Abstract
PRDM16 is a transcriptional coregulator involved in translocations in acute myeloblastic leukemia (AML), myelodysplastic syndromes, and T acute lymphoblastic leukemia that is highly expressed in and required for the maintenance of hematopoietic stem cells (HSCs), and can be aberrantly expressed in AML. Prdm16 is expressed as full-length (fPrdm16) and short (sPrdm16) isoforms, the latter lacking the N-terminal PR domain. The role of both isoforms in normal and malignant hematopoiesis is unclear. We show here that fPrdm16 was critical for HSC maintenance, induced multiple genes involved in GTPase signaling, and repressed inflammation, while sPrdm16 supported B cell development biased toward marginal zone B cells and induced an inflammatory signature. In a mouse model of human MLL-AF9 leukemia, fPrdm16 extended latency, while sPrdm16 shortened latency and induced a strong inflammatory signature, including several cytokines and chemokines that are associated with myelodysplasia and with a worse prognosis in human AML. Finally, in human NPM1-mutant and in MLL-translocated AML, high expression of PRDM16, which negatively impacts outcome, was associated with inflammatory gene expression, thus corroborating the mouse data. Our observations demonstrate distinct roles for Prdm16 isoforms in normal HSCs and AML, and identify sPrdm16 as one of the drivers of prognostically adverse inflammation in leukemia.
Keywords: Hematology, Stem cells
Keywords: Adult stem cells, Bone marrow, Leukemias
Introduction
Many genes mutated or translocated in leukemia play a role in the function or development of hematopoietic stem cells (HSCs), mostly quiescent cells that reside in the bone marrow (BM), can self-renew, and generate all cells of the hematopoietic system (1). One such gene is PRD-BF1 and RIZ homology (PR) domain–containing 16 (PRDM16), a highly conserved (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI99862DS1) 140-kDa zinc finger transcriptional coregulator that is a fusion partner in t(1;3)(p36;q21) and t(1;21)(p36;q22) translocations in human acute myeloblastic leukemia (AML) (2–4). Similar translocations were found in myelodysplasia (5, 6) and in adult T cell lymphoblastic leukemia (7). Prdm16 is selectively expressed in HSCs, and required for their maintenance (8, 9). Prdm16 also plays important roles in nonhematopoietic tissues, as it is critical for brown fat (10, 11), craniofacial (12–15), and cardiac (16) development and for the maintenance of subventricular gray zone neural stem cells (9).
PRDM16 belongs to the PRDM protein family. In addition to PRDM16, several PRDM family members are involved in malignancy (17, 18), most notably MDS1/EVI1 (PRDM3), which is translocated in AML; BLIMP1 (PRDM1), which is often silenced in diffuse large B cell lymphoma (19); and PRDM2, PRDM5, and PRDM10, which are silenced in several solid tumors (17). Many members of the PRDM family, including PRDM16, are expressed as 2 distinct isoforms. Full-length proteins contain an N-terminal PR domain, with homology to SET domains, which catalyze protein lysine methylation. However, in all PRDM proteins, the most conserved region of the SET domain, responsible for its histone methyltransferase (HMT) activity, is absent (17). Full-length PRDM16 (fPRDM16) may have H3K4 or H3K9 methylation activity, however (20, 21). The 3 N-terminal exons of fPRDM16 are absent in the short isoform (sPRDM16) in both humans and mice, which therefore lacks the PR domain (Supplemental Figure 1). Potential transcription start sites (TSSs) for sPRDM16 have been suggested in exon 1, in cotranscription with fPRDM16, in exon 2, and in intron 3 (7, 22).
While deletion of Prdm16 severely impairs HSC function (8, 9), the role of the individual isoforms in HSC regulation is unclear. We have previously shown that sPrdm16 maintains elongated mitochondria in HSCs through induction of mitofusin 2 (Mfn2). Mfn2 is required for the maintenance of HSCs with extensive lymphoid potential. Expression of Mfn2 in Prdm16–/– HSCs did not rescue function, however (23). The role of PRDM16 isoforms in hematological malignancies has also not been defined. It has been proposed that the long isoforms of several PRDM family members may be tumor suppressors in human malignancies (17, 18). This notion is based on the fact that many tumors show deletion or inactivation of a long isoform, while its overexpression induces apoptosis or cell cycle arrest. This has been demonstrated, among others, for PRDM1 (19), PRDM2 (24), and PRDM5 (25). On the other hand, PRDM14 appears to function as an oncogene in lymphoid malignancies (26). A recent study showed that fPrdm16 inhibits MLL-AF9–mediated leukemogenesis in mice through induction of Gfi1b, which in turn represses Hoxa genes (21). This effect required H3K4 methyltransferase activity of the PR domain. In these studies, no biological role could be discerned for a methyltransferase-dead mutant, suggesting that the PR-deleted isoform of PRDM16 has no biological function. Taken together, these findings suggest that fPRDM16 is a suppressor of leukemia. However, in karyotypically normal leukemias, particularly those with nucleophosmin 1 (NPM1) mutations, both PRDM16 isoforms are overexpressed to varying degrees (27), and high expression of PRDM16 in AML is associated with worse overall survival (28–31), suggesting that although fPRDM16 is a tumor suppressor, sPRDM16 may promote leukemogenesis or leukemia progression. Several lines of evidence support a role for sPRDM16 in leukemia. In translocations involving PRDM16, the PR domain is often deleted (6, 7, 18, 22, 27), and sometimes only sPRDM16 is expressed (27). These leukemias show dysplastic features and are associated with poor survival (31–33). Similarly, leukemic translocations involving the closely related family member PRDM3 (MDS1/EVI1) delete the PR domain (17, 18). The 5′ region of PRDM16 is also a frequent target of retroviral insertional mutagenesis leading to immortalization (34) and leukemia (35) in mice. While these findings could be ascribed to deletion of a full-length tumor suppressor protein, overexpression of sPrdm16, but not of fPrdm16, in progenitor cells from Trp53–/– mice induced leukemic transformation (27). Consistent with these findings, forced expression of sPrdm16 promoted leukemic transformation during HOXB4-mediated immortalization of HSCs (36). Collectively, these findings point toward a role for sPRDM16 in leukemia.
We therefore examined the role of both Prdm16 isoforms in normal HSCs and in a mouse model of human MLL-AF9 leukemia. We show here that fPrdm16 is required for normal HSC function, while sPrdm16 expression in HSCs induces inflammation and promotes the generation of a specific marginal zone–biased lymphoid progenitor population. Furthermore, we show that sPrdm16 drives a prognostically adverse inflammatory signature in AML. In contrast, while physiological expression of fPrdm16 in HSCs does not affect leukemogenesis, aberrantly expressed fPrdm16 in leukemic cells has tumor-suppressive effects.
Results
The hematopoietic phenotype of mice with conditional Prdm16 deletion.
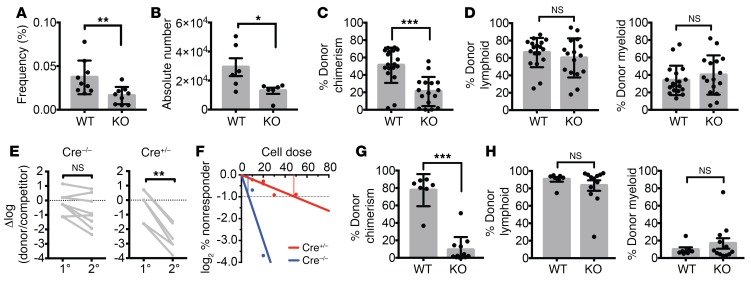
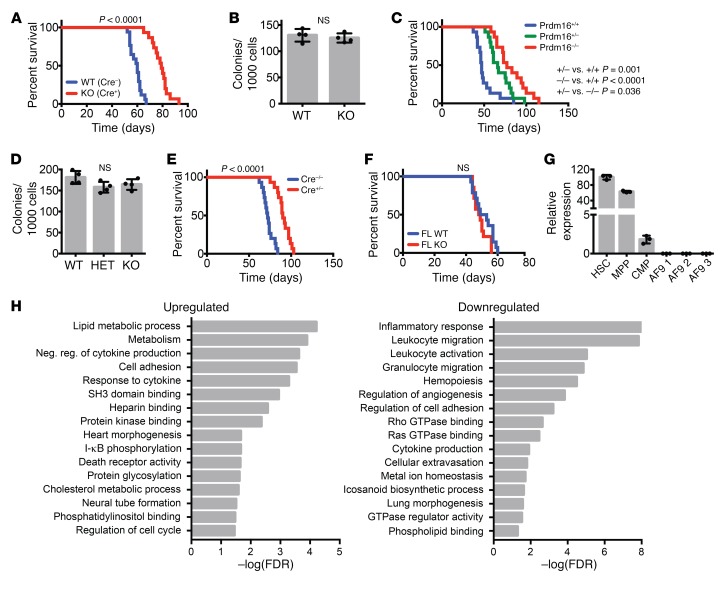
As germline-deleted Prdm16–/– mice die perinatally (8, 9), we generated Prdm16fl/fl mice and crossed these with Vav-Cre mice (37) (Prdm16fl/fl.Vav-Cre) to determine the role of isolated deletion of Prdm16 in the hematopoietic system (Supplemental Figure 2, A and B). Prdm16fl/fl.Vav-Cre mice were born in Mendelian ratios (not shown). Similarly to fetal liver (FL) HSCs from Prdm16–/– mice, the frequency and absolute number of phenotypically defined BM HSCs (Lin–Sca1+Kit+ (LSK) Flt3–CD48–CD150+; see Supplemental Figure 2C for representative analysis gates) were reduced (Figure 1, A and B), while BM cellularity was similar (not shown). Peripheral white cell counts (Supplemental Figure 2D), platelets, and hemoglobin (not shown) were similar. Competitive repopulation studies, however, revealed a profound, multilineage long-term repopulation defect (Figure 1, C and D) that became even more severe after serial transplantation (Figure 1E). Limiting-dilution competitive transplantation using purified HSCs revealed a decrease in functional HSC frequency in Prdm16fl/fl.Vav-Cre mice compared with WT littermates (1/47 vs. 1/8, respectively, P = 0.0006) (Figure 1F). Deletion of Prdm16 therefore not only decreased HSC number, but also impaired function of individual HSCs. The reconstitution defect in BM Prdm16fl/fl.Vav-Cre HSCs, however, appeared less severe than that in FL HSCs from germline-deleted Prdm16–/– mice we reported previously (8). Competitive repopulation studies using Prdm16fl/fl.Vav-Cre FL cells showed a more severe multilineage reconstitution defect similar to that previously reported by us (8) in Prdm16–/– FL HSCs, however (Figure 1, G and H). Cycling and apoptosis (Supplemental Figure 2, E and F) in adult BM HSCs, which were slightly but statistically significantly increased in Prdm16–/– FL HSCs (8), were marginally increased in Prdm16fl/fl.Vav-Cre HSCs, but this difference did not reach statistical significance. Similarly to germline Prdm16–/– FL cells, there was no difference in homing of donor cells to the BM after 24 hours in Prdm16fl/fl.Vav-Cre and WT littermate BM cells (Supplemental Figure 2G). Conditional deletion of Prdm16 within the hematopoietic system therefore recapitulates the effect of germline deletion.
Figure 1. Prdm16 supports normal HSC function.
(A and B) Frequency (n = 9) (A) and absolute number (n = 6) (B) of HSCs (Lin–cKit+Sca1+Flt3–CD48–CD150+) in BM of adult Vav-Cre–/– Prdm16fl/fl (WT) and Vav-Cre+/– Prdm16fl/fl (KO) mice. (C) Peripheral blood (PB) donor chimerism of WT or KO BM HSCs in competitive transplants with CD45.1 BM 16 weeks after transplantation (n = 16–18 mice, 3 independent transplants). (D) Percentage lymphoid (CD19+ or CD3+) and myeloid (Gr1+ or Mac1+) donor cells from C (n = 16–18 mice). (E) Change in donor/competitor ratio (log10 scale) in primary competitive transplantation recipients and secondary recipients (n = 8 mice, 2 independent transplants). (F) Limiting-dilution assay of WT versus KO BM HSCs. (G) PB donor chimerism of WT or KO FL HSCs 16 weeks after competitive transplantation (n = 8–10 mice, 2 independent transplants). (H) Percentage of donor lymphoid or myeloid donor cells from G (n = 8–10 mice). Mean ± SEM. NS, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test.
Genome-wide expression indicates regulation of GTPase signaling and mitochondrial metabolism by Prdm16.
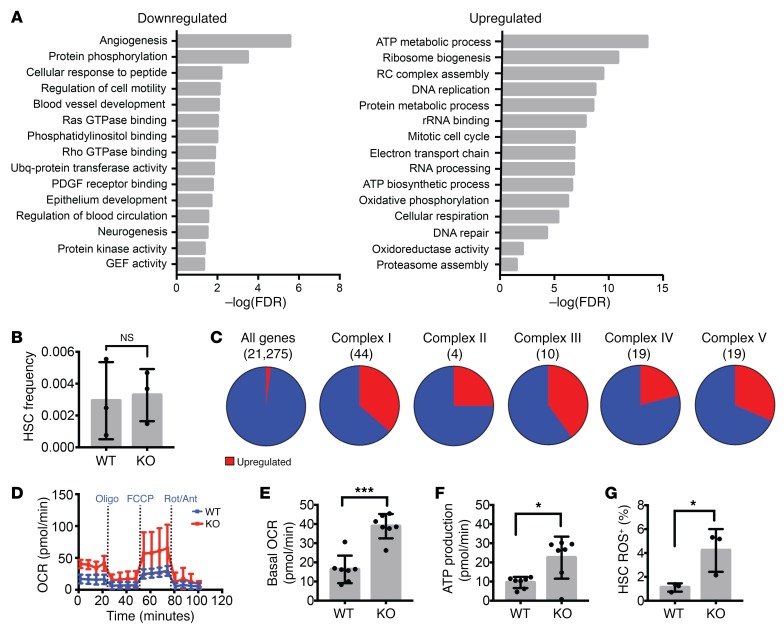
We performed genome-wide expression analysis on RNA isolated and amplified from purified LSK CD150+CD48+Flt3– HSCs (3 independent biological replicates; Supplemental Figure 3, A and B). RNA-Seq data in this publication are accessible through GEO Series accession number GSE112860 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112860). Five hundred sixty-one genes were significantly downregulated, while 411 genes were upregulated. As expected, Prdm16 mRNA was reduced in Prdm16fl/fl.Vav-Cre HSCs, with complete absence of exons 6 and 7, leading to a frameshift and a premature stop codon (Supplemental Figure 3, C and D). Pathway analysis using PANTHER (38, 39) with a 0.1 false discovery rate (FDR) cutoff showed that Prdm16 directly or indirectly regulates a broad set of pathways (Figure 2A). Rho and Ras GTPase signal transduction pathways and genes regulating cell migration and vascular development, in which Rho GTPases are involved, were significantly downregulated. As Rho signaling plays a major role in HSC homing and mobilization (40), we assessed the frequency of phenotypically defined HSCs in the peripheral blood (PB). No differences were observed between Prdm16fl/fl.Vav-Cre mice and WT littermates, however (Figure 2B). On the other hand, pathways related to mitochondrial respiration were upregulated (Figure 2A). Thirty-one of 96 electron transport chain genes were overexpressed, significantly more than expected (4/96, P < 0.0001) (Figure 2C). Measurement of metabolism using a Seahorse metabolic flux analyzer (Figure 2D) revealed elevated basal oxygen consumption (Figure 2E) and respiratory ATP production (Figure 2F) in HSCs from Prdm16fl/fl.Vav-Cre mice compared with Prdm16fl/fl littermate HSCs. Mitochondrial ROS production was also increased (Figure 2G). This effect was cell type–specific, as we did not observe enhanced respiration in Prdm16–/– mouse embryonic fibroblasts (Supplemental Figure 3E).
Figure 2. Increased respiration in adult Prdm16-deficient HSCs.
(A) GO pathways significantly up- or downregulated in Prdm16-deficient HSCs. Values expressed as –log10 of the P value, determined by PANTHER analysis. (B) HSC frequency in PB of Vav-Cre–/– Prdm16fl/fl (WT) and Vav-Cre+/– Prdm16fl/fl (KO) mice (n = 3). (C) Fraction of genes upregulated (red) in Prdm16-deficient HSCs among all genes and the 5 respiratory complexes. (D) Extracellular metabolic flux analysis of WT and KO BM HSCs (n = 3 experiments in duplicate, 5 mice per experiment). Rot, rotenone; Ant, antimycin; Oligo, oligomycin. (E and F) Basal oxygen consumption rate (OCR) (E) and respiratory ATP production (F) measured from D (n = 3). (G) ROS measured by the percentage of CellROX Deep Red–positive WT or KO BM HSCs (n = 3). Mean ± SEM. NS, P > 0.05; *P < 0.05; ***P < 0.001; Student’s t test, and χ2 test for observed vs. expected values.
We also compared FL Prdm16fl/fl.Vav-Cre and WT littermate HSCs. Similar pathways (Rho and Ras GTPase signaling, blood vessel development, and cell migration) were downregulated in Prdm16fl/fl.Vav-Cre HSCs (Supplemental Figure 3F). No significant differences were observed in mitochondrial respiration, however, indicating that regulation of respiration by Prdm16 is specific to adult HSCs. A possible explanation is that FL stem and progenitor cells are overall more oxidative than their adult counterparts (41).
Generation of fPrdm16–/– mice.
To identify specific roles of each isoform, we used CRISPR/Cas9 by pronuclear injection of guide RNA (gRNA) into fertilized C57BL/6 embryos (42). To target sPrdm16, a 500-bp region in intron 3 corresponding to a putative TSS was deleted (7) (Supplemental Figure 4A). However, using subtractive quantitative PCR (qPCR) (Supplemental Figure 4B), we did not observe reduced expression of sPrdm16 (Supplemental Figure 4C), although sequencing revealed that the putative TSS was deleted (Supplemental Figure 4D). sPrdm16 expression in HSCs therefore likely does not depend on the TSS in intron 3. Mice were born in Mendelian ratios, developed normally (not shown), and did not show HSC defects (Supplemental Figure 4, E and F). Targeting the sPrdm16 start codon would also mutate a methionine (Met-186) in fPRDM16, thus complicating the assignment of any phenotype to disruption of individual PRDM16 isoforms. This strategy was therefore not pursued.
Targeting exon 2 (Supplemental Figure 5A) yielded mouse strains with 47-bp (Δ47-fPrdm16–/–) and 13-bp (Δ13-fPrdm16–/–) frameshift deletions, respectively, leading to premature stop codons (Supplemental Figure 5B). Subtractive qPCR (Supplemental Figure 5C) and exon mapping (Supplemental Figure 5D) of RNA-Seq data in FL HSCs showed selective absence of fPrdm16 mRNA. The small amount of fPrdm16 mRNA detected would not translate to fPRDM16 protein given the frameshift within exon 2. Similarly to Prdm16–/– mice, both Δ47-fPrdm16–/– and Δ13-fPrdm16–/– mice, which developed in Mendelian ratios (Supplemental Figure 5E), died perinatally.
Hematopoietic phenotype of fPrdm16–/– mice.
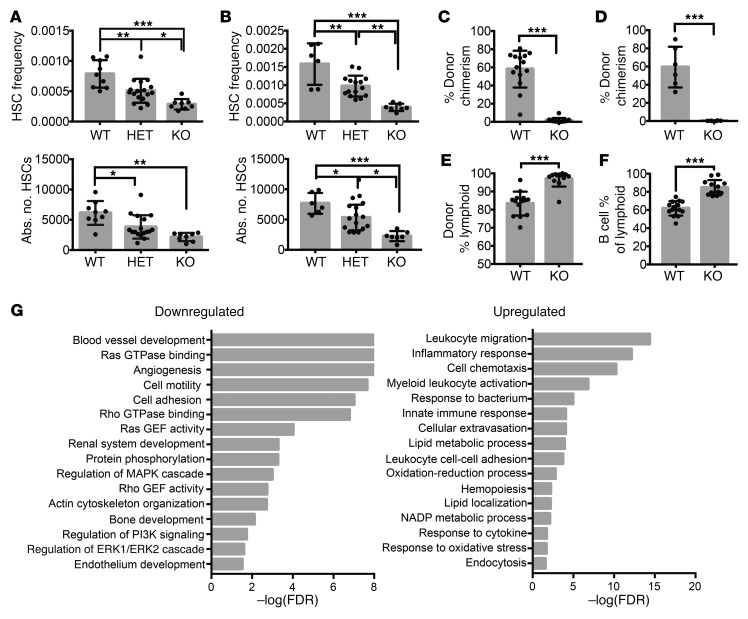
In E12.5–14.5 FL from both Δ47-fPrdm16–/– (Figure 3A) and Δ13-fPrdm16–/– mice (Figure 3B), the frequency and absolute number of HSCs were reduced, although the frequency of Lin–Sca1+Kit+ (LSK) cells was unchanged (Supplemental Figure 6A). Heterozygous mice displayed intermediate phenotypes, similar to Prdm16+/– mice. Given the similar phenotypes of Δ47-fPrdm16–/– and Δ13-fPrdm16–/– embryos, subsequent experiments were performed using Δ47-fPrdm16–/– embryos. The similar phenotypes of Δ47-fPrdm16–/– and Δ13-fPrdm16–/– embryos indicated that these were not caused by off-target effects. Furthermore, as any off-target indels would assort randomly after mating of Δ47-fPrdm16+/– heterozygotes and as WT littermates were used as controls, phenotypes described can be assigned with confidence to deletion of fPrdm16. This notion is further supported by the absence of a phenotype in mice in which the putative sPrdm16 TSS was targeted and in which fPrdm16 was intact, which can be considered nontargeting controls for fPrdm16. The fraction of cycling and apoptotic cells did not differ appreciably between Δ47-fPrdm16–/– and WT littermate FL HSCs (Supplemental Figure 6, B and C). In competitive transplantation studies, FL cells from Δ47-fPrdm16–/– mice showed a severe repopulation defect in PB (Figure 3C) and BM (Figure 3D) compared with FL cells from WT littermates. As in germline Prdm16–/– FL and Prdm16fl/fl.Vav-Cre BM cells, there was no defect in homing of donor cells to the BM after 24 hours in fPrdm16–/– FL cells (Supplemental Figure 5F). However, although FL HSCs typically show more pronounced lymphoid potential than adult BM HSCs, the Δ47-fPrdm16–/––derived donor cells displayed an even stronger and nearly absolute lymphoid bias (Figure 3E), and, within the lymphoid compartment, a bias toward B cells (Figure 3F). Such differentiation bias was not present in Prdm16–/– (8) or Prdm16fl/fl.Vav-Cre HSCs (Figure 1, D and H). Collectively, these data indicate that fPrdm16 is required for normal HSC function, but that sPrdm16 supports at least some lymphopoiesis.
Figure 3. Hematopoietic phenotype of fPrdm16-deficient mice.
(A) HSC frequency and absolute number (Lin–cKit+Sca+Mac1+CD48–CD150+) in FL of Δ47-fPrdm16–/– (KO), Δ47-fPrdm16+/– (HET), and WT littermate mice (n = 34 mice). (B) Analysis of Δ13-fPrdm16–/– mice, performed as in A (n = 28 mice). (C) PB donor chimerism 16 weeks after competitive transplantation of WT or KO FL HSCs (n = 12–14 mice, 3 independent experiments). (D) BM donor chimerism in recipient mice from C 16 weeks after transplantation (n = 6 mice). (E) Percentage lymphoid (CD19+ or CD3+) donor cells in PB from C (n = 12–14). (F) Percentage of B cells (CD19+) among lymphoid cells in E. (G) GO pathways significantly up- or downregulated in Δ47-fPrdm16–/– FL HSCs. Values expressed as –log10 of the P value, determined by PANTHER analysis. Mean ± SEM. NS, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; 1-way ANOVA for multiple comparisons or Student’s t test for single comparisons.
Genome-wide expression profiling of fPrdm16–/– HSCs.
RNA-Seq on purified WT and Δ47-fPrdm16–/– HSCs revealed 578 upregulated and 694 downregulated genes in Δ47-fPrdm16–/– HSCs. Top up- and downregulated genes and principal component analysis are shown in Supplemental Figure 7. As in Prdm16fl/fl.Vav-Cre FL and adult BM HSCs, pathways involving small GTPase signaling including cell motility, Rho and Ras GTPase binding and GEF activity, actin organization, vasculogenesis, and angiogenesis were downregulated in Δ47-fPrdm16–/– HSCs (Figure 3G), suggesting that these pathways are specifically induced by fPrdm16, and that their downregulation may contribute to the severe defects in Δ47-fPrdm16–/– and Prdm16fl/fl.Vav-Cre HSCs. A list of the top genes in these pathways reduced in both Δ47-fPrdm16–/– and Prdm16fl/fl.Vav-Cre HSCs is given in Supplemental Table 1. On the other hand, Δ47-fPrdm16–/– HSCs showed an increase in immune and inflammatory pathways. Although pathway analysis with rigorous multiple testing correction and an FDR cutoff of 0.1 did not reveal enhanced inflammation in WT compared with Prdm16fl/fl.Vav-Cre HSCs, an inflammatory signature was overrepresented in WT cells compared with Prdm16fl/fl.Vav-Cre HSCs at an FDR of 0.13. These findings indicate that sPrdm16 induces an inflammatory signature that is repressed by fPrdm16.
sPrdm16 supports the generation of Lin–Sca1+Kit+ lymphoid progenitors.
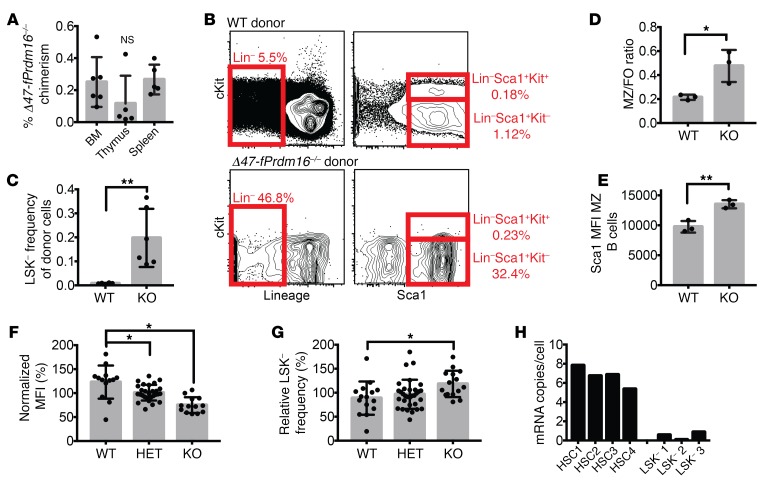
We further investigated the apparent lymphoid and B cell bias in recipients of Δ47-fPrdm16–/– cells. Donor repopulation in recipients of Δ47-fPrdm16–/– cells was reduced to a similar extent in spleen, thymus, and BM (Figure 4A). In BM, donor-derived LSK cells, common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs) were similarly reduced in recipients of Δ47-fPrdm16–/– cells and of WT littermate cells (Supplemental Figure 6D). However, a population of Lin–Sca1+Kit– (LSK–) cells was strikingly overrepresented relative to other donor populations in the BM of recipients of Δ47-fPrdm16–/– cells, and LSK– cells were, among Δ47-fPrdm16–/– donor cells, approximately 20-fold more frequent compared with donor LSK– cells in recipients of WT littermate FL cells (Figure 4, B and C).
Figure 4. sPrdm16 supports development of an LSK– B cell progenitor.
(A) Donor chimerism in BM, thymus, and spleen 16 weeks after competitive transplantation of Δ47-fPrdm16–/– FL HSCs (n = 6). (B) Representative flow cytometry plots showing gating of Lin–Sca1+Kit– (LSK– cells) in BM of recipients of Δ47-fPrdm16–/– (KO) and WT littermate FL cells (n = 6). (C) Donor LSK– frequency in recipients of WT and KO FL cells (n = 6). (D) Ratio of marginal zone (CD21hiCD23lo) to follicular B cells (CD23hiCD21lo) (MZ/FO) among donor splenic B cells (CD19+) in recipients of WT and KO FL cells (n = 3). (E) Sca1 mean fluorescence intensity (MFI) of donor MZ cells (n = 3). (F) Relative CD150 MFI of FL HSCs (n = 54 mice). (G) LSK– frequency (n = 73 mice) in FL from WT, Δ47-fPrdm16+/– (HET), and KO FL expressed as a percentage relative to litter average. (H) Prdm16 mRNA copies per cell in HSC and LSK– populations from 8-week-old WT mice (n = 3–4 mice, in triplicate). Mean ± SEM. NS, P > 0.05; *P < 0.05; **P < 0.01; 1-way ANOVA for multiple comparisons or Student’s t test for single comparisons.
Previous work from our group has shown that the LSK– population, which does not express cKit but expresses more Sca1 than LSK cells, contains a lymphoid progenitor distinct from CLPs, primarily possesses B cell potential, and displays a higher propensity to generate splenic marginal zone (MZ) B cells compared with CLPs. Furthermore, B cells generated from LSK– cells express more Sca1 than those derived from CLPs (43, 44). Analysis of the spleens of recipient mice revealed that the fraction of donor-derived MZ B cells was significantly higher in recipients of Δ47-fPrdm16–/– cells than in recipients of WT littermate cells (Figure 4D and Supplemental Figure 6E). Furthermore, Δ47-fPrdm16–/– MZ B cells expressed more Sca1 than WT littermate–derived MZ B cells (Figure 4E and Supplemental Figure 6F). These findings are consistent with B cell development that is predominantly derived from LSK– cells in Δ47-fPrdm16–/– FL cells. Further analysis of the stem and progenitor compartment in the FL of Δ47-fPrdm16–/– embryos showed lower CD150 mean fluorescence intensity (Figure 4F) compared with WT littermates. As low CD150 expression is associated with higher lymphoid potential (45, 46), these findings are consistent with the lymphoid bias of Δ47-fPrdm16–/– HSCs. Finally, although LSK– cells are rare in FL (43, 44), FL from Δ47-fPrdm16–/– embryos contained more LSK– cells compared with WT (Figure 4G). While highly expressed in HSCs as reported previously, Prdm16 mRNA was nearly undetectable in LSK– cells (Figure 4H). It is therefore most likely that sPrdm16 is required for the development of LSK– cells from HSCs, and not for their maintenance and differentiation into B cells.
Collectively, these results indicate that fPrdm16 promotes maintenance of HSCs and is not redundant with sPrdm16. Furthermore, both Prdm16 isoforms play distinct roles in HSCs as they drive different genome-wide expression signatures and as sPrdm16 is required for the development of LSK– lymphoid progenitors from HSCs.
Expression of sPrdm16 in HSCs is sufficient to enhance the progression of leukemia.
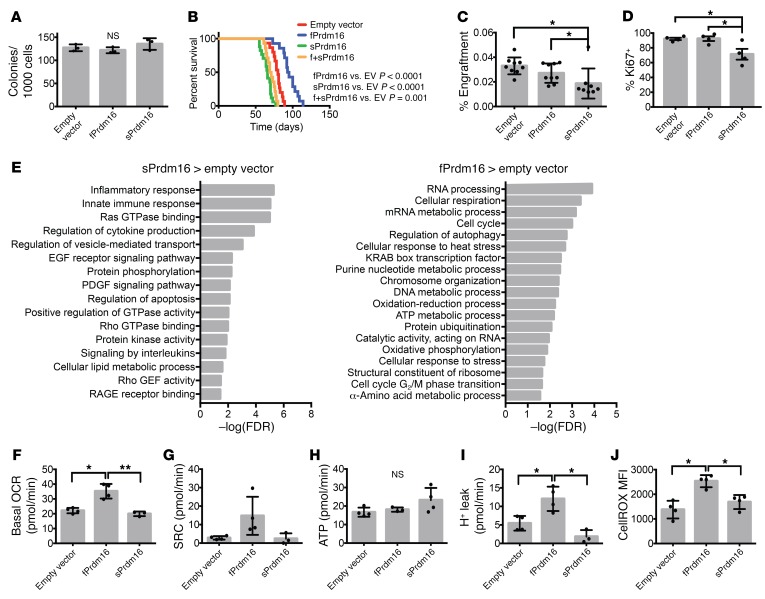
Next, we examined the role of Prdm16 in AML using retroviral transduction of HSCs with the MLL-AF9 fusion gene and an hNGFR reporter as a model (Supplemental Figure 8A) (47). Transduced purified adult BM Prdm16fl/fl.Vav-Cre, FL Prdm16–/–, FL Δ47-fPrdm16–/–, and appropriate WT littermate HSCs were expanded for 3–4 days, and 2 × 104 hNGFR+ cells were transplanted into irradiated recipient mice together with 2 × 105 supporting WT BM cells (Supplemental Figure 8, B and C). Multiple independent experiments from independent retroviral transductions were performed to avoid biological artifacts due to specific integration sites. Latency was significantly extended in recipients of immortalized Prdm16fl/fl.Vav-Cre compared with Prdm16fl/fl cells (Figure 5A). However, we observed no differences in AML-CFU formation and proliferation in vitro (Figure 5B). Leukemia was confirmed by accumulation of Mac1+Gr1+NGFR+ cells in PB (Supplemental Figure 8D) and H&E staining of PB (Supplemental Figure 8E). Similar data were obtained after transplantation of MLL-AF9–transduced FL HSCs from germline-deleted Prdm16–/– mice (Figure 5, C and D). Consistent with the haploinsufficiency with respect to HSC function in Prdm16+/– mice, leukemia latency in recipients of Prdm16+/– cells was also intermediate between that in recipients of WT and Prdm16–/– cells (Figure 5, C and D). Transduction of Lin–Sca1–Kit+ progenitors (a population containing GMPs, CMPs, and MEPs) followed by transplantation also yielded a longer latency in Prdm16-deficient cells (Figure 5E). Retroviral transduction of both sPrdm16 and fPrdm16 (Supplemental Figure 8F) partially restored latency to that of WT cells (Supplemental Figure 8G). In contrast, however, latency was similar in recipients of Δ47-fPrdm16–/– and WT littermate immortalized cells (Figure 5F). sPrdm16 is therefore sufficient to shorten latency to that observed in recipients of WT cells, while physiological expression of fPrdm16 in HSCs does not play a role in leukemogenesis.
Figure 5. sPrdm16 expression in HSCs shortens latency of MLL-AF9 leukemia.
(A) Survival of lethally irradiated mice transplanted with BM HSC-derived MLL-AF9 cells from Vav-Cre–/– Prdm16fl/fl (WT) and Vav-Cre+/– Prdm16fl/fl (KO) mice. (B) Colony-forming assays of MLL-AF9 cells from A (n = 4 independent assays in duplicate). (C) Survival of lethally irradiated mice transplanted with Prdm16+/+ (WT), Prdm16+/– (HET), or Prdm16–/– (KO) FL HSC-derived MLL-AF9 cells. (D) Colony-forming assays of MLL-AF9 cells from C (n = 4 independent assays in duplicate). (E) Survival of lethally irradiated mice transplanted with MLL-AF9 cells generated from BM Lin–Sca1–Kit+ cells from Vav-Cre–/– Prdm16fl/fl (WT) and Vav-Cre+/– Prdm16fl/fl (KO) mice. (F) Survival of lethally irradiated mice transplanted with FL HSC-derived MLL-AF9 cells from Δ47-fPrdm16–/– (KO) or WT littermate mice. (G) Expression of Prdm16 relative to HSC controls in stem and progenitor cells and in MLL-AF9 leukemic cells (n = 3, in triplicate). MPP, multipotent progenitor. (H) GO pathways significantly up- or downregulated in KO relative to WT MLL-AF9 cells from A. Values expressed as –log10 of the P value, determined by PANTHER analysis. Mean ± SEM. NS, P > 0.05; 1-way ANOVA for multiple comparisons, Gehan-Breslow-Wilcoxon test for comparison of survival curves; n = 13–15 recipients from 3 independently derived MLL-AF9 lines for each of the survival experiments in A, C, E, and F.
To determine whether expression of Prdm16 in leukemic cells or in the cell of origin was critical, we examined Prdm16 mRNA expression. Prdm16 mRNA was undetectable in leukemic cells (Figure 5G). Expression of sPrdm16 in the cell of origin was therefore likely the determinant of latency. An inheritable, epigenetic, leukemia-promoting effect of sPrdm16 is therefore plausible.
RNA-Seq (3 independent experiments) revealed a broad array of differentially regulated pathways. Eight hundred seventeen genes were significantly upregulated and 708 genes were downregulated in Prdm16fl/fl.Vav-Cre MLL-AF9 cells. Most strikingly, inflammatory and GTPase pathways were downregulated in Prdm16fl/fl.Vav-Cre leukemic cells (Figure 5H). This finding is consistent with the previously described expression signature of Prdm16fl/fl.Vav-Cre HSCs.
Distinct roles of Prdm16 isoforms expressed in leukemic cells.
Although Prdm16 was undetectable in MLL-AF9 leukemic cells, aberrant expression of PRDM16 is frequently observed in human AML and is associated with poor prognosis (27–31). We therefore examined the effect of forced expression of each isoform in leukemic cells. To avoid the confounding effect of endogenous expression of Prdm16 in HSCs, which promotes leukemogenesis (Figure 5), we transduced each isoform separately or together in MLL-AF9 immortalized HSCs from Prdm16fl/fl.Vav-Cre mice (Supplemental Figure 8, A and F). Similar GFP fluorescence indicated similar expression of each isoform in the respective lines (Supplemental Figure 8H). If the longer cDNA of fPrdm16 would impair transcription or translation compared with sPrdm16, GFP fluorescence should be lower in cells transduced with fPrdm16, as GFP is expressed off the IRES sequence. As with Prdm16 deletion, there were no differences in AML-CFU or overall growth in vitro among MLL-AF9 cells expressing either isoform (Figure 6A). However, sPrdm16 shortened latency while fPrdm16 further increased latency in recipients of Prdm16fl/fl.Vav-Cre MLL-AF9 cells (Figure 6B). Latency after coexpression of both isoforms was in between those extremes. These differences in latency were not caused by changes in engraftment, as 24-hour engraftment experiments showed that, in fact, fPrdm16-expressing cells engrafted more efficiently than sPrdm16-expressing cells (Figure 6C). Cytological analysis showed increased fragmented nuclei in cells expressing sPrdm16 (Supplemental Figure 8, I and J), a finding consistent with the dysplastic changes observed in AML with Prdm16 translocations (5, 6, 18, 28, 32, 33), where the PR domain is deleted. sPrdm16-expressing cells also included fewer cycling cells than those expressing fPrdm16 or empty vector (Figure 6D). These findings suggest an oncogenic role for sPrdm16 and a tumor suppressor role for fPrdm16 when expressed in leukemic cells that is not directly related to cycling or engraftment potential.
Figure 6. Distinct roles of Prdm16 isoforms in MLL-AF9 leukemia.
(A) Colony-forming assays of Prdm16-deficient Vav-Cre+/– Prdm16fl/fl (KO) MLL-AF9 cells expressing empty vector, fPrdm16, or sPrdm16 (n = 3 independent assays in duplicate). (B) Survival of lethally irradiated mice transplanted with MLL-AF9 cells expressing empty vector, fPrdm16, sPrdm16, or both (n = 14–15 recipients from 3 independent experiments). (C) Percentage of MLL-AF9 cells in BM of recipient mice 24 hours after transplantation (n = 9 recipients from 3 transplants). (D) Percentage Ki67+ cells among MLL-AF9 cells of leukemic mice (n = 4 recipients). (E) GO pathways significantly upregulated in sPrdm16- or fPrdm16-expressing MLL-AF9 cells isolated from leukemic mice. Values expressed as –log10 of the P value, determined by PANTHER analysis. (F) Basal oxygen consumption rate (OCR). (G) Spare respiratory capacity (SRC) (P[EV/fPrdm16] = 0.07, P[fPrdm16/sPrdm16] = 0.08). (H) Respiratory ATP production. (I) Proton leak in MLL-AF9 cells from leukemic mice. n = 4 recipients (F–I). (J) ROS measured by MFI of CellROX Deep Red in MLL-AF9 cells (n = 4). Mean ± SEM. NS, P > 0.05; *P < 0.05; **P < 0.01; 1-way ANOVA for multiple comparisons, Gehan-Breslow-Wilcoxon test for comparison of survival curves.
Enhanced inflammation induced by sPrdm16 in leukemic cells.
Expression profiles obtained by RNA-Seq from leukemic cells isolated from moribund mice in each cohort were compared with that of empty vector–transduced cells. Three hundred ninety-eight genes were significantly upregulated and 760 genes were significantly downregulated in fPrdm16-expressing Prdm16fl/fl.Vav-Cre MLL-AF9 cells. A much larger set of genes was regulated by sPrdm16: 1,608 genes were upregulated and 1,924 genes were downregulated. The top 50 genes up- and downregulated in each cohort, as well principal component analysis, are shown in Supplemental Figure 9, A and B. Differentially regulated pathways are shown in Figure 6E and Supplemental Figure 9C. fPrdm16, but not sPrdm16, upregulated respiration and oxidative phosphorylation pathways. Metabolic flux analysis showed that fPrdm16-expressing leukemic cells displayed higher basal oxygen consumption (Figure 6F) while higher spare respiratory capacity just failed to reach significance (Figure 6G) in comparison with control or sPrdm16-expressing cells. However, respiratory ATP production was similar in all groups (Figure 6H), while fPrdm16-expressing cells displayed increased proton leak (Figure 6I) and ROS production (Figure 6J). While enhanced uncoupling, as observed in brown fat, might play a role, no induction of Ucp1 was observed (not shown). On the other hand, sPrdm16 strikingly induced immune and inflammatory pathways, a signature we also found associated with expression of sPrdm16 in HSCs and in MLL-AF9 leukemia derived from WT compared with Prdm16fl/fl.Vav-Cre HSCs. Of the 418 genes in the Gene Ontology (GO) term “inflammatory process,” 151 were upregulated in at least 1 of these data sets. Of those, 56 (37%) were upregulated in at least 2, and 13 (9%) in all 3 data sets (Supplemental Table 2). sPrdm16 also induced several GO terms related to GTPase signaling (Figure 6E). As similar pathways were found to depend on the presence of fPrdm16 in normal HSCs, these data indicate that sPrdm16 also has context-dependent effects on gene expression signatures. Finally, as a previous report suggested that fPrdm16 but not a mutant without HMT activity induces Gfi1b and in doing so represses Hoxa genes to prevent leukemogenesis (21), we specifically analyzed expression of Gfi1b and Hoxa cluster genes. We found, however, that Gfi1b expression was very low and that fPrdm16 did not repress any Hoxa genes (Supplemental Table 3).
Association between PRDM16 and inflammation in NPM1-mutant and MLL leukemias.
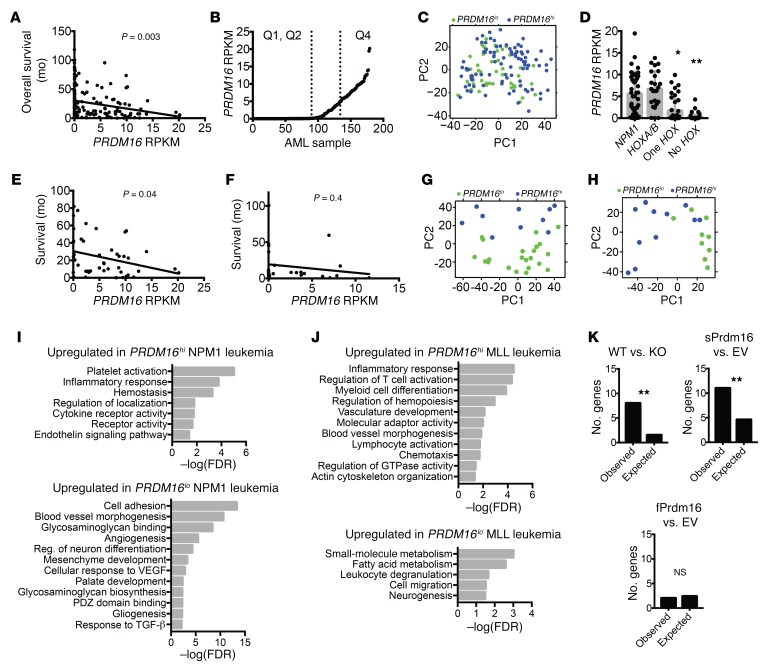
To explore the relation between PRDM16, inflammation, and leukemia progression in human AML, we used publicly available gene expression data from the Cancer Genome Atlas (48). Among the 179 AML samples, PRDM16 expression (as calculated by reads per kilobase of transcript per million mapped reads [RPKM]) correlated negatively with overall survival (Figure 7A), confirming multiple reports of PRDM16 expression as a negative prognostic factor (28–31). To detect a specific impact of sPRDM16 on prognosis, we calculated the RPKM values of exons 1–3 (fPRDM16 only) and exons 4–17 (total PRDM16), using the difference between those to estimate sPRDM16. Expression of fPRDM16 and expression of sPRDM16 were correlated, indicating that both isoforms are expressed in most PRDM16-expressing leukemias and confirming previously published findings (27). sPRDM16 expression, however, had a stronger negative prognostic value and a more negative correlation coefficient with survival than fPRDM16 (Supplemental Figure 10, A–C). These data are consistent with the shortening of latency after forced expression of sPrdm16 but not fPrdm16 in the MLL-AF9 mouse model, and support a primary negative prognostic role for sPRDM16. Notably, PRDM16 expression was not correlated with expression of EVI1/PRDM3 (Supplemental Figure 10D).
Figure 7. PRDM16 is associated with an inflammatory signature in a subset of human AML.
(A) Correlation between PRDM16 RPKM and overall survival in all 179 human AML samples from the Cancer Genome Atlas (CGA) (n = 179). (B) PRDM16 RPKM for all samples from A, ranked by RPKM, illustrating Q1/Q2 (PRDM16lo) and Q4 (PRDM16hi). (C) Principal component (PC) analysis of cohorts described in B. (D) PRDM16 RPKM compared within 4 mutually exclusive groups from the CGA AML cohort: NPM1 mutated, NPM1wt HOXA9/B4 double-positive (HOXA/B), HOXA9 or HOXB4 single-positive (One HOX), and HOXA9/HOXB4 double-negative (No HOX) (n = 179). (E and F) Correlation between PRDM16 RPKM and overall survival among NPM1-mutated AML samples (n = 47) (E) and MLL-rearranged AML samples (F) in the CGA (n = 21). (G and H) Principal component analysis of NPM1-mutated (G) and MLL-rearranged (H) AML cases from the CGA, comparing PRDM16hi and PRDM16lo cohorts. (I and J) Representative list of GO pathways upregulated in PRDM16hi or PRDM16lo cohorts of NPM1-mutated (I) or MLL-rearranged (J) AML cases in the CGA. Values expressed as –log10 of the P value, determined by PANTHER analysis. (K) χ2 analysis of observed versus expected number of dysregulated MDS-related genes in common with genes from our RNA-Seq analysis in Figure 5H and Figure 6E. Data represent mean ± SEM. NS, P > 0.05; *P < 0.05; **P < 0.01; Pearson’s test for linear correlations, 1-way ANOVA for multiple comparisons, χ2 test for comparing observed vs. expected values.
We next divided the cohort into 4 quartiles based on PRDM16 RPKM, and compared samples with low expression (Q1 and Q2, PRDM16lo with RPKM <0.1) and high expression (Q4, PRDM16hi with RPKM >5.0) of PRDM16 (Figure 7B). Principal component analysis showed no discernible clustering (Figure 7C). However, among differentially expressed genes between both groups, HOX genes appeared overrepresented (Supplemental Table 4). Thirteen of 40 HOX genes were upregulated in the PRDM16hi cohort, significantly more than expected (0.53/40, P < 0.001). We compared expression of PRDM16 after stratification based on HOX cluster expression, using HOXA9 and HOXB3 as representative genes. Quite strikingly, in HOX-negative AML and in cases in which only 1 HOX cluster was upregulated, PRDM16 expression was low to undetectable. Of the 66 HOX-negative samples, none had a PRDM16 RPKM greater than 5, and only 4 of 66 cases (6%) had a PRDM16 RPKM greater than 1. In contrast, in HOXA/B–double-positive AML and NPM1-mutated AML, in which both HOXA and HOXB genes are upregulated (49), the mean PRDM16 RPKM was greater than 5 (Figure 7D). As in the MLL-AF9 mouse model, no repression of HOX genes could be discerned, the association between PRDM16 and HOX cluster gene expression in human AML indicates that PRDM16 does not, as has been suggested (21), repress HOX genes, but rather that HOX genes may induce PRDM16.
We next focused on 2 specific AML subsets: karyotypically normal, NPM1-mutated AML (47 samples), because PRDM16 is frequently overexpressed in these leukemias (27), and MLL-rearranged leukemias (21 samples), as we found a role for Prdm16 in the MLL-AF9 mouse model and as Prdm16 downregulation has been reported to be required for pathogenesis in this model (21). As with the total AML cohort, PRDM16 expression negatively correlated with overall survival in NPM1-mutant AML (Figure 7E). These effects appeared to be independent of FLT3 or DNMT3A comutations, as a negative correlation was noted in both FLT3/DNMT3A mutant and WT populations, although sample size was likely too small to achieve statistical significance (Supplemental Figure 10, E and F). A similar trend was also present in the MLL cohort, but results were not significant (Figure 7F), again possibly owing to the smaller sample size.
We next divided both AML subsets into PRDM16hi and PRDM16lo groups, again using quartiles (Q1/Q2 vs. Q4). Principal component analysis clearly separated PRDM16hi and PRDM16lo groups both in NPM1-mutant (Figure 7G) and in MLL-translocated leukemias (Figure 7H). PANTHER pathway analysis showed that in both AML subsets PRDM16hi leukemias were associated with an upregulated inflammatory signature compared with PRDM16lo leukemias (Figure 7, I and J). That this association could not be detected in the overall AML cohort may indicate specificity to the NPM1 and MLL AML subsets, or be a result of the use of more homogenous leukemic cohorts.
Collectively, these findings indicate that PRDM16 is associated with a worse prognosis overall, and, at least within the NPM1-mutant and MLL-translocated leukemias, with an inflammatory expression signature, consistent with the inflammatory signature induced by sPrdm16 in the MLL-AF9 mouse model.
A core set of inflammatory genes induced by sPrdm16 are associated with myelodysplastic syndromes.
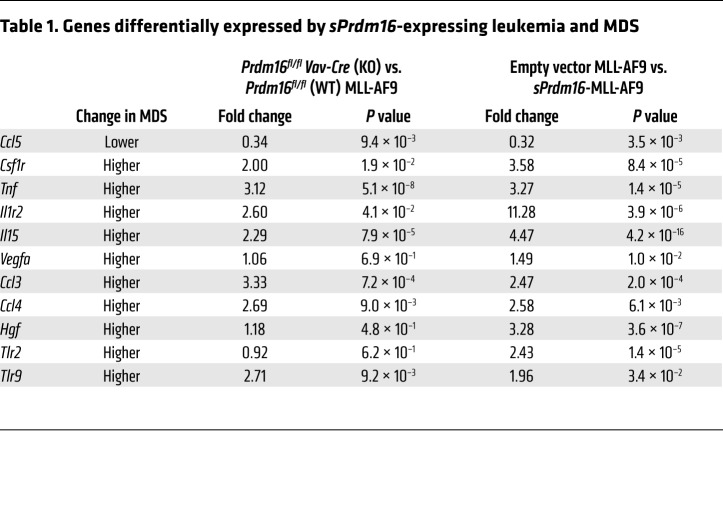
Myelodysplastic syndromes (MDS) are characterized by ineffective hematopoiesis by a dominant clone displaying enhanced proliferation and cell death, while normal hematopoiesis is suppressed. Inflammation is a key feature of MDS (50, 51). As sPrdm16-expressing MLL-AF9 cells display dysplastic features and express an inflammatory signature (Supplemental Figure 8, I and J), we extracted all genes from the GO term “inflammation” that were upregulated in sPrdm16-overexpressing compared with empty vector control cells and in WT compared with Prdm16–/– MLL-AF9 leukemic cells, and cross-referenced these with a consensus list of inflammatory genes frequently dysregulated in MDS (50). Of this MDS signature, a much higher fraction than expected through random association was also regulated by sPrdm16 (Figure 7K and Table 1). sPrdm16 therefore induces an inflammatory signature that overlaps with that observed in MDS.
Table 1. Genes differentially expressed by sPrdm16-expressing leukemia and MDS.
Discussion
We report here that the two PRDM16 isoforms, sPRDM16 and fPRDM16, have distinct roles in the maintenance of HSCs and in AML.
Prdm16 is required for normal HSC function (8, 9), and conditional deletion within the hematopoietic system indicates that this effect is cell-autonomous. We note that of the 14 genes found differentially regulated in germline Prdm16–/– HSCs by Fluidigm qPCR (8), only 2 (Mmi1 and Cdkn1c) were also found in the RNA-Seq experiments in Prdm16fl/fl.Vav-Cre and Δ47-fPrdm16–/– mice described here. The reasons may be technical. It is also possible, however, that the fact that we performed RNA-Seq on 3 independent replicates, thus decreasing statistical noise but increasing the probability of missing truly differentially expressed genes, may play a role. Regulation of pathways related to GTPase-mediated signaling, as well as pathways involved in cell motility and vasculogenesis, in which small GTPases play key roles, could be ascribed to expression of fPrdm16, which is required for HSC maintenance. These pathways play important roles in HSC function, cycling, homing, and mobilization (40, 52–55). Many genes downregulated in both Prdm16fl/fl.Vav-Cre and Δ47-fPrdm16–/– HSCs are involved in vesicle trafficking (Als2cl, ref. 56; Arhgef10, ref. 57; Rab11fip3, ref. 58) and/or are guanine exchange factors (Arhgef10; Fgd5, ref. 59; Itsn1, Obscn, Rgl1) (Supplemental Table 1). fPrdm16 may regulate endosomal trafficking in HSCs, which may play thus far underappreciated roles in HSC function. Increased mitochondrial respiration as observed in adult, but not in fetal, Prdm16fl/fl.Vav-Cre HSCs is associated with enhanced cycling and HSC exhaustion (60–65). Cycling was only slightly enhanced in Prdm16-deficient HSCs, though just shy of statistical significance. The effect of Prdm16 on respiration could not be assigned specifically to fPrdm16, as this effect was not present in FL HSCs and only FL HSCs could be assessed in Δ47-fPrdm16–/– mice. It is possible that the absence of increased respiration in Prdm16fl/fl.Vav-Cre FL HSCs is due to the already more oxidative nature of WT FL stem and progenitor cells compared with their adult counterparts (41).
Although, unfortunately, we did not succeed in generating an sPrdm16–/– mouse, at least some of the effects of sPrdm16 can be inferred from differences between the phenotypes resulting from deletion of both isoforms simultaneously and from deletion of fPrdm16 alone. We have previously shown, using overexpression studies and chromatin immunoprecipitation, that sPrdm16 induces Mfn2 in HSCs (23). Our current studies indicate a broader role for sPrdm16 in hematopoiesis. sPrdm16 is required for the generation of an LSK– lymphoid progenitor we described previously (43, 44), and most notably induces inflammatory pathways, both in HSCs and in leukemic cells. As inflammation is involved in HSC activation but is detrimental to HSC function when chronic (66), it is possible that specific deletion of sPrdm16 would also reveal a critical role for this isoform and that normal HSC function hinges on the balance between both isoforms.
In contrast to the role of both Prdm16 isoforms in the regulation of normal HSCs, we find only sPrdm16 to be relevant to leukemogenesis from HSCs, as physiological expression of sPrdm16, as in Δ47-fPrdm16–/– cells, is sufficient to restore the increased latency observed in Prdm16fl/fl.Vav-Cre MLL-AF9 leukemia to that of WT. Because in these experiments Prdm16 was not expressed in the leukemic cells, but only in the HSCs from which these were derived, it is plausible that this effect of sPrdm16 is at least in part epigenetic, a notion consistent with previous observations suggesting that epigenetics within the cell of origin has a pronounced effect on prognosis of leukemia (67).
Consistent with the induction of an inflammatory signature in normal HSCs, sPrdm16 also drives inflammation in MLL-AF9–induced leukemia in mice, both when expressed in HSCs and subsequently downregulated during leukemogenesis and after forced expression in leukemic cells. Multiple reports have indicated an association between inflammation and outcome in both AML and MDS, as well as in myeloproliferative neoplasms (MPNs) and MDS/MPN (50, 51, 68–73). Antiinflammatory and immune-suppressive treatments are currently being explored in MDS (50, 51). It is unclear, however, which genes drive inflammation in hematological malignancies, to what extent preexisting inflammation predisposes to the development of hematological malignancy, and whether the malignancy itself directly or indirectly promotes inflammation. The frequent association between preexisting autoimmune disease and the subsequent development of MDS suggests that, at least in MDS, inflammation plays a role in disease predisposition, although a role for inflammation caused by or emanating from the abnormal clone in disease progression has been suggested as well (74). Our observations indicate that at least in a subset of AML, the expression of sPRDM16 underlies an inflammatory process that originates in the leukemic clone.
The inflammatory signature induced by sPrdm16 in leukemic cells overlapped with that observed in MDS (50, 51). A core set of genes regulated by sPrdm16 and dysregulated in MDS (50) include cytokines and chemokines associated with prognosis in AML. High expression of hepatocyte growth factor (HGF) (71), vascular endothelial growth factor A (VEGFA) (70, 75), and tumor necrosis factor (TNF) (73), which are all increased in MLL-AF9 cells expressing sPrdm16, is associated with a worse prognosis. On the other hand, sPrdm16 repressed Ccl5 expression. Lower levels of CCL5 are observed in MDS (50) and adversely affect prognosis in AML (72). The inflammatory signature induced by sPRDM16 might explain why translocations involving PRDM16 can also be associated with MDS (5, 6), while AML with PRDM16 translocations displays dysplastic changes (31–33). Furthermore, we observed increased frequency of abnormal nuclei in MLL-AF9 cells expressing sPrdm16. Inflammatory cytokines and chemokines may suppress normal hematopoiesis, thus providing a competitive advantage to leukemic cells overexpressing sPRDM16, despite their lower proliferation and lower engraftment capacity. CCL3, for example, is also induced by sPRDM16, is associated with MDS (50), and inhibits leukemic proliferation and megakaryocyte/erythroid progenitors (76). A recent report lends additional support to this idea (77). AML cells in endosteal zones of the BM express inflammatory mediators and remodel endothelial cells, thus suppressing normal hematopoiesis (77). Taken together, our observations are consistent with a model wherein sPRDM16 induces inflammatory genes, and in particular secreted inflammatory cytokines, in leukemic cells and in doing so suppresses normal hematopoiesis, thus explaining its deleterious impact on outcome. This mechanism would also explain the absence of any effect of Prdm16 on proliferation and colony formation of leukemic cells in vitro.
For many PRDM genes involved in malignancy, the long form functions as a tumor suppressor (19, 24, 25). Our observations suggest that physiologically expressed fPRDM16 in HSCs does not suppress leukemogenesis, as leukemic cells from fPrdm16-deficient mice have similar latency to WT. Forced expression in leukemic cells, however, was associated with increased respiration, enhanced oxidative stress, and extended latency, suggesting tumor-suppressive activity that may counterbalance the leukemia-promoting effects of sPRDM16 in leukemias expressing PRDM16. The PRDM16 gene therefore encodes both an oncogene and a tumor suppressor. Given the negative prognostic impact of PRDM16 and in particular of sPRDM16 in human AML, the tumor-promoting effect of sPRDM16 appears to generally prevail over any tumor-suppressive actions of fPRDM16.
A previous report indicated that fPrdm16 is a tumor suppressor, but that sPrdm16 did not promote leukemia. These authors showed that the PR domain of PRDM16 harbors H3K4 methyltransferase activity (21). Using the same MLL-AF9 model, they found that fPrdm16, but not a mutant (mutPrdm16) without H3K4 activity, induced Gfi1b, which in turn reduced expression of Hoxa cluster genes and completely prevented leukemogenesis. Furthermore, knockdown of Prdm16 shortened latency. While we concur with Zhou et al. (21) that Prdm16 is downregulated during MLL-AF9–induced immortalization of HSPCs, our data are at variance with most of these findings. First of all, we do not find that fPrdm16 blocks leukemogenesis, and, in contrast to Zhou et al., we could also not detect any inhibitory effect of fPrdm16 on proliferation in vitro. Second, we find that sPrdm16, which, similarly to the mutant used by Zhou et al., cannot have any H3K4 methyltransferase activity, is biologically active, while Zhou et al. could not identify any role of mutPrdm16. Third, we could not find repression of Hoxa genes by either Prdm16 isoform. We did observe an association between PRDM16 and HOX expression in human AML, however. As Prdm16 did not affect Hoxa gene expression in the MLL-AF9 mouse model, PRDM16 is likely downstream and not upstream of HOX genes in leukemia. Indeed, Yu et al. showed that Hoxa9 and Hoxa10 induced Prdm16 (36), thus lending further support to the idea that HOX genes regulate Prdm16. There are several methodological differences between our work and that of Zhou et al. Zhou et al. evaluated the effect of fPrdm16 and mutPrdm16 through knockdown and forced expression approaches in a WT background. It is not clear whether or to what extent the knockdown of Prdm16 selectively affected one or the other isoform. Furthermore, forced expression of one isoform may induce homeostatic mechanisms aimed at balancing the expression of the respective isoforms (17). In contrast, we used mice with deletion of Prdm16 or with a selective ablation of fPrdm16, and in addition performed overexpression experiments in Prdm16-deleted leukemic cells to distinguish the biological effects of each isoform.
The mechanisms of action of PRDM16 are unclear. The largely distinct expression signatures indicate distinct mechanisms and targets for each isoform. Furthermore, at least some of the expression signatures, such as mitochondrial respiration, are context-dependent. PRDM proteins act through multiple downstream mechanisms. Although some, including PRDM16 (20, 21), may harbor HMT activity, most act by recruiting other HMTs, histone deactylases, and corepressors through zinc finger and proline-rich domains. To drive brown fat development, PRDM16 binds C/EBPα. Furthermore, they can bind DNA directly as well. To what extent any of these mechanisms are used by the long and short isoforms is unknown.
In summary, we showed here that sPrdm16 and fPrdm16 play distinct roles in the regulation of normal HSCs, and that sPrdm16 is a driver of a prognostically adverse inflammatory signature in leukemia. The association between expression of sPRDM16, inflammation, and progression of AML, and the similarities to MDS, might foster the exploration of antiinflammatory or immune-suppressive interventions that, while unlikely to be curative, might improve prognosis of a subset of AML expressing sPRDM16 and associated with inflammation.
Methods
Mice.
C57BL/6J mice (CD45.2) and competitor B6.SJL-PtprcaPep3b/BoyJ (CD45.1) mice were purchased from The Jackson Laboratory.
Prdm16fl/fl mice were generated by Ingenious Targeted Laboratory Inc. A neomycin resistance cassette flanked by FRT and a 3′-loxP site were inserted upstream of exon 6 and another loxP site was inserted downstream of exon 7 in a 10.2-kb fragment from a C57NL/6 BAC clone, extending from intron 5 through exon 9. The modified BAC clone was electroporated into mouse embryonic stem cells, and Neo-resistant clones were expanded, screened for retention of the Neo cassette and second loxP site, and injected into B6 blastocysts to generate chimeric mice. Removal of the Neo cassette was accomplished by crossing to FLP heterozygous mice, resulting in mutant, Prdm16fl/fl mice with loxP sites flanking exons 6 and 7.
Isoform-specific Prdm16-knockout mice were generated using CRISPR/Cas9. PX330-based plasmids (200 ng/μl; Addgene), containing CRISPR/Cas9 expression cassettes and specific gRNAs (Supplemental Table 5), were injected into fertilized B6 blastocysts. Mice chimeric for CRISPR-initiated mutations were bred to WT C57BL/6J. Heterozygous siblings were mated to generate a homozygous mutant. All primers and gRNA constructs were purchased from Integrated DNA Technologies. Prdm16–/– mice were obtained from Lexicon Genetics, and Vav-iCre (Vav-Cre) mice were purchased from The Jackson Laboratory.
MLL-AF9 transduction and cell culture.
pMSCV-FPRDM16-IRES-GFP/RFP and pMSCV-SPRDM16-IRES-GFP/RFP plasmids were subcloned by XhoI/EcoRI insertion of fPRDM16 or sPRDM16 cDNA into a backbone pMSCV-IRES-GFP/RFP plasmid (Addgene). pMSCV-MLL-AF9-IRES-hNGFR was subcloned by replacement of GFP from a pMSCV-MLL-AF9-IRES-GFP plasmid (Addgene) with hNGFR cDNA. Retroviral particles were produced by seeding of PlatE cells (Cell Biolabs) at 7 × 105 per cubic centimeter overnight followed by transfection of each packaging and expression construct (1:1:1) using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. Media were pooled after 48 hours, clarified and concentrated by ultracentrifugation (100,000 g), resuspended in RPMI media (Corning/Cellgro), and stored at –80°C. MLL-AF9 cells were generated by transduction of sorted HSCs (Lin–cKit+Sca1+Flt3–) or committed myeloid/erythroid progenitors (Lin–cKit+Sca1–CD16/32+) with an MSCV-MLL-AF9-hNGFR retroviral construct by spinfection at 750 g, 22°C for 90 minutes. hNGFR+ cells were sorted and expanded in RPMI media (Corning/Cellgro) containing 15% FBS (Atlanta Biologicals), 1× Glutamax (Gibco), penicillin/streptomycin (Gibco), 1× MEM Non-Essential Amino Acids (Gibco), stem cell factor (50 ng/ml), IL-6 (20 ng/ml), and IL-3 (10 ng/ml) (peptides from Peprotech). For colony-forming assays, 1,000 cells were plated in 1 ml media of the same composition containing 1.5% methylcellulose (Sigma-Aldrich), and colony number was counted after 7 days.
MLL-AF9 leukemia and HSC transplantation.
Two hundred purified HSCs, 2 × 105 unsorted BM or FL cells, or 2 × 104 purified MLL-AF9 immortalized cells, as indicated, were injected together with 2 × 105 competitor BM cells into lethally irradiated mice (2 doses of 475 cGy using a Rad Source RS-200 X-ray irradiator) by tail vein injection. Survival in leukemia experiments was determined as the number of days after transplant before mice became moribund.
Quantitative reverse transcriptase PCR.
After lysis (Trizol, Invitrogen), RNA was isolated according to the manufacturer’s instructions, using a chloroform/isopropanol extraction and 70% ethanol wash. cDNA was prepared using SuperScript III Reverse Transcriptase (Invitrogen). PCR was performed using the Viia7 Real-Time PCR System (Applied Biosystems), using either inventoried or custom TaqMan probes, or using custom primers and SYBR Green enzyme (Supplemental Table 5). Relative mRNA content was determined by the ΔΔCT method normalized to GAPDH-VIC or 18S-VIC housekeeping genes. To determine copy number of Prdm16 isoforms, an fPrdm16-specific (exon 2/3 junction) and total Prdm16 (tPrdm16) probe (exon 14/15 junction) were used. Copy number was calculated from a linear regression of serial dilutions of a Prdm16-containing plasmid, and sPrdm16 copy number was calculated via subtraction (sPrdm16 = tPrdm16 – fPrdm16).
Flow cytometry and cell staining.
Cells were analyzed on a BD LSRII or BD Fortessa flow cytometer (Becton Dickinson). Cells were sorted using a BD Influx cell sorter. For BM analysis, bones were crushed, lysed with ACK lysis buffer (Gibco, Thermo Fisher Scientific), and filtered through a 0.22-μm filter before staining. For FL analysis, livers were isolated from E13–15 embryos, passed through a 0.22-μm filter, and lysed with ACK buffer before staining. For PB analysis, blood obtained by submandibular bleeding was lysed twice with ACK buffer before staining. Staining was performed by incubation of cells with an antibody cocktail in FACS buffer for 30 minutes at 4°C and washing in FACS buffer before analysis. Antibodies are listed in Supplemental Table 5. All data were analyzed using FlowJo 9.6 (Tree Star Inc.). BM and PB from select moribund mice were stained with H&E (Vector Laboratories). Undiluted PB or BM diluted in 50 μl per femur was smeared on slides, fixed with methanol, and stained with H&E according to the manufacturer’s protocol.
Metabolic flux analysis.
Respiratory oxygen consumption rate was determined using a Seahorse XFe96 or XFp metabolic flux analyzer (Seahorse Bioscience). MLL-AF9 cells (1 × 106 cells per well) or enriched HSCs (Lin–cKit+Sca1+Flt3–, 7.5 × 104 cells per well) were suspended in unbuffered medium, equilibrated at 37°C in a CO2-free incubator, transferred to the Seahorse bioanalyzer, and assayed. The Mitochondrial Stress Test (Seahorse) was used to measure the respiratory properties of the cells.
Measurement of ROS.
Cells were suspended in 500 μl of PBS (1 × 105 cells/ml) at room temperature with 1 μl CellROX Deep Red (Thermo Fisher Scientific) for 45 minutes, washed, resuspended in FACS buffer, and analyzed by flow cytometry in the APC channel. Cells treated with both CellROX and tert-butyl hydroperoxide were positive controls.
RNA-Seq analysis.
mRNA from total RNA samples (400 ng per sample) was enriched by poly(A) pulldown. Libraries were prepared using TruSeq RNA prep kit (Illumina) and sequenced using Illumina HiSeq2000 at the Columbia Genome Center. Samples were multiplexed in each lane. Base calling was performed using RTA (Illumina). BCL and bcl2fastq programs were used to convert BCL to FASTQ format, coupled with adaptor trimming. Reads were mapped to a reference genome (UCSC/mm9) using TopHat with 4 mismatches and 10 maximum multiple hits. Binary alignment map (BAM) files were generated by TopHat to map reads to annotated genes, and converted to an annotated count matrix, using the Rsamtools and GenomicAlignments R packages. Data were then analyzed using DESeq to obtain differential expression analysis and principal component analysis. Pathway analysis was performed using PANTHER, with pathway data from the GO gene ontology database.
Statistics.
Statistical analysis was performed using GraphPad PRISM software. Primary statistical tests include 2-tailed Student’s t test for single comparisons of normally distributed data, 1-way ANOVA for multiple comparisons, Pearson’s correlation test for comparisons of PRDM16 RPKM versus survival, and Gehan-Breslow-Wilcoxon log-rank tests to compare survival of recipient mice after MLL-AF9 transplantation. A P value less than 0.05 was considered significant.
For RNA-Seq analysis, statistical P values for individual genes were calculated from the DESeq package in R using a binomial test accounting for size factors and intragene sample variance, and principal component analysis was also performed with this software. An FDR of less than 10% was used as the basis for a positive “hit.” RNA-Seq pathway analysis was performed using the PANTHER statistical overrepresentation test, with a Bonferroni correction for multiple testing. Chi-squared tests were used to test whether the observed frequency of selected “hits” from RNA-Seq analysis was larger than the expected frequency.
Study approval.
Experiments and animal care were performed in accordance with the Columbia University Institutional Animal Care and Use Committee, under the approved mouse protocol AC-AAAM4750, under which all contributing authors are approved.
Author contributions
HWS conceived the study and served as the primary author of the paper. DJC planned, performed, and analyzed the majority of the experiments, and helped to prepare the manuscript and figures. LLL developed the Prdm16fl/fl.Vav-Cre mice, planned, performed, and analyzed experiments shown in Figure 1, C–F, and contributed to experiments using the Seahorse bioanalyzer. LJW planned, performed, and analyzed experiments shown in Figure 1, G and H, and helped in the analysis of RNA-Seq data from these mice. MJDA planned, performed, and executed experiments shown in Figure 1, A and B. AS performed CuffDiff and exon analysis from Prdm16fl/fl.Vav-Cre RNA-Seq and analyzed exons using the IGV viewer as shown in Supplemental Figure 2C.
Supplementary Material
Acknowledgments
This work was supported by grants R01 CA167289, R01 HL135039, and R01 AG055910 (to HWS), as well as the Ruth L. Kirschstein award F31 CA196045 (to DJC). Research reported in this publication was performed in the Columbia Center for Translational Immunology Flow Cytometry Core, supported in part by the Office of the Director, NIH, under awards S10RR027050 and S10OD020056. This research was supported by the New York Stem Cell Foundation. AS is a New York Stem Cell Foundation–Druckenmiller Fellow. We thank Victor Lin from the Columbia Transgenic Mouse Shared Resource for advice and blastocyst injections of CRISPR/Cas9/gRNA. We also thank the JP Sulzberger Columbia Genome Center for performing RNA-Seq.
Version 1. 06/07/2018
In-Press Preview
Version 2. 07/23/2018
Electronic publication
Version 3. 08/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(8):3250–3264.https://doi.org/10.1172/JCI99862.
Contributor Information
David J. Corrigan, Email: djc2174@cumc.columbia.edu.
Larry L. Luchsinger, Email: lll2145@columbia.edu.
Mariana Justino de Almeida, Email: mj2569@columbia.edu.
Linda J. Williams, Email: lw2251@columbia.edu.
Alexandros Strikoudis, Email: as5109@cumc.columbia.edu.
Hans-Willem Snoeck, Email: hs2680@columbia.edu.
References
- 1.Rossi L, et al. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11(3):302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mochizuki N, et al. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96(9):3209–3214. [PubMed] [Google Scholar]
- 3.Sakai I, et al. Novel RUNX1-PRDM16 fusion transcripts in a patient with acute myeloid leukemia showing t(1;21)(p36;q22) Genes Chromosomes Cancer. 2005;44(3):265–270. doi: 10.1002/gcc.20241. [DOI] [PubMed] [Google Scholar]
- 4.Stevens-Kroef MJ, et al. Identification of truncated RUNX1 and RUNX1-PRDM16 fusion transcripts in a case of t(1;21)(p36;q22)-positive therapy-related AML. Leukemia. 2006;20(6):1187–1189. doi: 10.1038/sj.leu.2404210. [DOI] [PubMed] [Google Scholar]
- 5.Moir DJ, Jones PA, Pearson J, Duncan JR, Cook P, Buckle VJ. A new translocation, t(1;3) (p36;q21), in myelodysplastic disorders. Blood. 1984;64(2):553–555. [PubMed] [Google Scholar]
- 6.Xiao Z, Zhang M, Liu X, Zhang Y, Yang L, Hao Y. MEL1S, not MEL1, is overexpressed in myelodysplastic syndromes patients with t(1;3)(p36;q21) Leuk Res. 2006;30(3):332–334. doi: 10.1016/j.leukres.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida M, Nosaka K, Yasunaga J, Nishikata I, Morishita K, Matsuoka M. Aberrant expression of the MEL1S gene identified in association with hypomethylation in adult T-cell leukemia cells. Blood. 2004;103(7):2753–2760. doi: 10.1182/blood-2003-07-2482. [DOI] [PubMed] [Google Scholar]
- 8.Aguilo F, et al. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood. 2011;117(19):5057–5066. doi: 10.1182/blood-2010-08-300145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol. 2010;12(10):999–1006. doi: 10.1038/ncb2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6(1):38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet. 2010;19(5):774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaffer JR, et al. Genome-wide association study reveals multiple loci influencing normal human facial morphology. PLoS Genet. 2016;12(8):e1006149. doi: 10.1371/journal.pgen.1006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding HL, Clouthier DE, Artinger KB. Redundant roles of PRDM family members in zebrafish craniofacial development. Dev Dyn. 2013;242(1):67–79. doi: 10.1002/dvdy.23895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu F, et al. A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 2012;8(9):e1002932. doi: 10.1371/journal.pgen.1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arndt AK, et al. Fine mapping of the 1p36 deletion syndrome identifies mutation of PRDM16 as a cause of cardiomyopathy. Am J Hum Genet. 2013;93(1):67–77. doi: 10.1016/j.ajhg.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mzoughi S, Tan YX, Low D, Guccione E. The role of PRDMs in cancer: one family, two sides. Curr Opin Genet Dev. 2016;36:83–91. doi: 10.1016/j.gde.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Morishita K. Leukemogenesis of the EVI1/MEL1 gene family. Int J Hematol. 2007;85(4):279–286. doi: 10.1532/IJH97.06174. [DOI] [PubMed] [Google Scholar]
- 19.Pasqualucci L, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203(2):311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinheiro I, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150(5):948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 21.Zhou B, et al. PRDM16 suppresses MLL1r leukemia via intrinsic histone methyltransferase activity. Mol Cell. 2016;62(2):222–236. doi: 10.1016/j.molcel.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishikata I, et al. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood. 2003;102(9):3323–3332. doi: 10.1182/blood-2002-12-3944. [DOI] [PubMed] [Google Scholar]
- 23.Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M, Snoeck HW. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529(7587):528–531. doi: 10.1038/nature16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, et al. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58(19):4238–4244. [PubMed] [Google Scholar]
- 25.Deng Q, Huang S. PRDM5 is silenced in human cancers and has growth suppressive activities. Oncogene. 2004;23(28):4903–4910. doi: 10.1038/sj.onc.1207615. [DOI] [PubMed] [Google Scholar]
- 26.Dettman EJ, et al. Prdm14 initiates lymphoblastic leukemia after expanding a population of cells resembling common lymphoid progenitors. Oncogene. 2011;30(25):2859–2873. doi: 10.1038/onc.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shing DC, et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J Clin Invest. 2007;117(12):3696–3707. doi: 10.1172/JCI32390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamato G, et al. Clinical features and prognostic impact of PRDM16 expression in adult acute myeloid leukemia. Genes Chromosomes Cancer. 2017;56(11):800–809. doi: 10.1002/gcc.22483. [DOI] [PubMed] [Google Scholar]
- 29.Shiba N, et al. High PRDM16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT3-ITD, KMT2A-PTD, and NUP98-NSD1: the results of the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 trial. Br J Haematol. 2016;172(4):581–591. doi: 10.1111/bjh.13869. [DOI] [PubMed] [Google Scholar]
- 30.Jo A, et al. High expression of EVI1 and MEL1 is a compelling poor prognostic marker of pediatric AML. Leukemia. 2015;29(5):1076–1083. doi: 10.1038/leu.2015.5. [DOI] [PubMed] [Google Scholar]
- 31.Duhoux FP, et al. PRDM16 (1p36) translocations define a distinct entity of myeloid malignancies with poor prognosis but may also occur in lymphoid malignancies. Br J Haematol. 2012;156(1):76–88. doi: 10.1111/j.1365-2141.2011.08918.x. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo H, Goyama S, Kamikubo Y, Adachi S. The subtype-specific features of EVI1 and PRDM16 in acute myeloid leukemia. Haematologica. 2015;100(3):e116–e117. doi: 10.3324/haematol.2015.124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eveillard M, et al. The closely related rare and severe acute myeloid leukemias carrying EVI1 or PRDM16 rearrangements share singular biological features. Haematologica. 2015;100(3):e114–e115. doi: 10.3324/haematol.2014.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106(12):3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modlich U, et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22(8):1519–1528. doi: 10.1038/leu.2008.118. [DOI] [PubMed] [Google Scholar]
- 36.Yu H, et al. Downregulation of Prdm16 mRNA is a specific antileukemic mechanism during HOXB4-mediated HSC expansion in vivo. Blood. 2014;124(11):1737–1747. doi: 10.1182/blood-2013-10-534735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33(2):314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 38.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nayak RC, Chang KH, Vaitinadin NS, Cancelas JA. Rho GTPases control specific cytoskeleton-dependent functions of hematopoietic stem cells. Immunol Rev. 2013;256(1):255–268. doi: 10.1111/imr.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manesia JK, et al. Highly proliferative primitive fetal liver hematopoietic stem cells are fueled by oxidative metabolic pathways. Stem Cell Res. 2015;15(3):715–721. doi: 10.1016/j.scr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar R, Fossati V, Israel M, Snoeck HW. Lin-Sca1+kit- bone marrow cells contain early lymphoid-committed precursors that are distinct from common lymphoid progenitors. J Immunol. 2008;181(11):7507–7513. doi: 10.4049/jimmunol.181.11.7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fossati V, Kumar R, Snoeck HW. Progenitor cell origin plays a role in fate choices of mature B cells. J Immunol. 2010;184(3):1251–1260. doi: 10.4049/jimmunol.0901922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beerman I, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A. 2010;107(12):5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weksberg DC, Chambers SM, Boles NC, Goodell MA. CD150- side population cells represent a functionally distinct population of long-term hematopoietic stem cells. Blood. 2008;111(4):2444–2451. doi: 10.1182/blood-2007-09-115006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milne TA. Mouse models of MLL leukemia: recapitulating the human disease. Blood. 2017;129(16):2217–2223. doi: 10.1182/blood-2016-10-691428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research Network, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spencer DH, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015;29(6):1279–1289. doi: 10.1038/leu.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gañán-Gómez I, et al. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia. 2015;29(7):1458–1469. doi: 10.1038/leu.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cull AH, Rauh MJ. Success in bone marrow failure? Novel therapeutic directions based on the immune environment of myelodysplastic syndromes. J Leukoc Biol. 2017;102(2):209–219. doi: 10.1189/jlb.5RI0317-083R. [DOI] [PubMed] [Google Scholar]
- 52.Gu Y, et al. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 2003;302(5644):445–449. doi: 10.1126/science.1088485. [DOI] [PubMed] [Google Scholar]
- 53.Yang FC, et al. Rac and Cdc42 GTPases control hematopoietic stem cell shape, adhesion, migration, and mobilization. Proc Natl Acad Sci U S A. 2001;98(10):5614–5618. doi: 10.1073/pnas.101546898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X, et al. RhoA GTPase controls cytokinesis and programmed necrosis of hematopoietic progenitors. J Exp Med. 2013;210(11):2371–2385. doi: 10.1084/jem.20122348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Florian MC, et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell. 2012;10(5):520–530. doi: 10.1016/j.stem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki-Utsunomiya K, et al. ALS2CL, a novel ALS2-interactor, modulates ALS2-mediated endosome dynamics. Biochem Biophys Res Commun. 2007;354(2):491–497. doi: 10.1016/j.bbrc.2006.12.229. [DOI] [PubMed] [Google Scholar]
- 57.Shibata S, Teshima Y, Niimi K, Inagaki S. Involvement of ARHGEF10, GEF for RhoA, in Rab6/Rab8-mediating membrane traffic. Small GTPases. doi: 10.1080/21541248.2017.1302550. doi: 10.1080/21541248.2017.1302550. [published online ahead of print April 27, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Deretic D. The Arf and Rab11 effector FIP3 acts synergistically with ASAP1 to direct Rabin8 in ciliary receptor targeting. J Cell Sci. 2015;128(7):1375–1385. doi: 10.1242/jcs.162925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Citi S, Guerrera D, Spadaro D, Shah J. Epithelial junctions and Rho family GTPases: the zonular signalosome. Small GTPases. 2014;5(4):1–15. doi: 10.4161/21541248.2014.973760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen C, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205(10):2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohrin M, et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347(6228):1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maryanovich M, et al. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat Commun. 2015;6:7901. doi: 10.1038/ncomms8901. [DOI] [PubMed] [Google Scholar]
- 63.Takubo K, et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12(1):49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Halvarsson C, Eliasson P, Jönsson JI. Pyruvate dehydrogenase kinase 1 is essential for transplantable mouse bone marrow hematopoietic stem cell and progenitor function. PLoS One. 2017;12(2):e0171714. doi: 10.1371/journal.pone.0171714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho TT, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543(7644):205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.King KY, Goodell MA. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol. 2011;11(10):685–692. doi: 10.1038/nri3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krivtsov AV, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27(4):852–860. doi: 10.1038/leu.2012.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemmati S, Haque T, Gritsman K. Inflammatory signaling pathways in preleukemic and leukemic stem cells. Front Oncol. 2017;7:265. doi: 10.3389/fonc.2017.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deininger MWN, Tyner JW, Solary E. Turning the tide in myelodysplastic/myeloproliferative neoplasms. Nat Rev Cancer. 2017;17(7):425–440. doi: 10.1038/nrc.2017.40. [DOI] [PubMed] [Google Scholar]
- 70.Aguayo A, et al. Plasma vascular endothelial growth factor levels have prognostic significance in patients with acute myeloid leukemia but not in patients with myelodysplastic syndromes. Cancer. 2002;95(9):1923–1930. doi: 10.1002/cncr.10900. [DOI] [PubMed] [Google Scholar]
- 71.Kim JG, et al. Clinical implications of angiogenic factors in patients with acute or chronic leukemia: hepatocyte growth factor levels have prognostic impact, especially in patients with acute myeloid leukemia. Leuk Lymphoma. 2005;46(6):885–891. doi: 10.1080/10428190500054491. [DOI] [PubMed] [Google Scholar]
- 72.Kornblau SM, McCue D, Singh N, Chen W, Estrov Z, Coombes KR. Recurrent expression signatures of cytokines and chemokines are present and are independently prognostic in acute myelogenous leukemia and myelodysplasia. Blood. 2010;116(20):4251–4261. doi: 10.1182/blood-2010-01-262071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsimberidou AM, et al. The prognostic significance of cytokine levels in newly diagnosed acute myeloid leukemia and high-risk myelodysplastic syndromes. Cancer. 2008;113(7):1605–1613. doi: 10.1002/cncr.23785. [DOI] [PubMed] [Google Scholar]
- 74.Patnaik MM, et al. Flow cytometry based monocyte subset analysis accurately distinguishes chronic myelomonocytic leukemia from myeloproliferative neoplasms with associated monocytosis. Blood Cancer J. 2017;7(7):e584. doi: 10.1038/bcj.2017.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aguayo A, et al. Cellular vascular endothelial growth factor is a predictor of outcome in patients with acute myeloid leukemia. Blood. 1999;94(11):3717–3721. [PubMed] [Google Scholar]
- 76.Wang Y, et al. Leukemia cell infiltration causes defective erythropoiesis partially through MIP-1α/CCL3. Leukemia. 2016;30(9):1897–1908. doi: 10.1038/leu.2016.81. [DOI] [PubMed] [Google Scholar]
- 77.Duarte D, et al. Inhibition of endosteal vascular niche remodeling rescues hematopoietic stem cell loss in AML. Cell Stem Cell. 2018;22(1):64–77.e6. doi: 10.1016/j.stem.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.