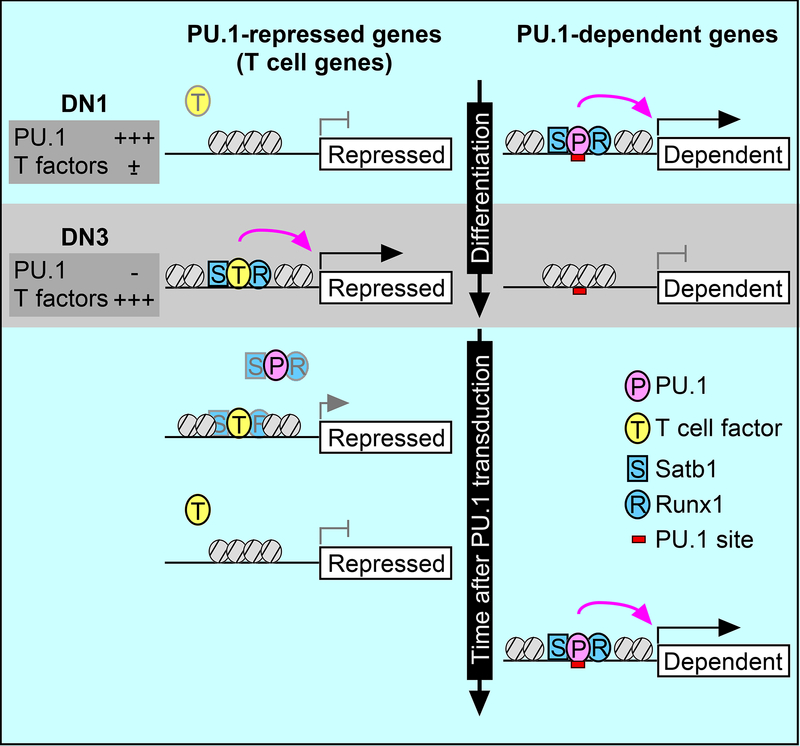
SUMMARY
Transcription factors normally regulate gene expression through their action at sites where they bind to DNA. However, the balance of activating and repressive functions that a transcription factor can mediate is not completely understood. Here, we showed that the transcription factor PU.1 regulated gene expression in early T cell development both by recruiting partner transcription factors to its own binding sites and by depleting them from the binding sites that they preferred when PU.1 was absent. The removal of partner factors Satb1 and Runx1 occurred primarily from sites where PU.1 itself did not bind. Genes linked to sites of partner factor ‘theft’ were enriched for genes that PU.1 represses despite lack of binding, both in a model cell line system and in normal T cell development. Thus, system-level competitive recruitment dynamics permit PU.1 to affect gene expression both through its own target sites and through action at a distance.
Keywords: Spi1, Runx1, Satb1, DNA accessibility, repression
eTOC blurb
Transcription factors regulate target genes via sequence-specific DNA binding. They may collaborate when bound together, but are assumed to be independent at sites where they bind alone. Hosokawa, Ungerbäck et al. show that PU.1 broadly shifts the genome-wide site choice of Runx1 DNA binding, enabling PU.1 to repress some target genes at a distance.
Graphical Abstract
INTRODUCTION
Cell-type identity in hematopoiesis is defined by actions of combinations of transcription factors that are both complex and dosage-sensitive. The same factors in modestly different ratios can promote differentiation or survival of very different blood cell types (Dahl et al., 2003; Taghon et al., 2007), and hematopoietic regulatory genes frequently show abnormal phenotypes if one allele is mutated (Cai et al., 2000; Carotta et al., 2010; Klein Wolterink et al., 2013; Lacaud et al., 2004; Lukin et al., 2010; Prasad et al., 2015; Rodrigues et al., 2005; Sun and Downing, 2004; Talebian et al., 2007). A central question is what kinds of regulatory factor mechanisms could explain this high dosage sensitivity. The program of transcription factor changes during T cell differentiation (Rothenberg et al., 2016; Yui and Rothenberg, 2014), is a highly accessible system in which to examine this question. Here, analysis of the T cell system revealed that developmentally dynamic transcription factors could affect cell fate not only by positive and negative actions at sites of their own specific DNA binding, but also by an ability to titrate other factors competitively, altering the genomic profile of the other factors’ preferred binding sites.
In T-lymphoid development, cells traverse at least three major phases of transcription factor action before they are fully programmed, when the global pattern of gene regulation shifts dramatically across the genome (Yui and Rothenberg, 2014). The first transition, commitment, separates the earliest “Early T cell Precursors” (or Kit-high double-negative 1 “DN1”) and “DN2a” stages from “DN2b” and “DN3” stages. This is when initially multipotent precursors lose access to other options and become fully committed to a T cell fate. One factor that occupies a major fraction of all open regulatory sites in the genomes of pre-commitment (DN1 and DN2a) cells is PU.1 (Ungerbäck, Hosokawa, Wang, Strid, Williams, Sigvardsson, Rothenberg, submitted). This is a signature factor of myeloid cells, dendritic cells, and B cells, which is potent in reprogramming various other cell types into myeloid cells (Feng et al., 2008; Iwasaki et al., 2006; Laiosa et al., 2006; Nerlov and Graf, 1998). Yet it is also initially well-expressed in virtually all DN1 and DN2a T-lineage progenitors, and is silenced only during commitment (Yui et al., 2010). Either gain or loss of PU.1 in experimental contexts can appear to push pro-T cells “backwards” or “forwards”, respectively, relative to the normal developmental program (Champhekar et al., 2015; Del Real and Rothenberg, 2013), implicating this factor in the control of developmental progression, while further elevation of PU.1 expression in early T cell progenitors activates genes important for the myeloid program.
Altered transcription factor activity potently affects cellular identity, as, for example, in induced pluripotent stem (iPS) cell reprogramming (Takahashi and Yamanaka, 2016). Introduced transcription factors not only turn on their own positively regulated target genes but also silence expression of genes associated with any program that the cells were following before transformation. In normal T cell development a kind of reprogramming also occurs naturally at commitment, when a gene expression program highly related to that of hematopoietic multipotent progenitors is silenced and replaced by a T cell specific program. This shift can be partially reversed if PU.1 is added back after commitment (Ungerbäck et al., op. cit.)(Del Real and Rothenberg, 2013). As in iPS cell reprogramming (Chronis et al., 2017; Koche et al., 2011) and artificial hematopoietic cell lineage trans-differentiation (Laiosa et al., 2006; van Oevelen et al., 2013; Xie et al., 2004), both the natural forward transition and the experimentally inducible reverse transition involve both repression and activation of target genes. In general, the repression is not as well understood as activation.
Hematopoietic lineage-determining transcription factors are often bifunctional in their direct binding effects, repressing as well as activating (de la Rica et al., 2013; Huang et al., 2008; McManus et al., 2011; Nechanitzky et al., 2013; Revilla-i-Domingo et al., 2012; Treiber et al., 2010). PU.1 activates and represses approximately equal numbers of genes in pro-T cells. However, its impact on genes actually linked to its binding sites is disproportionately activating (Zhang et al., 2012)(Ungerbäck et al., op. cit.), and evidence suggests that much of its negative regulation is mediated indirectly (Champhekar et al., 2015). One indirect way that PU.1 can antagonize T cell specific gene expression is by raising the threshold for Notch signaling that drives T-lineage specification (Del Real and Rothenberg, 2013; Franco et al., 2006). However, PU.1 still works to control differentiation speed even in strong Notch signaling conditions (Champhekar et al., 2015). Research on reprogramming has shown an additional way that factors can repress as well as activate, by “enhancer decommissioning”. Several recent reports document hit-and-run transcription factor binding that leads to the competitive eviction of previously bound factors and closure of regulatory sites (Chronis et al., 2017; Krishnakumar et al., 2016; Respuela et al., 2016; van Oevelen et al., 2013). However, such decommissioning could be seen as a temporally extended consequence of direct repression, as it is still based on local DNA binding.
Here, we showed that yet another mechanism contributes to the switch-like potency of PU.1 in both the normal “forward” T cell developmental process and the experimental reversal or redirection of this process. We confirmed that PU.1 activates its positive regulatory targets through efficient co-recruitment of other, stably expressed factors. Although the common collaborators of PU.1, i.e. IRF4 or IRF8 (Marecki and Fenton, 2000) and C/EBPα or C/EBPα (Heinz et al., 2013; Natoli et al., 2011), are absent in most pro-T cells, we showed that other partners were recruited for PU.1-mediated positive regulation. However, we also showed that this recruitment of collaborators had another side as those factors were quantitatively depleted from alternative sites, causing widespread reduced expression of many genes linked to sites that PU.1 itself does not bind at all.
Results
Satb1 and Runx1 interact with PU.1 in early T cells
In DN1-DN2b cells PU.1 acts as a stage-specific pioneer factor, able both to select its binding sites based on its own DNA-site affinity criteria and to induce opening (or maintain accessibility) of sites in facultatively closed chromatin (Ungerbäck et al., op. cit.). However, the ability of full-length PU.1 to establish occupancy in closed chromatin was found to depend not only on its own DNA binding domain but also on its other protein-interaction domains. This raised the question of what protein partners could be important for PU.1 binding and regulatory function. To dissect the molecular mechanisms involved, we took advantage of a highly tractable DN3-like pro-T cell line, Scid.adh.2c2, that normally lacks PU.1. Retroviral transduction of PU.1 into these cells causes loss of T cell marker CD25 expression while activating myeloid marker CD11b expression and shifting the cells to a globally altered regulatory state (Fig. S1A)(Del Real and Rothenberg, 2013; Dionne et al., 2005)(Ungerbäck et al., op. cit.).
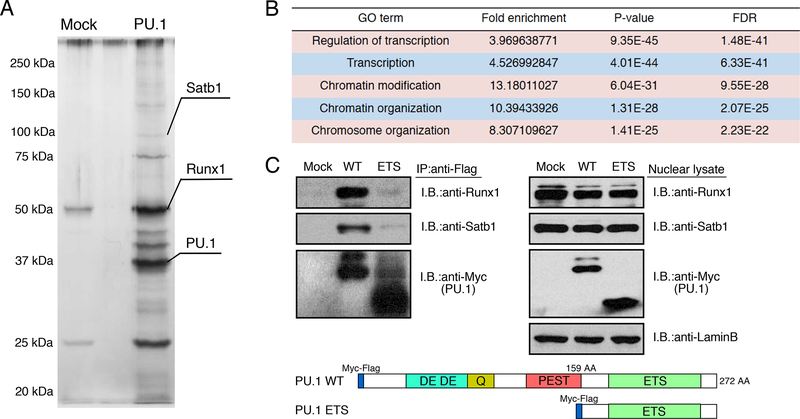
To identify the functional components of PU.1-containing complexes that mediate these effects, extracts from the Scid.adh.2c2 cells expressing a Myc-Flag-tagged construct of PU.1 were subjected to two-step affinity purification followed by SDS-PAGE and silver staining (Fig. 1A). Mass spectrometry identified more than 200 molecules with supra-threshold enrichment (Table S1). Gene ontology analysis of PU.1 interacting molecules showed that proteins involved in transcriptional regulation and chromatin remodeling were most highly enriched (Fig. 1B). Two sequence-specific transcription factors, Satb1 and Runx1, which are known to have important roles in hematopoiesis and T cell development (Alvarez et al., 2000; Huang et al., 2008; Satoh et al., 2013), were repeatedly detected in our mass spectrometry analyses; Runx1 signals were among the highest seen (Table S1). Their association with full-length PU.1WT was validated by co-immune precipitation (Fig. 1C, WT). These protein associations were lost with the truncated PU.1ETS mutant (Fig. 1C, ETS), which was seen to bind stably to sites in open but not in closed chromatin (Ungerbäck et al., op. cit.). Both Runx1 and Satb1 are expressed in DN1 cells which also highly express PU.1 protein, as well as through later DN stages (Fig. S1B). Together with the Runx motif enrichment prominent at many PU.1 sites (Ungerbäck et al., op. cit.), these findings suggest that Runx1 or Satb1 association might play roles in PU.1-mediated gene regulation in early T cells.
Figure 1. Satb1 and Runx1 are PU.1 interacting molecules in early T cells.
(A), Total extracts from Myc-Flag-PU.1-expressing Scid.adh.2c2 cells were subjected to two- step affinity purification followed by SDS-PAGE and silver staining. Mass spectrometry identified several specific polypeptides for both Satb1 and Runx1.
(B), Top five gene ontology (GO) terms for PU.1 interacting molecules.
(C), Total extracts from Scid.adh.2c2 cells transduced with Myc-Flag-PU.1 WT or ETS were subjected to immune precipitation (IP) with anti-Flag mAb followed by immunoblotting (IB) with anti-Runx1, anti-Satb1 or anti-Myc Abs (left). IB from total nuclear lysates are also shown (right). Schematic representation of the Myc-Flag-tagged PU.1 WT and ETS constructs are shown (bottom). Two independent experiments were performed with similar results (A, C)(cf. Fig. S1, Table S1).
PU.1 redirects transcription factor ensembles across the genome
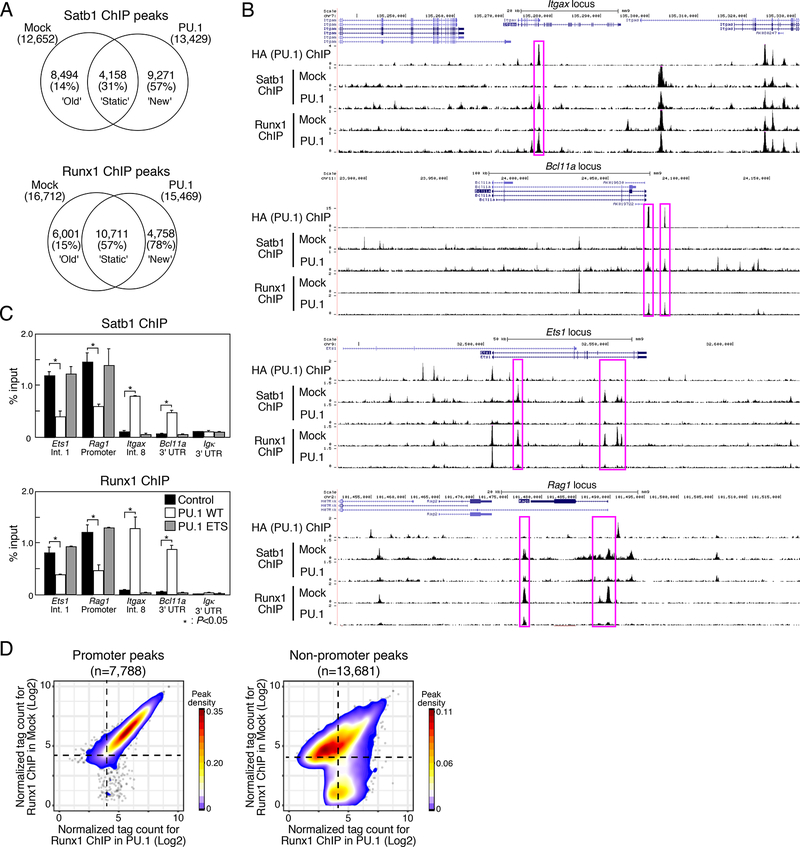
Chromatin immunoprecipitation (ChIP) seq analysis using mock- or PU.1-transduced Scid.adh.2c2 cells revealed that PU.1 not only shared binding regions with Satb1 and Runx1, but indeed recruited these factors de novo to PU.1 binding sites across the genome (Figs. 2A and B). Introduction of PU.1 into these cells did not repress Satb1 or Runx1 expression (Fig. 1C, right), and total numbers of Satb1 and Runx1 ChIP-seq peaks were very similar with and without PU.1 (Fig. 2A). However, PU.1 introduction both induced many ‘new’ Satb1 and Runx1 peaks and also caused a large fraction of ‘old’ peaks to disappear (Fig. 2A). Many ‘new’ Satb1 and Runx1 peaks were found around genes that are directly bound and activated by PU.1 in vivo (Champhekar et al., 2015; Del Real and Rothenberg, 2013; Ghisletti et al., 2010; Kamath et al., 2008)(Ungerbäck et al., op. cit.). For example, Itgax (encoding CD11c) and Bcl11a have sites that exogenous PU.1 can occupy when introduced into Scid.adh.2c2 cells. Although these are not sites occupied by Runx1 and Satb1 in Scid.adh.2c2 cells normally, PU.1 efficiently co-recruited Runx1 and Satb1 to these binding sites (Fig. 2B, upper), as PU.1 co-recruits different factors in myeloid cells (Ghisletti et al., 2010; Heinz et al., 2010). On the other hand, ‘old’ Runx1 and Satb1 sites, prominent in control Scid.adh.2c2 cells, were often linked to T cell program genes like Etsl and Ragl. A specific subset of Runx1 and Satb1 peaks at the Ets1 and Rag1 loci were examples of ‘old’ sites that disappeared upon introduction of PU.1 (Fig. 2B, lower).
Figure 2. PU.1 introduction induces redirection of Satb1 and Runx1 ChIP peaks.
(A), Satb1 and Runx1 ChIP-seq analyses of mock- or PU.1-introduced Scid.adh.2c2 cells. Venn diagrams show the number of ChIP peaks, with percentages of the peaks overlapping with PU.1 sites in parentheses.
(B), Binding patterns of PU.1, Runx1 and Satb1 at the Itgax, Bcl11a, Ets1 and Rag1 loci in mock- and PU.1-introduced Scid.adh.2c2 cells. Representative of two independent experiments.
(C), Scid.adh.2c2 cells were transduced with empty vector (mock), PU.1 WT or PU.1 ETS. The binding of Runx1 and Satb1 at the Etsl (intron1), Rag1 (promoter), Itgax (intron8), Bcllla (3’UTR) and Igk(3’UTR) loci were determined by ChIP assay with qPCR analysis. P-values were determined by Student t-test. Representative of three independent experiments.
(D), Contour display of Runx1 tag counts (log2 counts per 107 reads) in Scid.adh.2c2 cells transduced with PU.1 or control vector. Data in A, D are based on reproducible ChIP-seq peaks in two replicate samples.
This major loss of binding had two key features. First, no direct binding of PU.1 appeared in most sites that lost ‘old’ Satb1 or Runx1 peaks (<14% in Fig. 2A; Fig. 2B, lower), arguing against a competitive displacement mechanism. Second, the PU.1 ETS construct, consisting only of the DNA binding domain, both failed to recruit ‘new’ Satb1 and Runx1 peaks at PU.1 sites and also failed to induce loss of ‘old’ Runx1 and Satb1 peaks (Fig. 2C). This implied that the emptying of the ‘old’ sites was functionally linked with the ability of PU.1 to recruit these factors elsewhere. Genome-wide (Fig. 2D), PU.1 did not affect Runx1 binding to promoter sites. However, as PU.1 induced new Runx1 binding to non-promoter sites, it specifically reduced the effective binding of Runx1 to about half the non-promoter Runx1 sites seen in control cells, leaving the other half unchanged (Fig. 2D).
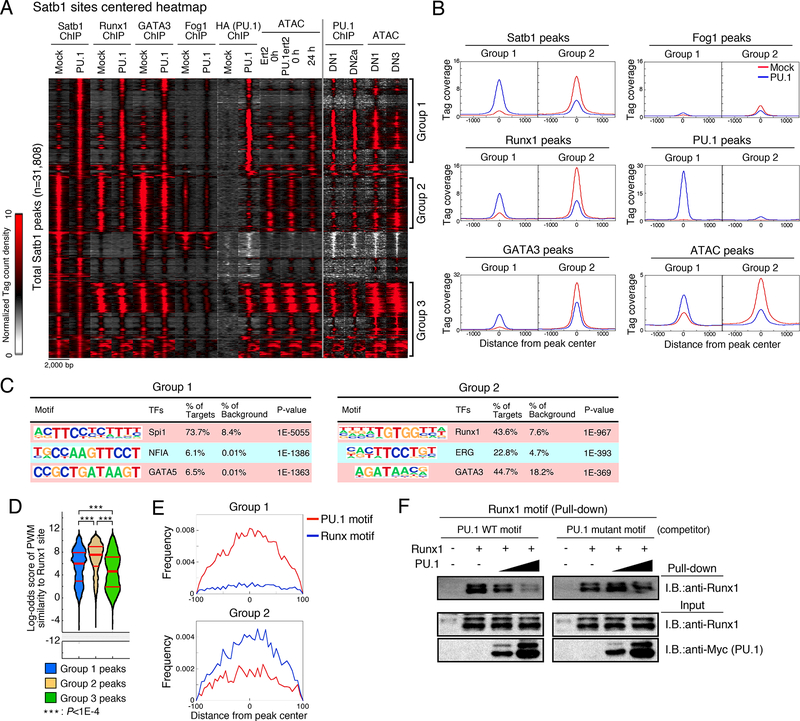
Distinctive properties of the shifted sites were shown in a clustered heatmap of all the ChIP-seq tags around Satb1 peaks in Scid.adh.2c2 with or without added PU.1 (Fig. 3A). This identified two distinct groups of sites: Group 1 sites associated with ‘new’ Satb1 and Runx1 peaks, induced by PU.1, and Group 2 sites associated with ‘old’ Satb1 and Runx1 peaks, diminished by PU.1. Group 1 sites were strongly associated with PU.1 occupancy peaks, and an assay for transposase-accessible chromatin (ATAC) signals showed that the chromatin in these regions is opened up by PU.1 within 24h or less (Fig. 3A and B). Importantly, many peaks in Group 1 were also bound by endogenous PU.1 in primary DN1 and DN2a cells ex vivo, and reduced their ATAC accessibility naturally as the PU.1 levels decline between DN1 and DN3 stages (Fig. 3A). In Group 2, in contrast, minimal PU.1 binding was detected in any conditions, yet the chromatin structure was closed down by PU.1 introduction (Figs. 3A and B). These Group 2 sites also lacked PU.1 in normal pro-T cells and naturally opened during the transition from DN1 to DN3. A third group of Satb1 sites, Group 3, was constitutively open, highly enriched for promoter sites, and minimally affected by PU.1 (Fig. 3A). Corresponding patterns were seen with heatmaps built on all Runx1 binding peaks (Fig. S1C, Groups 4–6) or all PU.1 peaks in the transduced cells (Fig. S1D).
Figure 3. PU.1 redirects transcription factor ensembles.
(A), Peak group classification: tag count distributions for Satb1, Runx1, GATA3, Fog1 and PU.1 ChIP, and ATAC signal around Satb1 peaks using mock- or PU.1-introduced Scid.adh.2c2 cells are shown. PU.1 ChIP in primary DN1 and DN2a, and ATAC signal in DN1 and DN3 are also indicated.
(B), Average tag count distributions in Group 1 and Group 2.
(C), Top three enriched sequence motifs in Group 1 and Group 2.
(D), Distribution of motif log-odds similarity scores for Groups 1, 2, and 3 against position weight matrix (PWM) for Runx1 sites in DN1 and DN3 cells (Fig S1E). Median, 25% and 75% percentiles are also shown. P-values determined by Kruskal-Wallis statistical test.
(E), PU.1 and Runx motif distributions around Group 1 and 2 peaks. Data are based on reproducible ChIP and ATAC signals in two replicate samples for (A, B, C, D, E). Scid.adh.2c2 cells were transduced with Runxl and/or Myc-Flag-PU.1 as indicated, and nuclear lysates subjected to pulldown assay using biotinylated Runx1 motif oligonucleotides with or without competitor oligonucleotides containing a canonical or mutant PU.1 motif. Precipitated proteins were eluted by SDS sample buffer and subjected to IB. Representative of two independent experiments. See also Fig. S1, S2 and Table S2.
Peaks in Group 1 were highly enriched for PU.1 (Spi1) motifs, whereas motifs for canonical T cell factors,, e.g. GATA3, were enriched at Group 2 sites (Fig. 3C). Group 2 peaks in fact showed enrichment for GATA3 binding in ChIP-seq analysis of control Scid.adh.2c2 cells. Like Satb1 and Runx1, GATA3 was also partially dissociated from many Group 2 sites by PU.1 introduction, and was partially recruited to Group 1 sites by PU.1 as well (Figs. 3A and B). A GATA cofactor, FOG1 (Zfpm1), is known to be repressed by PU.1 (Del Real and Rothenberg, 2013), and we considered whether effects via FOG1 might alter the stability of complexes binding Group 2 sites. However, over these time scales, introduction of PU.1 minimally reduced FOG1 binding (Figs. 3A and B). Thus, PU.1 introduction caused redirection of transcription factor ensembles not only by recruiting them to its own target sites but also by “stealing” Satb1, Runx1, and possibly GATA3 from T cell factor binding sites.
We asked whether the Satb1 and Runx1 binding shifted by PU.1 was specifically limited to lower affinity sites. Although the consensus for Satb1 is not well defined, that for Runx1 is known (Fig. S1E). Close analysis of the motifs represented in the Group 1 and Group 2 Runx1 sites showed that there was indeed an affinity difference between sites where Runx1 binds with and without PU.1, but not as expected. Using Runx1 occupancy sites in primary pro-T cells (see below), we calculated a position weight matrix (PWM, Table S2) for site preferences (see Methods; Fig. S1E) and used this as a quality metric to compare different classes of Runx1 occupancy. The promoter-enriched Group 3 and Group 6 (static) sites showed generally poor matches to the Runx PWM, implying low-affinity binding (Fig. 3D; Fig. S1F). The ‘old’ (Group 2, Group 5) sites in PU.1-transduced Scid.adh.2c2 cells showed much better matches to the Runx1 PWM. However, comparing ‘old’ and ‘new’ (Group 1, Group 4) sites, the results implied that PU.1 often recruits Runx1 to bind to significantly lower-quality Runx1 sites than it would otherwise occupy (Fig. 3D, Fig. S1F). In fact, Group 1 ‘new’ sites often lacked a canonical Runx motif (Fig. 3E; Table S2).
Binding of Satb1 and Runx1 to ‘new’ sites when PU.1 was introduced resulted from an association with PU.1 itself, not from passive exploration of sites opened in the chromatin via indirect effects (Fig. S2). We divided the sites that became newly ATAC-accessible in the chromatin of PU.1-transduced Scid.adh.2c2 cells into sites with and without strong local occupancy by PU.1 (Fig. S2A). Satb1 and Runx1 were substantially better recruited to those newly opened sites where PU.1 itself was present (Fig. S2B,C). Co-binding with PU.1 in a ternary complex could indeed compete Runx1 away from a superior Runx binding site, as shown by in vitro DNA binding using a pull-down assay (Fig. 3F). Runx1 was tested for its ability to bind to a canonical site (from Rag1) pulldown probe in the presence of a competing oligonucleotide containing a weaker ‘new’ PU.1-Runx site (from Itgax). Binding to the canonical Runx site probe was indeed competed away by titrating in increasing amounts of PU.1 protein, but only if the PU.1 could form a complex on the competing oligonucleotide. If the PU.1 binding site was mutated in the competitor, PU.1 could no longer remove Runx1 from the pulldown target (Fig. 3F). Taken together, these experiments show that PU.1 itself caused Runx1 to vacate sites of higher quality in favor of co-binding to DNA with PU.1 at sites of lower quality.
Satb1 and Runx1 binding predicts PU.1-mediated gene regulation in Scid.adh.2c2 cells
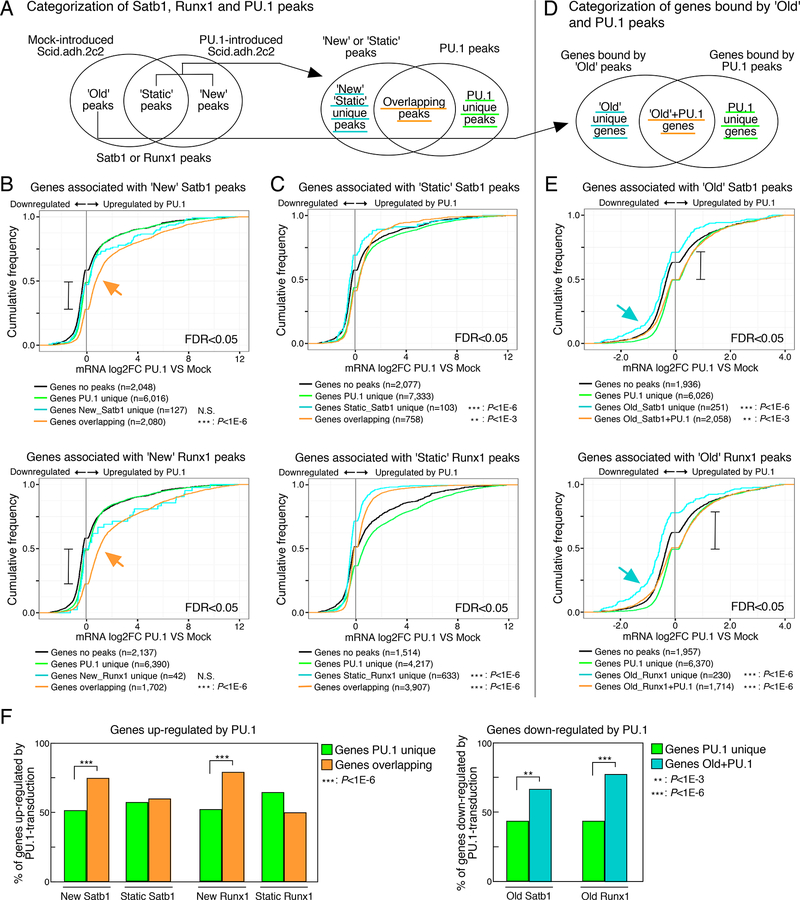
These results raised the question whether Satb1 and Runx1 redirections could underlie PU.1-mediated positive or negative gene regulation. First, we used ChIP-seq data to categorize genes associated with the sites bound by different combinations of Satb1, Runx1, and PU.1 in Scid.adh.2c2 control cells as well as after PU.1 introduction. We then tested these linked genes for differential responses to the addition of PU.1. Genes associated with ‘new’ peaks of Satb1 or Runx1 overlapping with PU.1 co-occupancy, PU.1 unique peaks, and any ‘new’ Satb1 or Runx1 unique peaks without PU.1, were identified as shown in Fig. 4A. We then measured the overall expression trends in these genesets caused by PU.1 (Fig. 4B, false discovery rate (FDR) <0.05; and Table S3). Empirical cumulative distribution frequency (ECDF) plots show that differentially expressed genes linked to PU.1 binding sites usually increased expression in response to PU.1, but that genes bound by PU.1 peaks overlapping with ‘new’ Satb1 or Runx1 peaks increased expression even more strongly than genes bound by PU.1 alone (Fig. 4B, orange arrows; 75% vs. ~40% activated, brackets in Fig. 4B). The expression of genes associated with ‘static’ Satb1 or Runx1 peaks tended to change little upon PU.1 introduction (Fig. 4C, blue & orange curves). In contrast, genes bound by ‘old’ Satb1 or Runx1 peaks, i.e. peaks that lost binding when PU.1 was introduced, but without direct binding of PU.1 (Fig. 4D), significantly lost expression when PU.1 was introduced (Fig. 4E, blue arrows). This seemed unlikely to reflect direct repression by PU.1, for the negative effect was much weaker on genes that had both ‘old’ Satb1 or Runx1 sites and additional sites of PU.1 binding (Fig. 4E, Old R1-S1 + PU.1, orange curves). Indeed, among negatively regulated genes, loss of Satb1 or Runx1 from ‘old’ sites without local PU.1 binding was a better predictor of repression than presence of PU.1 (70–75% repression vs. ~45% repression; brackets in Fig. 4E). These differences are summarized in Fig. 4F. Taken together, these data show that cofactor recruitment could be involved in direct activation of the PU.1 target genes, whereas removing ‘Old’ Satb1 and Runx1 peaks from other loci could be a mechanism enabling PU.1 to repress genes indirectly.
Figure 4. PU.1-mediated gene regulation is correlated with the redirection of Satb1 and Runx1.
(A), Categorization of Satb1, Runx1 and PU.1 peaks.
(B and C), Cumulative distributions of expression changes by PU.1 introduction for four groups of genes bound by ‘New’ (B) and ‘Static’ (C) peaks depicted in (A) and differentially expressed in PU.1-introduced Scid.adh.2c2 (FDR<0.05). Number of genes in each group and p-values (K-S tests relative to ‘Genes PU.1 unique’) are shown.
(D), Categorization of genes bound by ‘Old’ peaks only (‘Old unique’) and those with PU.1 peaks.
(E), Cumulative distributions for four groups of genes bound by ‘Old’ peaks depicted in (D) and differentially expressed in PU.1-introduced Scid.adh.2c2 (FDR<0.05).
(F), Summary, percentage of genes in each category up-regulated by PU.1-transduction (from B, C)(left), and down-regulated by PU.1 transduction (from E) (right). P-values are determined by Fisher’s exact test. Data are based on reproducible ChIP-seq peaks in two replicate samples and two replicates for RNA-seq results. See also Table S3.
Functional importance of Satb1 and Runx1 in gene regulation by PU.1
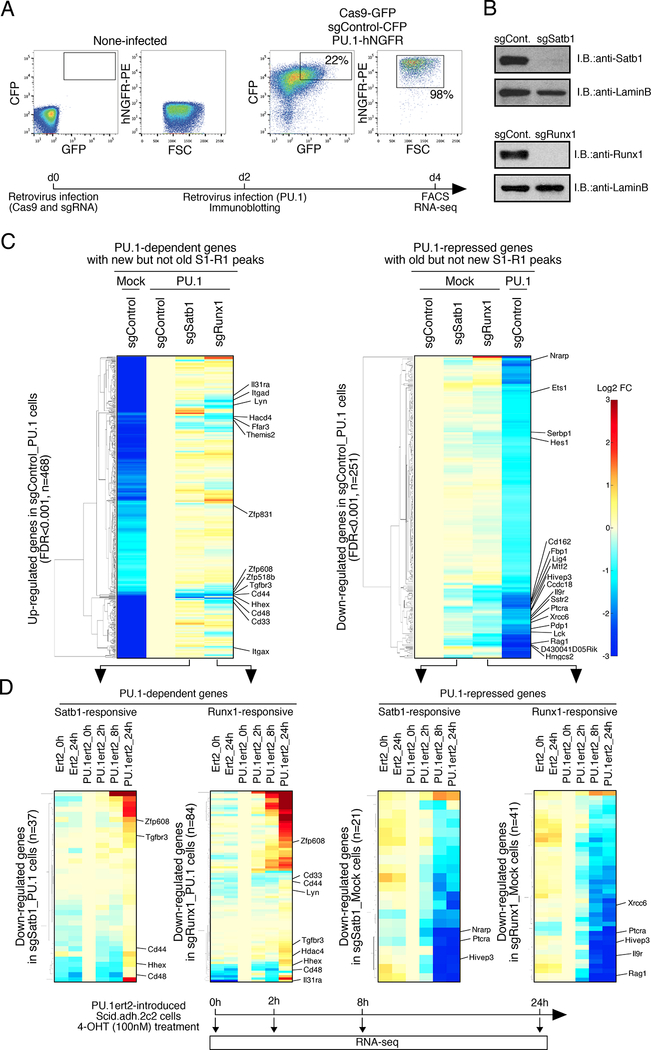
To test whether Satb1 and Runx1 themselves are functionally important for the effects of PU.1 on gene regulation, we disrupted Satb1 or Runx1 in Scid.adh.2c2 cells acutely, using a retroviral transduction method for CRISPR-Cas9 mutagenesis that is feasible in this highly permissive cell line (Fig. 5A). Two days after co-transduction with Cas9 and Satb1- or Runx1-specific guide RNAs (sgRNA), Satb1 and Runx1 proteins almost completely disappeared (Fig. 5B). Then, the cells were infected with a third retroviral vector encoding PU.1, and their responses analyzed two days later (Fig. 5A; Table S3). Fig. 5C(left) shows the effects of these mutations on the genes that were positively regulated by PU.1 (PU.1-dependent) and associated with peaks of PU.1 and ‘New’ Satb1 or Runx1 co-occupancy (‘overlapping new’ peaks, Fig. 4A). Among 468 PU.1- dependent genes, a subset including Il31ra, Itgad, Lyn, Cd44, Hhex, Cd48, Cd33, and Itgax were less activated if the cells were made Satb1- or Runx1-deficient before addition of PU.1 (Fig. 5C, left; Table S3). Satb1 and Runx1 seemed to have different effects on these genes, even though PU.1 caused most of the ‘new’ Satb1 and Runx1 binding peaks to move to the same sites (Figs. 3A vs 5C, left). Thus, Satb1 and/or Runx1 collaborate with PU.1 functionally but distinctly on many positive regulation targets. A majority of these genes that were PU.1-activated and Satb1- or Runx1-responsive in Scid.adh.2c2 cells also responded positively to PU.1 if it was overexpressed in primary DN2 cells (Ungerbäck et al., op. cit.), linking this functional collaboration to normal pro-T gene regulation (Fig. S3A, top).
Figure 5. Satb1 and Runx1 play roles in PU.1-mediated gene regulation in Scid.adh.2c2 cells.
(A), For CRISPR-Cas9-mediated deletion of Satb1 and Runx1 in Scid.adh.2c2 cells, cells were infected with Cas9-GFP and sgRNA-CFP retroviruses. Two days after 1st infection, they had 2nd retrovirus infection for empty vector or PU.1-hNGFR. Flow cytometry analyses of transduced Scid.adh.2c2 cells shows sort gates. Experimental scheme is also indicated (lower)
(B), Protein levels of Satb1 and Runx1 in sgRNA transduced Scid.adh.2c2 cells are shown. Two days after retrovirus infection, nuclear lysates of the Cas9 and sgRNA transduced cells were prepared and subjected to IB with anti-Satb1, anti-Runx1 and anti-LaminB Abs.
(C), Gene expression profiles of PU.1-dependent and -repressed genes in Scid.adh.2c2 cells (see A) were determined by RNA-seq analysis. Genes differentially expressed in PU.1-introduced Scid.adh.2c2 cells (FDR<0.001) were hierarchically clustered by expression patterns. The ‘PU.1- dependent’ (up-regulated by PU.1) genes shown were also selected for binding by ‘new’ Satb1 or Runx1 peaks overlapping with PU.1 peaks, but not by ‘old’ Satb1 or Runx1 unique peaks. The ‘PU.1-repressed’ (down-regulated by PU.1) genes were also selected for binding by ‘old’ Satb1 or Runx1 unique peaks but not by any ‘new’ Satb1 or Runx1 peaks overlapping with PU.1.
(D), The time course of activation or repression of Satb1- and Runx1-responsive genes in response to 4-OHT-driven mobilization of PU.1ert2 is shown. Satb1- or Runx1-responsive (sgSatb1 or sgRunx1/sgControl<0.75) genes were selected from PU.1-dependent and PU.1-repressed gene sets in (C). Hierarchical clustering analyses show expression of these genes over 24 h response to PU.1ert2 or empty ert2 control as indicated (upper). Heat map color scale is as in (C). Experimental scheme is also indicated (lower). Three independent experiments were performed with similar results for (A, B). Results in (C, D) are from two replicate RNA-seq datasets. See also Fig. S3, S4, Table S3 and S4.
Of more interest was whether PU.1-induced loss of Satb1 or Runx1 from ‘old’ sites could itself be responsible for the repression observed at linked genes. To minimize alternative explanations, we focused on genes associated only with Satb1 or Runx1 peaks in Group 2 (‘old’ Satb1 or Runx1 unique genes, Fig. 4D) without ‘new’ sites: 251 of these genes were down-regulated in expression by PU.1 (PU.1-repressed)(Fig. 5C, right; FDR<0.001). Even without PU.1 introduction, Satb1 or Runx1 disruption alone reduced expression of many of these genes, which include functionally important T cell genes including Ets1, Hes1, Ptcra, Lck and Rag1 (Fig. 5C right; Table S3). Responses to Satb1 or Runx1 disruption were much more prevalent among both positively and negatively PU.1-regulated genes with ‘new’ or ‘old’ sites than among expressed genes generally (Fig. S3B,C).
While effects of acute Runx1 gene disruption were stronger than those of Satb1 disruption, they were weaker than responses to PU.1 itself. Therefore we used a complementary approach to determine whether removal of Runx1 inputs really affected these genes. Scid.adh.2c2 cells were retrovirally transduced with a truncated competitive inhibitor form of Runx1 that includes the DNA binding and nuclear localization domains but lacks transactivation and repression domains (Runx1DN)(Jacobs et al., 2013; Telfer et al., 2004). Fig. S4 shows that even genes with weak responses to Cas9-mediated Runx1 disruption responded much more strongly (most FDR<1E- 10), and in the same direction, to forced expression of this RunxDN (Table S4). The genes repressed by Runx blockade also overlapped substantially with genes that were repressed by PU.1. Thus association with PU.1-sensitive ‘old’ Satb1 and Runx1 sites defined a distinct subset of PU.1 repressed targets that were enriched for functional dependence on Satb1 and/or Runx1.
Repression is not preceded by transient PU.1 binding to ‘old’ Runx1 sites
PU.1-mediated repression began rapidly. As described in our related study (Ungerbäck et al., op. cit.), we used a tamoxifen (4-OHT)-mobilized form of PU.1 (Pu.1ert2) to determine the kinetics of responses to PU.1 nuclear entry (Fig. 5C). Focusing on those PU.1-repressed genes that also showed a detectable Satb1 or Runx1-response (sgSatb1 or sgRunx1/sgControl<0.75), we found that the great majority began reducing expression within 2 h of 4-OHT mobilization of PU.1 and were clearly repressed within 8 h (Fig. 5D, right). This time course made it possible to test whether PU.1 might first transiently bind to these ‘old’ sites and evict the factors by displacement or whether Runx1 or Satb1 removal from ‘old’ sites really occurred by competitive recruitment elsewhere. PU.1 in these cells bound to 21,000–24,000 non-promoter sites at timepoints from 2–24 h (Ungerbäck et al., op. cit.), of which 21–25% overlapped with Runx1 sites as defined in control cells. As candidates for transient sites that might initiate the repressive mechanism, 6880 sites were bound by PU.1 at 2 h but not at 24 h. However, these early transient sites in fact had substantially less overlap with Runx1 sites (8.6%) than sites occupied by PU.1 at 24 h but not at 2 h (19%), after repression was complete. Thus, the mechanism of removal of Runx1 from ‘old’ sites for gene repression did not depend on expulsion by transiently bound PU.1.
PU.1 association shifts Satb1 and Runx1 binding patterns in primary DN cells
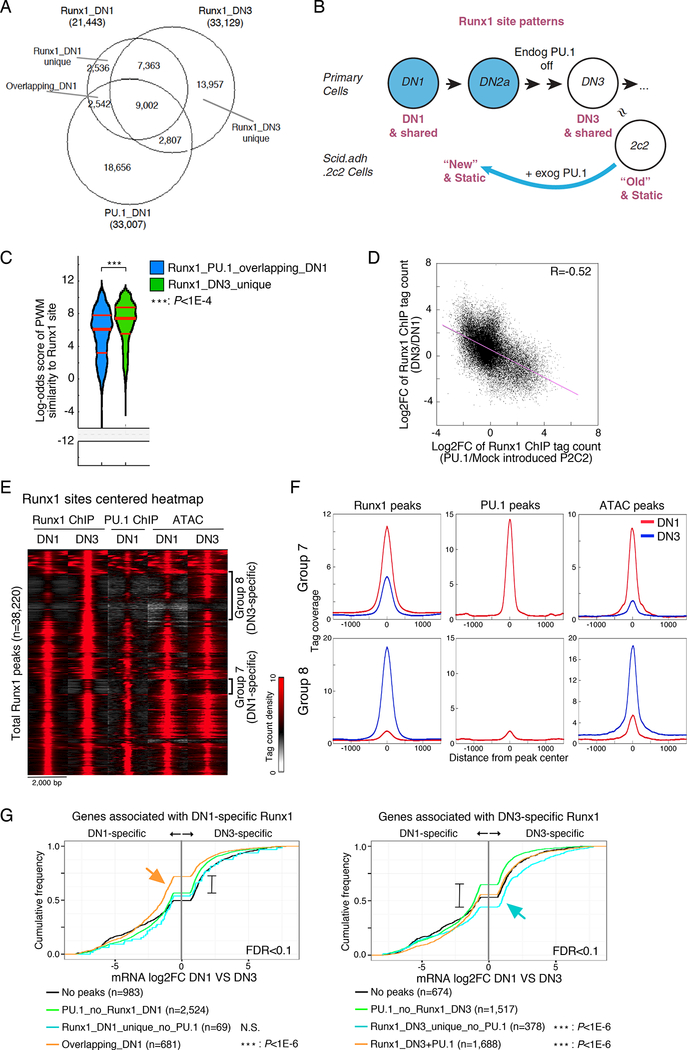
Runx1 and Satb1 normally increase expression from the DN1 to DN3 stages of pro-T cell development, but change expression less precipitously than PU.1 itself does (Fig. S1B). If the mode of repression by PU.1 shown above in PU.1-transfected Scid.adh.2c2 cells were relevant to the normal developmental timing of T cell gene expression, then Runx1 and Satb1 should occupy different genomic sites in the DN1 and DN2a stages, when PU.1 expression is naturally high, than in DN3 stage when PU.1 is largely absent. It was also important to test this prediction in the context of natural endogenous PU.1 expression levels. We focused on Runx1, which becomes essential for developing T cells around the time when PU.1 is downregulated (Egawa et al., 2007; Kawazu et al., 2005). To test whether endogenous PU.1 affects Runx1 deployment in natural developing DN cells, we compared primary DN1 and DN3 pro-T cells by Runx1 ChIP-seq (Fig. 6A,B). Consistent with the slight increase in Runx1 expression from DN1 to DN3, the number of Runx1 peaks in DN3 pro-T cells overall (~33K) was higher than in DN1 cells (~21K). However, we also found many DN1-specific Runx1 peaks that were vacated when the cells progressed to DN3, and half of these had been co-occupied by PU.1 in DN1 cells (Fig. 6A). Despite the lower level of Runx1 in DN1 cells, the DN1-specific sites occupied by Runx1, and especially those that were co-occupied with PU.1, were markedly lower in Runx site quality than the DN3-specific sites occupied when Runx1 levels were higher (Fig. 6C; Fig. S5A, B), similar to the results seen in PU. l -transduced Scid.adh.2c2 cells. Thus, even in primary DN1 cells, endogenous PU.1 appears to recruit Runx1 binding to a distinctive, lower-quality set of sites.
Figure 6. Redirection of Runx1 ChIP peaks in developing pro-T cells.
(A), Runx1 ChIP-seq analyses of bone marrow-derived primary DN1 and DN3 cells. Overlaps are shown among each set of Runx1 ChIP peaks and PU.1 ChIP peaks from DN1 cells.,
(B), Schematic view of Runx1 site patterns in primary DN cells and Scid.adh.2c2 cells with or without PU.1 introduction, relating effects of normal differentiation to the retrograde-like differentiation induced by exogenous PU.1 in Scid.adh.2c2 cells.,
(C), Distribution of motif log-odds similarity scores of Runx1_PU.1_overlapping_DN1 and Runx1_DN3_unique peaks in (A) against PWM for Runx1 motif. Median, 25% and 75% percentiles are also shown. P-values from Kruskal-Wallis test.,
(D), Scatter plot comparing redirection of the Runx1 peaks in primary DN cell development and in PU.1-transduced Scid.adh.2c2 cells. Points show log2 FC for Runx1 peak values for Mock vs PU.1-introduced Scid.adh.2c2 (x-axis) vs log2 FC in primary DN1 vs DN3 (y-axis). R, Pearson’s correlation coefficient.
(E), Tag count distributions for Runx1 and PU.1 ChIP signals and ATAC signals around Runx1 peaks in DN1 and DN3 cells.
(F), Association between redirection of Runx1 and chromatin accessibility in DN1 and DN3 cells. Average tag count distributions in Group 7 and Group 8 in (E) are indicated.
(G), Association of DN1- and DN3-specific Runx1 peaks with DN1-specific and DN3-specific gene expression. Cumulative distributions of expression changes between DN1 and DN3 stages are shown for four groups of genes bound by DN1-specific (left) and DN3-specific (right) Runx1 peaks (from A) and differentially expressed between DN1 and DN3 (FDR<0.1). Number of genes in each group and p-values (K-S tests for comparisons with genes bound by ‘PU.1_no_Runx1’ peaks) are indicated. Data are based on reproducible ChIP-seq peaks in two replicate samples and two replicates of RNA-seq results. See also Fig. S5.
The genomic sites of Runx1 peaks that increased or decreased intensity from DN1 to DN3 were well correlated with the genomic sites of Runx1 peaks in Scid.adh.2c2 that shifted in the opposite directions when PU.1 was artificially introduced, to drive differentiation “backward” in that cell-line model (Figs. 6B, D; S5C-E)(endogenous or exogenous PU.1: blue in Fig. 6B). The changes in occupancy and accessibility at these sites were also statistically significant predictors of the expression patterns of the genes linked to them, similar to the way ‘new’ and ‘old’ peaks predicted gene expression responses to exogenous PU.1 in the Scid.adh.2c2 cell line. Among the Runx1 peaks in the primary cells were two groups of regions that altered with development, Group 7 and Group 8, among many sites that changed much less (Fig. 6E). Group 7 contained DN1-specific Runx1 peaks, and these were frequently co-occupied with PU.1 and showed an open chromatin structure in DN1 that became more closed in DN3 cells (Figs. 6E,F). In contrast, Group 8 consisted of DN3-specific Runx1 peaks that had minimal PU.1 binding in the DN1 stage and opened in accessibility between DN1 and DN3 (Figs. 6E,F). Analysis of the expression of these genes showed that genes bound by DN1-specific Runx1 peaks overlapping with PU.1 peaks were biased to DN1-specific gene expression, even more than genes bound by PU.1 peaks without DN1-specific Runx1 peaks (Fig. 6G, left, orange arrow). On the other hand, genes bound by DN3-specific Runx1 peaks, with no evidence of PU.1 binding around them in DN1 cells, tended toward DN3-specific expression (Fig. 6G, right, blue arrow). In fact, these differences also reflected differential responses to short-term introduction of PU.1 itself into primary DN2b- DN3 (newly-committed) cells themselves. As shown in ECDF plots (Fig. S5F), the genes affected by exogenous PU.1 within the T cell program (PU1WTHA25) (Ungerbäck et al., op.cit.) were experimentally up-regulated by PU.1 if they had DN1-specific Runx1 peaks but downregulated by PU.1 if they had primarily DN3-specific Runx1 peaks. Thus, the Group 7 primary cell Runx1 sites found in DN1 cells resembled the ‘new’ sites induced by forcing Scid.adh.2c2 cells to express exogenous PU.1, whereas the Group 8 primary cell Runx1 sites found in DN3 cells appeared to be natural correlates of the ‘old’ sites in Scid.adh.2c2 cells that could be disrupted if PU.1 was introduced (Fig. 6B).
Functional gene response to endogenous PU.1in pro-T cells is predicted by Satb1 and Runx1 binding dynamics
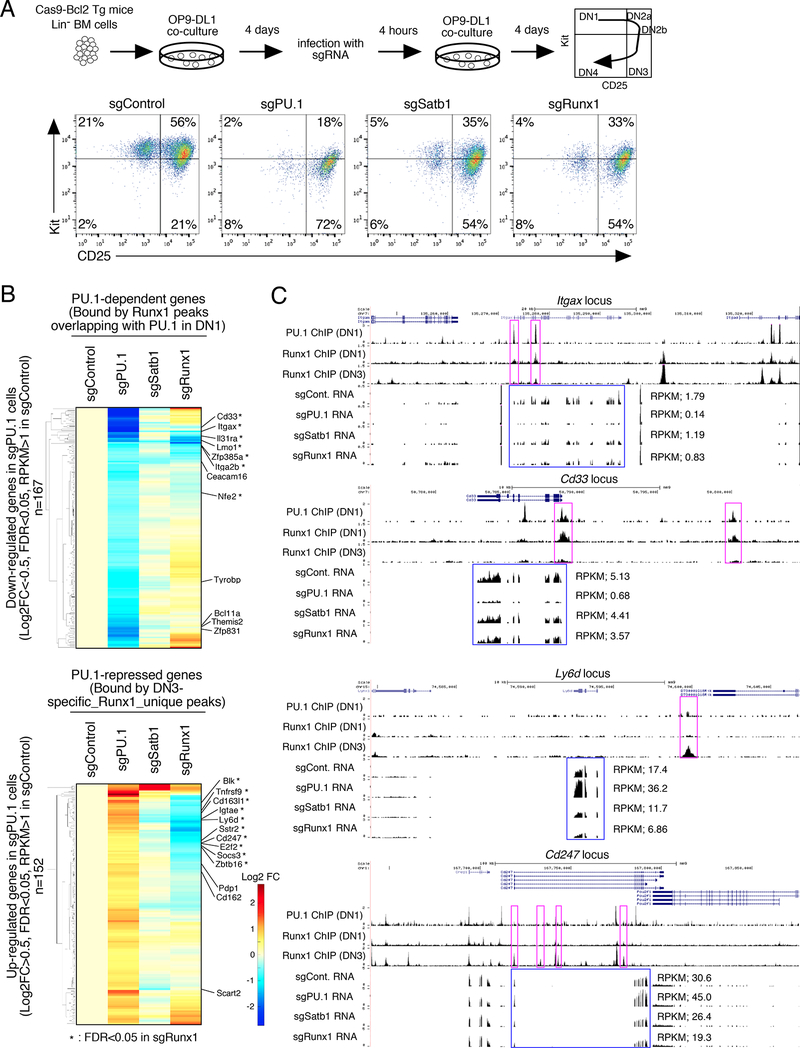
To test the functional importance of Satb1 and Runx1 for normal transcriptional control roles of endogenous PU.1 in primary pro-T cells, we obtained the cells from B6.ROSA26-Cas9 transgenic mice with a Bcl2 transgene. This genotype allowed us to induce acute disruption of PU.1, Satb1, or Runx1 by retroviral vector transduction of sgRNAs during in vitro differentiation when most cells were still in DN1 stage (Kit+ CD44+ CD25-, Fig. S6A). Four days later, transduced (CFP+) cells progressed developmentally but with gene-specific alterations (Fig. 7A, Fig. S6B,C), and the CFP+ transduced CD45+ CD25+ cells (Fig. S6B) were sorted for RNA-seq analysis. Those transduced with PU.1 sgRNA proliferated significantly less and progressed to the DN3 stage faster than control-transduced cells, consistent with Cre-dependent PU.1 deletion (Champhekar et al., 2015), and they had lost expression of PU.1 protein by day 8 (Fig. S6D). PU.1-disrupted cells reduced expression of a benchmark set of 500 PU.1 positively controlled genes (previously seen to be induced in DN2b cells with added PU.1), while increasing expression of 564 PU.1-repressed genes (inhibited by added PU.1 in DN2b cells; Ungerbäck et al., op. cit.)(Fig. 7B, Table S5). The presence of DN1- or DN3-specific Runx1 peaks at these genes correlated with their responses to deletion of endogenous PU.1, as shown in ECDF plots in Fig. S6E. Peaks of PU.1 overlapping with DN1-specific Runx1 peaks were significantly linked with genes that lost expression if PU.1 were deleted (Fig. S6E, left, orange arrow). In contrast, ~80% of genes bound by Runx1 at DN3-specific sites with no direct binding of PU.1 increased expression upon disruption of PU.1, even more than those with PU.1 sites as well as DN3- specific Runx1 sites (Fig. S6E, right, blue arrow). Thus, natural shifts in Runx1 occupancy not only correlated with endogenous PU.1, but also provided additional evidence needed to predict responses of linked genes to endogenous PU.1 activity.
Figure 7. Satb1 and Runx1 roles in PU.1-mediated gene regulation in primary DN cells.
(A), Flow cytometric analysis of primary pro-T cells after CRISPR-Cas9-mediated knockout of PU.1, Satb1 or Runx1. (Upper) Experimental scheme. (Lower) sgRNA transduced BM-derived precursors after 8 days of OP9-DL1 culture. Representative of three independent experiments.
(B), Effects of Satb1 and Runx1 disruption on PU.1-mediated gene regulation in DN cells, by RNA-seq of transduced CD25+ DN cells from (A). Hierarchical clustering of expression changes in response to different treatments are shown, among genes down-regulated (top) and up- regulated (bottom) in PU.1-deficient DN cells (|Log2FC|>0.5, FDR<0.05, RPKM>1 in sgControl). Based on RNA-seq from independent duplicates.
(C), The binding patterns of PU.1 (DN1) and Runx1 (DN1 and DN3), and RNA-seq tracks are shown for Spi1 (PU.1), Satb1 and Runx1-deficient DN cells at the Itgax and Cd33 PU.1- dependent loci and at the Ly6d and Cd247 PU.1-repressed loci. Representative of two independent experiments. See also Fig. S6 and Table S5
Recruitment of limiting Satb1 and Runx1 discriminates positive from negative effects of PU.1 in pro-T cells
To determine whether Satb1 and Runx1 truly work with PU.1 in primary cells to activate or repress target genes, we tested RNA expression in CD45+ CD25+ cells after acute deletion of Satb1 or Runx1, comparing it to expression after PU.1 deletion (Fig. 7A)(Table S5). Acute Runx1 disruption also reduced cell yield while Satb1 disruption caused less change in proliferation (Fig. S6C), and both caused less shift to DN2b/DN3 (Fig. 7A) than deletion of PU.1. We expected the interaction to be complex, because Runx1 is also needed for repression of PU.1 expression in DN cells (Huang et al., 2008; Zarnegar et al., 2010). Indeed, Runx1-deficient cells had 1.5x higher PU.1 expression than control cells, and some effects of Runx1 deletion phenocopied PU.1 overexpression as a result. However, distinct functional interactions emerged after separating the target genes that were positively and negatively regulated by PU.1 (Table S5).
Fig. 7B (top) focuses on the PU.1-dependent genes that were also linked with sites of co-binding of Runx1 and PU.1 in normal DN1 stage. Many of these genes also reduced expression upon deletion of Satb1 or Runx1 (Fig. 7B, top), consistent with positive PU.1-cofactor cooperation. For example, PU.1-dependent genes including Itgax and Cd33 have clear PU.1 peaks cooccupied with DN1-specific Runx1 peaks, and their expression was significantly decreased not only in PU.1-deficient but also in Runx1-deficient cells with intact PU.1 (Fig. 7C, top; FDR<0.05 in sgRunx1; Table S5). On the other hand, any genes repressed by PU.1 by the redeployment or “theft” mechanism should be genes that depend on positive regulation by Satb1 and Runx1 with T cell factors and are linked with sites that Runx1 binds better in DN3 cells when PU.1 is no longer present. Not only should their expression be increased in PU.1-deleted cells, but it should also be decreased in Satb1- or Runx1-deleted cells. As shown in Fig. 7B (bottom), in fact nearly half of these PU.1-repressed genes (68/152) were reduced in expression by sgRNA against Runx1 especially (Fig. 7B, bottom; Table S5). Fig. 7C also shows examples of T cell genes repressed by PU.1, Ly6d and Cd247, that were bound by DN3-specific Runx1 peaks and underwent significant downregulation of expression in Satb1- or Runx1-deficient cells (Fig. 7C, bottom; FDR<0.05 in sgRunx1). Importantly, these responses to Satb1 or Runx1 deletion were highly enriched in PU.1-induced and PU.1-repressed genes alike, as compared to all expressed genes in these primary DN cells (Fig. S6F).
Taken together, these results indicate that redeployment of Satb1 and Runx1 can play key collaborative roles in both positive and negative regulation of global gene expression by PU.1 in primary DN cells.
DISCUSSION
Here, we document a mechanism used by PU.1 in early T cells to regulate gene expression positively and negatively. It works not only by opening chromatin (Ungerbäck et al., op. cit.) and nucleating cooperative complexes with transcriptional cofactors at its binding sites, but also by competing to reallocate its cofactors, often removing them from sites where they could otherwise regulate different genes. Thus, PU.1 regulates gene expression in pro-T cells via two distinct mechanisms. PU.1 recruits Satb1 and Runx1 to new sites, and collaborates with them to activate expression of its direct target genes; but PU.1 also steals Satb1 and Runx1 from many genomic sites, thereby repressing expression of T cell genes indirectly. In this way, the loss of PU.1 expression that occurs at T cell commitment not only affects gene regulation through its own binding, but also through its impacts on the deployment of other factors. In support of this model, the pattern of Runx1 occupancy across the normal commitment transition implies that PU.1 indeed affects its deployment in natural development, and that these patterns of occupancy predict developmental regulation and acute responses of linked genes to PU.1. Thus, competitive recruitment of stably-expressed factors among distinct complexes can play a previously underappreciated, system-level role in the sharp differences between gene expression patterns across developmental transitions.
The mode of repression we have shown for PU.1 here is repression at a distance, without direct DNA binding at repressed sites, via interactions with a limiting pool of co-factors (Graphical abstract). It differs from other reports of “enhancer decommissioning” which are based on repression by binding, even if that binding is transient (Adam et al., 2015; Chronis et al., 2017; Respuela et al., 2016). PU.1 removes Runx1 from higher quality binding sites without PU.1 to lower quality sites that it can bind with PU.1, against its own concentration gradient. Such effects are often considered “squelching” artifacts (Zhao et al., 2003), confined to situations where transcription factors are ectopically expressed or in vast excess. Our data from primary cells, however, suggest that this mechanism is relevant to the natural switch-like gene regulatory transition that occurs as endogenous PU.1 levels decline normally in pro-T cells.
Some features of the mechanism remain to be clarified. The zero-sum nature of Runx1 and Satb1 distribution in the presence and absence of PU.1 makes it most likely that the pools of these factors in pro-T cells are limiting. However, proof that negative effects of PU.1 on DN3-specific genes might be reversed by increasing these factors must await better tools for level control, as Runx1 overexpression is itself toxic to pro-T cells (Telfer et al., 2004; Wong et al., 2010). Further analysis will also be needed to define how ‘theft’ occurs from some Runx1 sites but not others.
This is not the only mechanism that PU.1 can use to repress. The ‘theft’ mechanism may not explain the effect of PU.1 on the Notch signaling pathway that provides a crucial positive input into the T cell program (Del Real and Rothenberg, 2013). However, it could have pervasive impacts. Runx family transcription factor binding motifs are consistently enriched at active regulatory sites bound by diverse transcription factors in developing lymphocytes (Lin et al., 2010; Longabaugh et al., 2017; Miyazaki et al., 2011)(Ungerbäck et al., op. cit.)(Satb1 motifs remain less clearly defined). Furthermore, Runx1 itself plays a major role in the global organization of definitive stem-cell enhancer landscapes during the first wave of embryonic hematopoiesis (Lichtinger et al., 2012). Thus, if Runx protein levels are limited enough to constrain enhancer activity genome-wide, Runx redeployment could propagate through impacts on activity of many other transcription factors that collaborate with Runx factors at various enhancer classes, including such important T cell factors as GATA3, TCF7, and Bcl11b. PU.1 has little effect on the expression of any of these T cell factors under conditions that promote T cell development (Champhekar et al., 2015; Del Real and Rothenberg, 2013), which has left its broad impacts on T lineage developmental gene expression unexplained. However, if PU.1 binding with Runx1 could prevent Runx1 activity at sites where the actions of other T cell factors depend functionally on co-occupancy with Runx1, then PU.1 could affect not only its own direct targets, but also the function of multiple Runx-interacting factors in T lineage development.
This model has implications beyond the role of PU.1. It implies that each transcription factor may actually force other factors expressed at moderate levels to choose between alternative sites of engagement, with negative as well as positive consequences. This has not been widely examined elsewhere, but may be much more general. For example, it is known that ectopic expression of OCT3/4, SOX2, KLF4 and MYC in most cells generates iPSCs, but molecular mechanisms whereby these factors extinguish ongoing cell programs are still incompletely understood (Chronis et al., 2017; Takahashi and Yamanaka, 2016). It is attractive to consider that these factors may repress cell-type-specific gene expression indirectly via co-factor theft, as well as by recruiting co-factors to complete mobilization of pluripotency genes. The activity of PU.1 in early T cell precursors demonstrates that a transcription factor can work through systemwide redeployment of other factors and not only through sites that it binds itself.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ellen Rothenberg (evroth@its.caltech.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6, B6.Cg-Tg(BCL2)25Wehi/J (Bcl2-tg) and B6.Gt(ROSA)26Sortm1.1(CAG-cas9*,- EGFP)Fezh/J (Cas9) mice were purchased from the Jackson Laboratory. For CRISPR experiments in primary cells, we used F1 progeny of Bcl2-tg and Cas9 mice. No experiments were performed in vivo; mice were used only as a source of cells for experiments in vitro. Mice were used as a source of bone marrow at 2–3 months of age. Mice were used for thymocyte preparations at 4–6 weeks of age. Fetal mice were collected from pregnant females at embryonic day 13.5 for fetal liver cell collection. Both male and female mice were used, but in any one experiment comparing experimental and control groups, animals of the same age and sex were used for both groups. All animals were bred and maintained in the California Institute of Technology Laboratory Animal Facility, under specific pathogen free conditions, and the protocol supporting animal breeding for this work was reviewed and approved by the Institute Animal Care and Use Committee of the California Institute of Technology.
Cell line
Scid.adh.2c2 cells, a clonal DN3-like cell line developed in our laboratory (Dionne et al., 2005), were used for many experiments. Based on the presence of Y chromosome DNA sequences and the lack of Xist transcripts in genomic sequence tracks from these cells, these cells are male (data not shown). Cell culture details are provided under Method Details because they were varied according to the experiment.
METHOD DETAILS
Cell preparation and cell culture
All cell cultures were carried out at 37°C.
Bone marrow (BM) was removed from the femurs and tibiae of 2–3 month-old C57BL/6 or (Cas9 x Bcl2-tg)F1 mice. Suspensions of BM cells were prepared and stained for lineage markers using biotin-conjugated lineage antibodies (CD11b, CD11c, Gr1, TER-119, NK1.1, CD19, CD3ɛ, B220), then incubated with streptavidin-coated magnetic beads, and passed through a magnetic column. Lin- cells were eluted and cultured on OP9-DL1 monolayers using OP9 medium (α-MEM, 20% FBS, 50 µM β-ME, Pen-Step-Glutamine) supplemented with 10 ng/ml of IL-7 and 10 ng/ml of Flt3L. For deletion of PU.1, Satb1 and Runx1 genes, cultured cells were disaggregated on day 4, filtered through 40-µm nylon mesh, transferred onto RetroNectin-coated virus bound plates, and cultured with OP9 medium supplemented with 10 ng/ml of IL-7, Flt3L and SCF. Infected cells were cultured for an additional 4 days on OP9-DL1 and subjected to further analysis. For sorting, cells were stained with CD45, CD25, and a biotin-conjugated lineage cocktail (CD8α, CD11b, CD11c, Gr1, TER-119, NK1.1, CD19, TCRβ, TCRγδ), and were sorted for CD25+ infected cells (Lin-CD45+CD25+CFP+). For isolation of DN1 cells, Lin-CD45+KithiCD25- cells were sorted on day 5. For DN3 cells, cells were cultured on OP9-DL1 monolayers for 14 days and Lin-CD45+KitlowCD25+ cells were used.
To obtain primary thymocytes for ATAC-sequencing, thymocytes were stained with fluorescent antibodies against CD45, CD44, c-Kit, and CD25, and a biotin-conjugated lineage marker cocktail (CD11b, CD11c, Gr1, TER-119, NK1.1, CD19, CD3, CD8a,TCRγδ,TCRβ), and were sorted on a BD FACSAria™ to isolate DN1 (Lin-CD45+c-KithiCD44hiCD25-) or DN3 cells (Lin CD45+ c-Kit1o CD441o CD25+).
Fetal livers (FL) were dissected from E13.5 (day of plug, E0.5) C57BL/6 animals. Suspensions of FL cells were stained for lineage markers using biotin-conjugated lineage antibodies (CD11c, Gr1, TER-119, NK1.1, CD19, F4/80), incubated with streptavidin-coated magnetic beads (Miltenyi Biotec), and passed through a magnetic column (Miltenyi Biotec). Lin- precursor cells were used to initiate OP9-DL1 cultures using slight modifications of methods previously described (Champhekar et al., 2015) in OP9 medium supplemented with IL-7 and Flt3L (5 ng/ml each). On day 7, cells were harvested and enriched for CD25+ cells, and subjected to retrovirus infection for HA-PU.1. Two days after infection, retrovirus transduced cells were stained with antibodies against c-Kit, CD25, CD44 and CD45. Fractions sorted were CD45+CD25+ (“CD25+”) for samples transduced with EVGFP, PU1WTHA or CD45+ CD44+CD25- (“CD44+”) for samples transduced with PU1WTHA.
Scid.adh.2c2 cells (Dionne et al., 2005) were cultured in RPMI1640 with 10% fetal bovine serum (Sigma-Aldrich), sodium pyruvate, non-essential amino acids, Pen-Strep-Glutamine and 50 μM β-mercaptoethanol. For the PU.1 mobilization time course study, Lzrs-PU.1ert2- transduced Scid.adh.2c2 cells or cells transduced with an empty Lzrs-ert2 vector control were sorted for GFP positivity, and acute PU.1 mobilization was then induced for 0, 2, 8 or 24h with 0.1 μM 4-OHT (4-hydroxytamoxifen; Sigma-Aldrich). Controls were treated with 4-OHT in parallel. Additional details are provided in our related study (J. Ungerbäck, H. Hosokawa, X. Wang, T. Strid, B. A. Williams, M. Sigvardsson, E. V. Rothenberg: Pioneering, chromatin remodeling, and epigenetic constraint in early T cell gene regulation by PU.1, submitted). For analysis of RNA expression changes in response to Runx1DN (Fig. S4), Scid.adh.2c2 cells were transduced with Runx1DN or empty vector and sorted for vector expression after 48 h.
Acute Cas9-mediated deletion of genes
For CRISPR-Cas9-mediated deletion of Satbl and Runx1 in Scid.adh.2c2 cells, cells were first infected with Cas9-GFP and sgRNA-CFP retroviruses as indicated. Western blotting showed the efficient depletion of the proteins encoded by the targeted genes in doubly-infected cells if they were sorted two days after co-transduction with Cas9-GFP and sgRNA (Fig. 5B). At this point, to test the effect on responses to PU.1, the population could again be subjected to retroviral vector infection to introduce PU.1-hNGFR or an hNGFR empty vector control, and the triply infected cells were sorted two days later as shown in Fig. 5A. The high transducibility of Scid.adh.2c2 cells, usually well >80%, made it feasible to purify enough triply-transduced cells for RNA-seq or Western blotting experiments in a single sort, but the overall cell yield was limited by the lower infectivity of the Cas9-GFP construct as compared to the others, at least in part due to the large insert size. In other experiments where more cells were required, therefore, the protocol could be modified by transducing first with Cas9-GFP alone, then sorting GFP+ cells and only subsequently transducing them in two more rounds with the more efficient sgRNA and PU.1 expression vectors.
For Cas-mediated gene targeting in primary cells, we used BM precursors from F1 progeny of Cas9 transgenic and Bcl2 transgenic mice, as described under “Animals” above. These mice are healthy and show no obvious abnormalities of their T cell development. We initiated BM precursors from these Cas9-transgenic mice into T cell differentiation in vitro as described above using OP9-DL1 co-culture. After 4 days, we transduced them with retroviral vectors encoding sgRNA against PU.1, Satb1 or Runx1 with a Cyan Fluorescent Protein (CFP) reporter and returned them to OP9-DL1 culture. After four more days, transduced CFP+ cells and controls alike had progressed into DN2a or DN2b stages, as shown in Figs. 7A and S6B, and the cells were sorted for RNA analysis. Intracellular flow cytometric staining showed that endogenous PU.1 protein was extensively depleted from the cells transduced by PU.1 sgRNA at this timepoint.
Cloning
Myc-Flag tagged-PU.1 WT and ETS mutants were inserted into a multi-cloning site of the pMXs-IRES-hNGFR vector. cDNA for Cas9 was inserted into a multi-cloning site of the MSCV-IRES-GFP vector. The truncated Runx1 construct called “Runx1DN” in Fig. S4, also known as d190, was previously described (Jacobs et al., 2013; Telfer et al., 2004; Telfer and Rothenberg, 2001) and was also used in the MSCV-IRES-GFP vector. It contains the distal promoter sequence and extends through sequences coding for the DNA binding domain and nuclear localization sequences of Runx1, but not the repression, trans-activation, or nuclear matrix attachment regions. For sgRNA-expressing vectors (E42 dTet-CFP), we first made an empty sgRNA expression cassette with human U6 promoter and an mTurquoise2 reporter (brighter version of Cyan Fluorescent Protein with shorter EF-1a promoter), by modifying a pQCXIN backbone retroviral vector (Clontech) using Gibson cloning. 19-mer sgRNAs were designed using the CHOPCHOP web tool (https://chopchop.rc.fas.harvard.edu/) and inserted into the empty E42 dTet-CFP vector by PCR-based insertion. Three sgRNA-expression vectors were generated for each gene, and pooled retroviral plasmids were used to make retroviral supernatant.
Affinity purification of PU.1 complexes
PU.1 protein complexes were isolated by two-step affinity purification from the DN3-like cell line, Scid.adh.2c2. Myc-Flag-tagged cDNA for Spil (PU.1) was inserted into a multi-cloning site of the pMXs-IRES-hNGFR vector. The retrovirus-containing culture supernatant was prepared as described previously(Champhekar et al., 2015; Del Real and Rothenberg, 2013). Scid.adh.2c2 cells were infected with either mock control (pMXs-IRES-hNGFR) or Myc-Flag- PU.1-containing retrovirus. Three days after infection, Myc-Flag-tagged PU.1-infected Scid.adh.2c2 cells were solubilized with the following protease inhibitor-containing IP buffer:50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol, 0.1% Tween, 1 mM EDTA, 10 mM NaF, 1 mM DTT and a protease inhibitor cocktail (Roche Applied Science), and lysed on ice for 30 min with gentle shaking and sonicated on a Misonix S-4000 sonicator (Qsonica) for 3 cycles, amplitude 20 for 30 sec. followed by 30 sec. rest. The insoluble materials were removed by centrifugation and immunoprecipitation with anti-Flag M2 agarose (Sigma-Aldrich) was performed overnight at 4oC. Immune complexes were eluted from the agarose by 3xFlag peptide (Sigma-Aldrich), and the eluted PU.1 complexes were subjected 2nd immunoprecipitation with anti-Myc gel (MBL). Immune complexes were eluted from the gel with Myc peptide (MBL) and separated by SDS-PAGE. The bands were excised from the gel and subjected to a mass spectrometric analysis to identify corresponding proteins. The gel pieces were washed twice with 100 mM bicarbonate in acetonitrile and the proteins were digested with trypsin. After adding 0.1% formic acid to the supernatant, the peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with a LTQ Mass Spectrometer (Thermo Scientific). The resulting MS/MS data set was analyzed using the Mascot software program (Matrix Science,).
Immunoprecipitation and immunoblotting
Anti-Flag mAb (M2, Sigma-Aldrich) were used for immunoprecipitation. Protein extracts from Myc-Flag-tagged PU.1-infected Scid.adh.2c2 cells were prepared using RIPA buffer (1% NP- 40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 µg/ml of aprotinin, leupeptin and pepstatin, 1 mM Na3VO4, 1 mM NaF and Tris-HCl, 50mM, pH 7.4). For immunoprecipitation analyses, the cell lysates were subjected to a pre-clear process with control IgG coupled agarose (Sigma-Aldrich) at 4oC for 1 hr with rotation. The pre-cleared extracts were subjected to immunoprecipitation with an anti-Flag M2 agarose (Sigma-Aldrich) at 4oC for 16 hr, and then the immunocomplexes were eluted from the agarose by 3xFlag peptide, and the eluted PU.1 complexes were run on 10% polyacrylamide gels. After electrophoresis, the proteins were subjected to immunoblotting as described previously (Hosokawa et al., 2013). Nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). The antibodies used for the Immunoblot analyses were anti-Satb1 (ab109122, Abcam), anti-Runx1 (ab23980, Abcam), anti-Lamin B (sc-6217, Santa Cruz Biotechnology, Inc.), and anti-Myc (PL14, MBL).
Chromatin Immunoprecipitation (ChIP)
Ten million of BM-derived DN1, DN3 or Scid.adh.2c2 cells cells were fixed with 1 mg/ml DSG (Thermo Scientific) in PBS for 30 min at RT followed by additional 10 min with addition of formaldehyde up to 1%. The reaction was quenched by addition of 1/10 volume of 0.125M glycine and the cells were washed with HBSS (Gibco). Pelleted nucleus were dissolved in lysis buffer (0.5% SDS, 10 mM EDTA, 0.5 mM EGTA, 50 mM Tris-HCl (pH 8) and PIC) and sonicated on a Bioruptor (Diagenode) for 18 cycles of 30sec sonication followed by 30sec rest, with max power. Five µg per 107 cells of antibodies were hybridized to Dynabeads M-280 Sheep anti- Rabbit, Dynabeads M-280 Sheep anti- Mouse, or Dynabeads Protein A/G (Invitrogen) and added to the diluted chromatin complex. They were incubated over night at 4°C, then washed and eluted for 6hr at 65°C in ChIP elution buffer (20 mM Tris-HCl, pH 7.5, 5 mM EDTA 50 mM NaCl, 1% SDS, and 50 µg proteinase K). Precipitated chromatin fragments were cleaned up using Zymo ChIP DNA Clean & Concentrator and subjected to quantitative PCR on a 7900HT Fast Real-Time PCR System (Applied Biosystems) with SYBR® GreenER™ qPCR SuperMix (Invitrogen). Rabbit anti-Satb1 mAb (ab109122, Abcam), Rabbit anti-Runx1 Ab (ab23980, Abcam), Mouse anti-Gata3 mAbs (a mixture of sc-268, Santa Cruz Biotechnology and MAB26051, R&D Systems), Goat anti-Fog1 Ab (sc-9361, Santa Cruz Biotechnolog) and Rabbit HA-probe polyclonal IgG (sc-805x, Santa Cruz Biotechnology) were used. ChIP-seq libraries were constructed using NEBNext ChIP-Seq Library Preparation Kit (NEB #E6240) following manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq2500 in single read mode with the read length of 50 nt following manufacturer's instructions. Base calls were performed with RTA 1.13.48.0 followed by conversion to FASTQ with bcl2fastq 1.8.4 and produced approximately 30 million reads per sample.
mRNA and RNA-seq library preparation
Total RNA was isolated using an RNAeasy MicroKit (Qiagen) according to manufacturer’s recommendations. Libraries were constructed using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB #E7530) from ~1 µg of total RNA following manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq2500 in single read mode with the read length of 50 nt following manufacturer's instructions.
Pull-down assay
Interactions of Runx1 and Myc-Flag-PU.1 with Runx and PU.1 site oligonucleotides were tested by pulldown assays from total-cell lysates of Runx1- or Myc-Flag-PU.1-transduced Scid.adh.2c2 cells. The specific Runx1 motif oligonucleotide probes (from Rag1 promoter) were biotinylated and were tested for ability to be bound by Runx1 in the presence or absence of increasing amounts of PU.1 plus non-biotinylated competitor oligonucleotides. Competitor oligonucleotides (from Itgax int.8) contained a non-consensus Runx motif closely linked to a canonical PU.1 motif (WT) or a mutant PU.1 motif (TTCCµ → TAGC). Lysates of Scid.adh.2c2 cells transduced with Runx1 and/or Myc-Flag-PU.1 were incubated with 5 µg of poly(dl-dC) (Thermo Fisher), 250 nM of PU.1 motif oligonucleotides and 20µl of Dynabeads M280 Streptavidin (Invitrogen) carrying biotinylated Runx1 motif oligonucleotides. They were incubated for 2hr at 4°C and washed. Then, precipitated proteins were eluted by SDS sample buffer and subjected to immunoblotting. Sequences of the oligonucleotides were as follows: Runx1 motif (Rag1 promoter), 5’-CGGCTAACCACAGATGATG-3’; PU.1 canonical motif (Itgax int. 8), 5’- CCCACCACTTCCTCCTGTAA-3’; PU.1 mutant motif, 5’-CCCACCACTagCTCCTGTAA-3’.
Assay for Transposase Accessible Chromatin
Eighty thousand Scid.adh.2c2, 18x103 DN1 or 50x103 DN3 cells were subjected to Assay for Transposase Accessible Chromatin (ATAC)-sequencing library preparation as described in (Buenrostro et al., 2013). Briefly, cells were lysed with ATAC lysis buffer (10mM Tris-HCl pH7.5, 10mM NaCl, 0.1% IGEPAL CA-630, 3mM MgCl2), the lysed nuclei were immediately processed by tagmentation reaction mix including Nextera Tn5 transposase (Illumina) and incubated for 30min at 37°C, then cleaned up using a MinElute Kit(Qiagen). The library was constructed and barcoded using a Nextera library preparation kit (Illumina). For the primary cells, the final amplified libraries were purified and size-selected (x1.2 bead-to-DNA ratio for ~100–150bp cutoff DNA size) using SPRIselect-beads (Beckman-Coulter). Libraries were single-end sequenced on a HiSeq2500 (Illumina) (primary cells) or a NextSeq500 (Scid.adh.2c2) and produced approximately 30–50 million reads per sample.
QUANTIFICATION AND STATISTICAL ANALYSIS
ChIP-sequencing
ChIP-seq data were mapped to the mouse genome build NCBI37/mm9 using Bowtie (v1.1.1; http://bowtie-bio.sourceforge.net/index.shtml) with “-v 3 -k 11 -m 10 -t --best -strata” settings and HOMER tagdirectories were created with makeTagDirectory. ChIP peaks were identified with findPeaks.pl against a matched control sample using the settings “-P .1 -LP .1 -poisson .1 - style factor”. ChIP peak reproducibility was determined by a HOMER adaptation of the IDR (Irreproducibility Discovery Rate) package according to ENCODE guidelines (https://sites.google.com/site/anshulkundaie/proiects/idr). Only reproducible high quality peaks, with a normalized peak score ≥ 15, were considered for further analysis. Reproducible ChIP peaks were annotated to closest gene/transcriptional start site with proximity based annotation using HOMER annotatePeaks.pl (Heinz et al., 2010) (mm9 genome build). Tag density plots and heat maps were created with annotatePeaks.pl ( -hist or -hist & -ghist respectively) in a 2000 bp region surrounding indicated TF peak center, and by hierarchical clustering the tag count profiles in Cluster3 (de Hoon et al., 2004) with average linkage (if not stated) followed by TreeView visualization (Saldanha, 2004).
RNA-sequencing
Base calls were performed with RTA 1.13.48.0 followed by conversion to FASTQ with bcl2fastq 1.8.4 and produced approximately 30 million reads per sample. RNA-sequenced reads were trimmed with Trimmomatic (v.033; http://www.usadellab.org/cms/?page=trimmomatic) (Bolger et al., 2014) for removal of adapter -and low quality sequences (settings: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36). Resulting reads were then mapped onto the mouse genome build NCBI37/mm9 with STAR (v2.4.0) (Dobin et al., 2013) and post- processed with RSEM (v1.2.25; http://deweylab.github.io/RSEM/) (Li and Dewey, 2011) according to the settings in the ENCODE long-rna-seq-pipeline (https://github.com/ENCODE- DCC/long-rna-seq-pipeline/blob/master/DAC/STAR_RSEM.sh) with the minor modifications that settings “--output-genome-bam --sampling-for-bam” was added to rsem-calculate- expression. STAR and RSEM reference libraries were created from genome build NCBI37/mm9 together with the Ensembl gene model file Mus_musculus.NCBIM37.66.gtf. The resulting bam- files were used to create HOMER (Heinz et al., 2010) tag directories (makeTagDirectory with - keepAll setting). For analysis of statistical significance among differentially expressed genes the raw gene counts were derived from each tag directory with analyzeRepeats.pl with the -noadj - condenseGenes options followed by the getDiffExpression.pl command using EdgeR (v3.6.8; http://bioconductor.org/packages/release/bioc/html/edgeR.htmn (Robinson et al., 2010). For data visualization, rpkm normalized reads were derived using the analyzeRepeats.pl command with the options -count exons -condenseGenes -rpkm followed by log transformation. The resulting normalized datasets were hierarchically clustered in MatLab with “average” linkage. Results were visualized with the MatLab (clustergram).
Motif enrichment analysis
Motif enrichment analysis was performed with the findMotifsGenome.pl command in the HOMER package using a 200bp window. Overlapping peaks between samples were derived using mergePeaks.pl (default parameters). Annotation of peaks to genes and genomic regions (e.g. promoters, CG-rich regions, repeat regions) were performed with annotatePeaks.pl (default settings).
Runx site quality scoring
Position-weight matrix (PWM) log odds score analysis was performed with annotatePeaks.pl with the options -m {motif file} -mscore to derive the highest similarity score for each individual peak. Briefly, the top 12-mer motif was derived from a combined DN1 + DN3 combined Runx1 peak list (this paper), using findMotifsGenome.pl with default options. The top identified Runx- family motif PWM was provided as the motif files for the analysis. The highest reported log- odds similarity scores within each peak were imported into MATLAB (R2016a) and visualized as Violin distribution Plots (https://www.mathworks.com/matlabcentral/fileexchange/23661- violin-plots-for-plotting-multiple-distributions--distributionplot-m- /content/distributionPlot/distributionPlot.m) displaying 25,50 and 75 percentiles. Statistical significances of differences between groups were tested in GraphPad Prism 6 with a Kruskal- Wallis (K-W) non-parametric test (Dunn’s correction for multiple comparisons). Adjusted P- values ≤ 0.05 were considered as statistically significant.
Linking DNA-binding and gene expression
Enriched TF peaks were annotated to closest gene/transcriptional start site with proximity based annotation using HOMER annotatePeaks.pl (Heinz et al., 2010) (mm9 genome build). Entrez gene ID was then used to match gene expression tables (rpkm or DEG) with peak lists. Each gene category was assigned a unique value depending on the combination of features annotated to the genes. Empirical cumulative distribution (ECDF) plots were generated with the R package ggplot2 and statistical analysis was performed with Kolmogorov-Smirnov (K-S) two-sided tests.
ATAC-sequencing
ATAC-seq data were mapped to the mouse genome build NCBI37/mm9 using Bowtie (v1.1.1; http://bowtie-bio.sourceforge.net/index.shtml) with “-v 3 -k 11 -m 10 -t --best -strata” settings and HOMER tagdirectories were created with makeTagDirectory. Additionally, tag directories with reads mapped to the mitochondrial chromosome filtered out were created using the HOMER platform (Heinz et al., 2010) (makeTagDirectory). Abundance of ATAC-tags on transcription factor ChIP-seq peaks was analyzed in HOMER using the annotatePeaks.pl ( -hist or -hist & -ghist respectively) in a 2000 bp region surrounding indicated TF peak center.
UCSC Genome Browser bigwig visualization
BigWigs were generated from the aligned SAM or BED-file formats using Samtools (Li et al., 2009), Bedtools (Quinlan and Hall, 2010) and the UCSC genomeGoverageBed and bedGraphToBigWig and normalized to 1 million reads. For visualization of RNA-seq tracks, bamToBed and genomeCoverageBed were used with the “-split” setting enabled. BiGWig files were up-loaded to the UCSC-genome browser (http://genome.ucsc.edu) (Speir et al., 2016) for visualization.
Other statistical analyses
The statistical significance of differences between datasets was determined by Student’s t test, Kolmogorov-Smirnov (K-S) test or Pearson’s correlation coefficient using excel or the R package. Statistical details of experiments can be found in the figure legends.
DATA AND SOFTWARE AVATLABTLTTY
The accession numbers for the deep-sequencing data reported in this paper are GEO: GSE93755, GSE110020 and GSE103953. No new software was generated.
Supplementary Material
Highlights.
PU.1 interacts with Runx1 and Satb1 and recruits them to its DNA binding sites
Recruitment depletes partners from alternative sites causing repression by theft
Runx1 site choice in normal pro-T development follows PU.1 even to poor Runx sites
Theft-vulnerable Runx sites with lack of PU.1 sites predict PU.1 repression targets
ACKNOWLEDGMENTS
We thank Diana Perez, Jaime Tijerina, and Rochelle Diamond for cell sorting and advice, Ingrid Soto for mouse colony care, Vijaya Kumar for library preparation and sequencing, Henry Amrhein and Diane Trout for computational help, Igor Antoshechkin for sequencing management, and members of the Rothenberg group for valuable discussion and reagents. Fellowships from the Manpei Suzuki Diabetes Foundation (to HH) and from the Swedish Research Council (to JU) are gratefully acknowledged. This work was supported by grants from the USPHS to EVR (R01HD076915 and R01AI95943) and Grants-in-Aid for Advanced Research and Development Programs for Medical Innovation, the Takeda Science Foundation, and SENSHINE Medical Research Foundation to TT. This work was partly performed in the Collaborative Research Project Program of the Medical Institute of Bioregulation, Kyushu University, and was also supported by the L. A. Garfinkle Memorial Laboratory Fund and the Al Sherman Foundation, and by the Albert Billings Ruddock Professorship to EVR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
H.H., J.U. and E.V.R. designed the project, analyzed data, and wrote the manuscript. T.T. designed proteomic strategy, analyzed data, and reviewed the manuscript. H.H. and J.U. conducted most of the experiments. X.W. performed ATAC-seq experiments, S.M.C performed Runx1DN experiments, and M.M., K.I.N, and T.T. conducted proteomic analysis.
REFERENCES
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, et al. (2015). Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, and Kohwi-Shigematsu T. (2000). The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev 14, 521-535. [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, de Bruijn M, Ma X, Dortland B, Luteijn T, Downing RJ, and Dzierzak E. (2000). Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity 13, 423-431. [DOI] [PubMed] [Google Scholar]
- Carotta S, Dakic A, D'Amico A, Pang SH, Greig KT, Nutt SL, and Wu L. (2010). The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity 32, 628-641. [DOI] [PubMed] [Google Scholar]
- Champhekar A, Damle SS, Freedman G, Carotta S, Nutt SL, and Rothenberg EV. (2015). Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Dev 29, 832-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, and Plath K. (2017). Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, and Simon MC. (2003). Regulation of macrophage and neutrophil cell fates by the PU.1 :C/EBPα ratio and granulocyte colony-stimulating factor. NatImmunol 4, 1029-1036. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, and Miyano S. (2004). Open source clustering software. Bioinformatics 20, 1453-1454. [DOI] [PubMed] [Google Scholar]
- de la Rica L, Rodriguez-Ubreva J, Garcia M, Islam AB, Urquiza JM, Hernando H, Christensen J, Helin K, Gomez-Vaquero C, and Ballestar E. (2013). PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol 14, R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Real MM, and Rothenberg EV. (2013). Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development 140, 1207-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, and Rothenberg EV. (2005). Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol 280, 448-466. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, Tillman RE, Naoe Y, Taniuchi I, and Littman DR. (2007). The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 204, 1945-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, and Graf T. (2008). PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA 105, 6057-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, and Rothenberg EV. (2006). Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A 103, 11993-11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. (2010). Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317-328. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, and Glass CK. (2013). Effect of natural genetic variation on enhancer selection and function. Nature 503, 487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa H, Tanaka T, Suzuki Y, Iwamura C, Ohkubo S, Endoh K, Kato M, Endo Y, Onodera A, Tumes DJ, et al. (2013). Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc Natl Acad Sci U S A 110, 4691-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, et al. (2008). PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet 40, 51-60. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, and Akashi K. (2006). The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev 20, 3010-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs PT, Cao L, Samon JB, Kane CA, Hedblom EE, Bowcock A, and Telfer JC. (2013). Runx transcription factors repress human and murine c-Myc expression in a DNA- binding and C-terminally dependent manner. PLoS One 8, e69083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath MB, Houston IB, Janovski AJ, Zhu X, Gowrisankar S, Jegga AG, and DeKoter RP. (2008). Dose-dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B-cell identity. Leukemia 22, 1214-1225. [DOI] [PubMed] [Google Scholar]
- Kawazu M, Asai T, Ichikawa M, Yamamoto G, Saito T, Goyama S, Mitani K, Miyazono K, Chiba S, Ogawa S, et al. (2005). Functional domains of Runx1 are differentially required for CD4 repression, TCRb expression, and CD4/8 double-negative to CD4/8 double-positive transition in thymocyte development. JImmunol 174, 3526-3533. [DOI] [PubMed] [Google Scholar]
- Klein Wolterink RGJ, Serafini N, van Nimwegen M, Vosshenrich CA, de Bruijn MJ, Fonseca Pereira D, Veiga Fernandes H, Hendriks RW, and Di Santo JP. (2013). Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc Natl Acad Sci U S A 110, 10240-10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, and Meissner A. (2011). Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell 8, 96-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Chen AF, Pantovich MG, Danial M, Parchem RJ, Labosky PA, and Blelloch R. (2016). FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell 18, 104-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaud G, Kouskoff V, Trumble A, Schwantz S, and Keller G. (2004). Haploinsufficiency of Runx1 results in the acceleration of mesodermal development and hemangioblast specification upon in vitro differentiation of ES cells. Blood 103, 886-889. [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, and Graf T. (2006). Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25, 731-744. [DOI] [PubMed] [Google Scholar]
- Li B, and Dewey CN. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hannah R, Muller D, Clarke D, Assi SA, Lie ALM, Noailles L, Vijayabaskar MS, Wu M, et al. (2012). RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J 31, 4318-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. (2010). A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. NatImmunol 11, 635-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJR, Zeng W, Zhang JA, Hosokawa H, Jansen CS, Li L, Romero-Wolf M, Liu P, Kueh HY, Mortazavi A, et al. (2017). Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network. Proc Natl Acad Sci U S A 114, 5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukin K, Fields S, Lopez D, Cherrier M, Ternyak K, Ramirez J, Feeney AJ, and Hagman J. (2010). Compound haploinsufficiencies of Ebfl and Runxl genes impede B cell lineage progression. ProcNatlAcadSciUSA 107, 7869-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marecki S, and Fenton MJ. (2000). PU.l/Interferon Regulatory Factor interactions: mechanisms of transcriptional regulation. Cell BiochemBiophys 33, 127-148. [DOI] [PubMed] [Google Scholar]
- McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, Jaritz M, Tagoh H, and Busslinger M. (2011). The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J 30, 2388-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, and Murre C. (2011). The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol 12, 992-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Ghisletti S, and Barozzi I. (2011). The genomic landscapes of inflammation. Genes Dev 25, 101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, Diefenbach A, and Grosschedl R. (2013). Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol 14, 867-875. [DOI] [PubMed] [Google Scholar]
- Nerlov C, and Graf T. (1998). PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev 12, 2403-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad MA, Ungerback J, Ahsberg J, Somasundaram R, Strid T, Larsson M, Mansson R, De Paepe A, Lilljebjorn H, Fioretos T, et al. (2015). Ebf1 heterozygosity results in increased DNA damage in pro-B cells and their synergistic transformation by Pax5 haploinsufficiency. Blood 125, 4052-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respuela P, Nikolic M, Tan M, Frommolt P, Zhao Y, Wysocka J, and Rada-Iglesias A. (2016). Foxd3 promotes exit from naive pluripotency through enhancer decommissioning and inhibits germline specification. Cell Stem Cell 18, 118-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, Smeenk L, Trupke J, Sommer A, Jaritz M, et al. (2012). The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J 31, 3130-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, Enver T, Vyas P, and Scadden DT. (2005). Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 106, 477-484. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Ungerback J, and Champhekar A. (2016). Forging T-Lymphocyte Identity: Intersecting Networks of Transcriptional Control. Adv Immunol 129, 109-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. (2004). Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246-3248. [DOI] [PubMed] [Google Scholar]
- Satoh Y, Yokota T, Sudo T, Kondo M, Lai A, Kincade PW, Kouro T, Iida R, Kokame K, Miyata T, et al. (2013). The Satb1 protein directs hematopoietic stem cell differentiation toward lymphoid lineages. Immunity 38, 1105-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir ML, Zweig AS, Rosenbloom KR, Raney BJ, Paten B, Nejad P, Lee BT, Learned K, Karolchik D, Hinrichs AS, et al. (2016). The UCSC Genome Browser database: 2016 update. Nucleic acids research 44, D717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, and Downing JR. (2004). Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood 104, 3565-3572. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, and Rothenberg EV. (2007). Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol 8, 845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S. (2016). A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol 17, 183-193. [DOI] [PubMed] [Google Scholar]
- Talebian L, Li Z, Guo Y, Gaudet J, Speck ME, Sugiyama D, Kaur P, Pear WS, Maillard I, and Speck NA. (2007). T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFb dosage. Blood 109, 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer JC, Hedblom EE, Anderson MK, Laurent MN, and Rothenberg EV. (2004). Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. JImmunol 172, 4359-4370. [DOI] [PubMed] [Google Scholar]
- Telfer JC, and Rothenberg EV. (2001). Expression and function of a stem-cell promoter for the murine CBFa2 gene: distinct roles and regulation in natural killer and T cell development. DevelBiol 229, 363-382. [DOI] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET, and Grosschedl R. (2010). Early B cell Factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity 32, 714-725. [DOI] [PubMed] [Google Scholar]
- van Oevelen C, Kallin EM, and Graf T. (2013). Transcription factor-induced enhancer modulations during cell fate conversions. Curr Opin Genet Dev 23, 562-567. [DOI] [PubMed] [Google Scholar]
- Wong WF, Nakazato M, Watanabe T, Kohu K, Ogata T, Yoshida N, Sotomaru Y, Ito M, Araki K, Telfer J, et al. (2010). Over-expression of Runx1 transcription factor impairs the development of thymocytes from the double-negative to double-positive stages. Immunology 130, 243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, and Graf T. (2004). Stepwise reprogramming of B cells into macrophages. Cell 117, 663-676. [DOI] [PubMed] [Google Scholar]
- Yui MA, Feng N, and Rothenberg EV. (2010). Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol 185, 284-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, and Rothenberg EV. (2014). Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol 14, 529-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar MA, Chen J, and Rothenberg EV. (2010). Cell-type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol Cell Biol 30, 4922-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, and Rothenberg EV. (2012). Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149, 467-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, McCarrick-Walmsley R, Akerblad P, Sigvardsson M, and Kadesch T. (2003). Inhibition of p300/CBP by Early B-Cell Factor. Mol Cell Biol 23, 3837-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.