et al. 2007) using GAPDH as reference gene. Primer sequences for quantitative real-time PCR (qPCR) were:
BCL2A1 fwd GAATGGATAAGGCAAAACGGA
BCL2A1 rev TGTTGGCAATCGTTTCCATAT
GAPDH fwd TGCACCACCAACTGCTTA GC
GAPDH rev GGCATGGACTGTGGTCATGAG
Neutral comet assay
Microscopy slides were coated with 1.5% agarose in PBS; 10 µl of a cell suspension (from 1 × 106 NB4 cells/ml) was diluted in 120 µl of 0.5% low melting point agarose. This suspension was added to the slide, which was then covered and incubated for 5 min at 4 °C. After removal of cover slips, the slides were incubated for at least 1 h in lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 µM Tris pH 7.5, 1% sodium lauroyl sarcosinate) at 4 °C. Electrophoresis was carried out to allow comet formation. Slides were then fixed for 5 min in ethanol and dried for 2 h at room temperature; 50 µl of a 50 µg/ml solution of PI was added, and the slides were then analyzed microscopically; see (Nikolova et al. 2014) for further details.
Results
High affinity of HDACi confers robustness to cytoprotective effects of ATRA
We have previously shown that a pretreatment with ATRA protects NB4 cells from the induction of apoptosis by the class I HDACi MS-275 and VPA [Supplemental Fig. S1 and Table 1, see also (Bradner et al. 2010a, b)]. However, pretreatment with ATRA does not antagonize apoptosis that is caused by the pan-HDACi LBH589 (Hennig et al. 2015) (Table 1). To characterize this finding further, we tested the class I HDACi FK228 and SAHA (Table 1). SAHA inactivates class I HDACs and the class IIb deacetylase HDAC6 (Table 1), which is regarded as a crucial molecule in leukemic cells (Krämer et al. 2014).
Based on the finding that the pan-HDACi LBH589 can kill ATRA pre-treated NB4 cells (Hennig et al. 2015), we suspected that the broad-range HDACi SAHA would equally cause an ATRA-insensitive cell death program in NB4 cell cultures. Yet, a 24-h pretreatment with ATRA could abrogate the cytotoxic fragmentation of DNA in NB4 cells exposed to 1 µM SAHA (Fig. 1a). This unexpected outcome was not due to an inefficient inhibition of HDAC6. Western blot against acetylated tubulin, a prototypical HDAC6 target (Krämer et al. 2014), verified that our batch of SAHA efficiently blocked HDAC6 at 1 µM (Fig. 1b). At 2 µM, SAHA more effectively killed NB4 cells. The cytotoxic effect of 2 µM SAHA was significantly blocked by ATRA, but the induction of cell death remained significant too (Fig. 1a). The analysis of histone hyperacetylation, which prototypically indicates a suppression of class I HDACs (Mithraprabhu et al. 2013), revealed that the inhibition of these HDACs and not the hyperacetylation of tubulin indicates the cytotoxicity of SAHA (Fig. 1a, b).
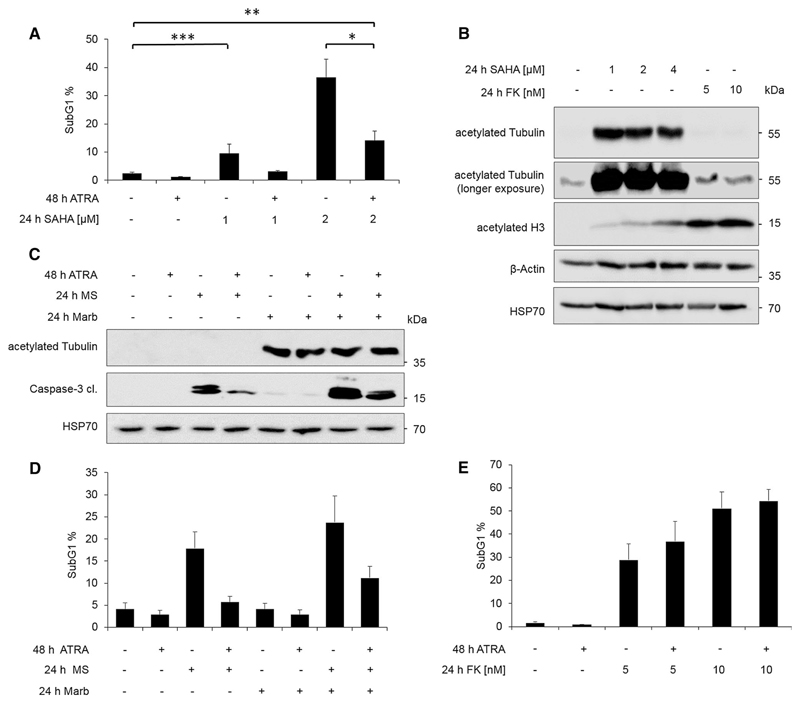
Fig. 1. Pre-treatment with ATRA protects NB4 cells from MS-275-but not from LBH589- or FK228-induced cell death.
a Flow cytometric analysis of NB4 cells treated for 48 h with 1 µM ATRA, and/or SAHA for 24 h with the indicated concentrations. Shown is the subG1 fraction of PI-stained cells (mean + SEM, n = 3–6). Statistics were carried out with one-way ANOVA *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005. b Immunoblot of cell lysates treated with indicated concentrations with SAHA or FK228 (FK) for 24 h; n = 3. c Immunoblot of NB4 cells treated for 48 h with 1 µM ATRA, and/or with 5 µM MS-275 (MS) and 1 µM Marbostat-100 (Marb) for 24 h; n = 5. Tubulin hyperacetylation indicates inhibition of HDAC6, and cleaved caspase-3 is the activated form of this apoptosis execution enzyme. d Flow cytometry analysis of PI-stained NB4 cells treated as described in c; n = 4. e Flow cytometry analysis of PI-stained NB4 cells treated with 1 µM ATRA for 48 h and/or 24 h FK228 (FK) with the indicated concentrations; n = 3; ATRA did not induce any significant cytoprotection
To further test the relevance of HDAC6 in our system, we used our newly developed HDAC6-specific drug Marbostat-100 (see “Materials and methods” for details). A dose of 1 µM Marbostat-100 evoked a strong accumulation of hyperacetylated tubulin in NB4 cells (Fig. 1c). However, Marbostat-100 alone could not promote DNA fragmentation or caspase-3 activation in NB4 cell cultures (Fig. 1c, d). Hence, we combined Marbostat-100 with MS-275 to see whether an additional inhibition of HDAC6 on top of class I inhibition by MS-275 (Table 1) could eliminate naïve and ATRA pre-treated NB4 cells. The combination of MS-275 and Marbostat-100 was slightly more effective than MS-275 alone (Fig. 1d). Consistent with our previous data (Hennig et al. 2015), a pre-treatment with ATRA ablated the cytotoxic effect of MS-275 (Fig. 1d). ATRA also reduced the cell death program that the MS-275/Marbostat-100 combination caused (Fig. 1d). Consistent with these results, we noted that ATRA suppressed the activation of the apoptotic enzyme caspase-3 in NB4 cells that were treated with MS-275 or MS-275 plus Marbostat-100 (Fig. 1c). These data disfavor that the activity of HDAC6 is primarily responsible for the cytoprotective effects that ATRA confers to NB4 cells.
To analyze the impact of HDACi on NB4 cells further, we used FK228 (Table 1). A main difference between MS-275 and FK228 is the about 1000-fold higher affinity of FK228 for class I HDACs (Table 1; the IC50 of FK228 for HDAC1–3 is 0.00005 µM and the IC50 of MS-275 for HDAC1–3 ranges from 0.04 to 0.8 µM). These divergent properties of the class I HDACi MS-275 and FK228 allowed us to identify the relevance of HDACi affinity for the killing of NB4 cells. FK228 was able to induce cell death in a concentration-dependent fashion as measured by PI staining followed by flow cytometry analysis (Fig. 1e). We further noted that a 24-h pretreatment with ATRA did not protect NB4 cells from FK228-induced cell death (Fig. 1e).
Compared to SAHA, FK228 more strongly induced the hyperacetylation of histone H3, a typical target of class I HDACs (Spange et al. 2009; Mithraprabhu et al. 2013; Krämer et al. 2014) (Fig. 1b). These data agree with the lower Ki and IC50 of FK228 for class I HDACs (Table 1; the IC50 of FK228 for HDAC1–3 is 0.00005 µM and the IC50 of SAHA for HDAC1–3 ranges from 0.001 to 0.004 µM; the IC50 of FK228 against HDAC1 is more than 200-fold lower than the IC50 of SAHA against HDAC1). Dependent on the test system and the presence of reducing agents, FK228 shows some activity against HDAC6 in vitro (Table 1), but FK228 minimally blocks HDAC6 in vivo (Mithraprabhu et al. 2013; Yurek-George et al. 2007; Hui and Chiang 2014; Furumai et al. 2002). Accordingly, FK228 hardly changed the acetylation status of the HDAC6 target tubulin (Fig. 1b).
From these data, we conclude that the affinity for class I HDACs is a novel readout for the potency of HDACi against naïve and ATRA-treated, maturated APL cells.
Inhibition of BCL proteins does not break resistance mechanisms evoked by ATRA
We next analyzed whether anti-apoptotic BCL proteins determine the sensitivity of NB4 cells to HDACi. We suspected a role of BCL proteins because LBH589 reduced NB4 cell survival as well as BCL-XL expression much stronger than MS-275 and VPA (Hennig et al. 2015). Furthermore, ATRA promotes the accumulation of the BCL2A1 mRNA, which is an ATRA-inducible NF-κB-dependent target in NB4 cells (Altucci et al. 2001; Edelstein et al. 2003), and VPA slightly accentuates the accumulation of BCL2A1 mRNA in NB4 cells (Fig. 2a). Therefore, it is conceivable that BCL2 proteins protect cells from HDACi.
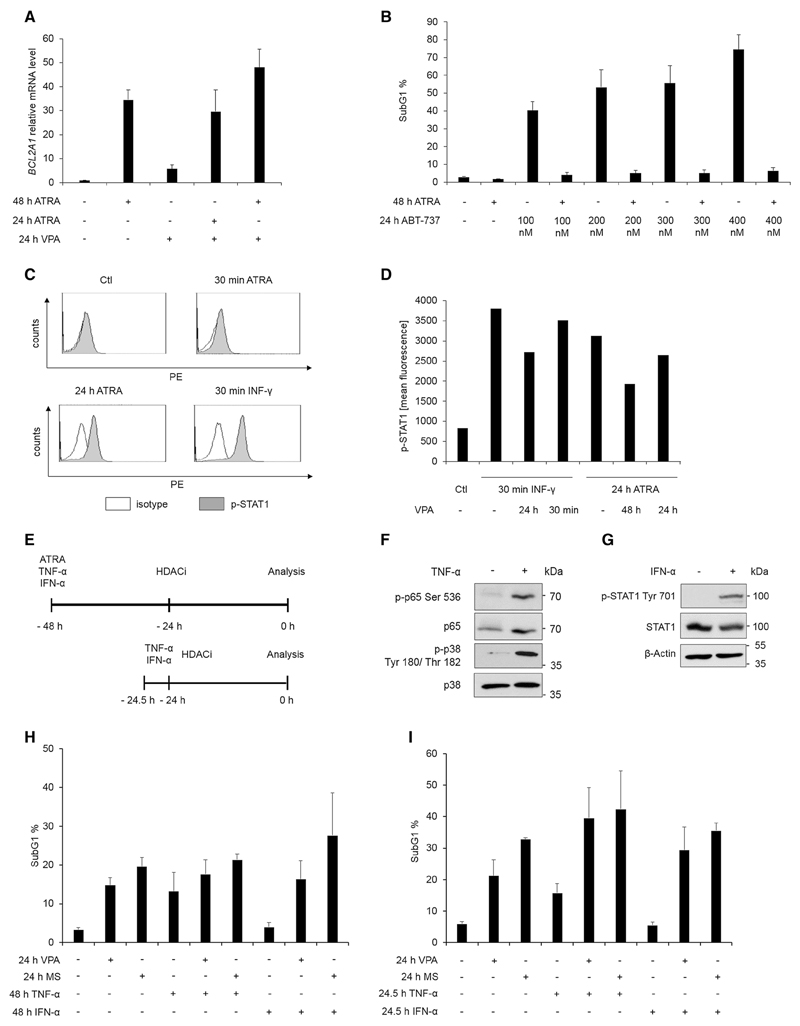
Fig. 2. Activation of NF-κB and STAT1 does not protect NB4 cells from HDACi.
a Cells were treated for the indicated times with 1 µM ATRA and/or 1.5 mM VPA; n = 3. Expression of Bcl2A1 mRNA was analyzed by quantitative real-time PCR. Relative expression relates to corresponding values for samples from untreated cells, set as 1 (mean ± SE). GAPDH, served as reference gene. b SubG1 phase of NB4 cells treated for 48 h with 1 µM ATRA and/or 100–400 nM ABT-737 for 24 h. DNA fragmentation was determined by flow cytometry after PI staining; n = 3. c NB4 cells were stimulated for indicated times with 1 µM ATRA or with 100 ng/ml INF-γ. Cells were incubated with an antibody against the Tyr701-phosphorylated form of STAT1 or an isotype control and analyzed by flow cytometry. Histogram overlays show intracellular p-Tyr 701-STAT1 and were calculated with the Weasel.jar software; n = 2. d NB4 cells were treated for the indicated times with 5 mM valproic acid (VPA), INF-γ (100 ng/ml for 30 min) or ATRA (1 µM for 24 h). Mean fluorescence of intracellular p-Tyr 701-STAT1 stain was quantified by flow cytometry analysis; see Supplemental Fig. S3A for a similar experimental outcome. e Treatment schedule for treatment with ATRA, TNF-α, IFN-α, and HDACi. Cells were either pre-treated for 24 h or for 30 min and then treated for 24 h with HDACi. f NB4 cells were treated for 30 min with 50 ng/ml TNF-α and stained with immunoblot for phosphorylation of NF-κB p65 and MAPK p38; n = 3. g NB4 cells were treated for 30 min with 1000 U IFN-α and analyzed by immunoblot for p-STAT1 and loading controls; n = 4. h Flow cytometry analysis of PI-stained NB4 cells treated as shown in 2e for the indicated times with cytokines (1000 U IFN-α or 50 ng/ml TNF-α) and HDACi [5 mM VPA or 5 µM MS-275 (MS)]; n = 3. i Same as in 2h but with a 30-min pre-incubation with cytokines
We used the BCL inhibitor ABT-737 (Oltersdorf et al. 2005) to challenge this idea. While treatment with ABT-737 led to cell death, pretreatment with ATRA inhibited this process (Fig. 2b). Similar data were collected for the BCL inhibitor Obatoclax (data not shown).
These observations disfavor the possibility that a loss of BCL proteins can break ATRA-mediated resistance mechanisms.
Activation of NF-κB and STAT1 does not protect NB4 cells from HDACi
ATRA activates a plethora of different pathways. For example, ATRA induces the transcription factors NF-κB and STAT1 (Altucci et al. 2001; Mathieu et al. 2005; Dimberg et al. 2000; Gianni et al. 1997). We noted that ATRA evoked the phosphorylation of STAT1 at Y701 in NB4 cells. Compared to IFN-γ-treated cells, this phosphorylation occurred slower and at quantitatively lower levels in ATRA-treated NB4 cells (Fig. 2c). Since HDACi can attenuate the phosphorylation of STAT1 at Y701 (Wieczorek et al. 2012; Moon et al. 2016; Kotla and Rao 2015), we asked whether HDACi have an impact on the ATRA-induced phosphorylation of STAT1. We found that a prolonged exposure of NB4 cells to VPA decreased the ATRA-dependent phosphorylation of STAT1 at Y701 (Fig. 2d, S3A). In order to analyze whether this effect is linked to altered protein stability, we performed Western blot analyses for STAT1. While the treatment with 1.5 mM VPA does not alter STAT1 levels, 5 mM VPA reduced STAT1 expression (Fig. S3B). These data agree with the accelerated turnover of STAT1 in apoptotic cells (Licht et al. 2014).
Based on these data we tested whether an activation of NF-κB and STAT1 may protect NB4 cells from HDACi. We stimulated the cells with typical inducers of these pathways: TNF-α to activate NF-κB and IFN-α to activate STAT1. To verify our batches of IFN-α and TNF-α, we stained immunoblots for phosphorylated STAT1 as well as for phosphorylated forms of NF-κB p65 and the kinase p38. After a 30-min incubation time with TNF-α or IFN-α, we found a clear phosphorylation of the above-mentioned marker proteins (Fig. 2f, g).
We then used the experimental setup seen in Fig. 2e. We focused on MS-275 and VPA in the following experiments because ATRA can abrogate the pro-apoptotic effects of these HDACi (Hennig et al. 2015, this work, and Supplemental Fig. S1). Incubation of NB4 cells with VPA and MS-275 for 24 h induced apoptosis, as evidenced by an increase in cells in the subG1 fraction and the appearance of cleaved caspase-3 in immunoblots (Fig. 2h, Supplemental Fig. S4). Incubation of NB4 cells with IFN-α alone for 48 h did not induce apoptosis, but TNF-α was effective against these cells (Fig. 2h). Flow cytometry analyses and immunoblots for active caspase-3 illustrated that the combined treatment with IFN-α or TNF-α did not rescue the cells from MS-275 or VPA (Fig. 2h, Supplemental Fig. S4).
To corroborate our results, we additionally chose a 30-min cytokine stimulation prior to the addition of MS-275 or VPA. Again, IFN-α and TNF-α were not able to inhibit the MS-275- and VPA-induced cell death (Fig. 2i).
We conclude that activation of NF-κB or STAT1 alone does not protect NB4 cells from HDACi. Hence, an induction of these pathways by ATRA unlikely renders NB4 cells resistant against HDACi.
Expression and relevance of C/EBP proteins for HDACi-induced cytotoxicity
ATRA induces myeloid differentiation that is causally associated with an increased expression of the C/EBP family members C/EBPα, C/EBPβ, and C/EBPε (Duprez et al. 2003; Park et al. 1999; Hashimoto et al. 2006; Hennig et al. 2015) and Fig. 3a. In contrast, stimulation with TNF-α or IFN-α did not promote the accumulation of these transcription factors (Fig. 3a). Therefore, we hypothesized that a myeloid differentiation process involving C/EBPs might contribute to cytoprotective effects of ATRA.
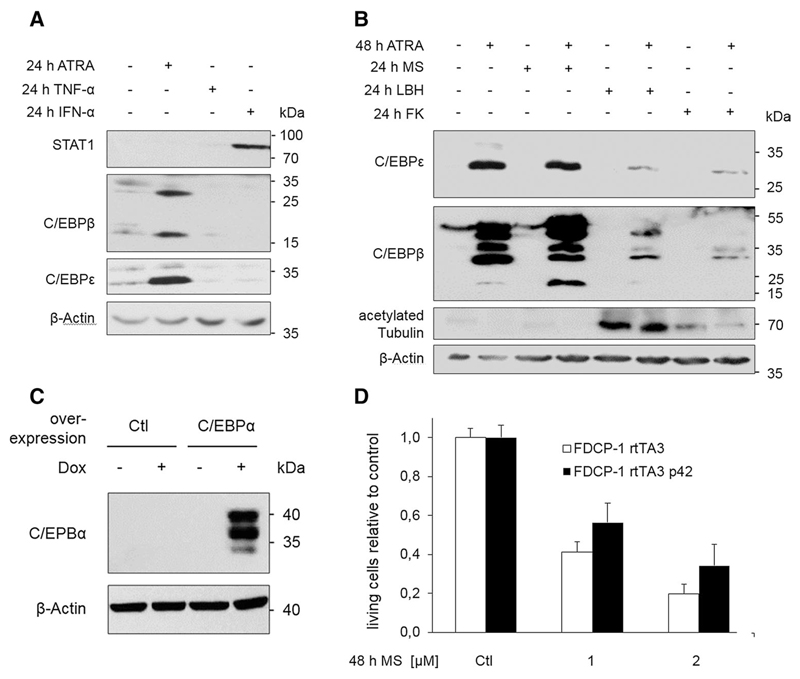
Fig. 3. Induction and role of C/EBP family members.
a NB4 cells were treated for 24 h with either 1 µM ATRA, 50 ng/ml TNF-α, or 1000 U IFN-α. Cells were subjected to immunoblot and probed with antibodies as indicated. Different membranes with the same samples were used; n = 3. b NB4 cells were stimulated for the indicated times with 1 µM ATRA, 5 µM MS-275, 100 nM LBH589 (LBH), or 10 nM FK228 (FK), lysed, and subjected to immunoblot. Different membranes with the same samples were used; n = 3. c FDCP-1 cells stably expressing rtTA3 or rtTA3 and C/EBPα p42 were treated with 1 µg/ml doxycycline for 48 h, and expression levels were detected by Immunoblot analysis with the indicated antibodies. d Protein expression was induced as in c, and FDCP-1 cells were simultaneously treated with the indicated concentrations of MS-275. After 48 h, the cells were harvested and living cells were determined by flow cytometry; n = 3
Since we found that the cytotoxicity of FK228 was insensitive to ATRA (Fig. 1e), we also tested whether this agent could attenuate the accumulation of C/EBP proteins in ATRA pre-treated NB4 cells; we previously noticed such an effect in LBH589-treated NB4 cells (Hennig et al. 2015). Like LBH589, FK228 largely suppressed the ATRA-induced accumulation of C/EBPε and C/EBPβ (Fig. 3b).
These findings encouraged us to exogenously express C/EBP proteins in NB4 cells, but such attempts failed because of a rapid induction of cell death upon transfection (n > 20; data not shown). Therefore, we alternatively analyzed the FDCP-1 cell line, which represents an established hematopoietic cell system for myeloid differentiation. Inducible expression of C/EBPα p42 activates C/EBPβ and C/EBPε and recapitulates principal aspects of myeloid maturation in these cells (Hashimoto et al. 2006; Welm et al. 1999; Friedman 2007; Grebien et al. 2015). We tested whether an ectopically induced overexpression of C/EBPα p42 rescued FDCP-1 cells from cell death induced by MS-275. FDCP-1 cells rTA3 were transduced with a retroviral construct allowing for doxycycline-inducible expression of C/EBPα p42. Expression of C/EBPα p42 was induced by 1 µg/ml doxycycline (Fig. 3c), and the cells were simultaneously exposed to MS-275 for 48 h (Fig. 3d). The anticipated C/EBPα-dependent induction of C/EBPβ and C/EBPε (Hashimoto et al. 2006; Welm et al. 1999; Friedman 2007) was confirmed (data not shown). Compared to mock-transduced cells, C/EBPα p42-overexpressing cells showed a trend toward a higher proportion of living cells in the presence of MS-275 (Fig. 3d). However, this effect cannot explain the abrogation of the MS-275-induced cell death in ATRA pre-treated NB4 cells (Hennig et al. 2015 and this work). Consequently, we addressed additional mechanisms, including the HDACi-evoked DNA damage.
MS-275 induces DNA damage
HDACi induce apoptosis by several ways, including the induction of ROS, mitochondrial damage (Rosato et al. 2003; Sanda et al. 2007), downregulation of anti-apoptotic proteins, upregulation of pro-apoptotic proteins (Bose et al. 2014), and DNA damage (Dasmahapatra et al. 2010; Gaymes et al. 2006). Based on these data, we investigated whether MS-275 can induce DNA damage in NB4 cells. We found a time-dependent increase in the phosphorylation of H2AX in MS-275-treated NB4 cells (Fig. 4a); such an accumulation of p-H2AX is a typical sign of DNA damage and replicative stress (Nikolova et al. 2014; Mah et al. 2010).
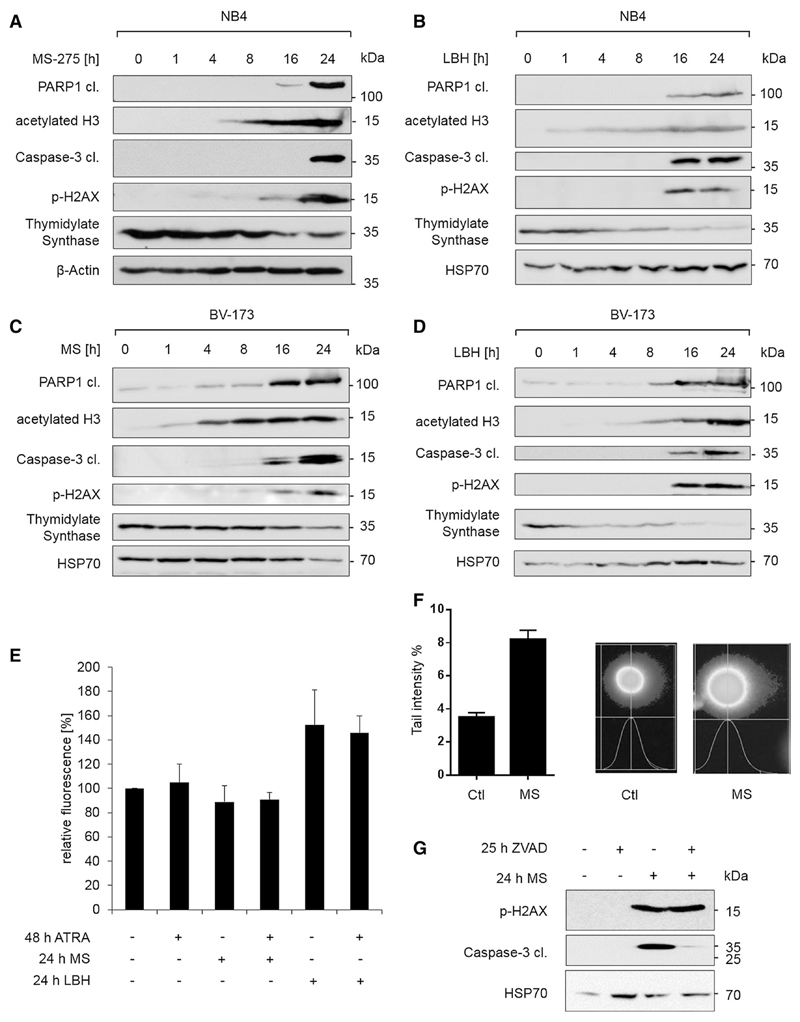
Fig. 4. HDACi induce DNA damage.
a–d NB4 and BV-173 cells were stimulated as indicated with 5 µM MS-275 (MS) or 100 nM LBH589 (LBH). Lysates were analyzed by immunoblot as indicated; n = 3. e NB4 cells were stimulated for the indicated times with 1 µM ATRA, 5 µM MS-275, or 100 nM LBH589 (LBH), and cytosolic ROS was measured. Unstained control was subtracted from each measurement, and data were normalized to control; n = 2. f NB4 cells were incubated for 12 h with 2 µM MS-275 and analyzed for DNA damage with comet assay; n = 2. g NB4 cells were treated as indicated and analyzed by immunoblot; 50 µM ZVAD-FMK (ZVAD) and 5 µM MS-275 were used. Different membranes with the same samples were used, n = 3
The accumulation of p-H2AX in MS-275-treated NB4 cells tied in with histone acetylation as well as with an activation of the apoptosis executioner caspase-3 and the cleavage of the DNA repair enzyme PARP1 (Fig. 4a). While we noted no signs of checkpoint kinase activation in MS-275-treated NB4 cells (data not shown), we saw a reduction in TdS (Fig. 4a), which supplies cells with nucleotides for S phase progression (Wilson et al. 2014).
LBH589 also induced the accumulation of p-H2AX, the cleavage of PARP1, and the reduction in TdS in NB4 cells (Fig. 4b). Moreover, the same effects occurred in another leukemia cell line, the BCR-ABL-transformed pre-B cell line BV-173 (Fig. 4c, d). Hence, our results are not restricted to a certain HDACi or cell type.
Since HDACi can induce apoptosis via elevated ROS levels (Sanda et al. 2007), these may cause DNA damage in HDACi-treated cells. Measurement of ROS levels in MS-275- and LBH589-exposed NB4 cells showed that ROS were elevated after treatment with LBH589, but surprisingly not after an exposure to MS-275 (Fig. 4e). The observation that ATRA has no discernable impact on ROS production further disfavors that ROS contribute to MS-275-induced DNA damage.
Assessing DNA damage with the neutral comet assay verified the ability of MS-275 to harm DNA integrity (Fig. 4f). However, apoptosis also leads to DNA fragmentation and H2AX phosphorylation (Rogakou et al. 2000). Therefore, we used the pan-caspase inhibitor ZVAD-FMK, to exclude that an apoptotic processing of DNA caused the phosphorylation of H2AX in HDACi-treated NB4 cells. Inhibition of caspases by ZVAD-FMK did not inhibit p-H2AX (Fig. 4g).
Hence, MS-275 appears as a causal inducer of DNA damage, which is associated with loss of PARP1 and TdS
Cell cycle progression promotes DNA damage and apoptosis of HDACi-treated NB4 cells
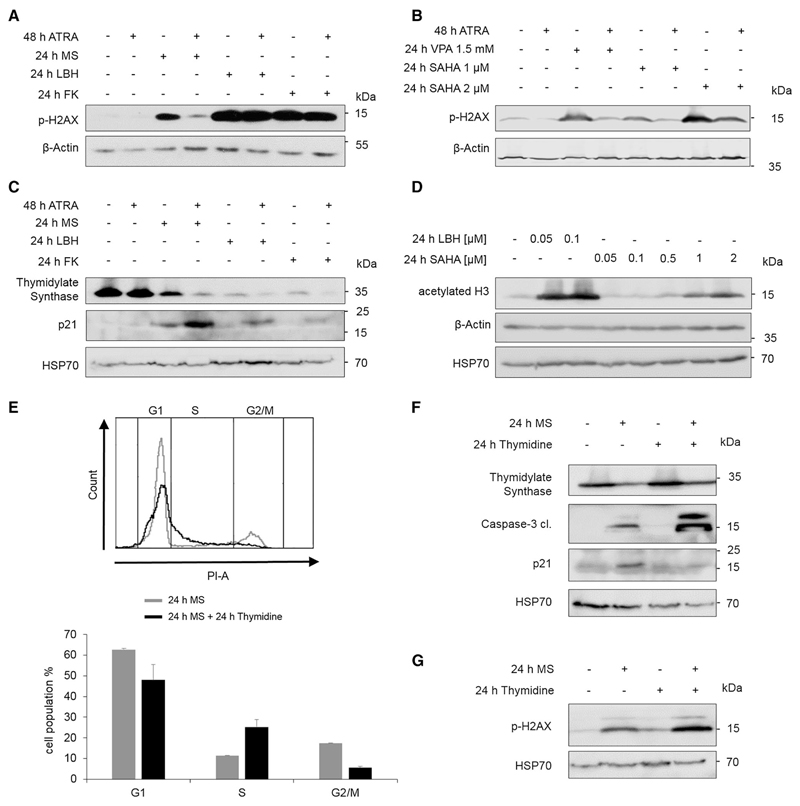
Like MS-275, LBH589 and FK228 caused an accumulation of p-H2AX in NB4 cells (Fig. 5a). Compared to NB4 cells that had been treated with MS-275, the amount of p-H2AX was higher in NB4 cells that had received LBH589 and FK228 (Fig. 5a). Thus, the extent of HDACi-induced apoptosis correlates with the levels of p-H2AX.
Fig. 5. Cell cycle distribution after HDACi and ATRA.
a–c NB4 cells were stimulated for the indicated times with 1 µM ATRA, 5 µM MS-275, 100 nM LBH589 (LBH),10 nM FK228 (FK) or 1.5 mM VPA or 1–2 µM SAHA, lysed, and subjected to immunoblot. Different membranes with the same samples were used; n = 3. d NB4 cells were incubated for 24 h with the indicated concentrations of LBH589 (LBH) or SAHA and subjected to immunoblot; n = 3. e NB4 cells were treated for 24 h with 5 µM MS-275 (gray) or with MS-275 + 200 µM thymidine (black), stained with PI and analyzed by flow cytometry; n = 2. Shown is a representative flow cytometry profile and a graphical evaluation of the two independent experiments. f–g NB4 cells were treated as described in eand analyzed by immunoblot; n = 2, experiments were done in duplicate
If the HDACi-evoked DNA damage was linked to cell death of NB4 cell cultures, ATRA should be able to impair the MS-275-induced accumulation of p-H2AX but should be ineffective against the accumulation of p-H2AX in response to LBH589 and FK228. Indeed, MS-275, LBH589, and FK228 induced DNA damage, but the pretreatment with ATRA only reduced the MS-275-induced accumulation of p-H2AX. The p-H2AX levels were equally high in naïve and ATRA pre-treated NB4 cells when they received LBH589 and FK228 (Fig. 5a).
To extent these analyses, we assessed how ATRA in combination with SAHA and VPA affected p-H2AX levels. Western blot experiments showed that ATRA prevented the induction of p-H2AX by 1.5 mM VPA and 1 µM SAHA (Fig. 5b). Congruently, ATRA protected cells from such concentrations of VPA (Hennig et al. 2015) and SAHA (Fig. 1a). For 2 µM SAHA, we found a reduction in p-H2AX, but ATRA did not prevent its accumulation completely (Fig. 5b). Figure 1a shows that ATRA protects the cells from 2 µM SAHA, but also not completely. Thus, the accumulation of p-H2AX and apoptosis correlate with each other.
We then asked whether ATRA was able to sustain the expression of TdS in MS-275-treated NB4 cells. We analyzed lysates from NB4 cells that had been incubated with ATRA, MS-275, LBH589, or FK228. Contrary to our expectation, HDACi reduced TdS even more strongly in combination with ATRA (Fig. 5c). Thus, the decrease in TdS can hardly be the sole determinant for the higher and ATRA-resistant levels of p-H2AX in NB4 cells that receive LBH589 and FK228.
Given that the impact of DNA damage on cellular fate can depend on the cell cycle phase (Karimian et al. 2016), we considered that different combinations of HDACi and ATRA might have different effects on the cell cycle of NB4 cells. The treatment with MS-275 and ATRA promoted an arrest of cells in the G1 phase more effectively than MS-275 alone (Hennig et al. 2015). This process involved an accumulation of the CDKI p21 (Fig. 5c, Supplemental S2). This molecule not only halts cells in the G1 phase but it is also a negative regulator of the MS-275-induced apoptosis in leukemic cells (Rosato et al. 2003). NB4 cell cultures that were treated with LBH589 and FK228 had lower numbers of cells in the G1 phase than cultures that were exposed to MS-275 or SAHA. Moreover, FK228 and LBH589 failed to induce an accumulation of p21 in NB4 cells (Fig. 5c, Supplemental Fig. S2, Hennig et al. 2015, and data not shown). These results suggest that a p21-associated arrest in the G1 phase partially protects against cytotoxic effects of HDACi.
We have deduced from Fig. 1 that the extent of class I HDAC inhibition, which we measured as histone hyperacetylation in response to FK228 and SAHA, is a readout for the robustness of HDACi-induced cytotoxic effects against ATRA. To corroborate this hypothesis, we compared the efficacy of LBH589 and SAHA side by side. We treated NB4 cells with 50–100 nM LBH589 or 50 nM to 2 µM SAHA. This experiment showed that LBH589 was about 500-fold more effective than SAHA in vivo (Fig. 5d). Congruent with Fig. 1, these data indicate that the inhibition of class I HDACs translates into HDACi-induced pro-apoptotic effects despite the presence of ATRA. Moreover, these data give further credit that very low Ki and IC50 values for class I HDAC inhibition (Table 1) render HDACi-induced cell death resistant against ATRA-induced cytoprotection.
We also considered that the different responses of NB4 cells to ATRA in combination with various HDACi rely on different modes of cell death, namely apoptosis or necrosis. We treated NB4 cells with different HDACi (MS-275, FK228, and LBH589) and stained them with Annexin V and PI to discriminate between early apoptotic (Annexin V+/PI−), late apoptotic (Annexin V+/PI+), and necrotic cells (Annexin V−/PI+). Flow cytometry analyses revealed that all three HDACi evoked a significant increase in apoptosis without a detectable induction of necrosis (Supplemental Figure S5).
The differential regulation of cell cycle arrest (Supplemental Fig. S2), together with the depletion of TdS in response to HDACi (Figs. 4a–d, 5c), let us speculate that we could drive MS-275-treated NB4 cells into S phase by supplementation with thymidine. Moreover, this may affect the HDACi-induced DNA damage. We treated NB4 cells with MS-275 or MS-275 plus thymidine. Cell cycle analysis of PI-stained cells by flow cytometry illustrated that a supplementation of MS-275-treated NB4 cells with thymidine decreased the number of cells in G1 phase and increased the amount of cells in early S phase (Fig. 5e). Immunoblots showed that this cell cycle progression was accompanied by reduced expression of p21 and an increase in cleaved caspase-3 (Fig. 5f). Furthermore, the combination of MS and thymidine enhanced the HDACi-evoked accumulation of p-H2AX (Fig. 5g).
We deduce that the abilities of ATRA to abrogate DNA damage and to arrest cells in the G1 phase of the cell cycle prevent cytotoxic effects of HDACi.
Discussion
Our previous data demonstrate that the inhibition of HDACs with MS-275 or VPA induces apoptosis of NB4 cells and that ATRA induces a differentiation program that renders NB4 cells resistant to such drugs (Hennig et al. 2015). We further analyzed the molecular mechanisms that promote the robustness of ATRA-treated NB4 cells against HDACi. Our data disfavor the inhibition of HDAC6 and the activation of NF-κB and STAT1 as key mediators of ATRA-mediated resistance. Myeloid differentiation induced by overexpression of C/EBPα has a slight protective effect on HDACi-treated hematopoietic cells. However, we find a clear association between avid HDAC inhibition and the induction of DNA damage and apoptosis. We discuss these findings and their implications as follows.
The stimulation of NB4 cells with ATRA, IFNs, or TNF-α activates STAT1 and NF-κB in NB4 cells (Fig. 2). These data are coherent with previous reports (Gianni et al. 1997; Altucci et al. 2001; Mathieu et al. 2005). Despite such similarities, IFN-α and TNF-α cannot protect NB4 cells from MS-275 and VPA; TNF-α rather acts in an additive manner with HDACi (Fig. 2h, i, Supplemental Fig. S4). Thus, we conclude that the activation of these transcription factors unlikely mediates the cytoprotective effect of ATRA against HDACi. We though cannot exclude that IFN-α and TNF-α cause additional effects that may override otherwise cytoprotective pathways. The finding that IFN-α has no effect on the HDACi-induced cell death (Fig. 2h, i) may be related to the processing of STAT1 by caspase-3 and 6 in HDACi-treated NB4 cells (Licht et al. 2014). This situation differs from melanoma cells, where the expression and acetylation of STAT1 increase in response to HDACi. In such cells, IFN-α and HDACi augment STAT1 levels and both agents combine favorably against melanoma cells (Krämer et al. 2006).
A striking difference between the stimulation of NB4 cells with ATRA or IFN-α/TNF-α is the induction of cell differentiation, i.e., the accumulation of C/EBPβ and C/EBPε (Fig. 3a), which are markers of myeloid cell maturation (Akagi et al. 2010; Schuster et al. 2003; Morosetti et al. 1997; Duprez et al. 2003; Truong et al. 2003). Therefore, we hypothesized that differentiation protects NB4 cells from HDACi. However, activation of C/EBPα p42, which induces maturation and a subsequent accumulation of C/EBPβ and C/EBPε (Hashimoto et al. 2006; Welm et al. 1999; Friedman 2007), only slightly attenuates apoptosis evoked by MS-275 (Fig. 3d). We are aware that C/EBPs may affect cell survival differentially in NB4 and FDCP-1 cells. Further studies are necessary to clarify this issue.
We rule out that HDAC6 is relevant for the survival of NB4 cells in the presence of ATRA and HDACi (Fig. 1d). The fact that apoptosis induction by VPA and SAHA, which block all four class I HDACs (Table 1), is susceptible to ATRA (Fig. 1a) also disfavors that HDAC8 is critical for the survival of HDACi- and ATRA-treated NB4 cells. Furthermore, an HDAC8-specific inhibitor induced apoptosis specifically in T cell tumor lines (Balasubramanian et al. 2008).
Our results suggest a radical model in which the avidity of an HDACi for the class I HDACs HDAC1–3, i.e., low Ki/IC50 values (Table 1), translates into its ability to eradicate APL cells at immature and differentiated stages. The accumulation of hyperacetylated histone H3 appears as a marker for the potency of HDACi against APL cells (Figs. 1, 5). A caveat of such highly potent HDACi may be cytotoxicity against normal cells. While a recent report raises caution about such undesired side effects (Wong et al. 2014), a large number of clinical studies corroborates tolerable effects of HDACi in patients suffering from cancer (Dokmanovic and Marks 2005; Müller and Krämer 2010; Ellis and Pili 2010; Lee et al. 2016a, b).
Since HDACi can evoke DNA damage in tumor cells (Lee et al. 2016a; Wells et al. 2013; Wang et al. 2012; Fukuda et al. 2015; Gaymes et al. 2006; Jiemjit et al. 2008), we asked whether apoptosis induced by HDACi was linked to a loss of DNA integrity in leukemic cells. Indeed, various HDACi induce DNA damage in NB4 and BV-173 cells (Figs. 4a–d, 5a, b). As PARP1, DNA-PK, and XRCC are differentially regulated upon monocytic maturation (Bauer et al. 2011), we also considered an impact of these molecules on the responses of ATRA-treated, granulocyte-like NB4 cells to different HDACi. However, we found no evidence for a regulation of XRCC by MS-275 in NB4 cells. Regarding PARP1, we found with flow cytometry that inhibition of PARP1 with rucaparib, simultaneously or 24 h prior to incubation with MS-275, had no influence on apoptosis after MS-275 alone and in combination with ATRA (data not shown). It appears plausible that these pathways are of different relevance for monocytic and granulocytic maturation and as well for primary and transformed cells.
A remaining question is the source of replicative stress in HDACi-treated cells. Previous data demonstrate that siRNA-mediated downregulation or selective pharmaceutical inhibition of HDAC3 reduces replicative fork velocity and induces replicative stress and chromosomal instability (Conti et al. 2010; Wells et al. 2013; Bhaskara et al. 2010). Our data agree with this notion, as MS-275, which only inactivates the class I HDACs HDAC1–3 (Table 1), suffices to promote the accumulation of p-H2AX. As HDAC1 and HDAC2 also control DNA damage responses and genomic stability (Wilting et al. 2010; Miller et al. 2010), these HDACs also represent critical targets for HDACi-induced DNA damage. Additional experiments are necessary to test such hypotheses.
We also see that MS-275, LBH589, and FK228 deplete the cellular TdS pool, which could equally promote replicative stress. The ability of HDACi to reduce TdS has also been reported by others (Chittur et al. 2008; Fazzone et al. 2009). As TdS levels strongly increase during S phase (Navalgund et al. 1980), it is possible that the reduction of S phase cells in the presence of HDACi is a reason for attenuated TdS levels. We are aware that others noted stable levels of TdS concomitant with a rapid occurrence of p-H2AX after a 2-h treatment with SAHA in breast cancer cells (Conti et al. 2010). Our results with leukemic cells rather suggest that the accumulation of p-H2AX occurs slowly and becomes detectable after 16 h and that TdS levels decrease simultaneously (Fig. 4a–d). Although such differences may be due to different cellular backgrounds (breast cancer vs. APL) and due to limitations in p-H2AX detection levels, they may reflect that HDACi-induced DNA damage can occur in several steps. There could be a first, rapid phase seen by Conti and colleagues and a further one that occurs later with the depletion of TdS and perhaps additional regulators of DNA metabolism and DNA repair. The latter hypothesis is congruent with the survival of MS-275/ATRA-treated NB4 cells that remain in the G1 phase. In such a setting, loss of TdS may even protect cells. Indeed, feeding MS-275-treated NB4 cells with thymidine promotes their entry into S phase (Fig. 5e), reduces p21 expression, increases p-H2AX, and activates caspase-3 (Fig. 5f, g). Obviously, the G1 phase is less susceptible to replicative stress as there is hardly DNA replication (Cheng et al. 2000). Moreover, a cytoprotective, pro-differentiating role of p21 in HDACi-treated AML cells is well documented (Wu et al. 2014; Rosato et al. 2003) and p21 protects cancer cells from apoptosis under replicative stress (Krämer et al. 2008a).
Taken together, our work shows that cytoprotective effects of ATRA against HDACi involve an attenuated HDACi-induced DNA damage and a p21-coupled cell cycle arrest. Dissociation constants for HDACi/HDAC complexes are predictive of whether an HDACi can eliminate ATRA-treated APL cells that have differentiated along the granulocytic lineage.
Supplementary Material
Electronic supplementary material The online version of this article (doi:10.1007/s00204-016-1878-5) contains supplementary material, which is available to authorized users.
Acknowledgements
We thank Anna Frumkina, ITOX Mainz, for help with the comet assay. This work was supported by the Federal Ministry of Education and Research (BMBF), Germany, FKZ: 01EO1002, the Wilhelm Sander-Foundation (#2010.078.2 to OHK), the Deutsche Krebshilfe (#110125 and #110909 to OHK), the Deutsche Forschungsgemeinschaft (#KR2291/4-1 and #KR2291/5-1 to OHK; MA2183/1-1 to SM), and startup grants from the UM Mainz and the NMFZ Mainz (to OHK). LS is the recipient of a DOC fellowship of the Austrian Academy of Sciences. Work in the laboratory of FG is supported by the ERC Starting Grant ONCOMECHAML.
Footnotes
Additional information The manuscript does not contain clinical studies or patient data. Archives of Toxicology complies with the recommendations of the Committee on Publishing Ethics (COPE).
Conflict of interest The authors declare that there are no conflicts of interest.
References
- Akagi T, Thoennissen NH, George A, et al. In vivo deficiency of both C/EBPβ and C/EBPε results in highly defective myeloid differentiation and lack of cytokine response. PLoS One. 2010;5:e15419. doi: 10.1371/journal.pone.0015419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altucci L, Gronemeyer H. The promise of retinoids to fight against cancer. Nat Rev Cancer. 2001;1:181–193. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- Altucci L, Rossin A, Raffelsberger W, et al. Retinoic acid-induced apoptosis in leukemia cells is mediated by paracrine action of tumor-selective death ligand TRAIL. Nat Med. 2001;7:680–686. doi: 10.1038/89050. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Ramos J, Luo W, et al. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22:1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- Bauer M, Goldstein M, Christmann M, et al. Human monocytes are severely impaired in base and DNA double-strand break repair that renders them vulnerable to oxidative stress. Proc Natl Acad Sci USA. 2011;108:21105–21110. doi: 10.1073/pnas.1111919109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigenzahn JW, Fauster A, Rebsamen M, et al. An inducible retroviral expression system for tandem affinity purification mass-spectrometry-based proteomics identifies mixed lineage kinase domain-like protein (MLKL) as an heat shock protein 90 (HSP90) client. Mol Cell Proteomics. 2016;15:1139–1150. doi: 10.1074/mcp.M115.055350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose P, Dai Y, Grant S. Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights. Pharmacol Ther. 2014;143:323–336. doi: 10.1016/j.pharmthera.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, Mak R, Tanguturi SK, et al. Chemical genetic strategy identifies histone deacetylase 1 (HDAC1) and HDAC2 as therapeutic targets in sickle cell disease. Proc Natl Acad Sci USA. 2010a;107:12617–12622. doi: 10.1073/pnas.1006774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010b;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman TR, Collins SJ, Keene BR. Terminal differentiation of human promyelocytic leukemic cells in primary culture in response to retinoic acid. Blood. 1981;57:1000–1004. [PubMed] [Google Scholar]
- Cai B, Lyu H, Huang J, et al. Combination of bendamustine and entinostat synergistically inhibits proliferation of multiple myeloma cells via induction of apoptosis and DNA damage response. Cancer Lett. 2013;335:343–350. doi: 10.1016/j.canlet.2013.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- Chittur SV, Sangster-Guity N, McCormick PJ. Histone deacetylase inhibitors: a new mode for inhibition of cholesterol metabolism. BMC Genom. 2008;9:507. doi: 10.1186/1471-2164-9-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov AM, Silla A, Williamson EA, Koeffler HP. Modulation of DNA binding properties of CCAAT/enhancer binding protein epsilon by heterodimer formation and interactions with NFkappaB pathway. Blood. 2007;109:4209–4219. doi: 10.1182/blood-2005-09-031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino G, Lo-Coco F, Fenu S, et al. Sequential valproic acid/all-trans retinoic acid treatment reprograms differentiation in refractory and high-risk acute myeloid leukemia. Cancer Res. 2006;66:8903–8911. doi: 10.1158/0008-5472.CAN-05-2726. [DOI] [PubMed] [Google Scholar]
- Conti C, Leo E, Eichler GS, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs CC, Tavakkoli M, Tallman MS. Acute promyelocytic leukemia: where did we start, where are we now, and the future. Blood Cancer J. 2015;5:e304. doi: 10.1038/bcj.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra G, Lembersky D, Kramer L, et al. The pan-HDAC inhibitor vorinostat potentiates the activity of the proteasome inhibitor carfilzomib in human DLBCL cells in vitro and in vivo. Blood. 2010;115:4478–4487. doi: 10.1182/blood-2009-12-257261. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- de Thé H, Le Bras M, Lallemand-Breitenbach V. The cell biology of disease: acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol. 2012;198:11–21. doi: 10.1083/jcb.201112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko YN, Kuehl WM. A critical role for the NFkB pathway in multiple myeloma. Oncotarget. 2010;1:59–68. doi: 10.18632/oncotarget.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Masi A, Leboffe L, De Marinis E, et al. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Mol Aspects Med. 2015;41:1–115. doi: 10.1016/j.mam.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Dimberg A, Nilsson K, Oberg F. Phosphorylation-deficient Stat1 inhibits retinoic acid-induced differentiation and cell cycle arrest in U-937 monoblasts. Blood. 2000;96:2870–2878. [PubMed] [Google Scholar]
- Dimberg A, Karlberg I, Nilsson K, Oberg F. Ser727/Tyr701-phosphorylated Stat1 is required for the regulation of c-Myc, cyclins, and p27Kip1 associated with ATRA-induced G0/G1 arrest of U-937 cells. Blood. 2003;102:254–261. doi: 10.1182/blood-2002-10-3149. [DOI] [PubMed] [Google Scholar]
- Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J Cell Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- Droin N, Guéry L, Benikhlef N, Solary E. Targeting apoptosis proteins in hematological malignancies. Cancer Lett. 2013;332:325–334. doi: 10.1016/j.canlet.2011.06.016. [DOI] [PubMed] [Google Scholar]
- Duprez E, Wagner K, Koch H, Tenen DG. C/EBPbeta: a major PML-RARA-responsive gene in retinoic acid-induced differentiation of APL cells. EMBO J. 2003;22:5806–5816. doi: 10.1093/emboj/cdg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein LC, Lagos L, Simmons M, et al. NF-kappa B-dependent assembly of an enhanceosome-like complex on the promoter region of apoptosis inhibitor Bfl-1/A1. Mol Cell Biol. 2003;23:2749–2761. doi: 10.1128/MCB.23.8.2749-2761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L, Pili R. Histone deacetylase inhibitors: advancing therapeutic strategies in hematological and solid malignancies. Pharmaceuticals (Basel) 2010;3:2411–2469. doi: 10.3390/ph3082441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chen S-JJ, Tong J-HH, et al. Treatment of acute promyelocytic leukemia with ATRA and As2O3: a model of molecular target-based cancer therapy. Cancer Biol Ther. 2002;1:614–620. doi: 10.4161/cbt.308. [DOI] [PubMed] [Google Scholar]
- Fang Y, Zhong L, Lin M, et al. MEK/ERK dependent activation of STAT1 mediates dasatinib-induced differentiation of acute myeloid leukemia. PLoS One. 2013;8:e66915. doi: 10.1371/journal.pone.0066915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzone W, Wilson PM, Labonte MJ, et al. Histone deacetylase inhibitors suppress thymidylate synthase gene expression and synergize with the fluoropyrimidines in colon cancer cells. Int J Cancer. 2009;125:463–473. doi: 10.1002/ijc.24403. [DOI] [PubMed] [Google Scholar]
- Fredly H, Gjertsen BTT, Bruserud O. Histone deacetylase inhibition in the treatment of acute myeloid leukemia: the effects of valproic acid on leukemic cells, and the clinical and experimental evidence for combining valproic acid with other antileukemic agents. Clin Epigenetics. 2013;5:12. doi: 10.1186/1868-7083-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Wu W, Okada M, et al. Class I histone deacetylase inhibitors inhibit the retention of BRCA1 and 53BP1 at the site of DNA damage. Cancer Sci. 2015;106:1050–1056. doi: 10.1111/cas.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumai R, Matsuyama A, Kobashi N, et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- Gaymes TJ, Padua RA, Pla M, et al. Histone deacetylase inhibitors (HDI) cause DNA damage in leukemia cells: a mechanism for leukemia-specific HDI-dependent apoptosis? Mol Cancer Res. 2006;4:563–573. doi: 10.1158/1541-7786.MCR-06-0111. [DOI] [PubMed] [Google Scholar]
- Gianni M, Terao M, Fortino I, et al. Stat1 is induced and activated by all-trans retinoic acid in acute promyelocytic leukemia cells. Blood. 1997;89:1001–1012. [PubMed] [Google Scholar]
- Göttlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebien F, Vedadi M, Getlik M, et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat Chem Biol. 2015;11:571–578. doi: 10.1038/nchembio.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignani F, De Matteis S, Nervi C, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Sonoda Y, Yamakado M, et al. C/EBPalpha inactivation in FAK-overexpressed HL-60 cells impairs cell differentiation. Cell Signal. 2006;18:955–963. doi: 10.1016/j.cellsig.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, et al. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig D, Müller S, Wichmann C, et al. Antagonism between granulocytic maturation and deacetylase inhibitor-induced apoptosis in acute promyelocytic leukaemia cells. Br J Cancer. 2015;112:329–337. doi: 10.1038/bjc.2014.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XT, Zuckerman KS. Role of cell cycle regulatory molecules in retinoic acid- and vitamin D3-induced differentiation of acute myeloid leukaemia cells. Cell Prolif. 2014;47:200–210. doi: 10.1111/cpr.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui KF, Chiang AK. Combination of proteasome and class I HDAC inhibitors induces apoptosis of NPC cells through an HDAC6-independent ER stress-induced mechanism. Int J Cancer. 2014;135:2950–2961. doi: 10.1002/ijc.28924. [DOI] [PubMed] [Google Scholar]
- Iriyama N, Yuan B, Yoshino Y, et al. Enhancement of differentiation induction and upregulation of CCAAT/enhancer-binding proteins and PU.1 in NB4 cells treated with combination of ATRA and valproic acid. Int J Oncol. 2014;44:865–873. doi: 10.3892/ijo.2013.2236. [DOI] [PubMed] [Google Scholar]
- Jiemjit A, Fandy TE, Carraway H, et al. p21(WAF1/CIP1) induction by 5-azacytosine nucleosides requires DNA damage. Oncogene. 2008;27:3615–3623. doi: 10.1038/sj.onc.1211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Kotla S, Rao GN. Reactive oxygen species (ROS) Mediate p300-dependent STAT1 protein interaction with peroxisome proliferator-activated receptor (PPAR)-γ in CD36 protein expression and foam cell formation. J Biol Chem. 2015;290:30306–30320. doi: 10.1074/jbc.M115.686865. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Krämer OH, Baus D, Knauer SK, et al. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer OH, Knauer SK, Zimmermann D, et al. Histone deacetylase inhibitors and hydroxyurea modulate the cell cycle and cooperatively induce apoptosis. Oncogene. 2008a;27:732–740. doi: 10.1038/sj.onc.1210677. [DOI] [PubMed] [Google Scholar]
- Krämer OH, Müller S, Buchwald M, et al. Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARalpha. FASEB J. 2008b;22:1369–1379. doi: 10.1096/fj.06-8050com. [DOI] [PubMed] [Google Scholar]
- Krämer OH, Stauber RH, Bug G, et al. SIAH proteins: critical roles in leukemogenesis. Leukemia. 2013;27:792–802. doi: 10.1038/leu.2012.284. [DOI] [PubMed] [Google Scholar]
- Krämer OH, Mahboobi S, Sellmer A. Drugging the HDAC6-HSP90 interplay in malignant cells. Trends Pharmacol Sci. 2014;35:501–509. doi: 10.1016/j.tips.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Kuendgen A, Schmid M, Schlenk R, et al. The histone deacetylase (HDAC) inhibitor valproic acid as monotherapy or in combination with all-trans retinoic acid in patients with acute myeloid leukemia. Cancer. 2006;106:112–119. doi: 10.1002/cncr.21552. [DOI] [PubMed] [Google Scholar]
- Lanotte M, Martin-Thouvenin V, Najman S, et al. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3) Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- Lee H-SS, Lee NC, Kouprina N, et al. Effects of anticancer drugs on chromosome instability and new clinical implications for tumor-suppressing therapies. Cancer Res. 2016a;76:902–911. doi: 10.1158/0008-5472.CAN-15-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Mark TM, Shah JJ. Practical approaches to the management of dual refractory multiple myeloma. Curr Hematol Malig Rep. 2016b;11:148–155. doi: 10.1007/s11899-016-0312-7. [DOI] [PubMed] [Google Scholar]
- Leiva M, Moretti S, Soilihi H, et al. Valproic acid induces differentiation and transient tumor regression, but spares leukemia-initiating activity in mouse models of APL. Leukemia. 2012;26:1630–1637. doi: 10.1038/leu.2012.39. [DOI] [PubMed] [Google Scholar]
- Licht V, Noack K, Schlott B, et al. Caspase-3 and caspase-6 cleave STAT1 in leukemic cells. Oncotarget. 2014;5:2305–2317. doi: 10.18632/oncotarget.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah L-JJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- Mahboobi S, Sellmer A, Pongratz H, Leonhardt M, Krämer O, Böhmer F-D, Kelter G. Preparation of fused heterocyclic compounds as HDAC6 inhibitors and their uses. PCT Int Appl WO 2016020369 A1. 2016
- Mathieu J, Besançon F. Arsenic trioxide represses NF-kappaB activation and increases apoptosis in ATRA-treated APL cells. Ann NY Acad Sci. 2006;1090:203–208. doi: 10.1196/annals.1378.022. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Giraudier S, Lanotte M, Besançon F. Retinoid-induced activation of NF-kappaB in APL cells is not essential for granulocytic differentiation, but prolongs the life span of mature cells. Oncogene. 2005;24:7145–7155. doi: 10.1038/sj.onc.1208889. [DOI] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithraprabhu S, Khong T, Jones SS, Spencer A. Histone deacetylase (HDAC) inhibitors as single agents induce multiple myeloma cell death principally through the inhibition of class I HDAC. Br J Haematol. 2013;162:559–562. doi: 10.1111/bjh.12388. [DOI] [PubMed] [Google Scholar]
- Moon J, Kaowinn S, Cho I-RR, et al. Hepatitis C virus core protein enhances hepatocellular carcinoma cells to be susceptible to oncolytic vesicular stomatitis virus through down-regulation of HDAC4. Biochem Biophys Res Commun. 2016;474:428–434. doi: 10.1016/j.bbrc.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Morosetti R, Park DJ, Chumakov AM, et al. A novel, myeloid transcription factor, C/EBP epsilon, is upregulated during granulocytic, but not monocytic, differentiation. Blood. 1997;90:2591–2600. [PubMed] [Google Scholar]
- Müller S, Krämer OH. Inhibitors of HDACs–effective drugs against cancer? Curr Cancer Drug Targets. 2010;10:210–228. doi: 10.2174/156800910791054149. [DOI] [PubMed] [Google Scholar]
- Nagel S, Ehrentraut S, Meyer C, et al. NFkB is activated by multiple mechanisms in hairy cell leukemia. Genes Chromosom Cancer. 2015;54:418–432. doi: 10.1002/gcc.22253. [DOI] [PubMed] [Google Scholar]
- Navalgund LG, Rossana C, Muench AJ, Johnson LF. Cell cycle regulation of thymidylate synthetase gene expression in cultured mouse fibroblasts. J Biol Chem. 1980;255:7386–7390. [PubMed] [Google Scholar]
- Nikolova T, Dvorak M, Jung F, et al. The γH2AX assay for genotoxic and nongenotoxic agents: comparison of H2AX phosphorylation with cell death response. Toxicol Sci. 2014;140:103–117. doi: 10.1093/toxsci/kfu066. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Park DJ, Chumakov AM, Vuong PT, et al. CCAAT/enhancer binding protein epsilon is a potential retinoid target gene in acute promyelocytic leukemia treatment. J Clin Invest. 1999;103:1399–1408. doi: 10.1172/JCI2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann K, Buchwald M, Müller S, et al. Differential regulation of PML–RARα stability by the ubiquitin ligases SIAH1/SIAH2 and TRIAD1. Int J Biochem Cell Biol. 2012;44:132–138. doi: 10.1016/j.biocel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Quintás-Cardama A, Santos FP, Garcia-Manero G. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia. 2011;25:226–235. doi: 10.1038/leu.2010.276. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Nieves-Neira W, Boon C, et al. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem. 2000;275:9390–9395. doi: 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- Rosato RR, Almenara JA, Grant S. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 1. Cancer Res. 2003;63:3637–3645. [PubMed] [Google Scholar]
- Samudio I, Konopleva M, Carter B, Andreeff M. Apoptosis in leukemias: regulation and therapeutic targeting. Cancer Treat Res. 2010;145:197–217. doi: 10.1007/978-0-387-69259-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gonzalez B, Yang H, Bueso-Ramos C, et al. Antileukemia activity of the combination of an anthracycline with a histone deacetylase inhibitor. Blood. 2006;108:1174–1182. doi: 10.1182/blood-2005-09-008086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda T, Okamoto T, Uchida Y, et al. Proteome analyses of the growth inhibitory effects of NCH-51, a novel histone deacetylase inhibitor, on lymphoid malignant cells. Leukemia. 2007;21:2344–2353. doi: 10.1038/sj.leu.2404902. [DOI] [PubMed] [Google Scholar]
- Sanford D, Lo-Coco F, Sanz MA, et al. Tamibarotene in patients with acute promyelocytic leukaemia relapsing after treatment with all-trans retinoic acid and arsenic trioxide. Br J Haematol. 2015;171:471–477. doi: 10.1111/bjh.13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C, Forster K, Dierks H, et al. The effects of Bcr-Abl on C/EBP transcription-factor regulation and neutrophilic differentiation are reversed by the Abl kinase inhibitor imatinib mesylate. Blood. 2003;101:655–663. doi: 10.1182/blood-2002-01-0043. [DOI] [PubMed] [Google Scholar]
- Serio KJ, Reddy KV, Bigby TD. Lipopolysaccharide induces 5-lipoxygenase-activating protein gene expression in THP-1 cells via a NF-kappaB and C/EBP-mediated mechanism. Am J Physiol Cell Physiol. 2005;288:C1125–C1133. doi: 10.1152/ajpcell.00296.2004. [DOI] [PubMed] [Google Scholar]
- Shang Y, Baumrucker CR, Green MH. The induction and activation of STAT1 by all-trans-retinoic acid are mediated by RAR beta signaling pathways in breast cancer cells. Oncogene. 1999;18:6725–6732. doi: 10.1038/sj.onc.1203084. [DOI] [PubMed] [Google Scholar]
- Spange S, Wagner T, Heinzel T, Krämer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Stein B, Cogswell PC, Baldwin AS. Functional and physical associations between NF-kappa B and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassara M, Döhner K, Brossart P, et al. Valproic acid in combination with all-trans retinoic acid and intensive therapy for acute myeloid leukemia in older patients. Blood. 2014;123:4027–4036. doi: 10.1182/blood-2013-12-546283. [DOI] [PubMed] [Google Scholar]
- Thirugnanam R, George B, Chendamarai E, et al. Comparison of clinical outcomes of patients with relapsed acute promyelocytic leukemia induced with arsenic trioxide and consolidated with either an autologous stem cell transplant or an arsenic trioxide-based regimen. Biol Blood Marrow Transpl. 2009;15:1479–1484. doi: 10.1016/j.bbmt.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Truong B-THT, Lee Y-JJ, Lodie TA, et al. CCAAT/enhancer binding proteins repress the leukemic phenotype of acute myeloid leukemia. Blood. 2003;101:1141–1148. doi: 10.1182/blood-2002-05-1374. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou W, Zheng Z, et al. The HDAC inhibitor depsipeptide transactivates the p53/p21 pathway by inducing DNA damage. DNA Repair (Amst) 2012;11:146–156. doi: 10.1016/j.dnarep.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Weber M, Sydlik C, Quirling M, et al. Transcriptional inhibition of interleukin-8 expression in tumor necrosis factor-tolerant cells: evidence for involvement of C/EBP beta. J Biol Chem. 2003;278:23586–23593. doi: 10.1074/jbc.M211646200. [DOI] [PubMed] [Google Scholar]
- Wells CE, Bhaskara S, Stengel KR, et al. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PLoS One. 2013;8:e68915. doi: 10.1371/journal.pone.0068915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm AL, Timchenko NA, Darlington GJ. C/EBPalpha regulates generation of C/EBPbeta isoforms through activation of specific proteolytic cleavage. Mol Cell Biol. 1999;19:1695–1704. doi: 10.1128/mcb.19.3.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Wieczorek M, Ginter T, Brand P, et al. Acetylation modulates the STAT signaling code. Cytokine Growth Factor Rev. 2012;23:293–305. doi: 10.1016/j.cytogfr.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Wilson PM, Danenberg PV, Johnston PG, et al. Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat Rev Clin Oncol. 2014;11:282–298. doi: 10.1038/nrclinonc.2014.51. [DOI] [PubMed] [Google Scholar]
- Wilting RH, Yanover E, Heideman MR, et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29:2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Rao A, Avramis E, et al. Exposure to a histone deacetylase inhibitor has detrimental effects on human lymphocyte viability and function. Cancer Immunol Res. 2014;2:459–468. doi: 10.1158/2326-6066.CIR-13-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang N, Zhou WH, et al. Up-regulation of P21 inhibits TRAIL-mediated extrinsic apoptosis, contributing resistance to SAHA in acute myeloid leukemia cells. Cell Physiol Biochem. 2014;34:506–518. doi: 10.1159/000363018. [DOI] [PubMed] [Google Scholar]
- Xia C, Cheshire JK, Patel H, Woo P. Cross-talk between transcription factors NF-kappa B and C/EBP in the transcriptional regulation of genes. Int J Biochem Cell Biol. 1997;29:1525–1539. doi: 10.1016/s1357-2725(97)00083-6. [DOI] [PubMed] [Google Scholar]
- Ying M, Zhou X, Zhong L, et al. Bortezomib sensitizes human acute myeloid leukemia cells to all-trans-retinoic acid-induced differentiation by modifying the RARα/STAT1 axis. Mol Cancer Ther. 2013;12:195–206. doi: 10.1158/1535-7163.MCT-12-0433. [DOI] [PubMed] [Google Scholar]
- Yurek-George A, Cecil AR, Mo AH, et al. The first biologically active synthetic analogues of FK228, the depsipeptide histone deacetylase inhibitor. J Med Chem. 2007;50:5720–5726. doi: 10.1021/jm0703800. [DOI] [PubMed] [Google Scholar]
- Zhang X-WW, Yan X-JJ, Zhou Z-RR, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwergal A, Quirling M, Saugel B, et al. C/EBP beta blocks p65 phosphorylation and thereby NF-kappa B-mediated transcription in TNF-tolerant cells. J Immunol. 2006;177:665–672. doi: 10.4049/jimmunol.177.1.665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.