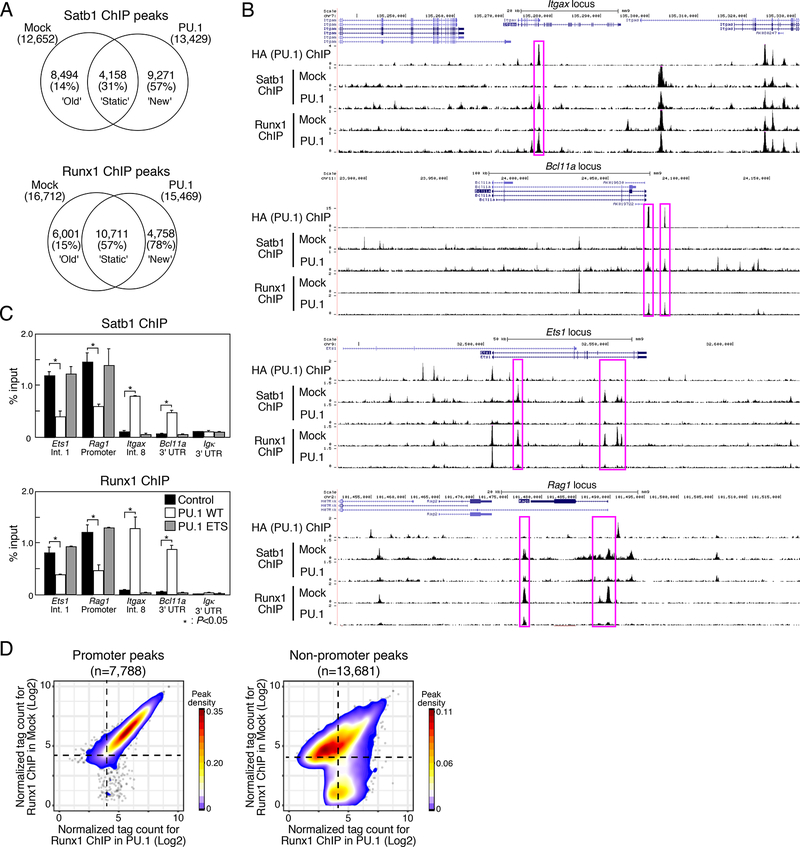
Figure 2. PU.1 introduction induces redirection of Satb1 and Runx1 ChIP peaks.
(A), Satb1 and Runx1 ChIP-seq analyses of mock- or PU.1-introduced Scid.adh.2c2 cells. Venn diagrams show the number of ChIP peaks, with percentages of the peaks overlapping with PU.1 sites in parentheses.
(B), Binding patterns of PU.1, Runx1 and Satb1 at the Itgax, Bcl11a, Ets1 and Rag1 loci in mock- and PU.1-introduced Scid.adh.2c2 cells. Representative of two independent experiments.
(C), Scid.adh.2c2 cells were transduced with empty vector (mock), PU.1 WT or PU.1 ETS. The binding of Runx1 and Satb1 at the Etsl (intron1), Rag1 (promoter), Itgax (intron8), Bcllla (3’UTR) and Igk(3’UTR) loci were determined by ChIP assay with qPCR analysis. P-values were determined by Student t-test. Representative of three independent experiments.
(D), Contour display of Runx1 tag counts (log2 counts per 107 reads) in Scid.adh.2c2 cells transduced with PU.1 or control vector. Data in A, D are based on reproducible ChIP-seq peaks in two replicate samples.