Abstract
Elucidation of the mechanistic relationships between drugs, their targets, and diseases is at the core of modern drug discovery research. Thousands of studies relevant to the drug-target-disease (DTD) triangle have been published and annotated in the Medline/PubMed database. Mining this database affords rapid identification of all published studies that confirm connections between vertices of this triangle or enable new inferences of such connections. To this end, we describe the development of Chemotext, a publicly-available Web server that mines the entire compendium of published literature in PubMed annotated by Medline Subject Heading (MeSH) terms. The goal of Chemotext is to identify all known drug-target-disease relationships and infer missing links between vertices of the DTD triangle. As a proof-of-concept, we show that Chemotext could be instrumental in generating new drug repurposing hypotheses or annotating clinical outcomes pathways for known drugs. The Chemotext Web server is freely-available at http://chemotext.mml.unc.edu.
Graphical Abstract
Introduction
The fundamental goal of small molecule drug discovery is the identification of bioactive compounds for the treatment of disease1. Many modern drug discovery projects start with the discovery of novel targets and then progress in the direction of finding ligands of these targets that are expected to affect the disease. Bioactivity data from drug repurposing/discovery campaigns are increasingly available in public databases such as PubChem2,3 and ChEMBL4. At the same time, much information about the biological underpinnings of disease, i.e., effector proteins and pathways, as well as drug targets are stored primarily in the biomedical literature. Thus, biomedically relevant relationships between drugs, biological targets, and diseases, which we call the DTD triangle, can be identified through mining the published biomedical literature.5,6
PubMed, the largest repository of published biomedical research, is a freely-accessible search engine maintained by the United States National Library of Medicine (NLM) at the National Institutes of Health (NIH)7. PubMed can be used to retrieve scientific articles containing specific search terms that are stored in the Medline bibliographic database. PubMed can also return a list of Medical Subject Headings (MeSH), or so-called MeSH terms8. The purpose of these MeSH terms is to index and categorize published studies by the subject matters discussed therein. As most drugs, biological targets, and diseases discussed in biomedical literature are captured by associated MeSH terms, relationships between terms in the DTD triangle (represented by edges of the triangle with vertices representing MESH terms) can be established based on their frequent co-occurrences within articles.
Indeed, such considerations led to the development of the Chemotext approach,9 which focused on the extraction of MeSH terms describing “chemicals”, “targets”, and “diseases”, i.e., the components of the DTD triangle, that were found to frequently co-occur in abstracts of papers annotated in PubMed. These co-occurrences were regarded as an indication of plausible assertions linking drugs, targets and diseases. Furthermore, Chemotext was conceived as an extension of Swanson’s ABC paradigm9–11 wherein “A” terms are chemical (drug) - related MeSH terms, “B” terms are so-called “target” MeSH terms, i.e., proteins and pathways, and “C” terms are MeSH terms for diseases (Figure 1). The underlying hypothesis generation starts with the observation that the name of drug “A” co-occurs in the same articles as the name of target “B” while the name of disease “C” co-occurs in the same or additional articles with the same target “B”. Thus, if drug “A” and disease “C” have not been mentioned together in the same article, an “A-C” connection mediated though target “B” can be inferred. This analysis leads to the identification of a new possible therapeutic use of drug “A”. This reasoning protocol illustrates one of possible uses of Chemotext for drug repurposing, which has emerged in the past decade as a boon to traditional drug discovery.12,13
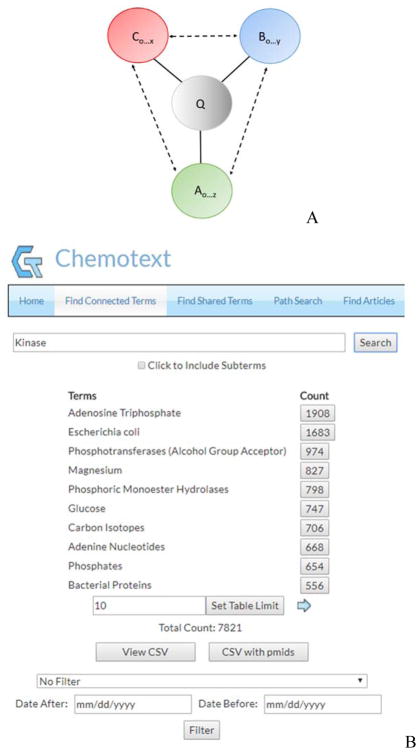
Figure 1. Swanson’s ABC paradigm used in Chemotext.
Chemical A is proposed to have an effect on Disease C since both terms are associated with Target B. Solid lines (edges) indicate an actual text-based relationship, while dashed lines (edges) indicate proposed connections.
Although efforts have been made to develop tools for text-mining of PubMed, such as “MeSHSim”,14 “pubmed.mineR”,15 and IBM-Watson,16 these current implementations are either available only as R-packages,14,15 which are not user-friendly, and/or proprietary.16 To this end, we have developed the publicly-available Chemotext Web server that mines published literature in PubMed in the form of MeSH terms. The goal of Chemotext is to establish text-based drug-target-disease relationships, which, as we show herein, can be used to generate novel drug repurposing hypotheses or elucidate clinical outcomes pathways that mechanistically connect drugs and diseases via intermediary, target-mediated biological effects of drug action. Similar to our Chembench Webportal,17 the Chemotext Web server is hosted by the Molecular Modeling Laboratory (MML) at the University of North Carolina – Chapel Hill and is freely-available at http://chemotext.mml.unc.edu/.
Methods
The Chemotext user interface is written in JavaScript with data retrieval through JQuery’s Asynchronous JavaScript and XML (AJAX) functionality.18 The data are stored in Neo4j, a graph database that uses nodes for articles and drug terminology. The server operates on Red Hat Linux and is hosted by the Longleaf computer cluster at UNC-Chapel Hill. MeSH term data were downloaded directly from the PubMed and Medline repository. Data were input into Neo4j using Cypher to parse the MeSH XML and to create the article and term nodes and relationships. Cypher queries allow for Neo4j to return sets of MeSH terms and article counts from the input term’s article relationships.
Data for calendar year 2016 were retrieved from the MEDLINE/PubMed Baseline Repository (MBR) in June 2017. Data are available at https://mbr.nlm.nih.gov/Downloads.shtml. Currently, the Chemotext database contains 19 282 732 articles and 78 758 882 connections between terms. Chemotext is currently fully functional only with the Google Chrome web browser on both PC and MAC operating systems.
Chemotext Environment
Chemotext generates text-based relationships via four modules described below: Find Connected Terms, Find Shared Terms, Path Search, and Find Articles. Within each module, there is a query bar that possesses the full dictionary of MeSH terms with an auto-complete function to facilitate searching. Each module can be executed separately or as part of a larger study design. On its homepage, Chemotext possesses direct link to the Medical Subject Headings search engine in order to facilitate the identification of correct MeSH terms for querying.
Find Connected Terms
In this module, every MeSH term that occurs in the same article as a query term is returned, and the total number of co-occurring terms and the associated article counts are reported. A schema of this module is presented in Figure 2A. To illustrate how this module is used, if “Kinase” is queried, 7 821 unique co-occurring MeSH terms are returned (Figure 2B), such as “Enzyme Inhibitors,” an A term with 333 article co-occurrences, and “Neoplasms”, a C term with 151 article co-occurrences (Table S1).
Figure 2. (A) Schema of the Find Connected Terms Module.
A query term (Q) is input and all co-occurring A, B, C terms connections are established and putative connections are proposed. Solid lines indicate actual text-based co-occurrences, while dashed lines indicate proposed connections. It should be noted that Q can be either an A, B, or C term. (B) Find Connected Terms Module Output. All A, B, and C terms (7 821 total) that co-occur in the same articles as the query term “Kinase” are returned with the associated article counts. Resultant terms can be filtered by sub-terms and date, and the results and PMIDs can be downloaded.
The resultant terms are rank-ordered by the number of unique articles in which the term co-occurs with the query. Thus, the article count serves as a proxy for the strength of the association between terms in the A-B-C paradigm. For each co-occurring term, the user can click on the article count and view all of the associated article PubMed Identification (PMID) numbers. These PMIDs are linked to PubMed, allowing the user to access and review the article(s) in which the two terms are mentioned together.
The full list of co-occurring terms can be filtered by MeSH term type, i.e, by “Chemical” terms, “Proteins-Pathways-Intermediaries-Other”, or by “Disease and Indication”, which correspond to A, B, and C terms, respectively (cf. Figure 1). Moreover, each MeSH term type (A, B, or C) has additional subtypes that facilitate further refinement of the co-occurring terms. For instance, Chemical (A) terms can be filtered by “Drug” terms, which allows the user to identify which FDA-approved drugs co-occur in the same articles as the query term. The full list of term subtypes for filtering is provided in the Supporting Information (Table S2). Aside from type, the co-occurring terms can be filtered by date of publication; thus, all terms appearing in articles published before or after a certain date can be retrieved.
Users are able to download two CSV files. First is a file of the co-occurring terms and the associated article counts, while the second is a file of co-occurring terms, the article counts, and the explicit PMIDs.
Find Shared Terms
In this module, two query terms are input, and co-occurring terms and the article counts that are shared between the queries are returned. A schema of this module is presented in Figure 3A.
Figure 3. (A) Schema of the Find Shared Terms Module.
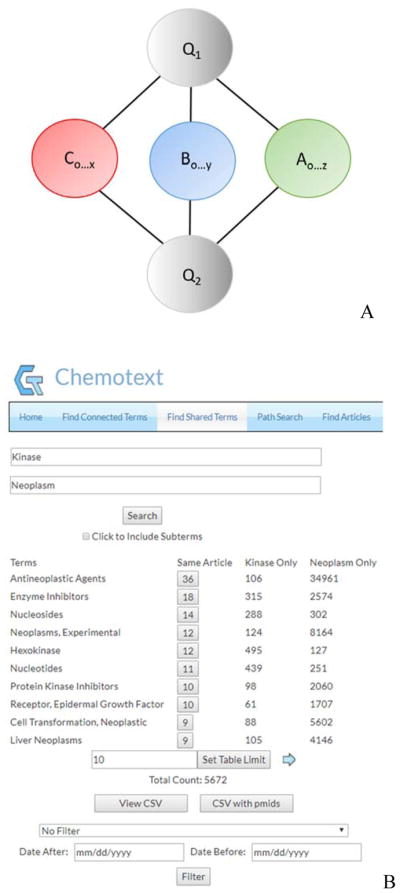
Two query terms, Q1 and Q2, representing any pair of A, B, and C terms, are input, and all co-occurring A, B, and C terms shared between the query terms are established. (B) Find Connected Terms Module Output. Two query terms, “Kinase” and “Neoplasm”, are input, and all co-occurring A, B, and C terms shared between the query terms are established (5 672 shared terms). Resultant terms can be filtered by sub-terms and date, and the results and PMIDs can be downloaded.
Thus, this type of search outputs the associated counts of co-occurrence for three instances: (i) when all three terms (query 1, query 2, and co-occurring term) are present in the same article, (ii) when the term co-occurs only in articles with query 1, and (iii) when the term co-occurs only in articles with query 2. For example, when “Kinase” and “Neoplasm” are queried together in this module (Figure 3B), the term “Antineoplastic Agents” co-occurs in 36 articles with both “Kinase” and “Neoplasm”, 106 articles with only “Kinase”, and 34 961 articles with only “Neoplasm” (Table S3). It should be noted, however, that if a term co-occurs with only one of the queries, then this co-occurring term is not returned in this module, as it does not occur with the other query. The term, therefore, is not shared between the two query terms.
The resultant terms are rank-ordered by the number of unique articles in which all three terms co-occur. Since all three terms occur in the same article(s), these associations are considered the strongest.
For each shared co-occurring term, the user can click on the article count and view all of the associated article PMID numbers when all three terms are present in the same article. As stated previously, these PMIDs are linked to PubMed. If for the case where the term co-occurs with query 1 and query 2, but are not necessarily present in the same articles, then the user can obtain these PMIDs and links to articles in the “Find Connected Terms” module. The same previously described filters and downloadable files are available in this module.
Path Search
In this module, complete text-based A-B-C connections can be made through co-occurring MeSH terms. The name of this module – “Path Search” – indicates that these A-B-C connections can be established through several “paths”, i.e., through multiple intermediary terms or through a single intermediary term. A schema of this module is presented in Figure 4A.
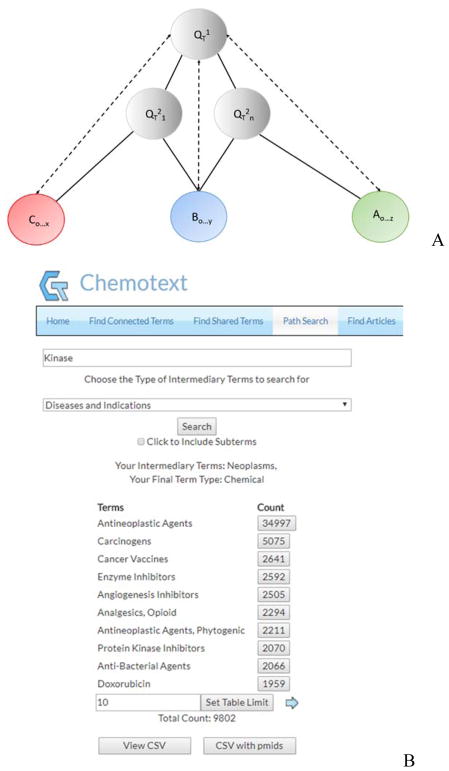
Figure 4. (A) Schema of the Path Search Module.
The first query term, QT1, is the input. Next, a second layer of query terms (QT2) that co-occur with QT1 are selected. The number of terms in the second query layer can range from one (QT21) to all associated terms (QT2n). Next, any category of MeSH term that co-occurs with QT2 are returned. Solid lines indicate actual text-based co-occurrences, while dashed lines indicate proposed connections. It should be noted that Q terms can be a combination A, B, or C terms. (B) Path Search Module Output. The first query term, “Kinase”, is the input. Co-occurring intermediary C terms, “Disease and Indications”, are returned. Within this group, “Neoplasms” is selected as the second query layer, and the 9 802 chemical terms that co-occur with that term are returned.
In the most complex and comprehensive path search, every possible A-B-C connection for a given query term can be established. For instance, if “Kinase” is queried and “Diseases and Indications” are chosen as the intermediary term, 1 242 unique MeSH terms are returned, representing 1 242 unique B-C connections. Examples of these unique B-C connections are as diverse as “Kinase-Neoplasms,” “Kinase-Gout,” and “Kinase-Leprosy.” Next, all 1 242 B-C connections can be queried for associated A-terms, thereby completing every possible A-B-C connection, i.e. DTD triangles. In this case, every chemical that can be associated with the B-term “Kinase” as mediated through the 1 242 C-terms is identified.
This path search can be simplified to identify more focused A-B-C connections through a single intermediary term. Using an above example, the single B-C connection of “Kinase-Neoplasms” can be queried for all co-occurring “Chemical” A-terms, resulting in 9802 unique A-B-C connections mediated through the “Kinase-Neoplasms” nodes (Figure 4B). Of these 9 802 unique A-B-C connection in this path search (Table S4), Chemotext retrieves 270 articles that establish the specific A-B-C connection of “Imatinib-Kinase-Neoplasms.” This connection represents a known drug-target-disease relationship, as the tyrosine kinase inhibitor imatinib is used to treat several cancers, including gastrointestinal stromal tumors (GIST) through the blockage of the receptor tyrosine kinase c-kit.19 In the Case Study, we will demonstrate that imatinib can also be repurposed as a treatment for asthma.
In the Path Search module, the intermediary term type can either be the MeSH term type, i.e., “Disease and Indication”, “Proteins-Pathways-Intermediaries-Other”, “Chemical” terms, or the MeSH term subtypes, such as “Viruses”, “Enzymes and Co-Enzymes”, and “Heterocyclic Compounds”. Regardless of the intermediary term type, resultant terms are ranked according to the highest co-occurring article count with the query term. One or more intermediary terms can be selected to complete the path search, and the final connection can either set as the MeSH term type or subtype. The resultant terms are again ranked by highest co-occurring article count with the intermediary terms. Once the path search has been completed, the user can access the articles associated with the final term via the PMID and can download the two previously described CSV files.
Find Articles
In this module, articles indexed in PubMed can be searched for using specific MeSH terms. Additionally, this module will allow the user to inspect the total number of articles associated with this term. For example, if the term “Neoplasms” is queried, 36 1190 unique hits are returned with direct links to the respective articles.
Case Study: Construction of a Clinical Outcome Pathway (COP) for a Drug-Disease Pair
In order to demonstrate the utility of Chemotext, we describe its application for finding the accurate solution of the recent National Center for Advancing Translational Science (NCATS) Biomedical Data Translator Challenge (https://ncats.nih.gov/translator/funding/not-tr-17-023). The task of this challenge was to construct a clinical outcome pathway (COP) for the drug-disease pair imatinib-asthma. It was stated that a clinical outcome pathway (COP) begins with (i) a molecule physically interacting with (ii) a biological target that affects (iii) a biological pathway relevant to (iv) a particular cell or tissue type that manifest as (v) a clinical phenotype and/or symptom which reflect (vi) a disease or condition. The challenge was to construct a COP for (i) imatinib that successfully reveals its (ii) biological target, (iii) the pathway affected by that target, (iv) the cell or tissue type, and (v) the manifested symptom germane to (vi) asthma in the form of relevant MeSH terms and associated article PMIDs for stages ii–v (Figure 5).
Figure 5.
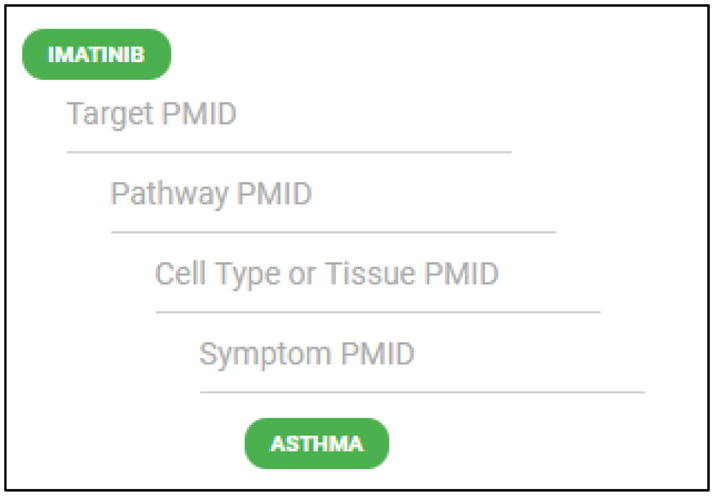
NCATS Biomedical Data Translator Challenge #5. The task was to successfully construct a COP connecting imatinib and asthma. Correct MeSH terms and associated article PMIDs had to be identified to solve the challenge.
In the first step of the solution-seeking algorithm, query terms “Imatinib” (i) and “Asthma” (vi) were searched in the Find Shared Terms module. The list of full associations was filtered by “Proteins-Pathways-Intermediaries-Other”. The MeSH term “Proto-Oncogene Proteins c-kit” was the fourth highest ranked shared term (two shared articles) selected as the potential biological target (ii) in the COP. The three more highly ranked terms, i.e., “Allergens”, “Stem Cell Factor”, and “Ovalbumin”, were deemed too broad or generic to be viable solutions. The two articles and their associated PMIDs related to “Proto-Oncogene Proteins c-kit” were then directly accessed through the Chemotext Web server. Both articles, upon visual inspection, confirmed the relevance of this DTD triangle. One article (PMID: 19722748)20, i.e., “Presence of c-KIT-positive mast cells in obliterative bronchiolitis from diverse causes”, was successfully chosen as the solution to stage (ii) of the COP, as later confirmed by the NCATS Challenge system.
To identify the biological pathway affected (iii) in this COP, query terms “Imatinib” (i) and “Proto-Oncogene Proteins c-kit” (ii) were searched in the Find Shared Terms module in the second step of the solution algorithm. The list of full associations was filtered by “Proteins-Pathways-Intermediaries-Other”. The MeSH terms and associated article counts were downloaded from Chemotext. Next, query terms “Proto-Oncogene Proteins c-kit” (ii) and “Asthma” (vi) were searched in the Find Shared Terms module and the same succeeding steps as above were performed. The intersection of the two lists, i.e., (i–ii) and (ii–vi) was obtained, and MeSH terms were sorted according to article count ranks (Table S5). The MeSH term “Phosphatidylinositol 3-Kinases” was one of the most highly ranked shared terms (22nd out of 928 terms). More highly ranked terms, such as “Biomarkers” and “Neoplasm Proteins”, were not selected because they were not relevant to the “Pathway” portion of this COP. Articles and their associated PMIDs related to “Phosphatidylinositol 3-Kinases” were then directly accessed through the Chemotext Web server. One article (PMID: 17546049)21, i.e., “KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway”, was chosen as the successful solution to stage (iii) of the COP.
In order to identify the cell or tissue type (iv) involved in this COP, “Imatinib” (i) and “Asthma” (vi) were again searched in the Find Shared Terms module. Co-occurring terms were then filtered by “Cells”. This resulted in the correct identification of “Mast Cells” (PMID: 16483568)22. Likewise, for the manifested symptom (v), the drug and the disease were queried in the Find Shared Terms module, and resultant connections were filtered by “Diseases and Indications”. The top co-occurring term was “Bronchial Hyperreactivity” (PMID: 24112389)23. Both the terms were later confirmed by the NCATS Challenge system as steps in the COP.
The full Imatinib-Asthma COP, as revealed by Chemotext and confirmed by the Challenge system, was: Imatinib (i) → Proto-Oncogene Proteins c-kit (ii) → Phosphatidylinositol 3-Kinases (iii) → Mast Cells (iv) → Bronchial Hyperreactivity (v) → Asthma (iv).
It should be emphasized that expert-based knowledge curation, in conjunction with the results of Chemotext, was key for the successful identification of terms and articles. For instance, in the first step of the solution algorithm, “Proto-Oncogene Proteins c-kit” was the correct target, but there were three more highly ranked terms, i.e., “Allergens”, “Stem Cell Factor”, and “Ovalbumin”. These terms were deemed too broad or generic to be viable solutions to this stage (ii) of the COP. Likewise, in the second step, “Phosphatidylinositol 3-Kinases” ranked 22nd out of 928 terms. More highly ranked terms, such as “Biomarkers” and “Neoplasm Proteins”, however, were not biologically relevant (not related to “Pathway”) for this COP and thus not investigated. This observation obligates that additional scoring functions - besides of article counts - should be considered to elucidate meaningful relationships.
Last, it should be noted that this COP may have many alternative plausible solutions that have not been investigated herein; we have described a single validated test case merely to illustrate Chemotext’s capabilities.
Conclusions
We have developed the Chemotext Web server to facilitate the identification of existing drug-target-disease (DTD) relationships and to generate hypotheses about novel relationships by mining of PubMed in the form of MeSH terms via four modules: Connected Terms, Find Shared Terms, Path Search, and Find Articles. In the Connected Terms module (Figure 2A), the user can query any type of MeSH terms, i.e., an A, B, or C term, and retrieve all MeSH terms that co-occur in the same articles as the query term. This module provides an overview of all text-based associations and makes connections between terms. In the Find Share Terms module (Figure 3A), two query terms are input, and co-occurring terms that are shared between the queries are returned. For instance, in this module the shared targets between two diseases or between a drug and disease can be identified. In the Path Search module (Figure 4A), full A-B-C connections can be established through intermediary MeSH terms. We provided an example of using Chemotext to generate drug repurposing candidates. Last, a focused search of PubMed via MeSH term keywords can be performed using the Find Articles module.
The Chemotext Web server was originally conceived of and developed as a text-mining tool for inferring new drug-disease associations9,24, i.e., drug repurposing; however, Chemotext can be used to establish DTD triangles or to mine any type of text-based relationships between biomedical terms or concepts. For example, Chemotext could be used to establish protein-protein interaction networks through co-occurring B-terms or to uncover correlations in disease progression through co-occurring C-terms. The potential number and types of relationships that can be generated with Chemotext are myriad and not limited to the A-B-C paradigm described herein. Indeed, in 2016, Alves et. al.25 used Chemotext outside of this paradigm to confirm the toxic effects of chemicals predicted as human skin sensitizers in a virtual screening campaign.
In spite of its obvious advantages, Chemotext in its current form has several limitations that must be addressed. First, the deposition of articles into PubMed is ever-growing. As per data availability in MBR, the database of terms that underlies Chemotext, must be updated regularly to capture these articles, and new literature-based connections between terms have to be generated. Additionally, from a functional aspect, relationships derived by Chemotext are limited to MeSH terms indexed in the abstracts of articles. Future implementations will seek to mine full articles, although this form of text-mining is orders of magnitude more difficult. In the same vein, Chemotext currently does not support natural language processing and provides no inference about the nature of the relationship between the terms (agonism vs. antagonism, cause vs. effect, mode of action vs. side effect, etc.). This may lead to a number of false positive hits that are not directly related to the desired effect. From a technical perspective, chemicals can be queried by multiple synonyms, i.e., aspirin vs. acetylsalicylic acid vs. dispril. The “Click to Include Subterms” feature of Chemotext ensures that all terms associated with a chemical will be investigated. On the other hand, chemicals will be returned only by the main MeSH term, i.e., aspirin. The user must be aware that the resultant chemical may be indexed by an unfamiliar construction, such as its IUPAC or generic name. Presently, the onus is placed on the user to then identify and investigate the chemical(s) of interest by the corresponding MeSH term. To address these issues and to improve the scope and functionality of Chemotext, regular updates and improvements are underway, such as improving its functionality on additional web browsers like Safari and Firefox and resolving chemical names.
The Chemotext Web server is freely-available at http://Chemotext.mml.unc.edu/index.html (currently fully operational via Google Chrome only). A user-friendly tutorial is also available at the site: http://chemotext.mml.unc.edu/ChemotextAppNote_Tutorial_v3.docx
Supplementary Material
Acknowledgments
The authors appreciate the financial support from NIH grant 1U01CA207160.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Associated content
Results of Chemotext queries described in the manuscript and other Chemotext related information (Tables S1–S6) including a user-friendly tutorial (ChemotextAppNote_Tutorial_v3.docx) are provided as Supporting Information. More specifically, Table S1 contains the results of querying connected term “kinase”; Table S2 –subterms for filtering the results; Table S3 – results of querying shared terms “kinase” and “neoplasm”; Table S4 – results of path search “Kinase – Neoplasms – Chemical”; Table S5 – results of querying “Find Shared Terms” and their overlap for case study of constructing imatinib-asthma clinical outcome pathway. These materials are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Frye S, Crosby M, Edwards T, Juliano R. US Academic Drug Discovery. Nat Rev Drug Discov. 2011;10:409–10. doi: 10.1038/nrd3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Suzek T, Zhang J, Wang J, He S, Cheng T, Shoemaker BA, Gindulyte A, Bryant SH. PubChem BioAssay: 2014 Update. Nucleic Acids Res. 2014;42:D1075–82. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. PubChem Substance and Compound Databases. Nucleic Acids Res. 2015;44:D1202–13. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012;40:D1100–7. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Przybyła P, Shardlow M, Aubin S, Bossy R, Eckart de Castilho R, Piperidis S, McNaught J, Ananiadou S. Text Mining Resources for the Life Sciences. J Biol databases curation. 2016;2016 doi: 10.1093/database/baw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei CH, Kao HY, Lu Z. PubTator: A Web-Based Text Mining Tool for Assisting Biocuration. Nucleic Acids Res. 2013;41:W518–W522. doi: 10.1093/nar/gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts RJ. PubMed Central: The GenBank of the Published Literature. Proc Natl Acad Sci U S A. 2001;98:381–2. doi: 10.1073/pnas.98.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J, DiCuccio M, Grigoryan V, Wilbur WJ. Navigating Information Spaces: A Case Study of Related Article Search in PubMed. Inf Process Manag. 2008;44:1771–1783. [Google Scholar]
- 9.Baker NC, Hemminger BM. Mining Connections between Chemicals, Proteins, and Diseases Extracted from Medline Annotations. J Biomed Inform. 2010;43:510–9. doi: 10.1016/j.jbi.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swanson DR. Fish Oil, Raynaud’s Syndrome, and Undiscovered Public Knowledge. Perspect Biol Med. 1986;30:7–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 11.Swanson DR. Migraine and Magnesium: Eleven Neglected Connections. Perspect Biol Med. 1988;31:526–57. doi: 10.1353/pbm.1988.0009. [DOI] [PubMed] [Google Scholar]
- 12.Nosengo N. Can You Teach Old Drugs New Tricks? Nature. 2016;534:314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 13.Blatt J, Farag S, Corey SJ, Sarrimanolis Z, Muratov E, Fourches D, Tropsha A, Janzen WP. Expanding the Scopre of Drug Repurposing in Pediatrics: The Children’s Pharmacy Collaborative. Drug Discov Today. 2014;19:1696–1698. doi: 10.1016/j.drudis.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J, Shui Y, Peng S, Li X, Mamitsuka H, Zhu S. MeSHSim: An R/Bioconductor Package for Measuring Semantic Similarity over MeSH Headings and MEDLINE Documents. J Bioinform Comput Biol. 2015;13:1542002. doi: 10.1142/S0219720015420020. [DOI] [PubMed] [Google Scholar]
- 15.Rani J, Shah ABR, Ramachandran S. pubmed.mineR: An R Package with Text-Mining Algorithms to Analyse PubMed Abstracts. J Biosci. 2015;40:671–82. doi: 10.1007/s12038-015-9552-2. [DOI] [PubMed] [Google Scholar]
- 16.Spangler S, Myers JN, Stanoi I, Kato L, Lelescu A, Labrie JJ, Parikh N, Lisewski AM, Donehower L, Chen Y, Lichtarge O, Wilkins AD, Bachman BJ, Nagarajan M, Dayaram T, Haas P, Regenbogen S, Pickering CR, Comer A. Automated Hypothesis Generation Based on Mining Scientific Literature. Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining - KDD ’14; New York, New York, USA: ACM Press; 2014. pp. 1877–1886. [Google Scholar]
- 17.Capuzzi SJ, Kim ISJ, Lam WI, Thornton TE, Muratov EN, Pozefsky D, Tropsha A. Chembench: A Publicly Accessible, Integrated Cheminformatics Portal. J Chem Inf Model. 2017;57:105–108. doi: 10.1021/acs.jcim.6b00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPherson S. [accessed Oct 1, 2017];JavaServer Pages: A Developer’s Perspective. http://www.oracle.com/technetwork/java/index.html.
- 19.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, Singer S, Fletcher CDM, Silberman S, Dimitrijevic S, Fletcher JA. Kinase Mutations and Imatinib Response in Patients with Metastatic Gastrointestinal Stromal Tumor. J Clin Oncol. 2003;21:4342–9. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 20.Fuehrer NE, Marchevsky AM, Jagirdar J. Presence of c-KIT-Positive Mast Cells in Obliterative Bronchiolitis from Diverse Causes. Arch Pathol Lab Med. 2009;133:1420–5. doi: 10.5858/133.9.1420. [DOI] [PubMed] [Google Scholar]
- 21.Bauer S, Duensing A, Demetri GD, Fletcher JA. KIT Oncogenic Signaling Mechanisms in Imatinib-Resistant Gastrointestinal Stromal Tumor: PI3-kinase/AKT Is a Crucial Survival Pathway. Oncogene. 2007;26:7560–7568. doi: 10.1038/sj.onc.1210558. [DOI] [PubMed] [Google Scholar]
- 22.Reber L, Da Silva CA, Frossard N. Stem Cell Factor and Its Receptor c-Kit as Targets for Inflammatory Diseases. Eur J Pharmacol. 2006;533:327–340. doi: 10.1016/j.ejphar.2005.12.067. [DOI] [PubMed] [Google Scholar]
- 23.Cleary RA, Wang R, Wang T, Tang DD. Role of Abl in Airway Hyperresponsiveness and Airway Remodeling. Respir Res. 2013;14:105. doi: 10.1186/1465-9921-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker NC, Fourches D, Tropsha A. Drug Side Effect Profiles as Molecular Descriptors for Predictive Modeling of Target Bioactivity. Mol Inform. 2015;34:160–170. doi: 10.1002/minf.201400134. [DOI] [PubMed] [Google Scholar]
- 25.Alves VM, Capuzzi SJ, Muratov EN, Braga RC, Thornton TE, Fourches D, Strickland J, Kleinstreuer N, Andrade CH, Tropsha A. QSAR Models of Human Data Can Enrich or Replace LLNA Testing for Human Skin Sensitization. Green Chem. 2016;18:6501–6515. doi: 10.1039/C6GC01836J. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.