Abstract
BACKGROUND. A common germline variant in HSD3B1(1245A>C) encodes for a hyperactive 3β-hydroxysteroid dehydrogenase 1 (3βHSD1) missense that increases metabolic flux from extragonadal precursor steroids to DHT synthesis in prostate cancer. Enabling of extragonadal DHT synthesis by HSD3B1(1245C) predicts for more rapid clinical resistance to castration and sensitivity to extragonadal androgen synthesis inhibition. HSD3B1(1245C) thus appears to define a subgroup of patients who benefit from blocking extragonadal androgens. However, abiraterone, which is administered to block extragonadal androgens, is a steroidal drug that is metabolized by 3βHSD1 to multiple steroidal metabolites, including 3-keto-5α-abiraterone, which stimulates the androgen receptor. Our objective was to determine if HSD3B1(1245C) inheritance is associated with increased 3-keto-5α-abiraterone synthesis in patients.
METHODS. First, we characterized the pharmacokinetics of 7 steroidal abiraterone metabolites in 15 healthy volunteers. Second, we determined the association between serum 3-keto-5α-abiraterone levels and HSD3B1 genotype in 30 patients treated with abiraterone acetate (AA) after correcting for the determined pharmacokinetics.
RESULTS. Patients who inherit 0, 1, and 2 copies of HSD3B1(1245C) have a stepwise increase in normalized 3-keto-5α-abiraterone (0.04 ng/ml, 2.60 ng/ml, and 2.70 ng/ml, respectively; P = 0.002).
CONCLUSION. Increased generation of 3-keto-5α-abiraterone in patients with HSD3B1(1245C) might partially negate abiraterone benefits in these patients who are otherwise more likely to benefit from CYP17A1 inhibition.
FUNDING. Prostate Cancer Foundation Challenge Award, National Cancer Institute.
Keywords: Endocrinology, Oncology
Keywords: Prostate cancer, Sex hormones
Introduction
Prostate cancer progression is highly dependent on androgen receptor (AR) stimulation by testosterone and its more potent 5α-reduced metabolite, 5α-dihydrotestosterone (DHT) (1, 2). The long-standing standard-of-care treatment for metastatic prostate cancer has therefore been androgen deprivation therapy (ADT) by way of medical or surgical castration (3). Frequent clinical responses to ADT are typically followed by the development of castration-resistant prostate cancer (CRPC), which is accompanied by intratumoral production of potent androgens (i.e., testosterone and DHT) from extragonadal precursor steroids (4). Synthesis of testosterone and/or DHT in CRPC from extragonadal precursors that originate from the adrenal gland (e.g., DHEA and DHEA-sulfate) and/or de novo synthesis from cholesterol both have an absolute requirement for steroid Δ5 → Δ4 isomerization and 3β-OH → 3-keto oxidation — reactions that are catalyzed by the enzyme 3β-hydroxysteroid dehydrogenase 1 (3βHSD1) (5, 6). The common (20%–35% allele frequency) germline variant HSD3B1(1245C) encodes for a protein degradation–resistant 3βHSD1, which effectively increases intratumoral testosterone and/or DHT synthesis from extragonadal precursor steroids (7). Patients with advanced prostate cancer who inherit the HSD3B1(1245C) variant have worse outcomes after initiation of ADT and more rapid development of CRPC because they have enabled enzymatic machinery that makes use of extragonadal precursor steroids (8–11). On the other hand, HSD3B1(1245C) inheritance may result in a tumor dependence on extragonadal precursors, as evidenced by its association with more sustainable clinical responses to the nonsteroidal CYP17A1 inhibitor, ketoconazole (12). Together, these data suggest that HSD3B1 inheritance is a prostate cancer determinant of relative gonadal versus extragonadal androgen dependence and that patients with HSD3B1(1245C) inheritance may benefit from earlier ablation of extragonadal androgen synthesis (13).
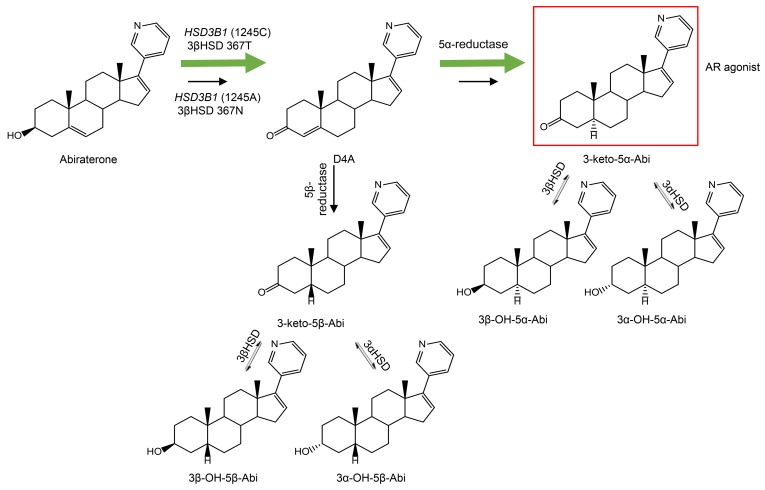
Abiraterone is a potent steroidal inhibitor of CYP17A1 (an enzyme required for extragonadal androgen synthesis) and when used in combination with prednisone it prolongs survival in the setting of CRPC (14, 15). More recently, abiraterone acetate (AA) plus prednisone has been shown to have a survival benefit that is more profound when administered up front along with ADT for the treatment of metastatic castration-sensitive prostate cancer (CSPC) (16, 17). However, the 3β-OH Δ5-steroidal structure of abiraterone makes it susceptible to catalysis by 3βHSD to Δ4-abiraterone (D4A) (18) and subsequently 5β-reductase and 5α-reductase, resulting in the generation of three 5β-reduced metabolites and three 5α-reduced metabolites in patients, including 3-keto-5α-abiraterone (3-keto-5α-Abi; Figure 1), which stimulates AR and tumor progression in mouse xenograft models (19, 20). Currently, it is not known if HSD3B1(1245C) inheritance increases 3-keto-5α-Abi synthesis, just as it hastens DHT synthesis from extragonadal precursor steroids. We hypothesized that HSD3B1(1245C) inheritance is associated with serum levels of 5α-reduced abiraterone metabolites, including 3-keto-5α-Abi. Here, we performed a 2-part study to test this hypothesis. First, we performed a pharmacokinetic (PK) study to define the temporal generation of steroidal abiraterone metabolites. Second, we determined the association between HSD3B1 genotype and steroidal abiraterone metabolites, including the three 5α-reduced metabolites (3-keto-5α-Abi, 3α-OH-5α-Abi, 3β-OH-5α-Abi) after normalizing for PK.
Figure 1. Schema of abiraterone metabolism by steroidogenic enzymes.
Abiraterone is metabolized by 3βHSD1 to D4A, which is then metabolized to 6 downstream metabolites, including the AR agonist 3-keto-5α-Abi. In patients who inherit the HSD3B1(1245A>C) variant, 3βHSD1 is resistant to degradation and metabolism is faster, leading to higher levels of 3-keto-5α-Abi. Green arrows denote increased metabolic flux with the 3βHSD1 variant enzyme. Red box denotes the metabolite with known AR agonist activity.
Results
Pharmacokinetics of biochemically active steroidal metabolites of abiraterone in healthy volunteers.
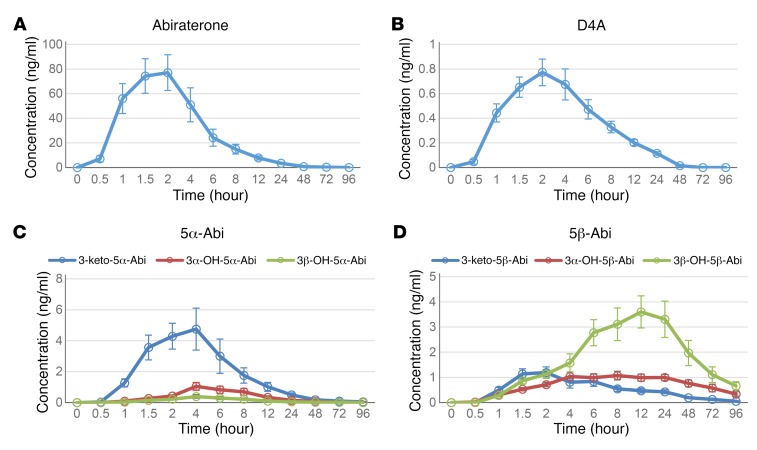
Fifteen healthy male volunteers received a single dose of AA (1,000 mg plus 240 mg apalutamide, an AR antagonist). Plasma samples were collected at –0.5 (predose), 0.5, 1, 1.5, 2, 4, 6, 8, 12, 24, 48, 72, and 96 hours, and analyzed for steroidal abiraterone metabolites by our published liquid chromatography–tandem mass spectrometry (LC-MS/MS) method (21). Using Phoenix WinNonlin 6.3 software, we performed a noncompartmental analysis to describe the disposition of each compound based on mechanisms of formation and degradation (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI98319DS1). The metabolites reached a maximum concentration 1.9–19.3 hours after administration. D4A and 3-keto-5α-Abi were readily formed, with a recorded mean time to reach the maximum concentration (tmax) of 2.07 hours and 2.70 hours, respectively. The 5β-abiraterone metabolites had longer tmax values: 16.8 hours for 3β-OH-5β-Abi and 19.33 hours for 3α-OH-5β-Abi. The mean time for the last detectable concentration (tlast) for D4A was 25.3 hours whereas the tlast value for 3-keto-5α-Abi was 62.4 hours. The mean maximum concentration (Cmax) was 90 ng/ml for abiraterone, 0.91 ng/ml for D4A, and 5.5 ng/ml for 3-keto-5α-Abi. Among the metabolites, 3-keto-5α-Abi had the highest Cmax, almost 6 times higher than D4A. As shown in the mean plasma concentration versus time plots (Figure 2, A–D), metabolism to D4A peaked the earliest and then rapidly declined. These data show that concentrations of the AR agonist 3-keto-5α-Abi are much higher than the AR antagonist D4A, even after a single dose of AA. These data also suggest that the elucidation of any link between genetics and the three 5α-reduced abiraterone metabolites that rapidly generate and decline probably requires an accounting of pharmacokinetics.
Figure 2. Mean plasma concentrations of abiraterone and steroidal metabolites after a single dose of AA and apalutamide in 15 healthy volunteers.
The mean plasma concentration at each time point was used to generate the plots for (A) abiraterone, (B) D4A, (C) 5α-Abi metabolites, and (D) 5β-Abi metabolites. Error bars represent SEM.
Association between HSD3B1 genotype and abiraterone metabolites in patients with CRPC.
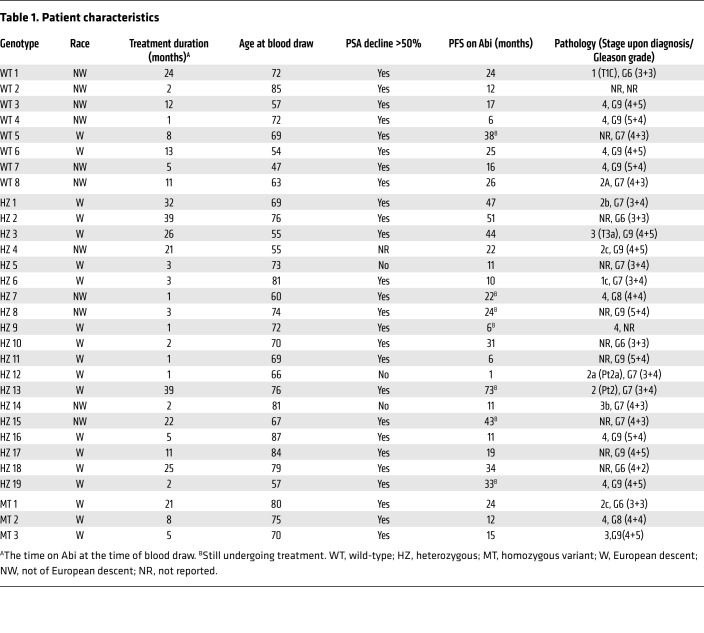
We next conducted a study in patients with metastatic CRPC who were receiving AA plus prednisone as standard-of-care treatment in order to determine if inheritance of the HSD3B1 genotype is associated with production of steroidal abiraterone metabolites that are regulated by the cognate 3βHSD1 enzyme (Table 1). In the context of low versus high 3βHSD1 enzymatic activity, robust 5α-reductase activity (which follows 3βHSD) would be expected to lead to low versus high levels of 5α-reduced steroid metabolites (22). We therefore hypothesized that inheritance of the HSD3B1(1245C) variant that encodes for a protein degradation–resistant enzyme would correlate with higher serum concentrations of 5α-abiraterone metabolites, including 3-keto-5α-Abi, which exhibits AR agonist activity (19). As the PK results in healthy volunteers show, the formation of 3-keto-5α-Abi starts early after AA administration, and the serum concentration is highly dependent on the time elapsed since administration. Patients received a daily dose of 1,000 mg AA, and blood was drawn during routine clinic visits. Time from prior AA dose to blood draw (1–16 hours) was recorded for each patient at the time of blood draw. Concentrations of abiraterone and its steroidal metabolites were measured in the sera using LC-MS/MS, as was done with the PK study in healthy volunteers (Supplemental Table 2) (21). To correct for variations between the last AA dose and blood draw among individual patients with CRPC, the abiraterone metabolite concentrations (D4A, 3-keto-5α-Abi, 3α-OH-5α-Abi, 3β-OH-5α-Abi, 3-keto-5β-Abi, 3α-OH-5β-Abi, and 3β-OH-5β-Abi) in these 30 patients were normalized to the 8-hour time point of the PK study in healthy controls to account for their respective PK parameters. All patients were genotyped for HSD3B1 using germline DNA from buffy coat and grouped by genotype. Of the 30 patients in this analysis, 8 (27%) were homozygous wild-type (WT), 19 (63%) were heterozygous (HZ), and 3 (10%) were homozygous variant (MT).
Table 1. Patient characteristics.
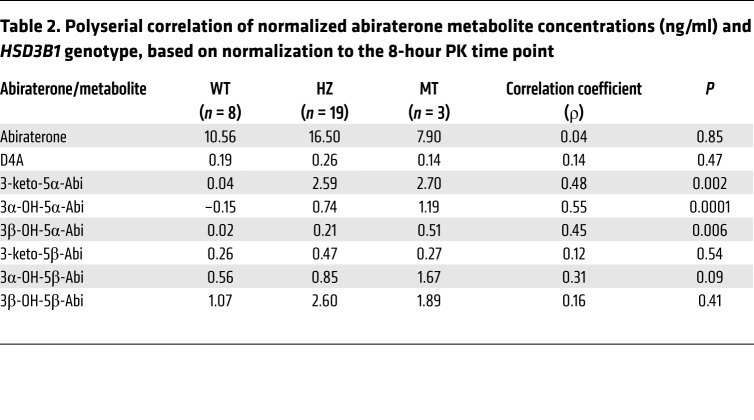
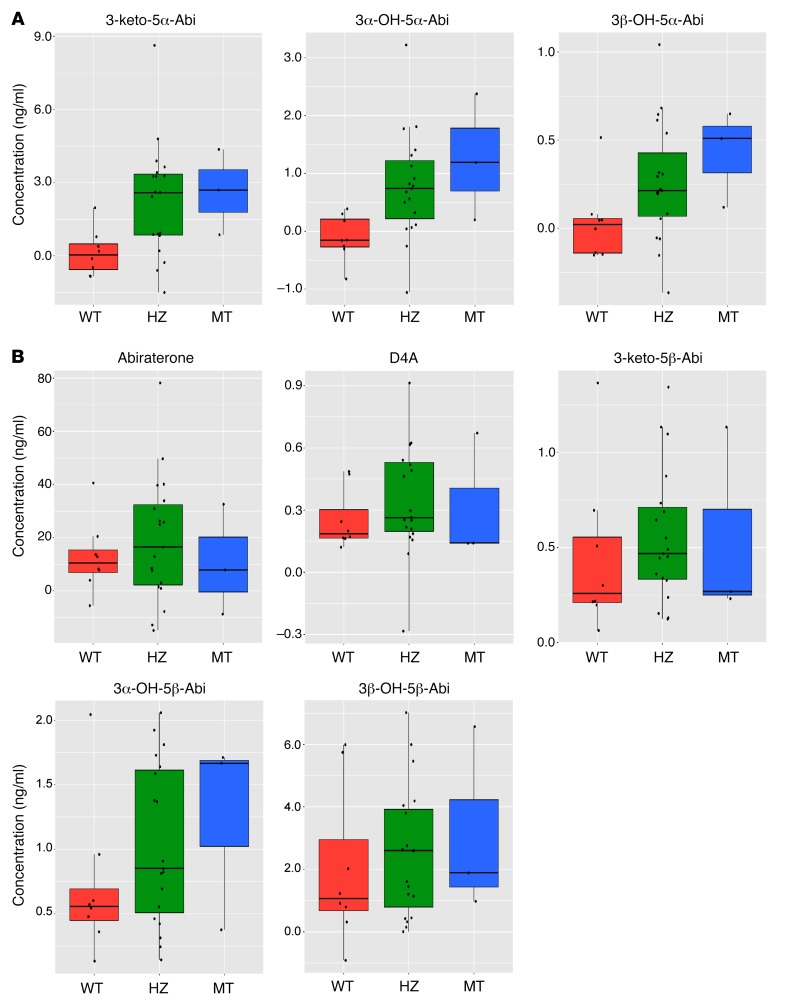
HSD3B1(1245C) variant allele inheritance was associated with a significant increase in the concentrations of all three 5α-reduced abiraterone metabolites, with the highest concentrations observed in patients with homozygous variant HSD3B1. The normalized values for 3-keto-5α-Abi, which is the specific metabolite with AR agonist activity (19), had a stepwise concentration increase of 0.04 ng/ml, 2.60 ng/ml, and 2.70 ng/ml (P = 0.002) for patients who inherited 0, 1, or 2 copies of the HSD3B1(1245C) variant, respectively (Table 2). Concentrations of 3α-OH-5α-Abi (P = 0.0001) and 3β-OH-5α-Abi (P = 0.006) similarly and significantly increased with the number of HSD3B1(1245C) variant alleles inherited, further imputing a mechanistic biochemical link between more active 3βHSD1 enzyme activity and downstream 5α-reduced steroidal metabolites. In contrast, there was no significant association between HSD3B1 variant allele inheritance and normalized concentrations of abiraterone, D4A, and all 5β-abiraterone metabolites (Table 2 and Figure 3). Alternative analysis using normalization to the 4-hour (instead of 8-hour) time point of the PK study (Supplemental Table 3) also yielded correlation coefficients that were statistically significant for all three 5α-reduced abiraterone metabolites but not the other metabolites. Comparisons with ANOVA, Tukey’s honest significant difference test (Supplemental Table 4), and corrections for false discovery rates (Supplemental Table 5) further support the association between HSD3B1 variant allele inheritance and the 5α-reduced abiraterone metabolites.
Table 2. Polyserial correlation of normalized abiraterone metabolite concentrations (ng/ml) and HSD3B1 genotype, based on normalization to the 8-hour PK time point.
Figure 3. HSD3B1(1245C) variant inheritance is associated with elevated generation of 3-keto-5α-Abi, 3α-OH-5α-Abi, and 3β-5α-Abi in 30 patients with CRPC treated with abiraterone.
(A) Higher levels of all three 5α-Abi metabolites were significantly present in patients with the HSD3B1(1245C) genotype. (B) Abiraterone, D4A, 3-keto-5β-Abi, 3α-OH-5β-Abi, and 3β-5β-Abi concentrations were not significantly correlated with HSD3B1 genotype. The levels of each metabolite were normalized to the 8-hour value in the PK study and the metabolite concentration for each patient at the fixed time point was predicted by a regression method. The significance of the difference among genotypes for each metabolite was determine by polyserial correlation analysis: 3-keto-5α-Abi, P = 0.002; 3α-OH-5α-Abi, P = 0.0001; 3β-5α-Abi, P = 0.006.
HSD3B1 (1245 A>C) and response to abiraterone acetate.
We performed Kaplan-Meier analysis for progression-free survival (PFS) on abiraterone acetate treatment, grouped by HSD3B1 genotype (Supplemental Figure 1). We also conducted the log-rank test and found no statistically significant difference between the 3 genotypes (P = 0.22). However, we note that this cohort is probably too small for meaningful evaluation.
Discussion
The standard of care for the upfront treatment of metastatic prostate cancer has recently progressed from the use of gonadal testosterone deprivation therapy (i.e., castration) to depletion of both gonadal and extragonadal androgens with the use of castration and AA plus prednisone (16, 17). However, the duration of response remains widely variable, suggesting frequent mismatches between the biology in individual patients and their respective treatments. The selection and application of frontline systemic therapy as well as the use of subsequent second-line therapies do not make use of genetic and biochemical information pertinent to the molecular and metabolic diversity of prostate cancers and the spectrum of disease-driving biological processes. Metabolic, genetic, and clinical data all show that the HSD3B1(1245C) variant encodes for a stable enzyme that increases DHT synthesis from extragonadal precursor steroids (7), leading to more rapid resistance to castration (8–11) and longer clinical responses to pharmacologic inhibition of extragonadal androgens (12). An open question was whether the HSD3B1(1245C) variant also engages in a confounding effect on pharmacologic therapy because it also utilizes abiraterone as a substrate, thereby converting it to abiraterone metabolites that directly interact with AR (18, 19).
The data presented here strongly suggest that germline HSD3B1 genotype inheritance is a determinant of abiraterone metabolism by steroidogenic enzymes. Of the 7 steroidal abiraterone metabolites downstream of 3βHSD1 tested, only the three 5α-reduced metabolites increased as a function of the number HSD3B1(1245C) alleles inherited. Notably, there was no increase in D4A, which is the immediate abiraterone metabolite downstream of 3βHSD1. This can be explained because the increased D4A that is generated with HSD3B1(1245C) inheritance is not allowed to accumulate due to rapid 5α-reduction to 3-keto-5α-Abi, 3α-OH-5α-Abi, and 3β-OH-5α-Abi. Although there is a trend for an increase in the concentration of 3α-OH-5β-Abi (P = 0.09) with HSD3B1(1245C) inheritance, there is no statistically significant increase in the concentration of any of the three 5β-reduced abiraterone metabolites, which are also downstream of D4A. However, the 5β-reduced metabolites exhibit a delayed rise and more sustained levels when compared with the 5α-reduced metabolites. Some individuals exhibit a rise after nadir 5β-reduced abiraterone metabolite levels (not shown) that are not apparent in the mean values. Together, these characteristics might be attributable in part to enterohepatic circulation, which is probably due in some part to the 5β-reduced structural commonality between these metabolites and bile acids, which are known to undergo enterohepatic circulation (23).
Although 3-keto-5α-Abi is a modest AR agonist that is substantially weaker than DHT, its presence in the near-complete absence of endogenous androgens in the setting of gonadal and extragonadal androgen deprivation may be deleterious. Furthermore, clinical outcomes data consistently show that HSD3B1(1245C) inheritance confers a switch to a decreased tumor dependence on gonadal androgens and increased dependence on extragonadal androgens (8–12), together suggesting that this genetically defined population that identifies a certain steroid physiology may have a disproportionate benefit from CYP17A1 blockade with a nonsteroidal CYP17A1 inhibitor (12). In contrast to nonsteroidal CYP17A1 inhibition, HSD3B1(1245C) inheritance does not appear to correlate with duration of response to AA therapy (24). Our data presented here offer an explanation for these contrasting findings and show that in the context of abiraterone therapy, HSD3B1(1245C) inheritance presents the specific conundrum of elevating synthesis of an AR agonist metabolite in a population that otherwise stands more to benefit from CYP17A1 inhibition. Although the clinical consequences of variations in steroidal abiraterone metabolite concentrations are not yet known, together these data suggest that patients with HSD3B1(1245C) inheritance may have a greater benefit from an alternative to abiraterone, such as a nonsteroidal agent that blocks extragonadal androgen synthesis or signaling.
A limitation of our work is that the PK studies in part 1 were done in noncastrated volunteers after a single dose of AA in combination with apalutamide, whereas the analysis in part 2 comparing HSD3B1 genotypes was done in castrated patients with continuous AA dosing who were also treated with prednisone. However, any differences attributable to single versus continuous dosing would be expected to confound the apparent correlation between HSD3B1 genotype and 5α-reduced abiraterone metabolites that we found and thus might be even stronger than our data suggest. Another limitation of our study is the small number (3 patients) in the homozygous variant group.
In conclusion, we find that inheritance of the HSD3B1(1245C) variant that encodes for a protein degradation–resistant 3βHSD1 enzyme is significantly associated with higher serum concentrations of 5α-reduced abiraterone metabolites, including the AR agonist 3-keto-5α-Abi. This pharmacogenetic effect may partially negate the benefit of abiraterone in a genetically defined subset of patients that is otherwise more likely to benefit from extragonadal androgen ablation. Alternative approaches to therapy should be explored in patients who harbor the HSD3B1(1245C) variant.
Methods
Subject recruitment and selection.
In part 1 of the PK study (conducted at Janssen Research & Development), 15 healthy male volunteers received a single oral dose of 1,000 mg AA plus 240 mg apalutamide under fasted conditions. The results from this study were used to generate reference data for abiraterone and the metabolites. In part 2, 30 patients with metastatic CRPC were recruited through the Department of Hematology and Oncology, Taussig Cancer Institute at the Cleveland Clinic, Cleveland, Ohio, USA. Patients were included if they were receiving treatment with AA plus prednisone. Patients with prior ketoconazole treatment or chemotherapy or who were currently receiving 5α-reductase inhibitors were excluded from the analysis. All subjects provided written informed consent before enrolling at the Cleveland Clinic under a protocol approved by the institutional review board (case 7813). Patient characteristics are listed in Table 1.
Determination of abiraterone metabolite pharmacokinetics in healthy male volunteers.
The PK sampling time points were –0.5 (predose), 0.5, 1, 1.5, 2, 4, 6, 8, 12, 24, 48, 72, and 96 hours. Samples were stored at –80°C for LC-MS/MS analysis. The plasma samples were delivered to the Cleveland Clinic and analyzed for the steroidal abiraterone metabolite concentrations using a validated LC–MS/MS method (21). The following PK parameters were determined using the noncompartment method and Phoenix WinNonlin 6.3 software: Cmax, tmax, time to reach half maximum concentration (t1/2), tlast, area under the plasma concentration – time curve from time 0 to time of the last quantifiable concentration (Clast) (AUClast), area under the plasma concentration time curve from time 0 to infinite time (AUC0–∞), elimination rate constant (Ke), mean residence time (MRT), and apparent volume of distribution during terminal phase after nonintravenous administration of a total plasma clearance of drug (CL/F) (Vz/F).
Determination of abiraterone metabolites in patients with CRPC.
In part 2, 30 patients with CRPC (47–87 years of age at time of blood draw) diagnosed with prostate cancer between January 1995 and May 2014 who began treatment with AA plus prednisone between December 2011 and October 2016 were available for study. Blood was collected between 1.5 hours and 16 hours after the 1,000 mg daily dose in Vacutainer Plus serum blood collection tubes (catalog BD367814, Becton Dickinson), and allowed to clot. Tubes were centrifuged at 3,740 g for 10 minutes. Serum aliquots were frozen at –80°C for LC-MS/MS analysis.
DNA extraction, amplification, and genotyping.
DNA was extracted from peripheral blood mononuclear cells using Qiagen’s DNEasy Blood & Tissue Kit. DNA concentration was quantified using a ThermoFisher NanoDrop 2000 Spectrophotometer as previously described (8). Genomic DNA was stored at 4°C. PCR was performed in 20 μl reactions (final volume) using the following for each reaction: 8 μl LightScanner Master Mix (Biofire; 2.5× Master Mix); 4 μl extracted genomic DNA (10 ng/μl); 1 μl forward primer (1 μM); 1 μl reverse primer (10 μM); and 1 μl unlabeled LNA6 probe (10 μM).
The PCR and the melting analyses were performed with an AB StepOnePlus Real-Time PCR System (Life Technologies) using the following conditions: initial denaturing for 2 minutes at 95°C, followed by 60 cycles of 94°C for 30 seconds, 66°C for 30 seconds, and 75°C for 30 seconds. The amplification cycles were followed by melting at 95°C for 30 seconds followed by cooling to 25°C for 60 seconds, followed by an increase to 50°C for 15 seconds, and a slow final denaturation to 95°C at a thermal ramp rate of 0.3%–1%. Data were analyzed using probe melting derivative peaks. Each validation run was performed with triplicate control samples representing the 3 different genotypes, and each sample was tested in triplicate during the study genotyping.
Liquid chromatography tandem mass spectrometry analysis.
Abiraterone and its steroidal metabolites were determined in the 2 cohorts using our previously published LC-MS/MS method (21). In brief, the analytes were extracted from the biological matrix following a liquid–liquid extraction protocol with methyl tert-butyl ether, and the analytes were separated applying reversed-phase chromatography.
Statistics.
Treating the HSD3B1 genotype as an ordinal variable (WT < HZ < MT) we used polyserial correlation to measure the correlation between the HSD3B1 genotype and each metabolite’s concentration value (25). We used polycor package in R (version 3.3.3) to compute the polyserial correlation coefficient ρ and its P value (the null hypothesis is ρ = 0).
Metabolite analysis.
Because samples were collected at multiple time points from multiple patients, we predicted all the patients’ metabolite concentration values at the same time point (8 hours) based on metabolite measurements from the 15 healthy volunteers. We first divided the entire time space into 6 time slots: (a) 0 to less than 2 hours, (b) 2 to less than 4 hours, (c) 4 to less than 6 hours, (d) 6 to less than 8 hours, (e) 8 to less than 12 hours, and (f) 12 hours or more. The patients were grouped based on time slot for time from last dose to blood draw.
For each time slot, we performed a z-transformation of the patients’ metabolite concentration values to obtain the same mean and variance as those of the healthy volunteers in the same time slot, then used the z-transformed value for analysis. To confirm our findings, we performed the analysis again and based it on the 4-hour prediction (Supplemental Table 3).
We then conducted ANOVA and Tukey’s honest significant difference (HSD) tests for each metabolite, which involves pairwise comparisons of mean metabolite concentration values between genotypes. This test provides the estimate of difference between 2 means and its confidence interval (Supplemental Table 4). We also added FDR corrected values for the P values (from polyserial correlation) using the Benjamini and Hochberg method (Supplemental Table 5).
Study approval.
The studies on human samples were reviewed and approved by the IRB at the Cleveland Clinic. The PK study was conducted at Janssen Research & Development and approved by the Chesapeake IRB (approval number Pro00015779). All subjects provided appropriate prior written informed consent for participation in the study.
Author contributions
MA and NS designed the studies and wrote the manuscript. MA performed the mass spectrometry metabolism studies and analyzed the data. HE, AT, VSK, PM, PG, BR, and JG coordinated clinical studies. SP, BH, and THH performed statistical analysis and correlations. JT and NA performed genotyping studies. MPB processed patient specimens. SU and RJA provided abiraterone metabolites. WDF and CJP generated the pharmacokinetic data analysis. CC designed and supervised the pharmacokinetic study. SP and THH modeled the pharmacokinetic fit. All authors analyzed and discussed the results and commented on the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by funding from a Prostate Cancer Foundation Challenge Award (to NS) and grants from the National Cancer Institute (R01CA168899, R01CA172382, and R01CA190289 to NS).
Version 1. 06/25/2018
Electronic publication
Version 2. 08/01/2018
Print issue publication
Footnotes
Conflict of interest: CC is an employee of Janssen Research & Development.
Reference information: J Clin Invest. 2018;128(8):3333–3340.https://doi.org/10.1172/JCI98319.
Contributor Information
Mohammad Alyamani, Email: alyamam@ccf.org.
Hamid Emamekhoo, Email: hemamekhoo@medicine.wisc.edu.
Sunho Park, Email: parks@ccf.org.
Jennifer Taylor, Email: taylorj16@ccf.org.
Nima Almassi, Email: almassn@ccf.org.
Sunil Upadhyay, Email: sunilku@med.umich.edu.
Allison Tyler, Email: tylera2@ccf.org.
Bo Hu, Email: hub@ccf.org.
Tae Hyun Hwang, Email: hwangt@ccf.org.
Caly Chien, Email: cchien@its.jnj.com.
Vadim S. Koshkin, Email: koshkiv@ccf.org.
Prateek Mendiratta, Email: mendirp@ccf.org.
Petros Grivas, Email: pgrivas@u.washington.edu.
Brian Rini, Email: rinib2@ccf.org.
Jorge Garcia, Email: garciaj4@ccf.org.
Nima Sharifi, Email: sharifn@ccf.org.
References
- 1.Attard G, et al. Prostate cancer. Lancet. 2016;387(10013):70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 2.Dai C, Heemers H, Sharifi N. Androgen signaling in prostate cancer. Cold Spring Harb Perspect Med. 2017;7(9):a030452. doi: 10.1101/cshperspect.a030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6(2):76–85. doi: 10.1038/ncpuro1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang KH, et al. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2011;108(33):13728–13733. doi: 10.1073/pnas.1107898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evaul K, Li R, Papari-Zareei M, Auchus RJ, Sharifi N. 3beta-hydroxysteroid dehydrogenase is a possible pharmacological target in the treatment of castration-resistant prostate cancer. Endocrinology. 2010;151(8):3514–3520. doi: 10.1210/en.2010-0138. [DOI] [PubMed] [Google Scholar]
- 6.Lorence MC, Murry BA, Trant JM, Mason JI. Human 3 beta-hydroxysteroid dehydrogenase/delta 5----4isomerase from placenta: expression in nonsteroidogenic cells of a protein that catalyzes the dehydrogenation/isomerization of C21 and C19 steroids. Endocrinology. 1990;126(5):2493–2498. doi: 10.1210/endo-126-5-2493. [DOI] [PubMed] [Google Scholar]
- 7.Chang KH, et al. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell. 2013;154(5):1074–1084. doi: 10.1016/j.cell.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hearn JWD, et al. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol. 2016;17(10):1435–1444. doi: 10.1016/S1470-2045(16)30227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hearn JWD, et al. Association of HSD3B1 genotype with response to androgen-deprivation therapy for biochemical recurrence after radiotherapy for localized prostate cancer. JAMA Oncol. 2018;4(4):558–562. doi: 10.1001/jamaoncol.2017.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agarwal N, Hahn AW, Gill DM, Farnham JM, Poole AI, Cannon-Albright L. Independent validation of effect of HSD3B1 genotype on response to androgen-deprivation therapy in prostate cancer. JAMA Oncol. 2017;3(6):856–857. doi: 10.1001/jamaoncol.2017.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiota M, et al. Independent validation of missense polymorphism in HSD3B1 in Japanese men treated with primary androgen-deprivation therapy for metastatic prostate cancer. J Clin Oncol. 2018;36(suppl_6):179 [Google Scholar]
- 12.Almassi N, et al. HSD3B1 and response to a nonsteroidal CYP17A1 inhibitor in castration-resistant prostate cancer. JAMA Oncol. 2018;4(4):554–557. doi: 10.1001/jamaoncol.2017.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hettel D, Sharifi N. HSD3B1 status as a biomarker of androgen deprivation resistance and implications for prostate cancer. Nat Rev Urol. 2018;15(3):191–196. doi: 10.1038/nrurol.2017.201. [DOI] [PubMed] [Google Scholar]
- 14.de Bono JS, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan CJ, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368(2):138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James ND, et al. Abiraterone for prostate cancer not previously treated with hormone therapy. N Engl J Med. 2017;377(4):338–351. doi: 10.1056/NEJMoa1702900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fizazi K, et al. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N Engl J Med. 2017;377(4):352–360. doi: 10.1056/NEJMoa1704174. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, et al. Conversion of abiraterone to D4A drives anti-tumour activity in prostate cancer. Nature. 2015;523(7560):347–351. doi: 10.1038/nature14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, et al. Redirecting abiraterone metabolism to fine-tune prostate cancer anti-androgen therapy. Nature. 2016;533(7604):547–551. doi: 10.1038/nature17954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claessens F, Moris L. Prostate cancer: The influence of steroid metabolism on CYP17A1 inhibitor activity. Nat Rev Urol. 2017;14(10):590–592. doi: 10.1038/nrurol.2017.132. [DOI] [PubMed] [Google Scholar]
- 21.Alyamani M, Li Z, Upadhyay SK, Anderson DJ, Auchus RJ, Sharifi N. Development and validation of a novel LC-MS/MS method for simultaneous determination of abiraterone and its seven steroidal metabolites in human serum: innovation in separation of diastereoisomers without use of a chiral column. J Steroid Biochem Mol Biol. 2017;172:231–239. doi: 10.1016/j.jsbmb.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alyamani M, et al. Steroidogenic metabolism of galeterone reveals a diversity of biochemical activities. Cell Chem Biol. 2017;24(7):825–832.e6. doi: 10.1016/j.chembiol.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell DW, Setchell KD. Bile acid biosynthesis. Biochemistry. 1992;31(20):4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 24.Hahn AW, et al. Germline variant in HSD3B1 (1245 A>C) and response to abiraterone acetate plus prednisone (AA) in men with new onset metastatic castration-resistant prostate cancer. Clin Genitourin Cancer. doi: 10.1016/j.clgc.2018.03.006. doi: 10.1016/j.clgc.2018.03.006. [published online ahead of print March 27, 2018]. [DOI] [PubMed] [Google Scholar]
- 25.Olsson U, Drasgow F, Dorans NJ. The polyserial correlation coefficient. Psychometrika. 1982;47(3):337–347. doi: 10.1007/BF02294164. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.