Abstract
Broad-spectrum antibiotics are widely used with patients in intensive care units (ICUs), many of whom develop hospital-acquired infections with Pseudomonas aeruginosa. Although preceding antimicrobial therapy is known as a major risk factor for P. aeruginosa–induced pneumonia, the underlying mechanisms remain incompletely understood. Here we demonstrate that depletion of the resident microbiota by broad-spectrum antibiotic treatment inhibited TLR-dependent production of a proliferation-inducing ligand (APRIL), resulting in a secondary IgA deficiency in the lung in mice and human ICU patients. Microbiota-dependent local IgA contributed to early antibacterial defense against P. aeruginosa. Consequently, P. aeruginosa–binding IgA purified from lamina propria culture or IgA hybridomas enhanced resistance of antibiotic-treated mice to P. aeruginosa infection after transnasal substitute. Our study provides a mechanistic explanation for the well-documented risk of P. aeruginosa infection following antimicrobial therapy, and we propose local administration of IgA as a novel prophylactic strategy.
Keywords: Infectious disease
Keywords: Bacterial infections
Introduction
National and international point-prevalence studies show that 64%–71% of all patients in intensive care units (ICUs) receive antibiotics (1, 2) and up to 10%–51% suffer from hospital-acquired infections (HAIs) (1–3). Recent estimates indicate that in Europe alone, 2.6 million cases of HAI occur each year, of which hospital-acquired pneumonia (HAP) accounts for the highest disease burden and number of deaths (4). HAP dramatically increases both the length of hospital stay and the healthcare costs, and is associated with an attributable mortality of up to 13% (5).
Pseudomonas aeruginosa represents an important opportunistic pathogen in critically ill patients and is a leading cause of HAP (6). The bacterium is intrinsically resistant to many antibiotics and rapidly acquires additional drug resistance–conferring genetic information (7). Rates of carbapenem resistance among P. aeruginosa isolates, for example, range from 10%–50% in numerous countries (8), demanding novel strategies to prevent and treat infections with P. aeruginosa.
Preceding antimicrobial therapy is a major independent risk factor for lung infections with P. aeruginosa (9–12). International guidelines therefore specifically recommend the use of P. aeruginosa–active antibiotics for empirical treatment of HAP in patients with prior antibiotic use (11, 13, 14). However, the mechanism underlying the enhanced P. aeruginosa infection susceptibility of patients receiving antimicrobial treatment is incompletely understood.
The microbiota mediates direct and indirect colonization resistance by outcompeting drug-resistant obligate or opportunistic pathogens and maintaining the local production of antibacterial peptides (15, 16). Moreover, recent studies indicate a beneficial influence of the microbiota on systemic and pulmonary immune responses during bacterial and viral infections. Accordingly, the microbiota calibrates the immune system by adjusting the release of antiinflammatory IL-10 (17), releasing short-chain fatty acids to stimulate infection resolution via the G protein–coupled receptor 43 (18), enhancing the antibacterial activity of neutrophils via NOD1 (19), priming inflammasome activities (20), and controlling responsiveness of mononuclear phagocytes to produce type I IFNs and other inflammatory cytokines (21, 22). Antimicrobial therapies cause dramatic and long-lasting collateral damage to the bacterial populations forming the patient’s microbiota (23, 24). Whether an impairment of aforementioned or other microbiota-dependent mechanisms explains the high susceptibility of individuals receiving antibiotics toward P. aeruginosa infection is currently unknown.
Here we examine the effect of preceding antimicrobial therapy on acute lung infection and identify a novel mechanism underlying the high risk of individuals receiving antibiotics to develop P. aeruginosa pneumonia.
Results
Antibiotic-mediated microbiota depletion enhances susceptibility to P. aeruginosa–induced pneumonia.
In order to assess the effect of the microbiota on P. aeruginosa–induced pneumonia, we first examined mice that received a cocktail of antibiotics to deplete the microbiota, as previously described (25). As expected, the antimicrobial treatment virtually depleted the respiratory and intestinal microbiota (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI97065DS1). Mice were intranasally infected with P. aeruginosa 4 to 5 days after ending antibiotic treatment. Microbiota-depleted mice showed an early defect in bacterial clearance in the lung (Figure 1, A and B), enhanced dissemination into the blood (Figure 1C), and reduced survival (Figure 1D) as compared with conventionally colonized mice. Similarly, animals born germ-free (GF) exhibited dramatically enhanced bacterial loads in the lung and blood compared with conventionally colonized mice (Supplemental Figure 2, A and B). The early inflammatory response, however, did not seem to be impaired by microbiota depletion. The production of several inflammatory mediators and recruitment of neutrophils were not inhibited by microbiota depletion, but rather were slightly enhanced, probably due to enhanced proliferation of P. aeruginosa (Figure 1, E–O). Microbiota depletion also did not generally increase vascular permeability, as detected by leakage of Evans blue dye (Figure 1P). These results imply an impact of microbiota depletion on a defense mechanism beyond the mechanisms analyzed as central for early P. aeruginosa resistance in the lung.
Figure 1. Antibiotic-mediated microbiota depletion impairs antibacterial defense against P. aeruginosa.
(A–D) Conventionally colonized mice (Conv) and mice that were treated with an antibiotic cocktail for 6–8 weeks (ABx) were infected with P. aeruginosa. (A) Bacterial loads in BALF (n = 9), (B) lung tissue (n = 11 for Conv mice; n = 9 for ABx mice), and (C) blood (n = 11 for Conv mice; n = 9 for ABx mice) were counted at the indicated time points, or (D) survival was monitored (n = 11 for Conv mice; n = 12 for ABx mice). (E–M) Mice were intranasally infected with P. aeruginosa or treated with PBS for 6 hours. Production of inflammatory mediators was measured by ELISA (n = 10 for uninfected Conv and ABx mice; n = 10 for infected Conv mice; n = 11 for infected ABx mice) and qRT-PCR (n = 8 for uninfected Conv and ABx mice; n = 10 for infected Conv mice; n = 9 for infected ABx mice). (N and O) Alveolar macrophages (n = 9 for uninfected Conv and ABx mice; n = 9 for infected Conv mice; n = 7 for infected ABx mice) and neutrophils (n = 9 for uninfected Conv and ABx mice; n = 9 for infected Conv mice; n = 8 for infected ABx mice) were counted by FACS analysis after staining of BALF samples. (P) Lung vascular permeability of control and microbiota-depleted animals was measured by Evans blue dye leakage in the lungs (n = 8 for Conv; n = 8 for ABx mice). Mann-Whitney U tests were applied to the data set, except for the survival curve (D), which was submitted to log-rank test. Values are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.005.
Microbiota-dependent IgA production contributes to control of P. aeruginosa infection.
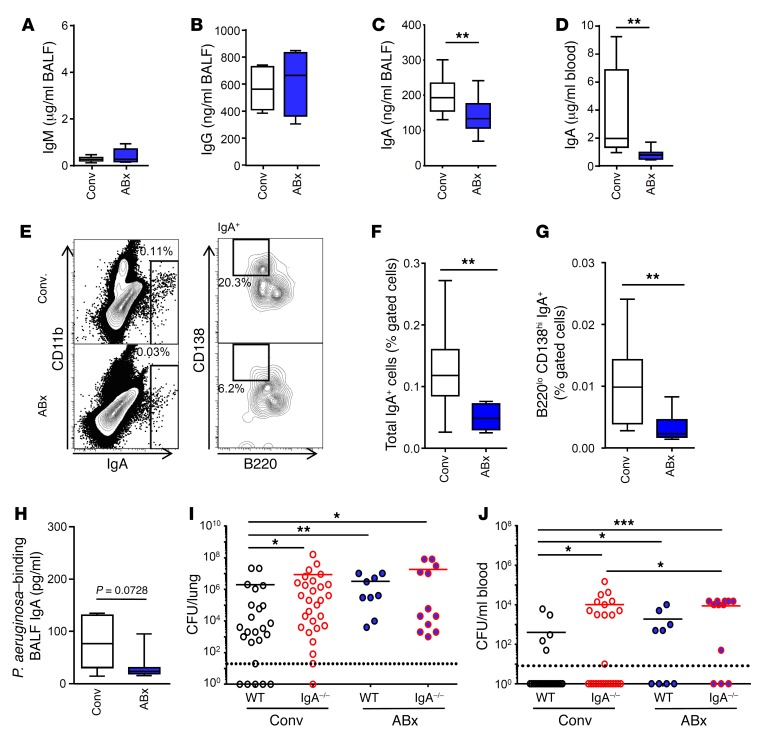
Considering humoral immune dysfunction as a potential mechanism underlying the high susceptibility of microbiota-depleted animals to P. aeruginosa infection (26), we next analyzed production of immunoglobulins. While IgM and IgG antibody titers were unaltered, we observed strongly reduced IgA levels in the lung and blood of antibiotic-treated animals as compared with control counterparts (Figure 2, A–D). Accordingly, microbiota depletion lowered the frequency of total IgA+ cells (probably consisting of IgA+ plasma cells, IgA+ plasmablasts, B cells, and cells expressing the polymeric immunoglobulin receptor carrying IgA; Figure 2, E and F) and IgA+ plasma cells (Figure 2, E and G) in the lung. Next, we coated ELISA plates with P. aeruginosa, incubated with bronchoalveolar lavage fluid (BALF) samples, quantified the bacteria-binding IgA, and found that levels of P. aeruginosa–binding IgA were almost significantly decreased in the BALF of antibiotic-treated animals as compared with samples from control mice (Figure 2H). In order to examine whether impaired IgA production is indeed responsible for the enhanced susceptibility of microbiota-depleted animals toward acute P. aeruginosa lung infection, we compared bacterial clearance in WT and IgA-deficient mice that either received antibiotics or were left untreated. We observed that both microbiota-depleted WT and conventionally colonized IgA–/– mice harbored higher bacterial loads in the lungs and blood as compared with control animals (Figure 2I). Of note, microbiota depletion did not further increase the bacterial burden in lungs of IgA-deficient animals. However, a slight effect of antibiotic treatment of IgA–/– animals suggests a minor contribution of an IgA-independent mechanism of the microbiota to restrict bacterial dissemination (Figure 2J). Taken together, these data reveal that IgA contributes to the early antibacterial defense during P. aeruginosa–induced pneumonia, and that antibiotic-mediated microbiota depletion following antibiotic administration impairs the resistance of mice by lowering IgA levels.
Figure 2. Microbiota depletion impairs antibacterial defense in the lung by decreasing pulmonary IgA production.
(A–D) Total levels of (A) IgM (n = 8 for Conv; n = 5 for ABx mice), (B) IgG (n = 5 for Conv; n = 4 for ABx mice), and (C) IgA (n = 14 for Conv; n = 14 for ABx mice) in the BALF or (D) in plasma (n = 9 for Conv; n = 7 for ABx mice) were measured. (E) Lung IgA+ plasma cells were identified by FACS as IgAhi (E, left) and B220lo CD138hi cells (E, right). (F) Total lung IgA+ cells and (G) IgA+ plasma cells were quantified by FACS (n = 10 for Conv and ABx mice). (H) Levels of P. aeruginosa–binding IgA were quantified by coating P. aeruginosa to ELISA plates and measuring bound IgA (n = 7 for Conv and ABx mice). (I, J) WT and IgA–/– mice were pretreated or not with antibiotics, and afterwards infected with 1 × 105 CFU per mouse P. aeruginosa. Bacterial loads in the (I) lungs and (J) blood were measured at 24 hours after infection (n = 24 for Conv WT; n = 28 for Conv IgA–/–; n = 9 for WT ABx; n = 12 for IgA–/– ABx mice). Mann-Whitney U tests were applied to the data set. Values are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.005.
IgA production in the lung is dependent on TLR and a proliferation-inducing ligand (APRIL).
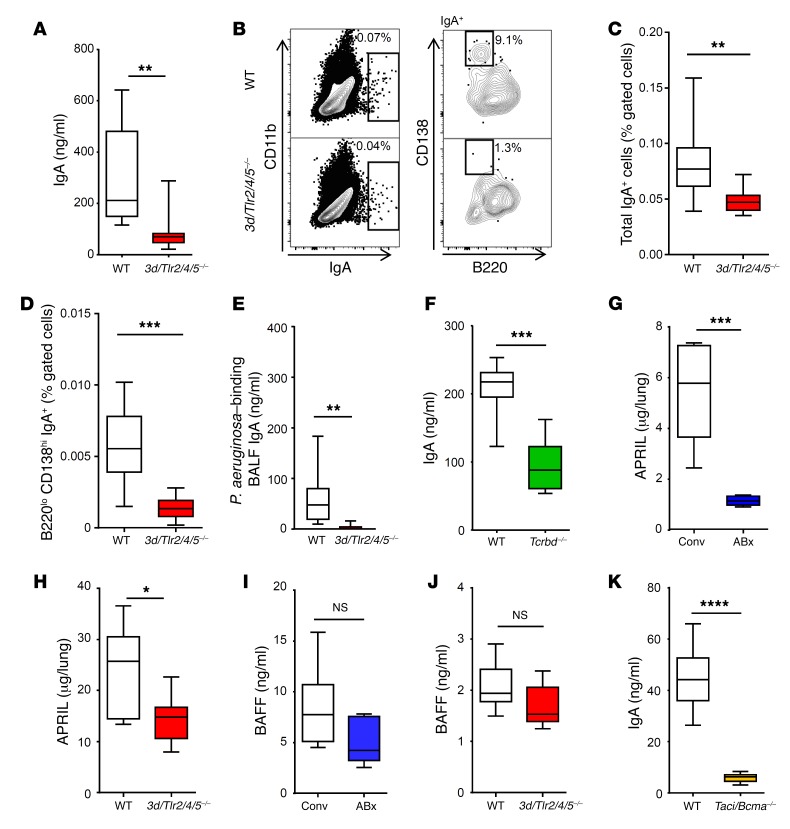
Next, we aimed to unravel the mechanism that drives IgA production in the lung in the presence of the microbiota. Innate immune stimulation mediated, for example, through the family of TLRs provides the costimulus required for an efficient adaptive immune response (27). We therefore analyzed mice devoid of TLR activities due to lack of expression of TLR2, TLR4, and TLR5 as well as the endosomal TLR chaperone UNC93B1 (3d/Tlr2/4/5–/–). In 3d/Tlr2/4/5–/– mice, production of IgA (Figure 3A), frequency of total IgA+ cells (Figure 3, B and C) and IgA+ plasma cells (Figure 3, B and D), and levels of P. aeruginosa–binding IgA (Figure 3E) were strongly reduced. In contrast, IgA levels were not affected by lack of TLR2 and TLR4 alone (Supplemental Figure 3, A and B). Accordingly, oral or intranasal application of the TLR4 agonist LPS did not affect IgA levels in antibiotic-treated animals (Supplemental Figure 3, C–F), although intranasal pretreatment with LPS increased (apparently IgA-independent) antibacterial defense in the lung against P. aeruginosa (Supplemental Figure 3, G–J). Moreover, IgA production was also reduced in T cell–deficient animals (Figure 3F).
Figure 3. IgA production is dependent on microbiota, TLRs, APRIL, and TACI/BCMA.
(A) IgA levels in BALF (n = 10 for WT; n = 7 for 3d/Tlr2/4/5–/– mice) and (B–D) total IgA+ cells and IgA+ plasma cells in whole lungs of WT and 3d/Tlr2/4/5–/– (comprehensive and specific lack of TLR function) mice were examined (n = 10 for each group). (E) Levels of P. aeruginosa–binding IgA were examined in BALFs of WT and 3d/Tlr2/4/5–/– mice (n = 10 for WT; n = 7 for 3d/Tlr2/4/5–/–). (F) IgA levels were measured in BALF of WT and Tcrbd–/– animals (n = 7 for WT; n = 10 for Tcrbd–/– mice). (G–J) Levels of APRIL and BAFF were measured in lung homogenate supernatants of animals as indicated (n = 10 for Conv mice; n = 5 for ABx mice; n = 10 for WT mice; n = 10 for 3d/Tlr2/4/5–/– mice). (K) IgA levels were assessed in BALFs of WT and Taci/Bcma–/– animals (n = 8 for WT; n = 9 for Taci/Bcma–/–). Mann-Whitney U tests were applied to the data set. Values are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001.
Production of intestinal IgA has been shown to depend on the TNF family cytokine APRIL and B cell–activating factor (BAFF) (28–30). Accordingly, the lungs of antibiotic-treated and 3d/Tlr2/4/5–/– animals contained lower levels of APRIL, and by trend of BAFF, as compared with lungs of respective control groups (Figure 3, G–J). Moreover, constitutive IgA production was virtually absent in mice lacking expression of the APRIL and BAFF receptors transmembrane activator and CAML interactor (TACI) and B cell maturation antigen (BCMA) (hereafter referred to as Taci/Bcma–/– mice) (Figure 3K). Together, our data indicate the requirement of microbiota-dependent activation of TLRs (probably involving members other than TLR2/4) for APRIL expression (and possibly BAFF expression) and IgA production in the lung.
Clinical antimicrobial therapies lower IgA levels in mice and human patients.
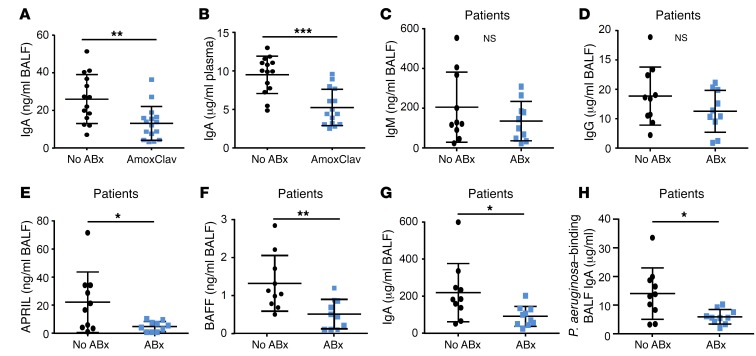
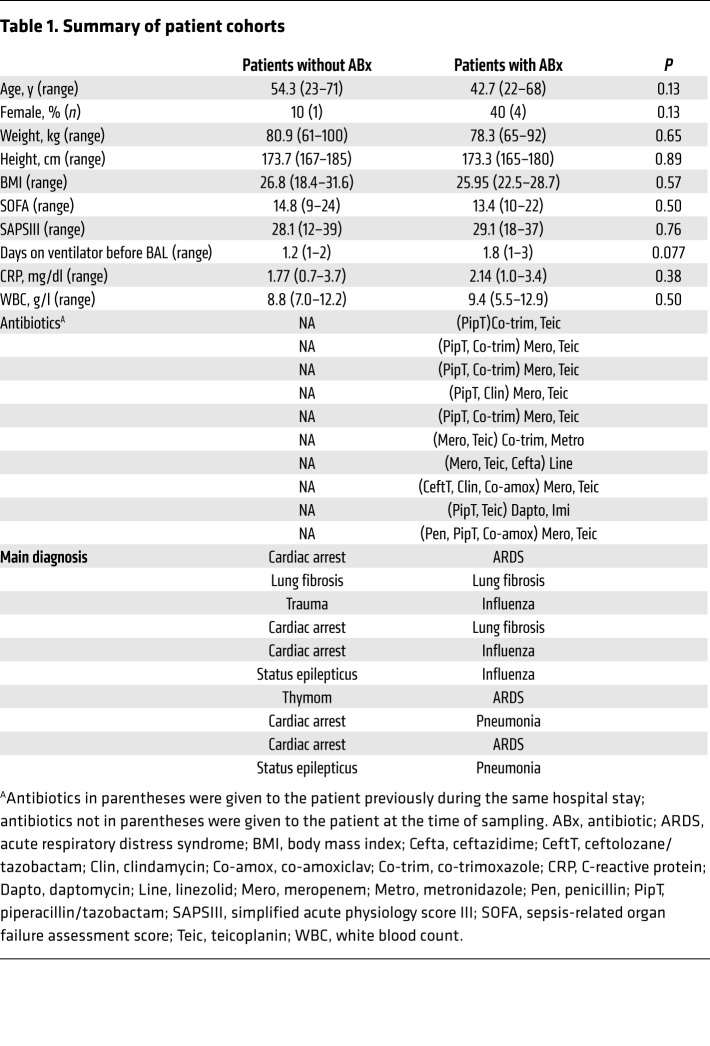
We next asked whether clinically relevant antimicrobial drugs exhibited a similar effect on IgA production. Indeed, we observed reduced levels of IgA (but not of IgM and IgG) in mice that were treated for 4 weeks with amoxicillin and clavulanate as compared with control mice (Figure 4, A and B, and our unpublished observations). Next, we examined the effect of antimicrobial therapies on pulmonary IgA production in human patients. We compared BALF samples from ICU patients who were either treated with antibiotics at the time of sampling (at least one broad-spectrum antibiotic for at least 2 weeks; for details refer to Table 1) or who did not receive antibiotic therapy for at least 3 months before sampling. Antibiotic-treated patients showed unaltered IgM and IgG levels (Figure 4, C and D), but significantly lower levels of APRIL, BAFF, and total IgA in their BALFs as compared with individuals who did not receive antimicrobial therapy (Figure 4, E–G). According to the results of a univariate analysis, APRIL and BAFF levels affected IgA BALF levels in antibiotic-treated patients trend-wise, since significance was not reached (P = 0.066). Moreover, levels of P. aeruginosa–binding IgA were reduced in the lungs of antibiotic-treated patients (Figure 4H). These results indicate that clinically relevant antimicrobial therapies reduce pulmonary IgA production. They suggest that the antibiotic-associated secondary IgA deficiency contributes to a high susceptibility of ICU patients toward P. aeruginosa (and possibly other bacterial) infections.
Figure 4. Production of IgA is impaired by antimicrobial therapy in mice and ICU patients.
(A and B) WT mice were treated with amoxicillin and clavulanate (AmoxClav) for 4 weeks and IgA levels in the BALF (A) and plasma (B) were examined (n = 14 for mice without AmoxClav; n = 16 or 15 for mice with AmoxClav). (C–H) Levels of IgM, IgG, APRIL, BAFF, total IgA, and P. aeruginosa–binding IgA were measured by ELISA from human BALF samples of ICU patients with or without antimicrobial therapy (n = 10 for patients without ABx; n = 10 for patients with ABx). Mann-Whitney U tests were applied to the data set. Values are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.005.
Table 1. Summary of patient cohorts.
Local application of IgA increases resistance of antibiotic-treated mice.
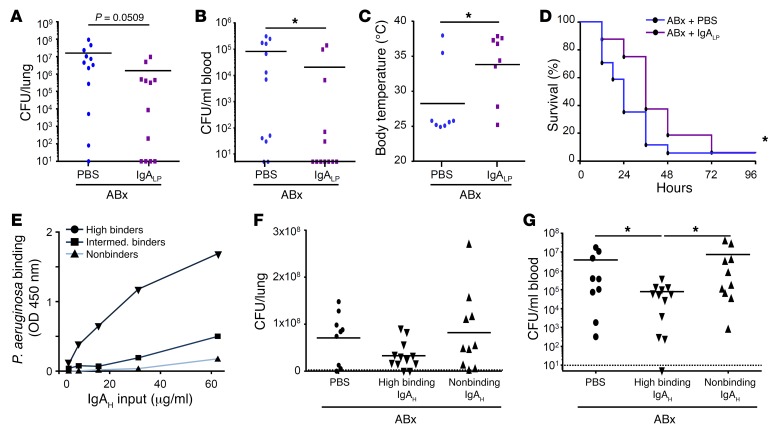
Finally, we tested whether local (i.e., pulmonary) application of IgA rescues the antibacterial defense in antibiotic-treated mice. First, polyclonal IgA was affinity purified from supernatants of ex vivo murine intestinal tissue cultures and found to bind to P. aeruginosa (our unpublished observation). Mouse polyclonal IgA of the source described above was further intranasally applied to antibiotic-pretreated animals 30 minutes prior to P. aeruginosa infection. Importantly, IgA-treated animals harbored fewer bacteria in their lungs and blood as compared with control mice (Figure 5, A and B). Accordingly, pretreatment of mice with a single dose of IgA reduced the infection-induced body temperature decrease (Figure 5C) and significantly prolonged the survival upon infection (Figure 5D). Next, we analyzed whether a P. aeruginosa–binding capacity of the administered IgA was required for its antibacterial effect in microbiota-depleted animals. IgA preparations from various IgA hybridomas were tested for their capacity to bind to P. aeruginosa, grouped and pooled into high P. aeruginosa binders, intermediate P. aeruginosa binders, and non–P. aeruginosa binders (Figure 5E). Intranasal application of high P. aeruginosa–binding IgA (pool of 5 hybridoma preparations) 30 minutes prior to infection lowered the bacterial load in the lung by trend and significantly reduced Pseudomonas dissemination into blood, as compared with PBS or nonbinding IgA (pool of 2 hybridoma preparations) (Figure 5, F and G). Collectively, these results indicate a substantial capacity of pretreatment with IgA to improve the antibacterial host defense and thus enhance the resistance of antibiotic-treated mice against P. aeruginosa.
Figure 5. Local application of IgA enhances the antibacterial defense against P. aeruginosa in antibiotic-treated animals.
(A–D) IgA was purified from ex vivo murine lamina propria culture, and purified lamina propria IgA (IgALP) or PBS was intranasally applied to animals pretreated with antibiotic 30 minutes prior to infection with P. aeruginosa. Bacterial loads (n = 12 for mice with and without IgA treatment) in (A) lung and (B) blood, and (C) body temperature (n = 8 for mice with and without IgA treatment) were measured 24 hours after infection, or survival was monitored over 4 days (n = 17 for mice with and without IgA treatment) (D). (E) Pooled hybridoma IgA (IgAH) preparations were tested in vitro for their binding to P. aeruginosa. (F and G) High and non–P. aeruginosa–binding IgA prepared from IgA hybridoma were administrated intranasally 30 minutes prior to infection and P. aeruginosa loads were measured 24 hours later in lung (F) and blood (G) (n = 9 mice for PBS group, n = 12 mice for high P. aeruginosa–binding IgA, n = 10 mice for non–P. aeruginosa–binding IgA). Values are mean ± SD, each dot corresponds to the data from one mouse. Mann-Whitney U tests were applied to the data set, except for survival and body temperature data. A t test with Welch’s correction was applied to body temperature data (see Methods). The Gehan-Breslow Wilcoxon test was applied to the survival data. *P < 0.05.
Discussion
Antimicrobial therapy constitutes a major risk factor for subsequent infection with P. aeruginosa and other antibiotic-resistant opportunistic pathogens (9–12). Our present study revealed that antibiotic-mediated inhibition of a microbiota-, TLR-, and APRIL-dependent pulmonary production of (probably cross-reactive) IgA enhances the susceptibility of mice toward P. aeruginosa lung infection. Likewise, we found reduced IgA levels in ICU patients treated with antibiotics. This suggests that antibiotic-associated secondary IgA deficiency is relevant in humans and might contribute to the high susceptibility of ICU patients toward infections with P. aeruginosa and possibly other bacteria. Finally, we demonstrate that local application of IgA partially restored the antibacterial defense against P. aeruginosa in antibiotic-pretreated mice, and that the capacity to bind Pseudomonas is required for the protective effect of IgA. We therefore hypothesize that ICU patients on broad-spectrum antimicrobial therapy might benefit from prophylactic and/or therapeutic pulmonary IgA administration or other novel strategies to enhance local IgA levels.
Primary IgA deficiency is the most common genetic immunodeficiency, with a prevalence of approximately 1:600 in the Western population (31). Recurrent pulmonary infections constitute the most prevalent comorbidity in IgA-deficient patients. However, many others remain asymptomatic, which could be explained by a compensatory increase in IgM production in individuals with primary IgA deficiency (32). Moreover, P. aeruginosa is poorly pathogenic in (otherwise) healthy individuals, and most persons outside the hospital are not colonized with this bacterium. This potentially explains why individuals with primary IgA deficiency are typically not at risk for P. aeruginosa–induced pneumonia.
On the contrary, hospitalized patients are frequently colonized with P. aeruginosa in the upper respiratory tract, which is the first step for subsequent lower respiratory tract infection (33). Our results suggest that the acquisition of antibiotic-dependent IgA deficiency (possibly in addition to the attenuation of other antibacterial immune pathways) in colonized patients allows aspirated bacteria to replicate and disseminate to cause pneumonia. This assumption is supported by a small prospective study that found an association between low bronchial IgA levels and onset of nosocomial pneumonia in ventilated ICU patients (34).
Several clinical trials indicated that the administration of probiotics to mechanically ventilated critically ill patients substantially decreases the incidence of ventilator-associated pneumonia (35, 36). Although IgA levels have, to our best knowledge, not been examined in these studies, the protective probiotic effect might be at least partially mediated by rescue of pulmonary IgA production. This assumption is supported by recent demonstration that oral application of bacterial extracts has the capability to enhance pulmonary IgA levels in mice (37).
It is well-established that the microbiota is critical for regulating intestinal IgA production, as IgA-secreting cells and IgA production are almost absent in the intestinal tract of GF mice (38). Microbial signals are known to activate TLRs on intestinal DCs and epithelial cells to induce production of APRIL and BAFF, which in turn promote IgA production by plasma cells (29, 30, 39, 40). Moreover, microbiota-derived short-chain fatty acids have recently been demonstrated to positively regulate IgA production (41, 42). Production of high-affinity and antigen-specific IgA in Peyer’s patches and mesentery lymph nodes has further been shown to be T cell dependent (43), whereas low-affinity polyreactive IgA responses were mainly induced in isolated lymphoid follicles and subepithelial B cells in a T cell–independent manner (44). Both types of IgA regulate the composition of intestinal microbiota by coating many of its bacterial components to maintain intestinal homeostasis (45, 46). In addition, IgA has long been known to mediate neutralizing of invasive intestinal pathogens by various mechanisms (47, 48).
Considerably less is known about the regulation of IgA production in the respiratory tract. A recent study demonstrated that the microbiota regulates the ability of specific lung DC subtypes to stimulate IgA class switch reaction and to induce gut homing molecules in B cells ex vivo (49). This was shown to depend on MyD88/TRIF-mediated TGF-β production as well as on retinoic acid, respectively. Moreover, this recent report revealed that the lung DC–dependent mechanism is critical for IgA-dependent gastrointestinal immune responses following intranasal immunization (49). Our study provides important new insight by showing that microbiota-dependent IgA production is required for antibacterial immunity during acute bacterial pneumonia. Moreover, we indicate that this mechanism is attenuated by antimicrobial therapies, and that the antibiotic-associated susceptibility to bacterial pneumonia can be reduced by local application of IgA.
Our data clearly show that attenuation of IgA production is causally linked to the enhanced susceptibility of antibiotic-treated animals to P. aeruginosa, and that application of IgA just 30 minutes before infection is sufficient to improve antibacterial defense. There are limitations to our study, however. First, our data do not exclude the possibility that regulation of additional antimicrobial pathways known to depend on microbiota signals (17, 19, 20, 50) might also play a role. Second, although our mouse experiments indicate a causal relationship among antibiotic-mediated microbiota depletion, attenuation of IgA production, and enhanced susceptibility to P. aeruginosa infection, the human data only show a correlation between antimicrobial therapy and low IgA levels. Prospective studies examining the impact of antibiotics on pulmonary IgA production and incidence of HAP are therefore required before interventional studies to increase IgA levels in high-risk ICU patients might be considered. Future studies are also necessary to define the specificity of the respiratory and/or intestinal microbiota being involved and how different classes of antibiotics and respective delivery routes (e.g., oral, inhaled, or parenteral) impact IgA-dependent immunity.
In summary, we here show that administration of broad-spectrum antibiotic inhibits microbiota-dependent IgA production in the lung and increases susceptibility toward subsequent infections with P. aeruginosa. Our data suggest that patients subjected to antimicrobial therapy might benefit from strategies to increase pulmonary IgA levels to decrease their risk of developing bacterial pneumonia.
Methods
Mice and antibiotic treatment.
Female C57BL/6 WT mice or KO mice on the same background were used for the experiments. To virtually deplete the intestinal microbiota, 6- to 8-week-old mice were transferred to sterile cages and treated by adding ampicillin (1 g/l; Ratiopharm), vancomycin (500 mg/l; Cell Pharm), ciprofloxacin (200 mg/l; Bayer Vital), imipenem (250 mg/l; MSD), and metronidazole (1 g/l; Fresenius) to the drinking water ad libitum for 6–8 weeks as described before (ABx mice) (25). Antibiotic-treated animals were monitored for fungal outgrowth. Mice in which fungal outgrowth was observed were eliminated from the study. Other 8-week-old animals were treated by adding amoxicillin and clavulanic acid (1 g/l Amox/142.5 mg/l Clav; 1A Pharma) to the drinking water ad libitum (renewed every 2 days, AmoxClav mice). Control mice for the ABx or AmoxClav groups were maintained in the same facilities during the same period of time as the animals undergoing antibiotic treatment. IgA–/– (Laboratory Animals Services Center), Taci/Bcma–/– (provided by Susan Kalled, Biogen) (51), Tlr2/4–/–, 3d/Tlr2/4/5–/– (52), and Tcrbd–/– mice (provided by Max Löhning, Charité Universitätsmedizin Berlin) on C57BL/6 background were housed under specific pathogen-free conditions and comparatively analyzed with WT control animals that were housed in the same facilities.
Intestinal microbiota analyses.
For molecular analysis of the intestinal microbiota, DNA was extracted from fecal samples as previously described (25). Briefly, DNA extracts and plasmids were quantified using Quant-iT PicoGreen reagent (Invitrogen) and adjusted to 1 ng/μl. Then, abundance of the main bacterial groups of murine intestinal microbiota was assessed by quantitative real-time PCR (qRT-PCR) with group-specific 16S rRNA gene primers (Tib MolBiol) as previously described (53, 54). The number of 16S rRNA gene copies per microgram DNA of each sample was determined and frequencies of respective bacterial groups were calculated proportionally to the eubacterial (V3) amplicon.
Respiratory tract microbiota analyses.
The protocol was adapted from ref. 55. BALF and upper respiratory lavage (URL) samples from 3 mice were pooled. After host cell removal by centrifugation, BAL or URL DNA was extracted by bead homogenization and isolated using the Qiagen DNeasy Blood & Tissue Kit. The control group (in triplicate) consisted of PBS that had been processed the same way as the BAL and URL samples. Total bacterial qPCR was performed using a 10:1 mixture of the universal forward primers 8FM (5′-AGAGTTTGATCMTGGCTCAG-3′) and 8FB (5′-AGGGTTCGATTCTGGCTCAG-3′), with reverse primer Bact515R (5′-TTACCGCGGCKGCTGGCAC-3′) and TaqMan probe Bact338K (5′-FAM/CCAKACTCCTACGGGAGGCAGCAG/TAMRA-3′) (56). Relative quantification was calculated by division with the control group.
Germ-free mice.
GF C57BL/6 mice were bred and kept in the animal facilities at the University of Marburg. GF mice were kept in plastic isolators (Metall and Plastik) with autoclaved food, bedding, and water. Sterility of animals was checked biweekly by culturing feces in thioglycollate medium under aerobic and anaerobic conditions for at least 10 days. All handling procedures for GF mice were conducted in a laminar flow hood under sterile conditions. All experiments were conducted according to German animal protection law.
Oral and intranasal LPS application in antibiotic-treated mice.
During the antibiotic treatment, mice received LPS (from Escherichia coli O111:B4; Sigma, catalog L2630) orally by adding 15 μg/ml LPS (57) to the drinking water ad libitum during the last 3 weeks of the antibiotic treatment (day –27 until day –4 or day –6 before infection), or mice received 2 μg LPS intranasally 3 consecutive times (days –22, –13, and –5 before infection)(20).
Pseudomonas aeruginosa infection.
P. aeruginosa strain PA103 was from ATCC (ATCC29260). PA103 was cultured in Luria Bertani (LB) or Tryptic Soy (TSB) broth to mid-logarithmic phase, washed twice in PBS, and inoculated into anesthetized mice (10 μl in each nostril) for an infection dose of 1 × 105 CFU per mouse. Control groups received PBS in an identical manner. At indicated time points, mice were sacrificed, blood was drawn, and whole lungs were harvested and homogenized in sterile PBS. Bacterial loads were quantified in lung homogenates, BALs, and blood by serial dilutions and plating on agar plates. The remaining lung homogenates were spun down (300 g for 15 minutes) to separate cells (for further FACS analysis or RNA extraction) from supernatants (for ELISA). BAL was performed 6 and 12 hours after infection for cell recruitment analysis by FACS and ELISA.
ELISAs.
Levels of prostaglandin E2 (PGE2) and the following cytokines and immunoglobulins were measured by ELISAs according to the manufacturers’ recommendations: mPGE2 (R&D Systems, catalog KGE004B), mIL-1α (eBioscience, catalog 50-112-5107), mIL-1β (eBioscience, catalog 88-7013-88), mTNF-α (eBioscience, catalog 88-7324-88), mIL-6 (eBioscience, catalog 88-7064-88), mCXCL1 (R&D Systems, catalog DY453-05), mCXCL2 (R&D Systems, catalog DY452-05), CCL2 (eBioscience, catalog 88-7391-22), mBAFF (R&D Systems, catalog MBLYS0), mAPRIL (LSBio, catalog LS-F7691), mIgM (eBioscience, catalog 88-50470-22), mIgG (eBioscience, catalog 88-50400), mIgA (eBioscience, catalog 88-50450-77, or direct ELISA with anti–mouse IgA/HRP cross-adsorbed from Thermo Fisher Scientific, catalog PA1-74397), hIgA (Thermo Fisher Scientific, catalog 88-50600-77), hIgM (eBioscience, catalog 88-50620-22), and hIgG (Thermo Fisher Scientific, catalog BMS2091TEN). For the IgA-binding ELISA, a mid-logarithmic phase PA103 suspension was washed twice with PBS and adjusted to an OD of 0.2, and then transferred onto a 96-well plate, spun-down (500 g for 10 minutes), and dried out after supernatant removal. After blocking, BAL samples were loaded onto the plate and incubated for 2 hours at room temperature. Binding IgA ELISAs and direct IgA ELISAs were detected by anti–mouse IgA/HRP (Thermo Fisher Scientific, catalog PA1-74397). Lung permeability was determined by quantifying Evans blue dye leakage into the lung.
qPCR.
RNA was extracted by Trizol from whole-lung homogenates. After reverse transcription using the High Capacity Reverse Transcription Kit (Applied Biosystems), targeted cDNA was amplified and measured by Taqman assay on an AB Applied Biosystems thermocycler. Primers used were IL-10 (Mm00439614, Thermo Fisher Scientific) and GAPDH (fwd: 5′-TGTGTCCGTCGTGGATCTGA-3′, rev: 5′-CCTGCTTCACCACCTTCTTGA-3′, probe: 5′-FAM-CCGCCTGGAGAAACCTGCCAAGTATG–TAMRA-3′). Relative quantification (RQ) values were calculated based on ΔΔCt, with conventionally colonized PBS-treated mice as a control group.
Flow cytometry analysis.
For recruited neutrophils and macrophages, cells from BAL were gathered by spinning down the BALF (400 g for 10 minutes RT). After incubation with blocking antibody (anti-CD16/32, BioLegend, catalog 101302), cells were stained using CD11c-FITC (N418, BioLegend), SiglecF-PE (E50-2440, BD Biosciences), F4/80-APC (BM8, BioLegend), CD45-Alexa700 (30-F11, BioLegend), Ly6G-BV421 (1A8, BioLegend), and CD11b-BV510 (M1/70, BioLegend). Beads for counting were used as recommended by the manufacturer (CountBright beads, Molecular Probes). For IgA-producing plasma cells, the lungs were mashed through a 100-μm filter. After incubation with blocking antibody (anti-CD16/32, BioLegend), cells were stained using IgA-FITC (mA-6E1, eBioscience, intracellular staining), B220-Alexa700 (RA3-6B2, BioLegend), and CD138-BV421 (281-2, BioLegend). Intracellular staining using BD Cytofix/Cytoperm solution (Thermo Fisher Scientific) was done according to the manufacturer’s recommendations. The samples were acquired on FACS CantoII (Becton Dickinson). Data were analyzed using the FlowJo data analysis software. Gating strategies were processed after exclusion of the doublets. Alveolar macrophages were defined as CD11c+ SiglecF+, neutrophils as CD11b+ Ly6G+, and IgA-producing plasma cells as B220lo, CD138hi, and IgAhi. Fluorescence minus one (FMO) was performed when necessary.
Purification of polyclonal murine IgA.
Intestines from C57Bl/6 mice were extensively washed in PBS/BSA containing penicillin and streptomycin and cultured in RPMI-1640 supplemented with 10% FCS/penicillin/streptomycin at 37°C and 5% CO2 in tissue flask. Tissues were kept in cultures for 2 weeks with complete medium replacement every 2 days. Harvested culture supernatants were centrifuged, filtered via 0.2 μm filters, dialyzed in a total of 6 l PBS, and preserved at −20°C for further purification. IgA was purified using protein L-Sepharose, and IgA concentration and purity were determined by gel electrophoresis and ELISA (amount of IgA was 50 μg/ml, whereas IgM and IgG levels were as low as 6 μg/ml or 1.2 μg/ml, respectively).
Generation of IgA hybridomas and purification of IgA.
Lamina propria cells were isolated by digesting C57Bl/6 intestines using 1 mg/ml collagenase D (Roche Diagnostics) and 1 mg/ml dispase II (Sigma-Aldrich). Cells were further fused with P3X63Ag8.653 myeloma cells (ATCC) using 50% polyethylene glycol (Mr ~1450 Da) and were cultured in RPMI-1640 containing 20% FBS, 1% penicillin and streptomycin, 100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine. Cloned hybridomas were tested for IgA by ELISA and homogeneity of IgA expression was examined by flow cytometry. Monoclonal IgA antibodies were purified from supernatants using Sepharose conjugated with anti–IgA antibody (Southern Biotech).
IgA treatment of microbiota-depleted animals.
Antibiotic-pretreated mice were anesthetized and purified IgA from lamina propria (1 μg per mouse) or hybridoma (1 μg per mouse) or PBS was given intranasally 30 minutes prior to infection with P. aeruginosa.
Human samples.
The patient cohorts were recruited from the tertiary care ICU of a university clinic from 2016–17. Patients gave their informed consent for sampling. Inclusion criteria comprised endotracheal intubation, 18 years and older, antibiotic therapy for at least 2 weeks prior, or no antibiotic therapy for at least 3 months prior. Exclusion criteria comprised active infection, pregnancy, and immunodeficiency. BALF was obtained by the same experienced physician to avoid interindividual variability. Patients received sedoanalgesia with propofol and/or midazolam by the physician in charge to minimize discomfort, as well as standard monitoring (ECG, heart rate, blood pressure, oximetry). After positioning the bronchoscope in a wedge position, 10 ml saline was instilled and extracted by suction. Samples were centrifuged (4,000 g for 10 minutes), supernatants were aliquoted in sterile tubes, and kept frozen at –80°C until further use.
Statistics.
Data are mean ± SD. For comparison of 2 groups, the Mann-Whitney U test (2-tailed) was used. When the normality test (D’Agostino & Pearson omnibus normality test) was passed, parametric statistical analyses were applied: unpaired t test with Welch’s correction (2-tailed). For all statistical analyses, P < 0.05 was considered significant with *P < 0.05, **P < 0.01, ***P < 0.005. To detect a possible influence of APRIL and BAFF on IgA levels in patients, univariate analysis was used.
Study approval.
All animal experiments were carried out in strict adherence to the Animal Protection Act under German law, following the approval of the corresponding institutional (Charité-Universitätsmedizin Berlin) and governmental (LAGeSo Berlin, approval ID G0367/11, G0178/14, and G0100/16) animal welfare authorities. The human part of the trial was approved by the institutional review board (Ethics Committee of the Medical University of Vienna, identifier 1364/2016) and was registered at the clinical trial registry (http://www.ClinicalTrials.gov, identifier NCT02912520). Written informed consent was received from all participants.
Author contributions
OHR, CC, MMH, SP, BG, KR, SB carried out most of the experimental work and analyzed the data under the guidance of LES, MW, NS, and BO. AAK and JN purified the monoclonal and polyclonal IgA preparations. HH, CJK, VM, MWH, JB, M. Suter, PS, M. Schnare, and US provided experimental tools. BO conceived and supervised the project. BO and CC wrote the manuscript. All authors approved and edited the manuscript.
Supplementary Material
Acknowledgments
We thank Sylvia Knapp, University of Vienna, for help with the Pseudomonas aeruginosa mouse model. We also thank Ursula Rüschendorf, Alexandra Bittroff-Leben, Ulrike Escher, Gernot Reifenberger, Ira Ekmekciu, Eliane von Klitzing, and the staff of the animal research facility of the Charité University Medicine Berlin for excellent technical assistance, animal breeding, and generation of secondary abiotic mice. Moreover, we are grateful to Berlin-Brandenburg Center for Regenerative Therapies for access to the FACS. This work was supported by the Deutsche Forschungsgemeinschaft (SFB-TR84 A5 to BO, CC, and MMH; C3/C6 to MW; SPP1656 to MWH) and by a Russian Science Foundation grant (IgA antibody generation, 17-74-20059 to AAK). OHR was temporally supported by a research fellowship of the Charité Universitätsmedizin Berlin. PS is supported by grants from the Swiss National Science Foundation.
Version 1. 05/17/2018
In-Press Preview
Version 2. 07/16/2018
Electronic publication
Version 3. 08/01/2018
Print issue publication
Funding Statement
to B.O., C.C., M.M.H., M.W., N.S.
to M.W.H.
to A.K.
to P.S.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(8):3535–3545.https://doi.org/10.1172/JCI97065.
See the related Commentary at Antibiotic therapy–induced collateral damage: IgA takes center stage in pulmonary host defense.
Contributor Information
Oliver H. Robak, Email: oliver.robak@charite.de.
Markus M. Heimesaat, Email: markus.heimesaat@charite.de.
Andrey A. Kruglov, Email: kruglov@drfz.de.
Sandra Prepens, Email: sandra.prepens@charite.de.
Justus Ninnemann, Email: justus.ninnemann@drfz.de.
Birgitt Gutbier, Email: birgitt.gutbier@charite.de.
Katrin Reppe, Email: Katrin.Reppe@charite.de.
Hubertus Hochrein, Email: hubertus.hochrein@bavarian-nordic.com.
Mark Suter, Email: Mark.Suter@bavarian-nordic.com.
Carsten J. Kirschning, Email: carsten.kirschning@uk-essen.de.
Veena Marathe, Email: veena.marathe@uk-essen.de.
Jan Buer, Email: jan.buer@uk-essen.de.
Mathias W. Hornef, Email: mhornef@ukaachen.de.
Markus Schnare, Email: schnarem@staff.uni-marburg.de.
Pascal Schneider, Email: Pascal.Schneider@unil.ch.
Martin Witzenrath, Email: martin.witzenrath@charite.de.
Stefan Bereswill, Email: stefan.bereswill@charite.de.
Ulrich Steinhoff, Email: ulrich.steinhoff@staff.uni-marburg.de.
Norbert Suttorp, Email: norbert.suttorp@charite.de.
Leif E. Sander, Email: Leif-Erik.Sander@charite.de.
Catherine Chaput, Email: catherine.chaput@charite.de.
Bastian Opitz, Email: bastian.opitz@charite.de.
References
- 1.Zarb P, et al. The European Centre for Disease Prevention and Control (ECDC) pilot point prevalence survey of healthcare-associated infections and antimicrobial use. Euro Surveill. 2012;17(46):20316. doi: 10.2807/ese.17.46.20316-en. [DOI] [PubMed] [Google Scholar]
- 2.Vincent JL, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302(21):2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 3.Magill SS, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassini A, et al. Burden of six healthcare-associated infections on European population health: estimating incidence-based disability-adjusted life years through a population prevalence-based modelling study. PLoS Med. 2016;13(10):e1002150. doi: 10.1371/journal.pmed.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melsen WG, et al. Attributable mortality of ventilator-associated pneumonia: a meta-analysis of individual patient data from randomised prevention studies. Lancet Infect Dis. 2013;13(8):665–671. doi: 10.1016/S1473-3099(13)70081-1. [DOI] [PubMed] [Google Scholar]
- 6.Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis. 2010;51 Suppl 1:S81–S87. doi: 10.1086/653053. [DOI] [PubMed] [Google Scholar]
- 7.Edelstein MV, et al. Spread of extensively resistant VIM-2-positive ST235 pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis. 2013;13(10):867–876. doi: 10.1016/S1473-3099(13)70168-3. [DOI] [PubMed] [Google Scholar]
- 8.Gniadek TJ, Carroll KC, Simner PJ. Carbapenem-resistant non-glucose-fermenting gram-negative bacilli: the missing piece to the puzzle. J Clin Microbiol. 2016;54(7):1700–1710. doi: 10.1128/JCM.03264-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metersky ML, Frei CR, Mortensen EM. Predictors of pseudomonas and methicillin-resistant staphylococcus aureus in hospitalized patients with healthcare-associated pneumonia. Respirology. 2016;21(1):157–163. doi: 10.1111/resp.12651. [DOI] [PubMed] [Google Scholar]
- 10.Shindo Y, et al. Risk factors for drug-resistant pathogens in community-acquired and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2013;188(8):985–995. doi: 10.1164/rccm.201301-0079OC. [DOI] [PubMed] [Google Scholar]
- 11.Woodhead M, et al. Guidelines for the management of adult lower respiratory tract infections—full version. Clin Microbiol Infect. 2011;17(Suppl 6):E1–59. doi: 10.1111/j.1469-0691.2011.03672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venier AG, et al. Identifying new risk factors for pseudomonas aeruginosa pneumonia in intensive care units: experience of the French national surveillance, REA-RAISIN. J Hosp Infect. 2011;79(1):44–48. doi: 10.1016/j.jhin.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Kalil AC, et al. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5):e61–e111. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalhoff K, et al. [Epidemiology, diagnosis and treatment of adult patients with nosocomial pneumonia. S-3 Guideline of the German Society for Anaesthesiology and Intensive Care Medicine, the German Society for Infectious Diseases, the German Society for Hygiene and Microbiology, the German Respiratory Society and the Paul-Ehrlich-Society for Chemotherapy] Pneumologie. 2012;66(12):707–765. doi: 10.1055/s-0032-1325924. [DOI] [PubMed] [Google Scholar]
- 15.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13(11):790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352(6285):535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souza DG, et al. The essential role of the intestinal microbiota in facilitating acute inflammatory responses. J Immunol. 2004;173(6):4137–4146. doi: 10.4049/jimmunol.173.6.4137. [DOI] [PubMed] [Google Scholar]
- 18.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37(1):171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 22.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaura E, et al. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. MBio. 2015;6(6):e01693–e01615. doi: 10.1128/mBio.01693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heimesaat MM, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol. 2006;177(12):8785–8795. doi: 10.4049/jimmunol.177.12.8785. [DOI] [PubMed] [Google Scholar]
- 26.Zeng MY, et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity. 2016;44(3):647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 28.Tezuka H, et al. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448(7156):929–933. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- 29.Litinskiy MB, et al. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3(9):822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He B, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26(6):812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Wang N, Hammarström L. IgA deficiency: what is new? Curr Opin Allergy Clin Immunol. 2012;12(6):602–608. doi: 10.1097/ACI.0b013e3283594219. [DOI] [PubMed] [Google Scholar]
- 32.Yazdani R, Azizi G, Abolhassani H, Aghamohammadi A. Selective IgA deficiency: epidemiology, pathogenesis, clinical phenotype, diagnosis, prognosis and management. Scand J Immunol. 2017;85(1):3–12. doi: 10.1111/sji.12499. [DOI] [PubMed] [Google Scholar]
- 33.Garrouste-Orgeas M, et al. Oropharyngeal or gastric colonization and nosocomial pneumonia in adult intensive care unit patients. A prospective study based on genomic DNA analysis. Am J Respir Crit Care Med. 1997;156(5):1647–1655. doi: 10.1164/ajrccm.156.5.96-04076. [DOI] [PubMed] [Google Scholar]
- 34.Annane D, et al. Immunoglobulin A levels in bronchial samples during mechanical ventilation and onset of nosocomial pneumonia in critically ill patients. Am J Respir Crit Care Med. 1996;153(5):1585–1590. doi: 10.1164/ajrccm.153.5.8630606. [DOI] [PubMed] [Google Scholar]
- 35.Petrof EO, Dhaliwal R, Manzanares W, Johnstone J, Cook D, Heyland DK. Probiotics in the critically ill: a systematic review of the randomized trial evidence. Crit Care Med. 2012;40(12):3290–3302. doi: 10.1097/CCM.0b013e318260cc33. [DOI] [PubMed] [Google Scholar]
- 36.Bo L, et al. Probiotics for preventing ventilator-associated pneumonia. Cochrane Database Syst Rev. 2014;10:CD009066. doi: 10.1002/14651858.CD009066.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasquali C, et al. Enhanced mucosal antibody production and protection against respiratory infections following an orally administered bacterial extract. Front Med (Lausanne) 2014;1:41. doi: 10.3389/fmed.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crabbé PA, Bazin H, Eyssen H, Heremans JF. The normal microbial flora as a major stimulus for proliferation of plasma cells synthesizing IgA in the gut. The germ-free intestinal tract. Int Arch Allergy Appl Immunol. 1968;34(4):362–375. doi: 10.1159/000230130. [DOI] [PubMed] [Google Scholar]
- 39.Tezuka H, et al. Prominent role for plasmacytoid dendritic cells in mucosal T cell-independent IgA induction. Immunity. 2011;34(2):247–257. doi: 10.1016/j.immuni.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy DD, et al. Mice overexpressing BAFF develop a commensal flora-dependent, IgA-associated nephropathy. J Clin Invest. 2011;121(10):3991–4002. doi: 10.1172/JCI45563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20(2):202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu W, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017;10(4):946–956. doi: 10.1038/mi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12(12):821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 44.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288(5474):2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 45.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2(5):328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101(7):1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langford TD, et al. Central importance of immunoglobulin A in host defense against Giardia spp. Infect Immun. 2002;70(1):11–18. doi: 10.1128/IAI.70.1.11-18.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmons CP, et al. Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium. Infect Immun. 2003;71(9):5077–5086. doi: 10.1128/IAI.71.9.5077-5086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruane D, et al. Microbiota regulate the ability of lung dendritic cells to induce IgA class-switch recombination and generate protective gastrointestinal immune responses. J Exp Med. 2016;213(1):53–73. doi: 10.1084/jem.20150567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuijt TJ, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65(4):575–583. doi: 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shulga-Morskaya S, et al. B cell-activating factor belonging to the TNF family acts through separate receptors to support B cell survival and T cell-independent antibody formation. J Immunol. 2004;173(4):2331–2341. doi: 10.4049/jimmunol.173.4.2331. [DOI] [PubMed] [Google Scholar]
- 52.Oldenburg M, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337(6098):1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- 53.Heimesaat MM, et al. MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut. 2010;59(8):1079–1087. doi: 10.1136/gut.2009.197434. [DOI] [PubMed] [Google Scholar]
- 54.Bereswill S, et al. Novel murine infection models provide deep insights into the “ménage à trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One. 2011;6(6):e20953. doi: 10.1371/journal.pone.0020953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dickson RP, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1(10):16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heimesaat MM, et al. Exacerbation of murine ileitis by Toll-like receptor 4 mediated sensing of lipopolysaccharide from commensal Escherichia coli. Gut. 2007;56(7):941–948. doi: 10.1136/gut.2006.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.