Abstract
[4 + 2]-Cycloadditions are increasingly being recognized in the biosynthetic pathways of many structurally complex natural products. A relatively small collection of enzymes from these pathways have been demonstrated to increase rates of cyclization and impose stereochemical constraints on the reactions. While mechanistic investigation of these enzymes is just beginning, recent studies have provided new insights with implications for understanding their biosynthetic roles, mechanisms of catalysis and evolutionary origin.
Graphical Abstract

1 Introduction and scope
This review is concerned with biocatalysts that have come into existence via the process of evolution by natural selection and have been or currently are under investigation as potential Diels-Alderases. A number of other biological catalysts have also been reported to accelerate a Diels-Alder reaction; however, these have been produced under various forms of artificial selection such as directed evolution,1,2 computational design2–6 and immunological selection7–9 and are not considered here. The identification and study of naturally selected Diels-Alderases has advanced significantly in the past five years, and as a testament to the current excitement in the field, there have been no less than half-a-dozen reviews of the subject in the past year alone.10–15 Furthermore, despite a large catalog of biosynthetic pathways hypothesized to involve an enzyme catalyzed Diels-Alder reaction,16–18 progress in the area has proven to be quite slow.
There are several challenges associated with the identification, isolation and characterization of enzymes as putative Diels-Alderases.19 Biosynthetic [4 + 2]-cycloadditions are frequently associated with natural products having complex structures. Therefore, even if the biosynthetic gene cluster can be identified, preparation of proposed substrates often proves difficult and time-consuming for the testing of activity and mechanistic study of the candidate enzymes. Furthermore, a number of these enzymes such as LovB, solanapyrone synthase and riboflavin synthase are multifunctional and appear to catalyze a priming reaction that increases the susceptibility of the substrate to a nonenzymatic intramolecular [4 + 2]-cycloaddition. This then complicates efforts to determine whether the enzyme indeed accelerates the cycloaddition or merely serves as a chiral template. Given the large number of recent reviews in the area, all of which are highly recommended to the interested reader, the present discussion will focus on providing an up-to-date survey and background for those enzymes that have had a significant impact on the search for a naturally selected Diels-Alderase. However, it is first necessary to clarify what is meant by a Diels-Alderase.
1.1 [4 + 2]-Cyclases vs. Diels-Alderases
Enzymes are often classified by the reactions that they bring more rapidly to thermodynamic equilibrium. For example, an alcohol dehydrogenase accelerates the specific equilibration of an alcohol and an oxidant (e.g., NAD) with the corresponding carbonyl and reductant (e.g., NADH) compared to what would be observed in the absence of the enzyme. A cycloaddition reaction has been defined by the International Union of Pure and Applied Chemistry (IUPAC) as20
“A reaction in which two or more unsaturated molecules (or parts of the same molecule) combine with the formation of a cyclic adduct in which there is a net reduction of the bond multiplicity… Cycloadditions may be pericyclic reactions or (non-concerted) stepwise reactions.”
It therefore stands to reason that a [4 + 2]-cyclase (or [4 + 2]-carbocyclase, when the cycloadduct is a cyclohexene ring) be regarded as an enzyme that accelerates the equilibration between a [4 + 2]-cycloadduct and the separated, diene and dienophile components regardless of mechanism. However, while there are a number of enzymes that can be so classified as [4 + 2]-cyclases, they are not all considered Diels-Alderases.
A Diels-Alder reaction is frequently treated as synonymous with a [4 + 2]-cycloaddition that proceeds stereoselectively in accord with the Woodward-Hoffmann rules.21–25 This in turn implies that either the reaction lacks an intermediate (i.e., is concerted) or the intermediate does not permit significant deviation from the requisite stereoselectivity.10,21,23,26 It has been concluded only relatively recently that most Diels-Alder reactions, including the reaction between ethylene and butadiene, is concerted and proceeds through a single, pericyclic transition state;10,23,27–29 however, examples abound where intermediates (e.g., biradicals) have been implicated.21,26,30 Nevertheless, as the case of macrophomate synthase discussed below shows, some intermediates evidently disqualify a stereospecific [4 + 2]-cycloaddition from being a Diels-Alder reaction.
Rather than attempting to parse which intermediates are allowed, we take a conservative approach in the following discussion and regard a Diels-Alder reaction as a [4 + 2]-cycloaddition where there is no local minimum in free energy (i.e., intermediate state) along the reaction coordinate connecting the minima corresponding to the reactant and product states. This definition is still admittedly incomplete (e.g., it still relies on a preconceived notion of a “reaction coordinate”); however, it intuitively describes an idealized pericyclic [4 + 2]-cycloaddition and serves as a useful point of reference. Therefore, a Diels-Alderase will be regarded here as a hypothetical enzyme that catalyzes a [4 + 2]-cycloaddition via a Diels-Alder reaction in its active site.
These working definitions are bound to raise objections by a number of readers—and rightly so! None of the enzymes discussed below can currently be said to unequivocally meet these criteria, and it is possible that no enzyme could be experimentally established to do so beyond all reasonable doubt. However, this is ultimately a distraction from the real point of interest, because the goal of enzymology is to understand enzymatic mechanisms of catalysis rather than argue the semantic implications of different monikers. For this reason, the following discussion emphasizes the chemistry of [4 + 2]-cyclases and the ways they approach or diverge from the idealized case, which for the sake of convenience is called a Diels-Alderase.
2 Macrophomate Synthase
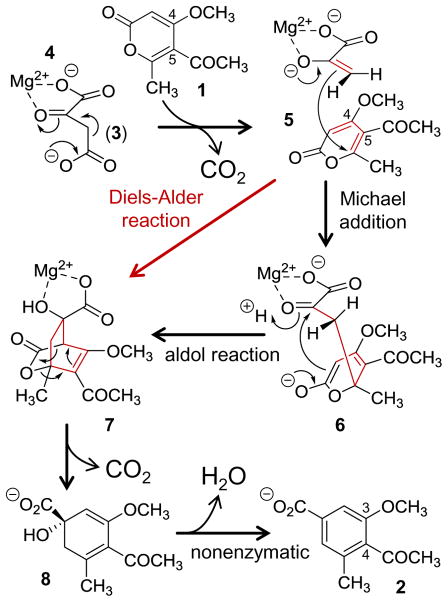
One of the best examples of the complications involved in attempting to label an enzyme as a Diels-Alderase is the case of macrophomate synthase. This enzyme is produced by the phytopathic fungus Macrophoma commelinae and is responsible for the conversion of 5-acetyl-4-methoxy-6-methyl-2-pyrone (1) to 4-acetyl-3-methoxy-5-methylbenzoic acid (i.e., macrophomic acid, 2).31–33 As it is currently understood (see Scheme 1), the catalytic cycle of macrophomate synthase begins with the binding of oxaloacetate (3) into the active site where it coordinates an active site Mg2+ ion via its C1 carboxylate and C2 carbonyl (4).33–35 The Mg2+ ion is required for activity and serves not only to orient 3 but likely also acts as a Lewis acid to promote the decarboxylation of oxaloacetate to form the enol tautomer of pyruvate (4→5).33,34,36 Preliminary kinetics suggest that the 2-pyrone substrate (1) then binds after decarboxylation of oxaloacetate,36,37 whereupon it is situated adjacent to the Mg2+-coordinated enolpyruvate (see Figure 1).35 Enolpyruvate next adds to the 2-pyrone to produce a bicyclo[2.2.2] adduct (5→[6]→7) that decomposes via decarboxylation (7→8).36 This essentially completes the catalytic cycle as the conversion of 8 to macrophomate (2) is likely not enzyme catalyzed.38 While a general mechanistic outline for macrophomate synthase has been largely established, the details regarding the individual steps and in particular formation of 7 have been the subject of considerable scrutiny.
Scheme 1.
Proposed mechanisms for the reaction catalyzed by macrophomate synthase.
Figure 1.
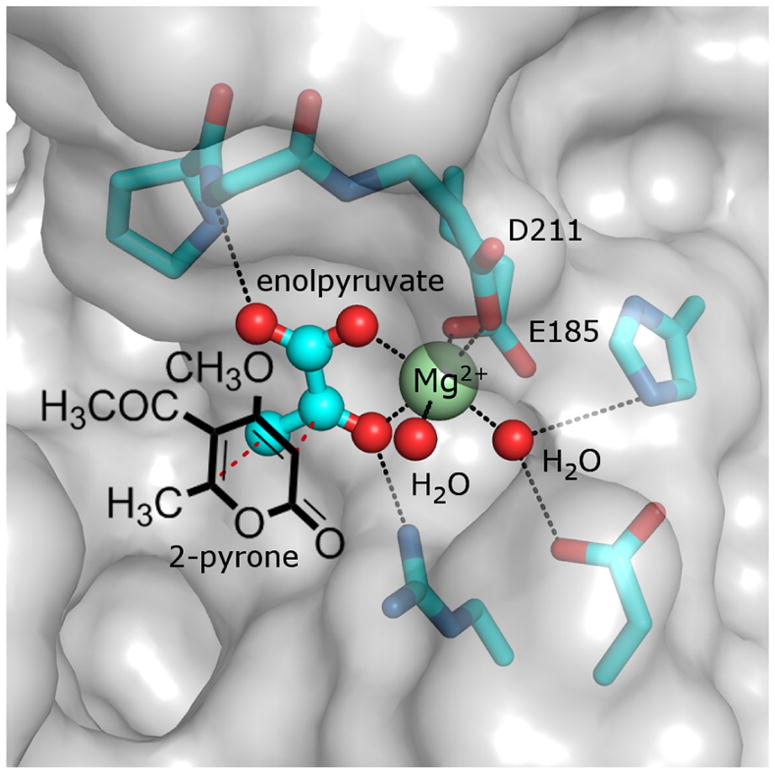
Structure of the active site of macrophomate synthase with enolpyruvate coordinated to the catalytic Mg2+ ion (PDB: 1IZC).35 The active site is located in a solvent-exposed cavity on the surface of the enzyme (grey) with the Mg2+ ion coordinated by the carboxylates of Asp211 (D211) and Glu185 (E185), two water ligands and enolpyruvate as shown. The crystal structure does not have the 2-pyrone substrate 1 bound; however, modeling simulations35 suggest that it may bind relative to enolpyruvate in a configuration conducive to a Diels-Alder reaction similar to that sketched here.
The existence of the bicyclic intermediate 7 was initially suggested by the Oikawa laboratory when they observed derivatives of methyl coumalate (9), which are analogs of 1, to react with macrophomate synthase and oxaloacetate and produce isolable bicyclo[3.2.1] intermediates (e.g., 11) rather than the predicted decarboxylation and dehydration products (see Scheme 2).36 The authors argued that such intermediates could be explained as arising from the rearrangement of strained bicyclo[2.2.2] intermediates (e.g., 10) similar to 7. This suggested that the enzyme does indeed catalyze the combination of two unsaturated carbon fragments (i.e., enolpyruvate and 1) to from a cycloadduct (7) with an overall net reduction in bond multiplicity. As this meets the IUPAC definition of a [4+2]-cycloaddition,20 macrophomate synthase can be regarded as exhibiting [4 + 2]-carbocyclase activity, though this is one of several steps in the overall reaction it catalyzes. The authors of the same study also observed stereospecific transfer of deuterium from oxaloacetate stereospecifically monodeuterated at C3 to either the bicyclo[3.2.1] adducts of a methyl coumalate derivative or macrophomate, though these studies were complicated by solvent exchange and racemization of the deuterated substrate.36 This was further supported when a 1.7 Å crystal structure of macrophomate synthase was reported with enolpyruvate bound and shown to accommodate the 2-pyrone substrate modeled into the active site in a configuration conducive to the formation of a pericyclic transition state as shown in Figure 1.35 Despite being circumstantial, these results led to the initial perception that this [4 + 2]-cycloaddition was in fact an enzyme catalyzed Diels-Alder reaction.
Scheme 2.

Proposal for the macrophomate synthase-catalyzed reaction between oxaloacetate and methyl coumalate.
Complications arose, however, when Jorgensen and coworkers took advantage of the crystal structure in an attempt to model the enzyme catalyzed [4 + 2]-cycloaddition using mixed quantum mechanical/molecular mechanical (QM/MM) computations.39 Using both the semiempirical PM3 as well as DFT computations for the QM part, the authors were only able to identify transition states for the stepwise formation of the bicylco[2.2.2] intermediate (7) from 5 in the enzyme active site. The authors were able to locate a structure suggestive of a concerted, Diels-Alder transition state; however, this was only possible at the PM3 level of theory and using a simplified model exclusive of the active site with restrictions placed on the two forming C—C bonds. Furthermore, this latter structure, which is not a true computed transition state, had a free energy 17.7 kcal/mol and 12.1 kcal/mol respectively greater than those for the Michael (5→6) and aldol (6→7) transition states calculated for the stepwise mechanism.39 Based on these observations, the authors suggested that the stepwise conversion was more energetically favorable thereby arguing against a concerted [4 + 2]-cycloaddition. This result also agreed with the observation that macrophomate synthase is structurally related to 2-dehydro-3-deoxygalactarate aldolase,35 which catalyzes an aldol reaction between pyruvate and tartronic semialdehyde.40 Furthermore, it was subsequently shown by the Hilvert laboratory that macrophomate synthase can also serve as an aldolase forming β-hydroxy phenylpropanoate from oxaloacetate and benzaldehyde with decarboxylation of oxaloacetate rate-limiting under saturating conditions.37
Based on these more recent investigations, macrophomate synthase is no longer considered to catalyze a Diels-Alder reaction during formation of the proposed bicyclic intermediate (7), and the Michael addition/aldol reaction is believed to be the more likely route.10,18,19,39 For this reason, it appears that interest in the enzyme has waned in recent years. This is rather unfortunate, however, given the complexity of its catalytic cycle. Furthermore, there has yet to be a direct experimental test of the concerted versus stepwise hypotheses or even a definitive demonstration of the proposed bicyclic intermediate.
Also of interest is the second decarboxylation reaction. If this decarboxylation takes place from the proposed bicyclo[2.2.2] intermediate (7→8) as shown in Scheme 1, then it would itself represent a cycloelimination reaction. Furthermore, if this elimination is concerted and accelerated by macrophomate synthase, then the enzyme would still catalyze a hetero-Diels-Alder reaction, albeit in reverse of the biosynthetic direction. Therefore, macrophomate synthase remains an interesting and important enzyme in the study of [4 + 2]-cyclases not only for its unusual chemistry but also the subtleties it exemplifies in the mechanistic characterization of these complex enzymes.
3 Riboflavin Synthase
Riboflavin synthase41 catalyzes the dismutation of two molecules of 6,7-dimethyl-8-ribityllumazine (12) to form riboflavin (13) (see Scheme 3)42 in a process that may involve an intermediary Diels-Alder reaction. The enzyme itself is a homotrimer with three active sites located at the interfaces of the three monomeric units.43,44 Each active site binds two molecules of 6,7-dimethyl-8-ribityllumazine (12) with one serving as the donor and the other as the acceptor of a four-carbon fragment to yield riboflavin and 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (14).43,44 The trimeric riboflavin synthase is further encapsulated in a large multimeric capsid made up of approximately sixty 6,7-dimethyl-8-ribityllumazine synthases that catalyze the condensation of the side product 14 and L-3,4-dihydroxy-2-butanone 4-phosphate (15) to regenerate one of the substrates for riboflavin synthase (14+15→12).45–47 Despite half a century of intense scientific investigation, many key aspects of the catalytic cycle of riboflavin synthase still remain largely unknown, and only relatively recently has new information come to light implicating a pericyclic [4 + 2]-cycloaddition.
Scheme 3.
Summary of abbreviated mechanisms proposed for the reaction catalyzed by riboflavin synthase.
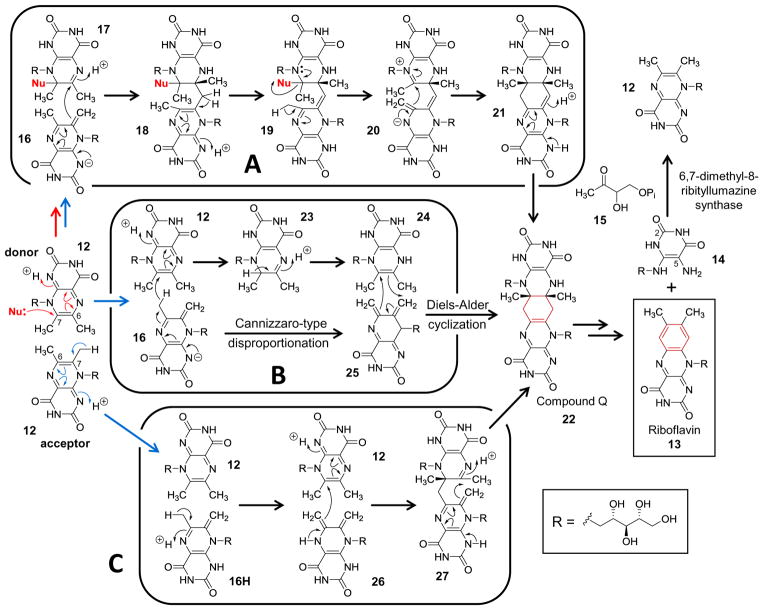
It was recognized early on that the nonenzymatic reaction to generate 13 and 14 also takes place at an appreciable rate when neutral to acidic aqueous solutions of 12 are heated.48–50 Early mechanistic hypotheses were thus guided by the notion that the enzymatic and nonenzymatic mechanisms would be similar, especially since both reactions proceed with the same regiospecific transfer of the isotopically labeled four-carbon fragment.51 This placed the focus on the formation of a covalent intermediate between the donor dimethyl ribityllumazine and either water or a nucleophilic side chain of the enzyme during the early steps of the catalytic cycle (12+12→16+17),42,52 though subsequent steps were much more ambiguous. A major breakthrough was achieved, however, when Illarionov, Bacher and coworkers discovered a catalytically competent, pentacyclic intermediate dubbed compound Q (22) in the catalytic cycle of riboflavin synthase.53–55 This was a significant discovery given that the central cyclohexene ring of the ionized compound Q implicated a possible Diels-Alder reaction in its formation.
Currently there are at least three main mechanistic hypotheses for the riboflavin synthase catalyzed generation of compound Q as shown in Scheme 3. The first (Scheme 3A) involves attack at the C7 carbon of the donor dimethyl ribityllumazine by an exogenous nucleophile and deprotonation of the acidic C7 methyl group (pKa ≈ 8) of the acceptor (12+12→16+17).56–60 Subsequent nucleophilic attack by the C7 methylene group of the acceptor at C6 of the donor is then the first step to formation of compound Q (16+17→18). Next, tautomerization and elimination of the activating nucleophile along with deprotonation of the acceptor C6 methyl (18→19→20) precedes nucleophilic attack at C7 of the donor to form a tautomer of compound Q (20→21). This reaction sequence is consistent with a primary deuterium isotope effect of approximately 5 on V/K when the C6 methyl of 12 is deuterated.61 This mechanism is also supported by the observation that the N-terminal fragment of E. coli riboflavin synthase, which bears the acceptor binding site,44 specifically binds the exomethylene anion of 6,7-dimethyl-8-ribityllumazine (16) that results from deprotonation of the C7 methyl of 12.59 Subsequent aromatization of 22/21 and elimination of 14 produces riboflavin (13) thereby completing the catalytic cycle.
A key objection to this hypothesis, however, is that structural analyses of the active site have failed to identify a suitable activating nucleophile.44,62,63 Therefore, an alternative mechanism was proposed59 that involves an initial Cannizzaro-type disproportionation of the acceptor and donor dimethyl ribityllumazines following formation of the exomethylene anion (16+12→23+25, see Scheme 3B). Tautomerization of the resulting intermediate (23→24) generates a diene-dienophile pair that can subsequently undergo a Diels-Alder reaction to produce compound Q (24+25→22). However, when this proposal was investigated computationally using density functional theory calculations, the transition state for the hydride transfer step (16+12→23+25) was found to be considerably higher in energy compared to that involving nucleophilic activation.60 Furthermore, no pericyclic transition state could be identified in the gas phase calculations using a CPCM solvation model, and instead only a stepwise mechanism was found involving a zwitterionic intermediate.60 As in the case of macrophomate synthase, these computations have thus cast doubt on the possibility that riboflavin synthase possesses Diels-Alderase activity.
The computational report also analyzed a third mechanistic possibility, which is abbreviated in Scheme 3C.60 This catalytic cycle likewise involves formation of a diene-dienophile pair (12+16H→12+26); however, the subsequent [4 + 2]-cycloaddition to form compound Q is stepwise rather than concerted (12+26→27→22). The computed free energy diagram for this reaction coordinate identified two transition states of roughly equal energy governing the first and second C—C bond formations. Furthermore, this pathway was not only lower in energy than that involving the hydride transfer and Diels-Alder reactions but also the pathway involving activation by an exogenous nucleophile.60
One drawback of this third mechanism, however, is that it does not fully explain the large primary deuterium isotope effect associated with deuteration of the C6 methyl group.60 The authors did not report the free energy of the transition state governing deprotonation of the C6 methyl group, which occurs prior to formation of the first C—C bond in a subsequent step. Therefore, unless the transition state for deprotonation is particularly energetic or deprotonation takes place in concert with C—C bond formation during the actual enzymatic reaction, only a much smaller α-secondary deuterium V/K isotope effect would have been expected.64–66 In any case, while a variation of the stepwise [4 + 2]-cycloaddition in Scheme 3 (bottom row) may presently be the most attractive for riboflavin synthase, questions still abound and await clarification through further experimental investigation.
4 LovB/LovC and the Biosynthesis of Statins
The statins are a class of compounds that inhibit hydroxymethylglutaryl coenzyme A (HMG-CoA) reductase and have become important drugs in the treatment of hyperlipi-demia as well as the prevention of cardiovascular disease (see Tobert67 for an excellent overview). The first two statins were discovered by Endo and coworkers as fungal natural products and have come to be known as mevastatin (or compactin)68 and lovastatin (also known as monacolin K or mevinolin, 28 see Scheme 4).69 Since their discovery, a number of synthetic and semisynthetic HMG-CoA reductase inhibitors have been identified; however, all of the natural product forms are characterized by a hexahydronaphthalene ring system that has been hypothesized to result from a biological [4 + 2]-cycloaddition.70,71 Since its initial discovery in Monascus ruber,69,72 lovastatin has also been identified in several other fungal species such as Aspergillus terreus73 and Pleurotus ostreatus.74 Lovastatin has since become the working system for understanding the biosynthesis of this clinically useful class of natural products and the possible involvement of a Diels-Alderase therein.
Scheme 4.
Biosynthetic pathway for lovastatin.
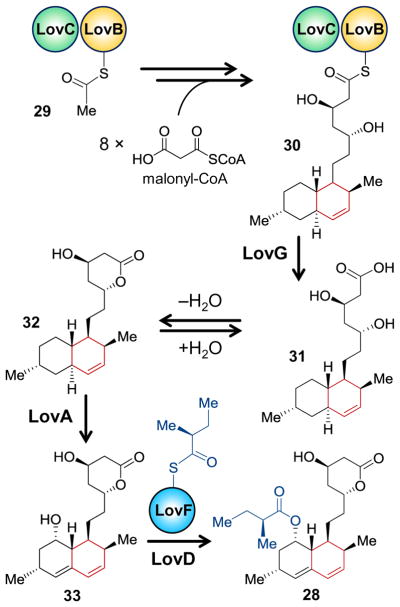
Lovastatin is a polyketide natural product generated via the linkage of a large non-aketide fragment containing the hexahydronaphthalene group with the diketide 2-methylbutyrate.70,71,75–79 The biosynthetic gene cluster for lovastatin was first described in Aspergillus terreus and found to encode two type I polyketide synthases dubbed LovB and LovF, which are responsible for the nonaketide and diketide fragments, respectively.80,81 LovB (also known as lovastatin nonaketide synthase or LNKS) works iteratively in a complex with the type II polyketide synthase LovC, which is an enoyl reductase also encoded in the cluster, to produce the intermediate dihydromonacolin L bound to LovB as a thioester (30, see Scheme 4).81–84 Dihydromonacolin L (31/32) is next hydrolyzed from the LovB-acyl carrier module in a reaction catalyzed by LovG.85 The freed intermediate is subsequently processed to the hexahydronapthalene-containing monacolin J (33) via post-PKS tailoring steps prior to coupling with the LovF-bound 2-methylbutyryl thioester to form lovastatin (33→28).81,86,87 The observation that 31 already possesses the cyclohexene-containing trans-decalin ring system strongly suggested that if a biological Diels-Alder reaction takes place during the biosynthetic pathway, then it would be among the nearly 40 chemical transformations catalyzed by the LovB/LovC complex.
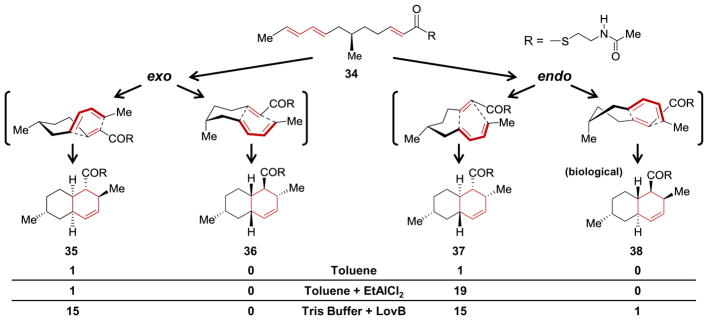
Evidence for the involvement of [4 + 2]-carbocyclase activity was provided even before the discovery of LovB/LovC. When Witter and Vederas prepared the N-acetylcysteamine hexaketide analog 34 of the predicted intermediate (i.e., 34 with R = LovB), they found that it cyclized when heated in toluene to give a one-to-one mixture of the cis and trans stereoisomers 35 and 37, respectively (see Figure 2).88 Furthermore, in the presence of a Lewis acid (i.e., EtAlCl2) the reaction was accelerated, and this ratio changed to favor the trans stereoisomer by 19 to 1, though neither of these product isomers matched the stereochemistry of the trans-decalin ring observed in dihydromonacolin L (compare 31 & 38).88 However, when Vederas and coworkers incubated 34 in Tris buffer with purified LovB, they again observed an increase in the rate of cyclization but this time with formation of the biological trans form 38, despite being the minor product (i.e., 38 : 37 : 35 ratios of 1 : 15 : 15).89 While the presence of a Lewis acid was also noted to significantly increase the rate of cyclization,88 these observations supported the hypothesis that LovB played a role in directing the stereochemistry of the [4 + 2]-cycloaddition if not also increasing the rate of cyclization.
Figure 2.
The hexaketide 34 is an analog of the predicted reactant for the [4 + 2]-cyclization during biosynthesis of dihydromonacolin L bound to LovB (30, see Scheme 4). This hex-aketide can undergo an intramolecular [4 + 2]-cycloaddition to produce four diastereomers in different relative proportions depending on the reaction conditions as shown.88,89
Further mechanistic investigation of the [4 + 2]-cycloaddition during the biosynthesis of dihydromonacolin L has been hampered by the multifunctionality of the LovB/LovC complex. Nevertheless, this polyketide megasynthase remains an active area of research with an emphasis on parsing and understanding the individual reactions and their control so that they can be adapted to the production of new pharmaceuticals and polyketides in general.90–92 As work proceeds, additional insights into the mechanism of the cycloaddition are sure to be revealed. In the meantime, however, new discoveries in the biosynthesis of spirotetramates (see below), which are also characterized by a trans-decalin ring system, are corroborating the hypothesis, albeit indirectly, that the [4 + 2]-cycloaddition during LovB/LovC catalysis may indeed be an enzyme catalyzed Diels-Alder reaction.
5 Solanapyrone synthase
The solanapyrones A through E represent a family of phytotoxins produced by the phytopathogenic fungi Alternaria solani and Ascochyta rabiei, which cause early blight disease in potato, tomato and chickpea.93–97 The solanapyrones are polyketides each characterized by a substituted pyrone connected to either a cis (A–C) or trans (D & E) decalin (see Scheme 5) and are thus structurally reminiscent of the lovastatin, spirotetronate and spirotetramate natural products. Moreover, a number of synthetic approaches to the solanapyrones have been reported, and each takes advantage of an intramolecular [4 + 2]-cycloaddition to generate the decalin moiety.94,98–100 For these reasons, the solanapyrone biosynthetic pathway has long been recognized as potentially harboring a Diels-Alderase.
Scheme 5.
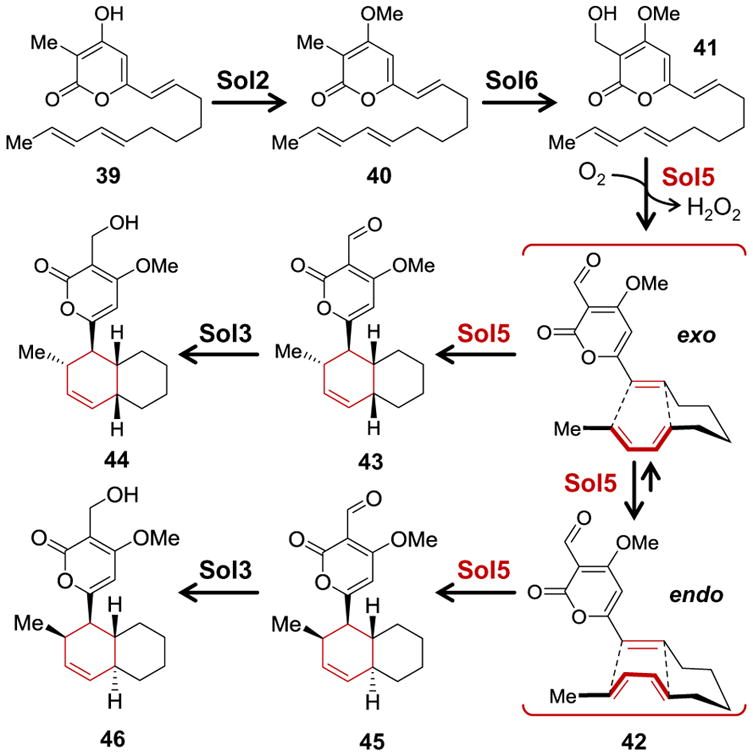
Biosynthetic pathway for solanapyrones A (43), B (44), D (45) and E (46). Desmethylprosolanapyrone I (39) is constructed from 8 acetate units and a methyl from SAM under the action of the polyketide synthase Sol1.
The solanapyrones are octaketides,101 and the sol gene cluster for their biosynthesis has been identified in Alternaria solani and Ascochyta rabiei.102,103 Biosynthesis begins with production of desmethylprosolanapyrone I (39) from acetate in a series of reactions catalyzed by the type I polyketide synthase Sol1. Post-PKS processing of 39 by the O-methyltransferase Sol2 results in prosolanapyrone I (40), which is monooxygenated by the cytochrome P450 enzyme Sol6 to yield the alcohol prosolanapyrone II (41, see Scheme 5). Prosolanapyrone II is relatively stable in aqueous solution undergoing nonenzymatic [4 + 2]-cycloaddition with a half-life of approximately 6 days at 30 °C.98,104 However, in the presence of molecular oxygen and the FAD-dependent oxidase Sol5, which is homologous to 6-hydroxy-nicotine oxidase,105 41 is converted to the aldehyde prosolanapyrone III (42) with hydrogen peroxide as a byproduct.102,106 Sol5 (solanapyrone synthase) also appears to accelerate the intramolecular [4 + 2]-cycloaddition of 42 to produce a mixture of solanapyrone A (43) and solanapyrone D (45).33,95,102,106,107 In the final step of the pathway, the dehydrogenase Sol3 reduces the aldehyde functionality of 43 and 45 to provide 44 and 46, respectively.102
Unlike prosolanapyrone II, the nonenyzamatic [4 + 2]-cycloaddition of prosolanapyrone III in aqueous solution is much more rapid with a half-life of around 1 to 2 h.98,104 This reaction results in a roughly 20- to 30-fold preference for the trans-decalin ring system of solanapyrone D (45) rather than the cis-decalin ring system of solanapyrone A (43).98,104 While this agrees with the well-known endo-preference for Diels-Alder reactions in aqueous media and is also seen with the homologs to the lovastatin precursor,88,108 it is not consistent with the predominance of solanapyrone A over D in biological extracts.96 However, Oikawa, Fujii and coworkers discovered that in the presence of Alternaria solani cell extracts or purified solanapyrone synthase, prosolanapyrone II and III cyclize with exo-preference to favor the cis-decalin ring system (i.e., 43) by 6 to 7-fold over the trans-decalin.98,102,104 As in the case of LovB/LovC, this stereochemical evidence implicated a role for the enzyme in controlling the stereochemistry of the cyclization. Furthermore, though the rate at which 42 cyclizes is only modestly increased in the presence of solanapyrone synthase, this rate does appear to increase in direct proportion to the concentration of the cellular extracts consistent with an active role in catalysis.104
Similar to LovB/LovC, solanapyrone synthase catalyzes a priming reaction to activate its substrate (42) for an intramolecular [4 + 2]-cycloaddition over which it also exerts some degree of stereochemical control. Whereas LovB/LovC is a polyketide megasynthase directly responsible for construction of the reactive triene, solanapyrone synthase polarizes the dienophile by bringing it into conjugation with a carbonyl. This can then serve to lower the energy of the LUMO for the dienophile and thereby promote a pericyclic cycloaddition. Given the modest increases in rate of cyclization caused by solanapyrone synthase, however, it is still not entirely clear whether the enzyme further activates the diene-dienophile pair for reaction or serves primarily as a template to prevent alternative stereochemical outcomes. Recent work on the spirotetronates/tetramates suggests the former hypothesis (see below); however, additional research into the mechanism of cyclization and catalysis by solanapyrone synthase is likely necessary before a definitive answer can be provided.
6 Thiopeptide Lynchpins
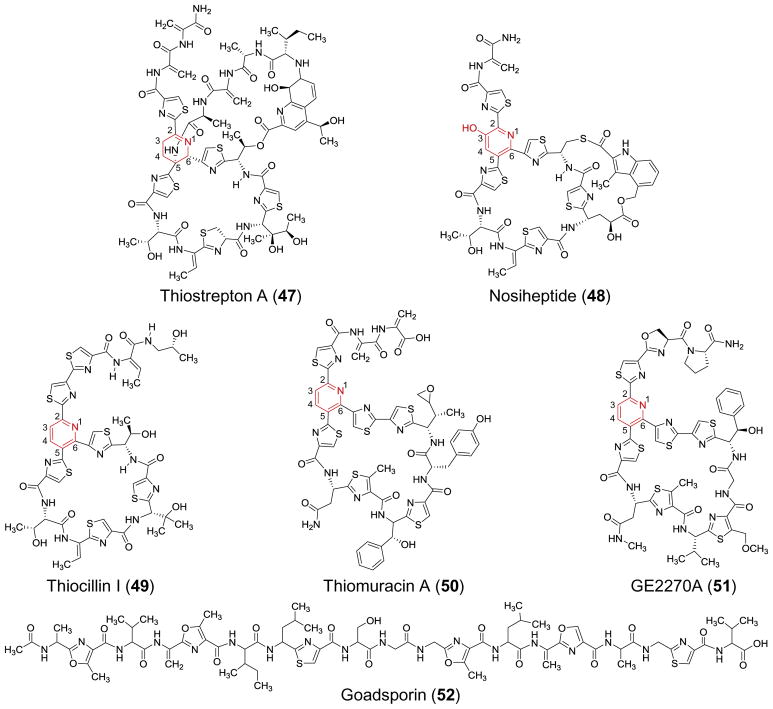
The thiopeptides (also known as thiazolyl peptides) are ribosomally synthesized, post-translationally modified peptides (RiPPs) frequently characterized by a macrocyclic structure internally linked via a lynchpin nitrogen-containing heterocycle (see Figure 3).109–113 Formation of the lynchpin is one of several features of thiopeptide biosynthesis to have drawn attention, because it has been hypothesized to result from a hetero-Diels-Alder reaction.
Figure 3.
Examples of thiopeptides that have been used to study lynchpin biosynthesis.
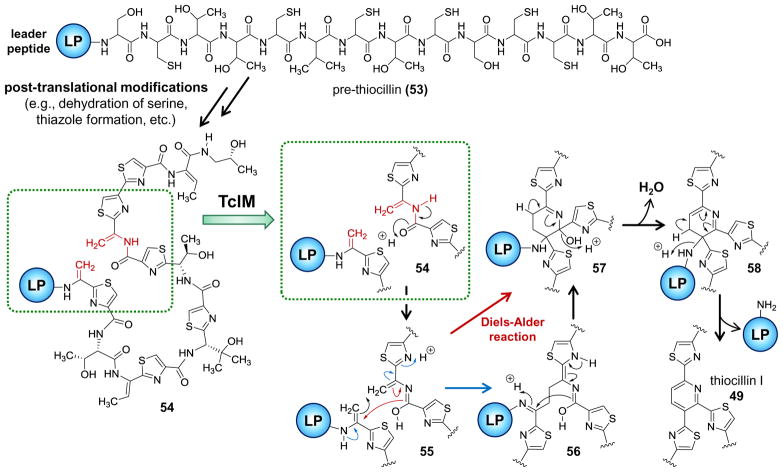
Collectively, the thiopeptides are produced by many Gram-positive bacteria and exhibit antibacterial activity resulting from their ability to inhibit protein synthesis.111 These natural products are biosynthesized via a series of post-translational modifications of a prepeptide precursor that is also encoded in the biosynthetic gene cluster.109,110,114 Following ribosomal translation, the prepeptide consists of an N-terminal 34 to 55 residue leader peptide followed by a 12 to 17 residue structural peptide at the C-terminus,109 which is rich in serine and cysteine residues (e.g., see 53 in Scheme 6). Foremost among the post-translational modifications is the dehydration of serine residues to form dehydroalanine residues along with the conversion of peptide bonds to thiazole linkages via the cyclization of cysteine residues (e.g., 53→54 in Scheme 6).109,110,114 The modified structural peptide is then converted to a macrocycle with the introduction of the lynchpin and removal of the leader peptide.
Scheme 6.
Current mechanistic hypotheses for the reactions catalyzed by TclM as a paradigm for the biosynthesis of lynchpin heterocycles in macrocyclic thiopetide antibiotics.114,122
Early evidence for the involvement of Diels-Alder chemistry during the final, macrocyclization phase was provided by following the incorporation of isotope tracers into the dehydropiperidine and hydroxypyridine lynchpins of thiostrepton (47) and nosiheptide (48), respectively. Upon supplementing cultures of the nosiheptide producing strain Streptomyces actuosus with 13C- and 2H-labeled isotopologs of serine, Floss and coworkers were able to show that the C3 and C4 carbons of the hydroxypyridine derive from the β-carbons of two serine residues.115–117 Furthermore, the C6 carbon of the hydroxypyridine in 48 originates from the carboxylate of an adjacent cysteine.117 Analogous labeling patterns were also obtained for the dehydropiperidine motif of thiostrepton.118 Together these labeling studies indicated that the lynchpin motif is generated from the intact carbon skeletons of two molecules of serine and one cysteine. This led to the hypothesis that a concerted [4 + 2]-cycloaddition involving dehydroalanine residues may ultimately be responsible for the formation of the heterocyclic lynchpin.118,119
Further insight into the biogenesis of thiopeptide lynchpins came with the identification of the biosynthetic gene clusters.109,111 By comparing the gene clusters for thiocillin (49), thiostrepton (47), nosiheptide (48) and GE2270A (51), Walsh and coworkers identified a collection of homologous genes that were not readily annotatable based on sequence alone and did not have a representative homolog in the gene cluster for the acyclic thiopeptide goadsporin (52, see Figure 3).120,121 Based on this observation, the authors identified the gene tclM as a prime candidate for encoding the enzyme responsible for introducing the pyridine lynchpin in thiocillin. When tclM was knocked out in the Bacillus cereus producing strain, only acyclic variants of thiocillin were recovered from cultures.120 Furthermore, reintroduction of tclM into the knockouts led to recovery of macrocyclic thiocillin with the aromatized pyridine lynchpin fully intact.120 Though indirect, these studies were the first to implicate TclM and its homologs in other gene clusters as [4 + 2]-heterocyclases.
Direct evidence for the [4 + 2]-heterocyclase activity of TclM was later provided when the Bowers laboratory heterologously overexpressed and purified the tclM gene product from E. coli.122 When isolated TclM was coincubated in vitro with an acyclic precursor to a thiocillin analog, not only was macrocyclization effected, but the pyridine lynchpin was introduced in its fully aromatized form and the leader peptide was cleaved. A similar conclusion was reached by van der Donk and coworkers when they partially reconstituted in vitro the biosynthetic pathway for thiomuracin (50) from Thermobispora bispora.123 Like thiocillin (49), thiomuracin also contains a pyridine lynchpin that is introduced in its fully aromatized form from the modified, acyclic prepetide precursor in a reaction catalyzed by TbtD, which is a homolog of TclM. Like TclM, TbtD is not only responsible for the macrocyclization of thiomuracin and lynchpin formation but also cleavage of the leader peptide.123 Moreover, TbtD could be replaced by the homologous PbtD gene product from the biosynthetic gene cluster for GE2270A (51), another pyridine-containing thiopeptide from Planobispora rosea ATCC3733.123,124 Together these works firmly established TclM, TbtD, PbtD and their homologs as not only [4 + 2]-heterocyclases but also the enzymes responsible for cleavage of the leader peptide and aromatization of pyridine lynchpins.
While the enzymes responsible for biogenesis of pyridine lynchpins have been identified, many questions regarding their enzymology remain. Of particular interest is the mechanism of cyclization that may very well involve a pericyclic cycloaddition as shown in Scheme 6. In this case, the cyclization would likely first involve the tautomerization of an amide linkage to a conjugated iminol (54→55) prior to concerted [4 + 2]-cycloaddition with an opposing dehydroalanine side chain (55→57). Subsequent dehydration (57→58) and elimination of the leader peptide (58→49) completes the aromatization and construction of the linkage.122 However, an alternative mechanism is possible whereby the cycloaddition takes place in a stepwise fashion with an adjacent thiazole moiety serving as an activating electron sink (55→56→57). Additionally, only enzymes responsible for installing pyridine lynchpins have so far been considered in any detail, and it remains to confirm that similar chemistry is at work in the biogenesis of dehydropyridine linkages as are found in thiostrepton125 and siomycin126,127 as well as hydroxypyridine linkages in the case of nosiheptide.128 Taken together, the thiopeptides represent an exciting area in the study of natural product biosynthesis and possibly the first examples of a biosynthetic pathway involving enzymes naturally selected to catalyze hetero-Diels-Alder reactions.
7 SpnF
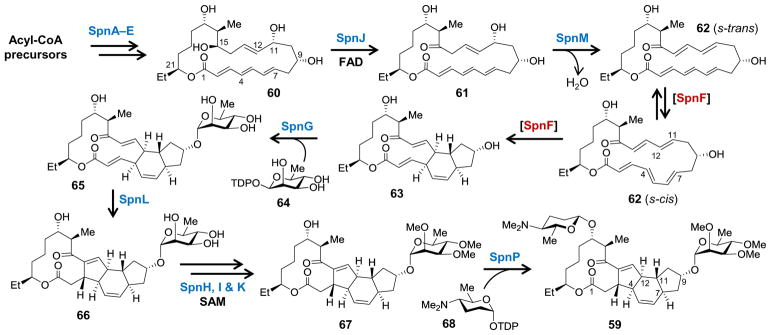
Spinosyn A (59) is a polyketide macrolide produced by the actinomycete bacterium Saccaropolyspora spinosa and represents the main active component of spinosad, which is used in various commercial insecticides.129 The aglycone core of spinosyn A is a tetracyclic macrolactone containing a perhydro-as-indacene ring system and is glycosylated at the C9 and C17 positions with the atypical sugars tri-O-methyl-D-rhamnose and D(+)-forosamine, respectively.129 Considerable attention has been focused on how the perhydro-as-indacene is constructed, because its central cyclohexene ring suggests the involvement of a biological Diels-Alder reaction. In fact, nonenzymatic total syntheses of spinosyn A have also relied on an intramolecular [4 + 2]-cycloaddition to install the cyclohexene,130,131 though other approaches have been utilized as well.132,133 Studies of the biosynthetic pathway for spinosyn A led to the identification of the spn gene cluster (see Figure 4)134 as well as the discovery of the enzyme SpnF, which was the first naturally selected enzyme found to specifically catalyze a [4 + 2]-cycloaddition.135,136
Figure 4.
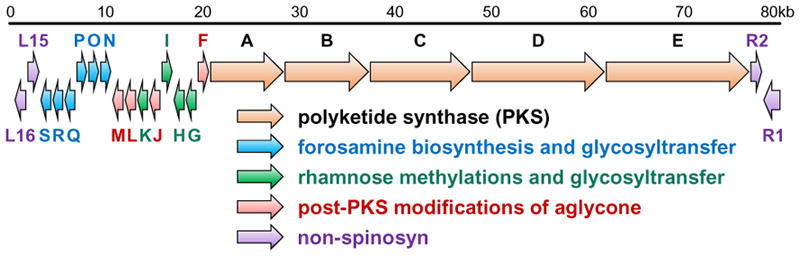
Biosynthetic gene cluster for spinosyn A from Saccaropolyspora spinosa.
The carbon backbone of spinosyn A is biosynthesized under the action of modular type I polyketide synthases (SpnA–E) encoded by the spn biosynthetic gene cluster (see Figure 5).134,137 Intramolecular transesterification of the nascent ACP-bound polyketide results in elimination of the 22-member macrolactone 60 that is subsequently oxidized by the FAD-dependent SpnJ to form the ketone 61.138 A second post-PKS tailoring step catalyzed by SpnM results in the 1,4-dehydration of 61 to produce species 62, which is susceptible to a transannular [4 + 2]-cycloaddition that results in the tricyclic product 63 with the same stereochemistry observed in the final product 59.135 The rate of the cycloaddition is independent of SpnM concentration indicating that it can take place nonenzymatically in aqueous solution with a half-time of approximately 25 min thereby excluding the possibility of SpnM playing a direct role in the cyclization. However, when SpnF was added to the reaction mixture, the rate at which 62 underwent cyclization to 63 was significantly increased and directly correlated with the SpnF concentration so that a rate enhancement (kcat/knon) of 500-fold could be measured. 135 The tricyclic product then undergoes rhamnosylation by SpnG (63→65) followed by a second cyclization reaction catalyzed by SpnL to complete the perhydro-as-indacene ring system (65→66) prior to SAM-dependent methylation (66→67) and the final glycosylation step by SpnP that ultimately results in the spinosyn A product (67→59).135,139–141
Figure 5.
Biosynthetic pathway for spinosyn A.
SpnF was unique at the time of its discovery, because all previously reported [4 + 2]-carbocyclases such as macrophomate synthase, LovB, salonapyrone synthase and riboflavin synthase are multifunctional. As discussed above, each of these latter enzymes also catalyze what amount to priming reactions to introduce an activated diene-dienophile pair that can subsequently undergo cyclization. This complicated efforts to deconvolute the roles the enzymes play in the actual cyclization reactions. In the case of spinosyn A biosynthesis, however, the priming reaction (61→62) is catalyzed by a separate enzyme (i.e., SpnM) such that acceleration of the [4 + 2]-cycloaddition (62→63) could be clearly attributed to SpnF. One question that remains open, however, is why SpnF is present in the first place, given that nonenzymatic cyclization can hardly be called slow in its absence and proceeds with the same stereochemistry. One hypothesis is that despite separable activities, SpnM and SpnF may work synergistically to prevent both unproductive side reactions during dehydration of 61 as well as attack on 62 by spectator nucleophiles.140
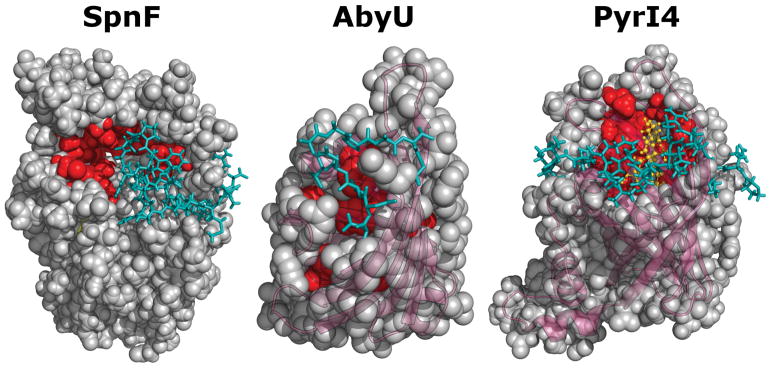
Experimental investigations of the SpnF mechanism of catalysis are currently underway with the first reported being the elucidation of the X-ray crystal structure (see Figure 7). The active site of SpnF is nestled in a β-sheet surrounded by α-helices that also encloses a molecule of S-adenosyl-homocysteine (SAH).142 The presence of SAH is consistent with the annotation of SpnF as an S-adenosyl-L-methionine (i.e., SAM) dependent methyltransferase.134 While SpnF is not known to catalyze any methyltransferase reaction nor needs SAM or SAH present in the reaction buffer to catalyze the cyclization of 62, site-directed mutagenesis of the enzyme has shown that SAM/SAH may serve as a necessary structural feature.135,142 The crystal structure does not have the SpnF substrate or an analog thereof bound; however, docking calculations indicate that it binds into the active site primarily via hydrophobic contacts and is not physically in contact with the bound SAH.142 Furthermore, the residue Thr196 may serve as a hydrogen-bond donor to the C15 carbonyl thereby activating the C11=C12 dienophile.142 Finally, a flexible loop composed of residues Arg181–Leu202 was discovered that can serve as a lid to fully enclose the bound substrate in the active site and trap it in a conformation conducive to an intramolecular [4 + 2]-cycloaddition (see Figure 7).142 Such an induced lidding/fit mechanism of constraining the bound substrate in a productive orientation has also been hypothesized based on the crystal structures of AbyU143 and PyrI4144 (see below) suggesting a more general paradigm in the operation of [4 + 2]-carbocyclases.
Figure 7.
Structures of the monomeric units of SpnF (PDB: 4PNE),142 AbyU (PDB: 5DYQ)143 and PyrI4 (PDB: 5BU3)144 showing the lidding regions (in teal) that occlude the active sites (in red) in the closed complexes. The structure of PyrI4 has the product species 97 bound in the active site and is rendered in gold. The β-barrel structure of AbyU and PyrI4 is highlighted in purple.
A number of computational studies on the cyclization of 62 have also been reported; however, they have focused primarily on models of the nonenzymatic reaction. The first of these studies was reported by Hess and Smentek and considered density functional theory calculations on a simplified model of the macrolactone 62 in the gas-phase.145 The investigators were able to locate a transition state in which the formation of the C7—C11 bond (bond length: 1.912 Å) is significantly more advanced compared to the C4—C12 bond (length: 2.997 Å). Intrinsic reaction coordinate (IRC) calculations likewise failed to identify a second transition state connecting the reactant and product states; however, what appear to be two inflection points forming a caldera-like region following the single transition state were also observed in the IRC plot.145 A later study by Gordeev and Ananikov helped to extend these results by including H-bonding interactions between modeled amino acids and the C9 and C17 hydroxyls as well as the C15 carbonyl.146 Their results also implicated a single transition state with respect to the [4 + 2]-cycloaddition while at the same time highlighting how both H-bonding and conformational preorganization of the substrate in the active site could account for the observed rate enhancement by SpnF. Together, these results implicated a highly asynchronous Diels-Alder reaction for the cyclization and provided a rationale for SpnF catalysis consistent with hypotheses based on crystal structures of other [4 + 2]-carbocyclases.142–144
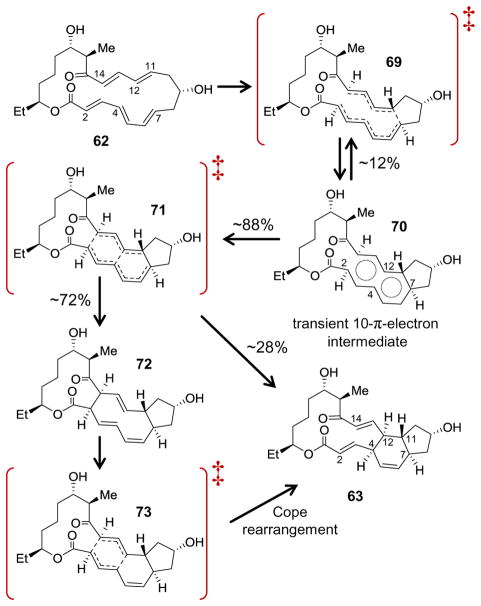
More recently, the Houk and Singleton laboratories released a detailed analysis of the nonenzymatic reaction that has emphasized the complications involved in trying to describe a reaction mechanism in terms of one-size-fits-all eponyms. Using density functional theory computations, these authors obtained results similar to those of Hess and Smentek;145 however, they also observed that the reaction coordinate leading away from the transition state (i.e., potential energy saddle point) in the forward direction could bifurcate resulting in either the observed product (63) or a [6 + 4]-cycloaddition product (72, see Scheme 7).30 This saddle point was therefore designated as an ambimodal transition state (represented by 69 in Scheme 7), because it can lead directly to two different products without an intermediary local minimum in potential energy.147–152 Furthermore, the steepest path of decent from the transition state led to 72, which calculations indicated30 would readily undergo a Cope rearrangement to generate 63 thereby explaining why the former compound is not observed experimentally.135,140 At face value, these computations suggested that the nonenzymatic reaction proceeds by both concerted and stepwise pathways.
Scheme 7.
Mechanism proposed for the nonenzymatic cyclization of 62.
Recognizing the deficiencies in interpreting reaction coordinates under conventional transition state theory alone, Houk, Singleton and coworkers also examined the reaction using molecular dynamics (MD). Simulations initiated forward from the ambimodal transition state (69) were found to result in one of three end points.30 Most MD trajectories (i.e., 63%) resulted in the [6 + 4] product while the remaining trajectories either led to the [4 + 2] product (25%) or completed formation of the C7—C11 bond before passing back through the ambimodal transition state to form the starting compound 62 (12%). Of equal interest was the observation that the MD trajectories remained for multiple bond vibrations in a region of configuration space about the caldera that had been observed in the previous IRC profiles.30,145 These configurations, which are represented by 70 in Scheme 7, were characterized by essentially complete formation of the C7—C11 bond and partial formation of both the C4—C12 and C2—C14 bonds (ca. 3.0 Å each). 30 This implied a (dynamical) bottleneck for trajectories that proceed beyond the ambimodal transition state (69) and led the authors to apply variational transition state theory (VTST) to characterize the free energy landscape along the minimum energy path (MEP) for the reaction.
In contrast to conventional transition state theory calculations, VTST does not restrict the surface corresponding to the transition state to cross the (potential energy) maximum along the reaction coordinate connecting reactant and product states.153,154 This permits identification of free energy maxima along the MEP, which are referred to as variational transition states. Two such states were found: one approximating the ambimodal transition state (69) and a second corresponding to the dynamical bottleneck (represented by 71 in Scheme 7) with a local minimum in the free energy residing in between. In other words, the region where trajectories were briefly trapped in the MD simulations (70) corresponds to a formal intermediate in what the authors described as a 10-π-electron aromatic state.30
Whether the cyclization of 62 proceeds through a [6 + 4]-cycloadduct (72) or not (at the time of this writing there is no experimental evidence for or against a [6 + 4]-cycloadduct), the computational results30 imply that the nonenzymatic [4 + 2]-cycloaddition of 62 is not concerted with respect to its free energy profile. Therefore, under the conservative definition previously discussed, this reaction would not be considered a Diels-Alder reaction, and if SpnF catalysis proceeds in a similar fashion, it would not be a Diels-Alderase. However, the reaction defies classification in terms of the standard alternatives (i.e., a biradical or zwitterionic intermediate) ultimately demonstrating that the chemistry underlying the cycloaddition as well as the enzymology of SpnF and other [4 + 2]-cyclases may be more complex and nuanced than previously appreciated.
8 Spirotetronates and Spirotetramates
8.1 Overview
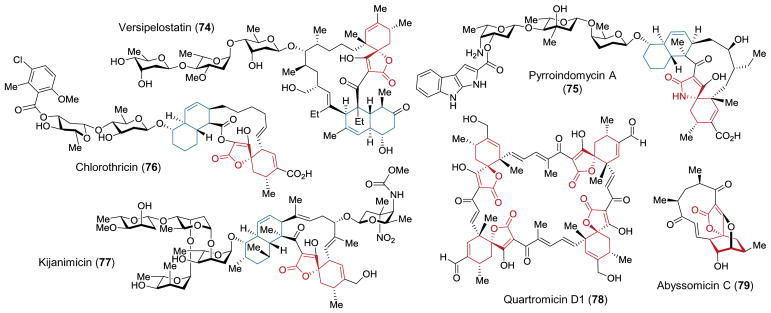
The tetronate antibiotics have been the focus of many recent biosynthetic research programs on account of their unusual polycyclic structures as well as their value as potential antimicrobial and antiproliferative agents.155–157 A subclass of this family of natural products has drawn particular interest, because the tetronate or tetramate namesake appears to have undergone an intramolecular Diels-Alder reaction with a 1,3-diene to form a cyclohexene ring with a tertiary carbon joint.12,157 Examples of such spirotetronates and spirotetramates are shown in Figure 6 and include versipelostatin (74),158–161 pyrroindomycin (75),162–166 chlorothricin (76),167–175 kijanimicin (77),176–181 the quartromicins (78),182–186 the abyssomicins (79),187–192 tetrocarcin,193–198 the lobophorins,199–205 nomimicin,206 maklamicin,207–209 and tetronothiodin.210–214 Furthermore, several of these compounds also possess a second, decalin ring system, the formation of which is highly reminiscent of the reactions catalyzed by LovB and solanapyrone synthase (see above). Therefore, the pathways responsible for the biosynthesis of the spirotetronates and spirotetramates are unique, because many may involve not one but two enzyme catalyzed Diels-Alder reactions.
Figure 6.
Selected examples of spirotetronate and spirotetramate natural products. The spirotetronate/tetramate heterocycles are highlighted in red, whereas the decalin ring systems are highlighted in blue.
The tetronate and tetramate natural products as a whole are polyketides typically produced by the action of modular type I polyketide synthases.155–157 The tetronate (i.e., 4-hydroxy-[5H]furan-2-one) moiety is generated during elimination of the nascent polyketide from the acyl carrier protein (ACP) (see Scheme 8).215 This reaction involves a transesterification between the ACP-bound polyketide and an ACP-bound D-glycerate thioester (80+81→82, X, Z = O) followed by a Dieckmann cyclization that releases the tetronate (82→83) from the second ACP.215 The tetronate formed is then acetylated in a post-PKS tailoring step (83→84), and elimination of acetate produces a methylene tetronate (84→85).186,216 The resulting methylene can then serve as the dienophile in an intramolecular Diels-Alder reaction with the polyunsaturated polyketide (85→86). Likewise, an ACP-bound cysteine (81, X = NH, Z = S) can substitute for the ACP-glycerate resulting in an amidation reaction prior to Dieckmann cyclization and thus elimination of a thiolated tetramate from the ACP.166 Formation of the methylene tetramate may follow an analogous path as described for the methylene tetronate. Interestingly, abiological syntheses of spirotetronate and spirotetramate antibiotics have expressly utilized a single [4 + 2]-cycloaddition to construct the whole spirotetronate or spirotetramate ring system217–219 and thus suggest that a similar, enzyme catalyzed reaction may also be at work in the corresponding biosynthetic pathways.
Scheme 8.
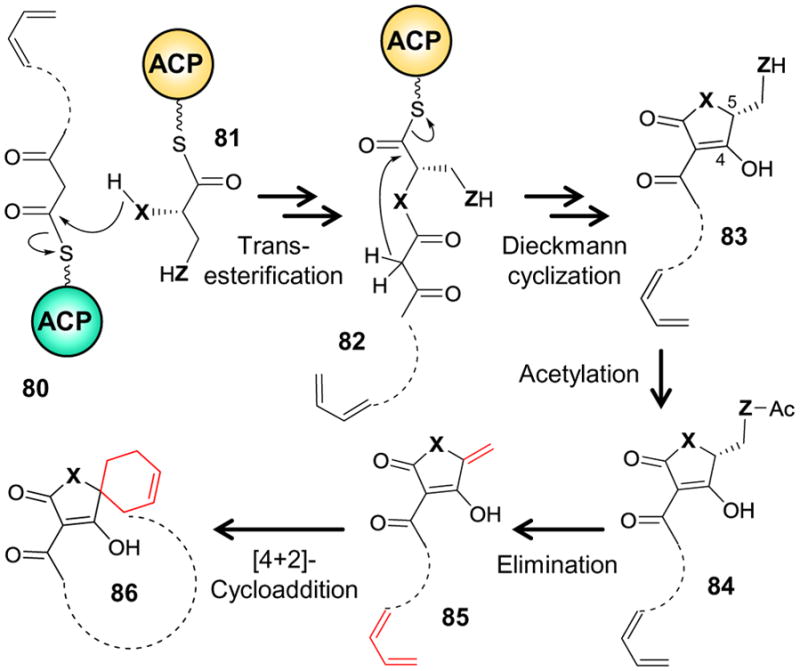
Biogenesis of the tetronate (X = O, Z = O) and tetramate (X = NH, Z = S) functionalities prior to cyclization to form the spirotetronate and spirotetramate ring systems, respectively.
8.2 Spirotetronates: VstJ and AbyU
One of the first investigations of this hypothesis focused on the biosynthesis of the spirotetronate versipelostatin (74, see Figure 6), which is produced by Streptomyces versipellis 4083-SVS6.158–160 Genome screening efforts by the Kuzuyama laboratory led to the discovery of the putative biosynthetic gene cluster vst, which was then confirmed when the authors transformed a nonproducing strain of Streptomyces albus into a producer via integration of the vst cluster into the S. albus genome.220 By comparing the vst cluster with previously reported spirotetronate biosynthetic gene clusters, these investigators went on to identify a small, highly conserved open reading frame of unknown function dubbed vstJ.220 A second S. albus transformant lacking only the vstJ gene was subsequently found to no longer produce versipelostatin and instead accumulated the acyclic intermediate 87 (see Scheme 9), which possesses both of the predicted diene and dienophile components.220 Moreover, intermediate 87 did not undergo nonenzymatic cyclization in vitro to form the spirotetronate 88 unless it was coincubated with VstJ, which had been heterologously overexpressed and purified as a His6-tagged conjugate from E. coli. 220
Scheme 9.

Reaction catalyzed by VstJ in the biosynthesis of versipelostatin (74).
Based on these observations, Kuzuyama and coworkers concluded that VstJ is a standalone [4 + 2]-carbocyclase tailoring enzyme that catalyzes the 87→88 reaction shown in Scheme 9. However, the discovery of VstJ was also important because it is absolutely required for the cycloaddition reaction unlike SpnF, solanapyrone synthase and LovB, where the cyclizations can also proceed nonenzymatically at appreciable rates (see previous discussion). This initial characterization also led to the annotation of several homologous genes such as chlL,174 tcaU4,198 lobU2202 and qmnH,185 as well as the newly identified open reading frames kijU/kijcyc181,220 and abyU/abycyc191,220 in the gene clusters for kijanimicin and abyssomicin C , respectively, as encoding putative [4 + 2]-carbocyclases involved in the biosynthesis of other spirotetronate-containing natural products.
This conjecture has been tested by careful study of the abyssomicin C (79, see Figure 6) biosynthetic pathway and in particular its VstJ homolog AbyU by Race and coworkers.143 These investigators cloned abyU from the abyssomicin C producing strain Verrucosispora maris AB-18-032 and heterologously overexpressed and purified the recombinant His6-tagged enzyme from E. coli. The enzyme was hypothesized to catalyze the cyclization of 89 to 91, and when the purified AbyU was coincubated with the putative substrate analog 90, cyclization to the spirotetronate 92 was indeed observed (see Scheme 10).143 The analog 90 can also undergo a nonenzymatic intramolecular [4 + 2]-cycloaddition to 92, and this has been utilized in synthetic preparations of abyssomicins.217,218 Race and coworkers were thus able to take advantage of this property to assign a rate enhancement (i.e., kcat/knon) of approximately 4 × 104 to AbyU with 90.143 Moreover, the synthetic precursor to 90 (i.e., 93) is not susceptible to measurable nonenzymatic cyclization presumably due to an increase in the energy of its LUMO versus that of 90 when the conjugated carbonyl is replaced with an alcohol. However, in the presence of AbyU, the 93 racemate is converted to two products consistent with the diastereomeric mixture of the corresponding spirotetronates (i.e., 94).143 These observations thus further established the class of VstJ homologous enzymes as [4 + 2]-carbocyclases responsible for spirotetronate formation.
Scheme 10.

Reactions catalyzed by AbyU.
8.3 Spirotetramates and trans-Decalins: PyrI4 and PyrE3
In tandem with the work on spirotetronates, the Liu laboratory published a detailed investigation of the biological origin of both the spirotetramate and transdecalin moieties of pyrroindomycin A (75), which is an antibiotic produced by Streptomyces rugosporus.162,163,166 Liu and coworkers began by using comparative genomics to identify PyrI4 as a homolog of VstJ implicating it the formation of the spirotetramate functionality.221 The PyrE3 gene product also drew attention given the absence of homologous genes in the clusters for abyssomicin C (79)191 and quartromicin D1 (78),185 both of which lack a decalin group (see Figure 6).221 A mutant strain of S. rugosporus without a functional copy of pyrI4 also failed to produce pyrroindomycin A or any identifiable intermediate in the pathway.221 Furthermore, knockout of the pyrE3 gene likewise did not result in the production of pyrroindomycin A, though in this case the linear tetramate 95 was found to accumulate instead.221 This led to the hypothesis that PyrE3 and PyrI4 are together responsible for the formation of the trans-decalin followed by the spirotetramate moieties in two sequential post-PKS tailoring reactions as shown in Scheme 11.
Scheme 11.

Sequential reactions catalyzed by PyrE3 and PyrI4 in the biosynthesis of pyrroindomycin A (75).
Confirmation of this hypothesis was provided when in vitro coincubation of 95 with heterologously expressed and purified PyrE3 resulted in the formation of the trans-decalin containing tetramate 96, while no reaction was observed between PyrI4 and 95.221 Similar to both 95 as well as the VstJ substrate 87, 96 was not susceptible to further cyclization to 97 unless PyrI4 was also included in the incubation.221 Having established PyrE3 and PyrI4 as sequential [4 + 2]-carbocyclases in the biosynthesis of pyrroindomycin, Liu and coworkers went on to demonstrate using a similar approach that the homologous enzymes ChlE3 and ChlL play respective roles in the chlorothricin (76) biosynthetic pathway.221 It should be noted that chlorothricin, unlike pyrroindomycin A, is a spirotetronate such that this work further extends the general class of VstJ homologs to also include those responsible for spirotetramate formation.
The enzyme PyrE3, and by extension ChlE3, is of particular note for two reasons. First, it catalyzes a reaction very similar to that of LovB (see above); however, unlike LovB, it is not a polyketide synthase and instead operates as a standalone [4 + 2]-carbocyclase. This provides evidence, albeit indirect, to suggest that LovB indeed also accelerates the [4 + 2]-cycloaddition during the biosynthesis of lovastatin—a hypothesis that has been previously difficult to test due to the multiple additional activities of this multienzyme complex. Second, PyrE3 is homologous to the flavin adenine dinucleotide (FAD)-dependent hydroxylases PgaE222,223 and OxyS224 from the biosynthetic pathways for angucycline and oxytetracycline, respectively.221 Consistent with this sequence homology, PyrE3 appears to require a bound FAD coenzyme for activity, despite the conversion of 95 to 96 being redox neutral and the observation that the enzyme does not require a redox cofactor such as NAD(P) for multiple turnovers.221 This suggests that the bound FAD is purely a structural, evolutionary remnant similar to SAM in the case of SpnF.135,142
8.4 Mechanistic studies
The studies discussed above established VstJ, AbyU and PyrI4 (and by extension their homologs) as the enzymes necessary for catalyzing the [4 + 2]-cycloadditions during spirotetronate/tetramate formation; however, they did not establish the mechanisms of catalysis. Currently only two studies have been published in this regard, which focus on protein structure elucidation and computational analyses. In the case of PyrI4, the crystal structure consists of a homodimer of two β-barrel cores each with a functionally essential lid-like N-terminal α-helix.144 Co-crystallization of PyrI4 and its substrate (96) resulted in a 1 : 1 structural complex with its product 97 alone bound in the β-barrel core (see Figure 7).144 This pointed to the site and configuration for the bound substrate, which when modeled computationally, implicated an exo-transition state for a concerted [4 + 2]-cycloaddition.144 The structure and characterization of several mutants also led to the identification of two H-bond networks in the proposed Michaelis complex that may serve to polarize the carbonyl and hydroxyl functionalities of the tetramate in order to make them better electron-withdrawing groups.144 Consequently, one mode of catalysis may involve decreasing the energy of the dienophile LUMO thereby facilitating a Diels-Alder reaction under normal demand.
Comparison of the PyrI4 crystal structures with and without 97 bound also demonstrated that the N-terminal α-helix is flexible and important for substrate binding as well as catalysis by essentially forming a partial lid over the active site (see Figure 7).144 The authors of this study thus suggested that a second mode of catalysis by PyrI4 involves constraint of the substrate in a conformation approximating the exo-transition state. Consistent with this hypothesis, disruption of the salt bridges necessary to stabilize the closed complex using site-directed mutagenesis led to enzyme inactivation thereby offering a mechanism to account for the reduction in entropy via enthalpic compensation upon forming the closed complex.144 Therefore, these observations suggest the hypothesis that PyrI4 catalyzes a Diels-Alder reaction by reducing both the enthalpy and entropy of activation for cyclization of the bound substrate.
The crystal structure of AbyU also demonstrates a homodimer of two β-barrel cores.143 Each β-barrel contains a hydrophobic active site with a flexible lid composed of residues Asp26–Gly36 (see Figure 7),143 which is reminiscent of the N-terminal α-helix of PyrI4.144 While a crystal structure of AbyU was obtained with HEPES rather than a substrate or product analog bound in the active site, docking simulations with the natural AbyU substrate (89) and product (91) revealed an active site tyrosine that may H-bond with the ring oxygens of the tetronate functionality as well as close proximity of both the C13 and C14 centers (3.6 Å) and the C10 and C15 centers (3.9 Å) of the substrate.143 This suggested that the active site is capable of stabilizing a pericyclic transition state, which was further supported by the identification of an asynchronous concerted reaction trajectory using QM/MM calculations.143 Unlike in the case of PyrI4, however, catalysis of the [4 + 2]-cycloaddition in the hydrophobic AbyU active site may rely more on preorganization of the substrate rather than reducing the enthalpy of activation.
At this time, the [4 + 2]-cyclases involved in the biosynthesis of spirotetronate/tetramates appear to be the best candidates for standalone Diels-Alderases. Current biochemical, structural and computational evidence is indeed highly suggestive of a single, pericyclic transition state along the reaction coordinate for both spirotetronate/tetramate as well as trans-decalin formation. However, additional thermochemical, computation and kinetic investigations will be required not only to carefully describe the reaction trajectories of these transformations but also to understand the mechanisms by which they are accelerated in the enzyme active sites.
9 Summary and Outlook
Despite the relatively sparse collection of [4 + 2]-cyclases that have been studied in isolation, a few general themes can be identified. In a number of cases, the cyclization follows an initial priming reaction that may be catalyzed by the cyclase itself (e.g., solanapy-rone synthase, LovB/LovC) or a separate enzyme (e.g., SpnM with SpnF being auxiliary or down-pathway) to generate a reactive intermediate that is predisposed towards nonenzymatic cyclization. This suggests that biosynthetic pathways may diversify to include [4 + 2]-cycloadditions in part by producing compounds that happen (by chance) to both undergo the reaction nonenzymatically and offer a reproductive advantage to the producing organism. This is consistent with known pathways where nonenzymatic [4 + 2]-cycloadditions are believed to be the operating biosynthetic transformation such as in the biosynthesis of paracaseolide A.225 Preorganization in the active sites of the priming, auxiliary or down-pathway enzymes may impose stereochemical constraints on the cyclization following the activation step and even accelerate the cyclization itself. Eventually, the priming activity of the enzyme may no longer be necessary and lost leaving the enzyme as a stand-alone cyclase like VstJ, PyrE3 and PyrI4. Similarly, auxiliary or down-pathway activities may no longer provide a selective advantage and be lost (e.g., methyl transferase activity in a precursor to SpnF). While entirely speculative, such a hypothesis seems apropos to comparisons of the FAD-binding enzymes solanapyrone synthase and PyrE3. Therefore, study of the [4 + 2]-cyclases may offer new opportunities to investigate the evolution and diversification of biosynthetic pathways.
Mechanistically, very little is known about how the [4 + 2]-cyclases operate as catalysts. Current structural investigations suggest that many of the enzymes (e.g., SpnF, PyrE3 and AbyU) bind substrate to form a closed-lidded Michaelis complex via induced-fit (see Figure 7).142–144 Situated in an enclosed, hydrophobic pocket within the enzyme, the diene and dienophile may be forced into a low-volume conformation that promotes cyclization in a manner analogous to the hydrophobic effect believed to accelerate Diels-Alder reactions in bulk aqueous solution.226,227 At the same time, electrostatic effects may contribute to polarization of the diene and dienophile and stabilization of charge redistributions in the transition state thereby lowering the enthalpy of activation. This mode of catalysis is observed in the bimolecular [4 + 2]-cycloaddition catalyzed by the catalytic antibody 1E9,228 and may also be important in many natural [4 + 2]-cyclases.
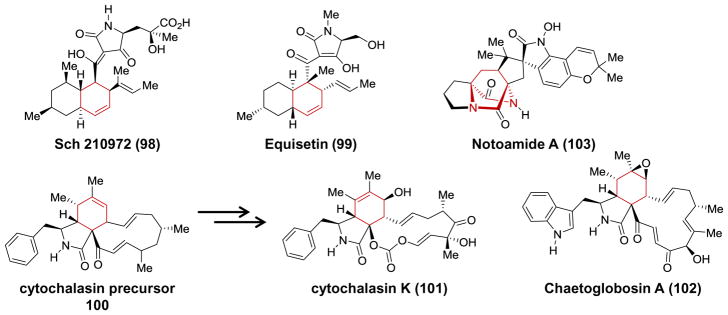
Finally, one should not be left with the impression that the number of natural [4 + 2]-cyclases is necessarily small. The biosynthetic pathways of an extensive collection of natural products are believed to involve [4 + 2]-cycloadditions that may be enzyme catalyzed. This can be appreciated from the catalogs compiled by Stocking and Williams as well as Oikawa and Tokiwano.16,17 Many of these pathways are currently under active investigation and already yielding new discoveries. For example, genes encoding potential [4 + 2]-cyclases have already been tested in vivo in the biosynthetic pathways of the decalin-containing natural products Sch210972 (98, cghA)229 and equisetin (99, fsa2)230 as shown in Figure 8. Likewise, the genes ccsF231 and cheA232 have been implicated as encoding enzymes that catalyze Knoevenagel condensations and possible [4 + 2]-cycloadditions to install the isoindolone rings of cytochalasin K (101) and chaetglobosins A (102) & C, respectively. Furthermore, notH, phqJ and malE have been identified as encoding enzymes likely necessary to construct the monooxopiperazine and dioxopiperazine ring systems in various bicyclo[2.2.2]diazaoctane indole alkaloids such as notoamide (103), paraherquamide and malbrancheamide, though the precise biosynthetic role of these gene products is still uncertain.10,233 In all of these cases, the biosynthetic [4 + 2]-cycloaddition is intramolecular; however, evidence has also been reported implicating bimolecular [4 + 2]-carbocyclase activity associated with cellular extracts from the fungus Hypocrea sp. AS 3.17108.234
Figure 8.
A partial list of natural products currently under investigation for the presence of a potential Diels-Alderase in the biosynthetic pathway.
Given the challenges in their study, experimental investigation of the [4 + 2]-cyclases is just beginning. Furthermore, the field appears to be quite broad with many open questions regarding their distribution, evolutionary origin and mechanism of action making this an exciting area for future research.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH GM040541) and the Welch Foundation (Welch F-1511).
References
- 1.Reetz MT. Artificial Metalloenzymes as Catalysts in Stereoselective Diels-Alder Reactions. Chemical Record. 2012;12:391–406. doi: 10.1002/tcr.201100043. [DOI] [PubMed] [Google Scholar]
- 2.Preiswerk N, Beck T, Schulz JD, Milovnik P, Mayer C, Siegel JB, Baker D, Hilvert D. Impact of Scaffold Rigidity on the Design and Evolution of an Artificial Diels-Alderase. Proc Natl Acad Sci US A. 2014;111:8013–8018. doi: 10.1073/pnas.1401073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kries H, Blomberg R, Hilvert D. De Novo Enzymes by Computational Design. Curr Opin Chem Biol. 2013;17:221–228. doi: 10.1016/j.cbpa.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Mak WS, Siegel JB. Computational Enzyme Design: Transitioning from Catalytic Proteins to Enzymes. Curr Opin Struct Biol. 2014;27:87–94. doi: 10.1016/j.sbi.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Tantillo DJ, Chen J, Houk KN. Theozymes and Compuzymes: Theoretical Models for Biological Catalysts. Curr Opin Chem Biol. 1998;2:743–750. doi: 10.1016/s1367-5931(98)80112-9. [DOI] [PubMed] [Google Scholar]
- 6.Eiben CB, Siegel JB, Bale JB, Cooper S, Khatib F, Shen BW, Players F, Stoddard BL, Popovic Z, Baker D. Increased Diels-Alderase Activity Through Backbone Remodeling Guided by Foldit Players. Nat Biotechnol. 2012;30:190–192. doi: 10.1038/nbt.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romesberg FE, Spiller B, Schultz PG, Stevens RC. Immunological Origins of Binding and Catalysis in a Diels-Alderase Antibody. Science. 1998;279:1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- 8.Hilvert D. Antibody Catalysis of Carbon-Carbon Bond Formation and Cleavage. Acc Chem Res. 1993;26:552–558. [Google Scholar]
- 9.Hilvert D, Hill KW, Nared KD, Auditor MTM. Antibody Catalysis of a Diels-Alder Reaction. J Am Chem Soc. 1989;111:9261–9262. [Google Scholar]
- 10.Klas K, Tsukamoto S, Sherman DH, Williams RM. Natural Diels-Alderases: Elusive and Irresistable. J Org Chem. 2015;80:11672–11685. doi: 10.1021/acs.joc.5b01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oikawa H. Nature’s Strategy for Catalyzing Diels-Alder Reaction. Cell Chem Biol. 2016;23:429–430. doi: 10.1016/j.chembiol.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Pang B, Zhong G, Tang Z, Liu W. Enzymatic [4 + 2] Cycloadditions in the Biosynthesis of Spirotetramates and Spirotetronates. Methods Enzymol. 2016;575:39–63. doi: 10.1016/bs.mie.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Q, Tian Z, Liu W. Recent Advances in Understanding the Enzymatic Reactions of [4 + 2] Cycloaddition and Spiroketalization. Curr Opin Chem Biol. 2016;31:95–102. doi: 10.1016/j.cbpa.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Minami A, Oikawa H. Recent Advances of Diels-Alderases Involved in Natural Product Biosynthesis. J Antibiot. 2016;69:500–506. doi: 10.1038/ja.2016.67. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto T, Kuzuyama T. Mechanistic Insights into Diels-Alder Reactions in Natural Product Biosynthesis. Curr Opin Chem Biol. doi: 10.1016/j.cbpa.2016.09.015. in press. [DOI] [PubMed] [Google Scholar]
- 16.Oikawa H, Tokiwano T. Enzymatic Catalysis of the Diels-Alder Reaction in the Biosynthesis of Natural Products. Nat Prod Rep. 2004;21:321–352. doi: 10.1039/b305068h. [DOI] [PubMed] [Google Scholar]
- 17.Stocking EM, Williams RM. Chemistry and Biology of Biosynthetic Diels-Alder Reactions. Angew Chem Int Ed. 2003;42:3078–3115. doi: 10.1002/anie.200200534. [DOI] [PubMed] [Google Scholar]
- 18.Kelly WL. Intramolecular Cyclizations of Polyketide Biosynthesis: Mining for a “Diels-Alderase”? Org BiomolChem. 2008;6:4483–4493. doi: 10.1039/b814552k. [DOI] [PubMed] [Google Scholar]
- 19.Kim HJ, Ruszczycky MW, Liu H-w. Current Developments and Challenges in the Search for a Naturally Selected Diels-Alderase. Curr Opin Chem Biol. 2012;16:124–131. doi: 10.1016/j.cbpa.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International Union of Pure and Applied Chemistry Organic Chemistry Division, Commission on Physical Organic Chemistry Glossary of Terms Used in Physical Organic Chemistry. Pure & Appl Chem. 1994;66:1077–1184. [Google Scholar]
- 21.Sauer J, Sustmann R. Mechanistic Aspects of Diels-Alder Reactions: A Critical Survey. Angew Chem Int Ed. 1980;19:779–807. [Google Scholar]
- 22.Berson JA. Discoveries Missed, Discoveries Made: Creativity, Influence, and Fame in Chemistry. Tetrahedron. 1992;48:3–17. [Google Scholar]
- 23.Houk KN, Gonzalez J, Li Y. Pericyclic Reaction Transition States: Passions and Punctilios, 1935–1995. Acc Chem Res. 1995;28:81–90. [Google Scholar]
- 24.Woodward RB, Hoffmann R. The Conservation of Orbital Symmetry. Angew Chem Int Ed. 1969;8:781–932. [Google Scholar]
- 25.Huisgen R. Cycloadditions—Definition, Classification, and Characterization. Angew Chem Int Ed. 1968;7:321–328. [Google Scholar]
- 26.Firestone RA. The Low Energy of Concert in Many Symmetry-Allowed Cycloaddi-tions Supports a Stepwise-Diradical Mechanism. Int J Chem Kinet. 2013;45:415–428. [Google Scholar]
- 27.Houk KN, Lin YT, Brown FK. Evidence for the Concerted Mechanism of the Diels-Alder Reaction of Butadiene with Ethylene. J Am Chem Soc. 1986;108:554–556. doi: 10.1021/ja00263a059. [DOI] [PubMed] [Google Scholar]
- 28.Storer JW, Raimondi L, Houk KN. Theoretical Secondary Kinetic Isotope Effects and the Interpretation of Transition State Geometries. 2. The Diels-Alder Reaction Transition State Geometry. J Am Chem Soc. 1994;116:9675–9683. [Google Scholar]
- 29.Beno BR, Houk KN, Singleton DA. Synchronous or Asynchronous? An “Experimental” Transition State from a Direct Comparison of Experimental and Theoretical Kinetic Isotope Effects for a Diels-Alder Reaction. J Am Chem Soc. 1996;118:9984–9985. [Google Scholar]
- 30.Patel A, Chen Z, Yang Z, Guteirez O, Liu H-w, Houk KN, Singleton DA. Dynamically Complex [6 + 4] and [4 + 2] Cycloadditions in the Biosynthesis of Spinosyn A. J Am Chem Soc. 2016;138:3631–3634. doi: 10.1021/jacs.6b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurai I, Suzuki H, Shimizu S, Yamamoto Y. Novel Biotransformation of a 2-Pyrone to a Substituted Benzoic Acid. Chem Pharm Bull. 1985;33:5141–5143. [Google Scholar]
- 32.Oikawa H, Yagi K, Watanabe K, Honma M, Ichihara A. Biosynthesis of Macrophomic Acid: Plausible Involvement of Intermolecular Diels-Alder Reaction. J Chem Soc, Chem Commun. 1997:97–98. [Google Scholar]
- 33.Oikawa H, Watanabe K, Yagi K, Ohashi S, Mie T, Ichihara AHM. Macrophomate Synthase: Unusual Enzyme Catalyzing Multiple Reactions from Pyrones to Benzoates. Tetrahedron Lett. 1999;40:6983–6986. [Google Scholar]
- 34.Watanabe K, Oikawa H, Yagi K, Ohashi S, Mie T, Ichihara A, Honma M. Macrophomate Synthase: Characterization, Sequence, and Expression in Escherichia coli of the Novel Enzyme Catalyzing Unusual Multistep Transformation of 2-Pyrones to Benzoates. J Biochem. 2000;127:467–473. doi: 10.1093/oxfordjournals.jbchem.a022629. [DOI] [PubMed] [Google Scholar]
- 35.Ose T, Watanabe K, Mie T, Honma M, Watanabe H, Yao M, Oikawa H, Tanaka I. Insight into a Natural Diels-Alder Reaction from the Structure of Macrophomate Synthase. Nature. 2003;422:185–189. doi: 10.1038/nature01454. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe K, Mie T, Ichihara A, Oikawa H, Honma M. Detailed Reaction Mechanism of Macrophomate Synthase. J Biol Chem. 2000;275:38393–38401. doi: 10.1074/jbc.M003119200. [DOI] [PubMed] [Google Scholar]
- 37.Serafimov JM, Gillingham D, Kuster S, Hilvert D. The Putative Diels-Alderase Macrophomate Synthase is an Efficient Aldolase. J Am Chem Soc. 2008;130:7798–7799. doi: 10.1021/ja8017994. [DOI] [PubMed] [Google Scholar]
- 38.Serafimov JM, Westfield T, Meier BH, Hilvert D. Trapping and Structural Elucidation of an Intermediate in the Macrophomate Synthase Reaction Pathway. J Am Chem Soc. 2007;129:9580–9581. doi: 10.1021/ja073087y. [DOI] [PubMed] [Google Scholar]
- 39.Guimaraes CRW, Udier-Blagovic M, Jorgensen WL. Macrophomate Synthase: QM/MM Simulations Address the Diels-Alder versus Michael-Aldol Reaction Mechanism. J Am Chem Soc. 2005;127:3577–3588. doi: 10.1021/ja043905b. [DOI] [PubMed] [Google Scholar]
- 40.Izard T, Blackwell NC. Crystal Structures of the Metal-Dependent 2-Dehydro-3-Deoxy-Galactarate Aldolase Suggest a Novel Reaction Mechanism. EMBO J. 2000;19:3849–3856. doi: 10.1093/emboj/19.15.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harvey RA, Plaut GWE. Riboflavin Synthase from Yeast. J Biol Chem. 1966;241:2120–2136. [PubMed] [Google Scholar]
- 42.Fischer M, Bacher A. Biosynthesis of Vitamin B2: Structure and Mechanism of Riboflavin Synthase. Arch Biochem Biophys. 2008;474:252–265. doi: 10.1016/j.abb.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Liao DI, Wawrzak Z, Calabrese JC, Viitanen PV, Jordan DB. Crystal Structure of Riboflavin Synthase. Structure. 2001;9:399–408. doi: 10.1016/s0969-2126(01)00600-1. [DOI] [PubMed] [Google Scholar]
- 44.Gerhardt S, Schott AK, Kairies N, Cushman M, Illarionov B, Eisenreich W, Bacher A, Huber R, Steinbacher S, Fischer M. Studies on the Reaction Mechanism of Riboflavin Synthase: X-Ray Crystal Structure of a Complex with 6-Carboxyethyl-7-Oxo-8-Ribityllumazine. Structure. 2002;10:1371–1381. doi: 10.1016/s0969-2126(02)00864-x. [DOI] [PubMed] [Google Scholar]
- 45.Schott K, Kellermann J, Lottspeich F, Bacher A. Riboflavin Synthases ofBacillus subtilis. J BiolChem. 1990;265:4202–4209. [PubMed] [Google Scholar]
- 46.Bacher A, Baur R, Eggers U, Harders HD, Otto MK, Schnepple H. Riboflavin Synthases ofBacillus subtilis. J BiolChem. 1980;255:632–637. [PubMed] [Google Scholar]
- 47.Ladenstein R, Fischer M, Bacher A. The Lumazine Synthase/Riboflavin Synthase Complex: Shapes and Functions of a Highly Variable Enzyme System. FEBS Journal. 2013;280:2537–2563. doi: 10.1111/febs.12255. [DOI] [PubMed] [Google Scholar]
- 48.Rowan T, Wood HCS. Biosynthesis of Riboflavin. Proc Chem Soc. 1963;1963:21–22. [Google Scholar]
- 49.Rowan T, Wood HCS. The Biosynthesis of Pteridines. Part V. The Synthesis of Riboflavin from Pteridine Precursors. J Chem Soc C. 1968:452–458. doi: 10.1039/j39680000452. [DOI] [PubMed] [Google Scholar]
- 50.Beach R, Plaut GWE. The Formation of Riboflavin from 6,7-Dimethyl-8-Ribityllumazine in Acid Media. Tetrahedron Lett. 1969;40:3489–3492. doi: 10.1016/s0040-4039(01)88428-8. [DOI] [PubMed] [Google Scholar]
- 51.Beach RL, Plaut GWE. Stereospecificity of the Enzymatic Synthesis of the o-Xylene Ring of Riboflavin. J Am Chem Soc. 1970;92:2913–2916. doi: 10.1021/ja00712a052. [DOI] [PubMed] [Google Scholar]
- 52.Bacher A, Eberhardt S, Fischer M, Kis K, Richter G. Biosynthesis of Vitamin B2 (Riboflavin) Annu Rev Nutr. 2000;20:153–167. doi: 10.1146/annurev.nutr.20.1.153. [DOI] [PubMed] [Google Scholar]
- 53.Illarionov B, Eisenreich W, Bacher A. A Pentacyclic Reaction Intermediate of Riboflavin Synthase. Proc Natl Acad Sci US A. 2001;98:7224–7229. doi: 10.1073/pnas.131610698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Illarionov B, Haase I, Bacher A, Fischer M, Schramek N. Presteady State Kinetic Analysis of Riboflavin Synthase. J Biol Chem. 2003;278:47700–47706. doi: 10.1074/jbc.M305050200. [DOI] [PubMed] [Google Scholar]
- 55.Illarionov B, Haase I, Fischer M, Bacher A, Schramek N. Pre-steady-state Kinetic Analysis of Riboflavin Synthase Using a Pentacyclic Reaction Intermediate as Substrate. Biol Chem. 2005;386:127–136. doi: 10.1515/BC.2005.016. [DOI] [PubMed] [Google Scholar]
- 56.Pfleiderer W, Hutzenlaub W. Pteridine, LVII. Synthesen und Eigenschaften von Lumazin-N-oxiden. Chemische Berichte. 1973;106:3149–3174. [Google Scholar]
- 57.Beach RL, Plaut GWE. Investigations of Structures of Substituted Lumazines by Deuterium Exchange and Nuclear Magnetic Resonance Spectroscopy. Biochemistry. 1970;9:760–770. doi: 10.1021/bi00806a009. [DOI] [PubMed] [Google Scholar]
- 58.Bown DH, Keller PJ, Floss HG, Sedlmaier H, Bacher A. Solution Structures of 6:7-Dimethyl-8-substituted-lumazines. 13C NMR Evidence for Intramolecular Ether Formation. J Org Chem. 1986;51:2461–2467. [Google Scholar]
- 59.Kim RR, Illarionov B, Joshi M, Cushman M, Lee CY, Eisenreich W, Fischer M, Bacher A. Mechanistic Insights on Riboflavin Synthase Inspired by Selective Binding of the 6,7-Dimethyl-8-ribityllumazine Exomethylene Anion. J Am Chem Soc. 2010;132:2983–2990. doi: 10.1021/ja908395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breugst M, Eschenmoser A, Houk KN. Theoretical Exploration of the Mechanism of Riboflavin Formation from 6,7-Dimethyl-8-ribityllumazine: Nucleophilic Catalysis, Hydride Transfer, Hydrogen Atom Transfer, or Nucleophilic Addition? J Am ChemSoc. 2013;135:6658–6668. doi: 10.1021/ja402099f. [DOI] [PubMed] [Google Scholar]
- 61.Plaut GWE, Beach RL, Aogaichi T. Studies on the Mechanism of Elimination of Protons from the Methyl Groups of 6,7-Dimethyl-8-ribityllumazine by Riboflavin Synthetase. Biochemistry. 1970;9:771–785. doi: 10.1021/bi00806a010. [DOI] [PubMed] [Google Scholar]
- 62.Meining W, Eberhardt S, Bacher A, Ladenstein R. The Structure of the N-Terminal Domain of Riboflavin Synthase in Complex with Riboflavin at 2.6 Å Resolution. J Mol Biol. 2003;331:1053–1063. doi: 10.1016/s0022-2836(03)00844-1. [DOI] [PubMed] [Google Scholar]
- 63.Truffault V, Coles M, Diercks T, Abelmann K, Eberhardt S, Luttgen H, Bacher A, Kessler H. The Solution Structure of the N-Terminal Domain of Riboflavin Synthase. J Mol Biol. 2001;309:949–960. doi: 10.1006/jmbi.2001.4683. [DOI] [PubMed] [Google Scholar]
- 64.Ray WJ., Jr Rate-Limiting Steps: A Quantitative Definition. Application to Steady-State Enzymic Reactions. Biochemistry. 1983;22:4625–4637. doi: 10.1021/bi00289a003. [DOI] [PubMed] [Google Scholar]
- 65.Ruszczycky MW, Anderson VE. Interpretation of V/K Isotope Effects for Enzymatic Reactions Exhibiting Multiple Isotopically Sensitive Steps. J Theor Biol. 2006;243:328–342. doi: 10.1016/j.jtbi.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 66.Tian G. Effective Rate Constants and General Isotope Effect Equations for Steady State Enzymatic Reactions with Multiple Isotope-Sensitive Steps. Bioorg Chem. 1992;20:95–106. [Google Scholar]
- 67.Tobert JA. Lovastatin and Beyond: The History of the HMG-CoA Reductase Inhibitors. Nat Rev Drug Discovery. 2003;2:517–526. doi: 10.1038/nrd1112. [DOI] [PubMed] [Google Scholar]
- 68.Endo A, Kuroda M, Tsujita Y. ML-236A, ML-236B, and ML-236C, New Inhibitors of Cholesterogenesis Produced byPencillium citrinum. JAntibiot. 1976;29:1346–1348. doi: 10.7164/antibiotics.29.1346. [DOI] [PubMed] [Google Scholar]
- 69.Endo A. Monacolin K, a New Hypocholesterolemic Agent Produced by a Monascus Species. J Antibiot. 1979;32:852–854. doi: 10.7164/antibiotics.32.852. [DOI] [PubMed] [Google Scholar]
- 70.Moore RN, Bigam G, Chan JK, Hogg AM, Nakashima TT, Vederas JC. Biosynthesis of the Hypocholesterolemic Agent Mevinolin by Aspergillus terreus. Determination of the Origin of Carbon, Hydrogen, and Oxygen Atoms by 13C NMR and Mass Spectrometry. J Am Chem Soc. 1985;107:3694–3701. [Google Scholar]
- 71.Yoshizawa Y, Witter DJ, Liu Y, Vederas JC. Revision of the Biosynthetic Origin of Oxygens in Mevinolin (Lovastatin), a Hypocholesterolemic Drug from Aspergillus terreus MF 4845. J Am Chem Soc. 1994;116:2693–2694. [Google Scholar]
- 72.Endo A. Monacolin K, a New Hypocholesterolemic Agent that Specifically Inhibits 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase. J Antibiot. 1980;33:334–336. doi: 10.7164/antibiotics.33.334. [DOI] [PubMed] [Google Scholar]
- 73.Alberts AW, et al. Mevinolin: a Highly Potent Competitive Inhibitor of Hydroxymethylglutaryl-coenzyme A Reductase and a Cholesterol-lowering Agent. Proc Natl Acad Sci U S A. 1980;77:3957–3961. doi: 10.1073/pnas.77.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alarcon J, Aguila S, Arancibia-Avila P, Fuentes O, Zamorano-Ponce E, Hernandez M. Production and Purification of Statins from Pleurotus ostreatus (Basidiomycetes) Strains. Z Naturforsch C. 2003;58:62–64. doi: 10.1515/znc-2003-1-211. [DOI] [PubMed] [Google Scholar]
- 75.Chan JK, Moore RN, Nakashima TT, Vederas JC. Biosynthesis of Mevinolin. Spectral Assignment by Double-Quantum Coherence NMR after High Carbon-13 Incorporation. J Am Chem Soc. 1983;105:3334–3336. [Google Scholar]
- 76.Wagschal K, Yoshizawa Y, Witter DJ, Liu Y, Vederas JC. Biosynthesis of ML-236C and the Hypocholesterolemic Agents Compactin by Penicillum aurantiogriseum and Lovastatin by Aspergillus terreus: Determination of the Origin of Carbon, Hydrogen and Oxygen Atoms by 13C NMR Spectrometry and Observation of Unusual Labelling of Acetate-Derived Oxygens by 18O2. J Chem Soc, Perkin Trans 1. 1996:2357–2363. [Google Scholar]
- 77.Kimura K, Komagata D, Murakawa S, Endo A. Biosynthesis of Monacolins: Conversion of Monacolin J to Monacolin K (Mevinolin) J Antibiot. 1990;43:1621–1622. doi: 10.7164/antibiotics.43.1621. [DOI] [PubMed] [Google Scholar]
- 78.Nakamura T, Komagata D, Murakawa S, Sakai K, Endo A. Isolation and Biosynthesis of 3α-Hydroxy-3,5-dihydromonacolin L. J Antibiot. 1990;43:1597–1600. doi: 10.7164/antibiotics.43.1597. [DOI] [PubMed] [Google Scholar]
- 79.Komogata D, Shimada H, Murakawa S, Endo A. Biosynthesis of Monacolins Conversion of Deshydroxytriol to Monacolin J by a Monooxygenase ofMonascus ruber. JAntibiot. 1989;42:407–412. doi: 10.7164/antibiotics.42.407. [DOI] [PubMed] [Google Scholar]
- 80.Hendrickson L, Davis CR, Roach C, Nguyen DK, Aldrich T, McAda PC, Reeves CD. Lovastatin Biosynthesis in Aspergillus terreus: Characterization of Blocked Mutants, Enzyme Actvities and a Multifunctional Polyketide Synthase Gene. Chem Biol. 1999;6:429–439. doi: 10.1016/s1074-5521(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 81.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of Polyketide Synthase Activity by Accessory Proteins During Lovastatin Biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 82.Ma SM, Li JWH, Choi JW, Zhou H, Lee KKM, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Complete Reconstitution of a Highly Reducing Iterative Polyketide Synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Auclair K, Kennedy J, Hutchinson CR, Vederas JC. Conversion of Cyclic Nonaketides to Lovastatin and Compactin by a lovC Deficient Mutant ofAspergillus terreus. Bioorg Med ChemLett. 2001;11:1527–1531. doi: 10.1016/s0960-894x(01)00290-6. [DOI] [PubMed] [Google Scholar]
- 84.Ames BD, Nguyen C, Bruegger J, Smith P, Xu W, Ma S, Wong E, Wong S, Xie X, Li JWH, Vederas JC, Tang Y, Tsai SC. Crystal Structure and Biochemical Studies of the Transacting Polyketide Enoyl Reductase LovC from Lovastatin Biosynthesis. Proc Natl Acad Sci U S A. 2012;109:11144–11149. doi: 10.1073/pnas.1113029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu W, Chooi YH, Choi JW, Li S, Vederas JC, Da Silva NA, Tang Y. LovG: The Thioesterase Required for Dihydromonacolin L Release and Lovastatin Nonaketide Synthase Turnover in Lovastatin Biosynthesis. Angew Chem Int Ed. 2013;52:6472–6475. doi: 10.1002/anie.201302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie X, Meehan MJ, Xu W, Dorrestein PC, Tang Y. Acyltransferase Mediated Polyketide Release from a Fungal Megasynthase. J Am Chem Soc. 2009;131:8388–8389. doi: 10.1021/ja903203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barriuso J, Nguyen DT, Li JWH, Roberts JN, MacNevin G, Chaytor JL, Marcus SL, Vederas JC, Ro DK. Double Oxidation of the Cyclic Nonaketide Dihydromonacolin L to Monacolin J by a Single Cytochrome P450 Monooxygenase, LovA. J Am Chem Soc. 2011;133:8078–8081. doi: 10.1021/ja201138v. [DOI] [PubMed] [Google Scholar]
- 88.Witter DJ, Vederas JC. Putative Diels-Alder-Catalyzed Cyclization during the Biosynthesis of Lovastatin. J Org Chem. 1996;61:2613–2623. doi: 10.1021/jo952117p. [DOI] [PubMed] [Google Scholar]
- 89.Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CJ, Vederas JC. Lovastatin Nonaketide Synthase Catalyzes an Intramolecular Diels-Alder Reaction of a Substrate Analogue. J Am Chem Soc. 2000;122:11519–11520. [Google Scholar]
- 90.Cacho RA, Thuss J, Xu W, Sanichar R, Gao Z, Nguyen A, Vederas JC, Tang Y. Understatnding Programming of Fungal Iterative Polyketide Synthases: The Biochemical Basis for Regioselectivity by the Methyltransferase Domain in the Lovastatin Megasynthase. J Am Chem Soc. 2015;137:15688–15691. doi: 10.1021/jacs.5b11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie X, Watanabe K, Wojcicki WA, Wang CCC, Tang Y. Biosynthesis of Lovastatin Analogs with a Broadly Specific Acyltransferase. Chem Biol. 2006;13:1161–1169. doi: 10.1016/j.chembiol.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 92.Ma S, Tang Y. Biochemical Characterization of the Minimal Polyketide Synthase Domains in the Lovastatin Nonaketide Synthase LovB. FEBS J. 2007;274:2854–2864. doi: 10.1111/j.1742-4658.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 93.Ichihara A, Tazaki H, Sakamura S. Solanapyrones A, B and C, Phytotoxic Metabolites from the FungusAlternaria solani. Tetraheron Lett. 1983;24:5373–5376. [Google Scholar]
- 94.Ichihara A, Miki M, Tazaki H, Sakamura S. Synthesis of (±)-Solanapyrone A. Tetraheron Lett. 1987;28:1175–1178. [Google Scholar]
- 95.Oikawa H, Suzuki Y, Naya A, Katayama K, Ichihara A. First Direct Evidence in Biological Diels-Alder Reaction of Incorporation of Diene-Dienophile Precursors in the Biosynthesis of Solanapyrones. J Am Chem Soc. 1994;116:3605–3606. [Google Scholar]
- 96.Oikawa H, Yokota T, Ichihara A, Sakamura S. Structure and Absolute Configuration of Solanapyrone D: A New Clue to the Occurrence of Biological Diels-Alder Reactions. J Chem Soc Chem Commun. 1989:1284–1285. [Google Scholar]
- 97.Alam SS, Bilton JN, Slawin AMZ, Williams DJ, Sheppard RN, Strange RN. Chickpea Blight: Production of the Phytotoxins Solanapyrones A and C byAscochyta rabiei. Phytochemistry. 1989;28:2627–2630. [Google Scholar]
- 98.Oikawa H, Kobayashi T, Katayama K, Suzuki Y, Ichihara A. Total Synthesis of (−)-Solanapyrone A via Enzymatic Diels-Alder Reaction of Prosolanapyrone. J Org Chem. 1998;63:8748–8756. [Google Scholar]
- 99.Lygo B, Bhatia M, Cooke JWB, Hirst DJ. Synthesis of (±)-Solanapyrones A and B. Tetrahedron Lett. 2003;44:2529–2532. [Google Scholar]
- 100.Wilson RM, Jen WS, MacMillan DWC. Enantioselective Organocatalytic Intramolecular Diels-Alder Reactions. The Asymmetric Synthesis of Solanapyrone D. J Am Chem Soc. 2005;127:11616–11617. doi: 10.1021/ja054008q. [DOI] [PubMed] [Google Scholar]
- 101.Oikawa H, Yokota T, Abe T, Ichihara A, Sakamura S, Yoshizawa Y, Vederas JC. Biosynthesis of Solanapyrone A, a Phytotoxin of Alternaria solani. J Chem Soc Chem Commun. 1989:1282–1284. [Google Scholar]
- 102.Kasahara K, Miyamoto T, Fujimoto T, Oguri H, Tokiwano T, Oikawa H, Ebizuka Y, Fujii I. Solanapyrone Synthase, a Possible Diels-Alderase and Iterative Type I Polyketide Synthase Encoded in a Biosynthetic Gene Cluster fromAlternaria solani. ChemBioChem. 2010;11:1245–1252. doi: 10.1002/cbic.201000173. [DOI] [PubMed] [Google Scholar]
- 103.Kim W, Park CM, Park JJ, Akamatsu HO, Peever TL, Xian M, Gang DR, Vandemark G, Chen W. Functional Analyses of the Diels-Alderase Gene sol5 of Ascochyta rabiei and Alternaria solani Indicate that the Solanapyrone Phytotoxins Are Not Required for Pathogenicity. MPMI. 2015;28:482–496. doi: 10.1094/MPMI-08-14-0234-R. [DOI] [PubMed] [Google Scholar]
- 104.Oikawa H, Katayama K, Suzuki Y, Ichihara A. Enzymatic Activity Catalyzing exo-Selective Diels-Alder Reaction in Solanapyrone Biosynthesis. J Chem Soc Chem Commun. 1995:1321–1322. [Google Scholar]
- 105.Koetter JWA, Schulz GE. Crystal Structure of 6-Hydroxy-D-nicotine Oxidase fromArthrobacter nicotinovorans. J MolBiol. 2005;352:418–428. doi: 10.1016/j.jmb.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 106.Katayama K, Kobayashi T, Chijimatsu M, Ichihara A, Oikawa H. Purification and N-Terminal Amino Acid Sequence of Solanapyrone Synthase, a Natural Diels-Alderase fromAlternaria solani. Biosci BiotechnolBiochem. 2008;72:604–607. doi: 10.1271/bbb.70600. [DOI] [PubMed] [Google Scholar]
- 107.Katayama K, Kobayashi T, Oikawa H, Honma M, Ichihara A. Enzymatic Activity and Partial Purification of Solanapyrone Synthase: First Enzyme Catalyzing Diels-Alder Reaction. Biochim Biophys Acta. 1998;1384:387–395. doi: 10.1016/s0167-4838(98)00040-5. [DOI] [PubMed] [Google Scholar]
- 108.Baldwin JE, Reddy VP. Stereochemistry of the Diels-Alder Reaction of Butadiene with Cyclopropene. J Org Chem. 1989;54:5264–5267. [Google Scholar]
- 109.Li C, Kelly WL. Recent Advances in Thiopeptide Antibiotic Biosynthesis. Nat Prod Rep. 2010;27:153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 110.Walsh CT, Acker MG, Bowers AA. Thiazolyl Peptide Antibiotic Biosynthesis: A Cascade of Post-Translational Modifications on Ribosomal Nascent Proteins. J Biol Chem. 2010;285:27525–275531. doi: 10.1074/jbc.R110.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide Antibiotics. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 112.Ardt HD, Schoof S, Lu JY. Thiopeptide antibiotic biosynthesis. Angew Chem Int Ed. 2009;48:6770–6773. doi: 10.1002/anie.200901808. [DOI] [PubMed] [Google Scholar]
- 113.Hughes RA, Moody CJ. From Amino Acids to HeteroaromaticsThiopeptide Antibiotics, Natures Heterocyclic Peptides. Angew Chem Int Ed. 2007;46:7930–7954. doi: 10.1002/anie.200700728. [DOI] [PubMed] [Google Scholar]
- 114.Brown LCW, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen Posttranslational Modifications Convert a 14-Residue Peptide into the Antibiotic Thiocillin. Proc Natl Acad Sci US A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Houck DR, Chen LC, Keller PJ, Beale JM, Floss HG. Biosynthesis of the Modified Peptide Antibiotic Nosiheptide inStreptomyces actuosus. J Am ChemSoc. 1987;109:1250–1252. [Google Scholar]
- 116.Houck DR, Chen LC, Keller PJ, Beale JM, Floss HG. Biosynthesis of the Modified Peptide Antibiotic Nosiheptide inStreptomyces actuosus. J Am ChemSoc. 1988;110:5800–5806. [Google Scholar]
- 117.Mocek U, Knaggs AR, Tsuchiya R, Nguyen T, Beale JM, Floss HG. Biosynthesis of the Modified Peptide Antibiotic Nosiheptide inStreptomyces actuosus. J Am ChemSoc. 1993;115:7557–7568. [Google Scholar]
- 118.Mocek U, Zeng Z, OHagan D, Zhou P, Fan LDG, Beale JM, Floss HG. Biosynthesis of the Modified Peptide Antibiotic Thiostrepton in Streptomyces azureus andStreptomyces laurentii. J Am ChemSoc. 1993;115:7992–8001. [Google Scholar]
- 119.Bycroft BW, Gowland MS. The Structures of Highly Modified Peptide Antibiotics Microccin P1 and P2. J Chem Soc Chem Comm. 1978:256–258. [Google Scholar]
- 120.Bowers AA, Walsh CT, Acker MG. Genetic Interception and Structural Characterization of Thiopeptide Cyclization Precursors fromBacillus cereus. J Am ChemSoc. 2010;132:12182–12184. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Onaka H, Tabata H, Igarashi Y, Sato Y, Furumai T. Goadsporin, a Chemical Substance which Promotes Secondary Metabolism and Morphogenesis in Streptomyces. I. Purification and Characterization. J Antibiot. 2001;54:1036–1044. doi: 10.7164/antibiotics.54.1036. [DOI] [PubMed] [Google Scholar]
- 122.Wever WJ, Bogart JW, Baccile JA, Chan AN, Schroeder FC, Bowers AA. Chemoenzymatic Synthesis of Thiazolyl Peptide Natural Products Featuring an Enzyme-Catalyzed Formal [4 + 2] Cycloaddition. J Am Chem Soc. 2015;137:3494–3497. doi: 10.1021/jacs.5b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hudson GA, Zhang Z, Tietz JI, Mitchell DA, van der Donk WA. In Vitro Biosynthesis of the Core Scaffold of the Thiopeptide Thiomuracin. J Am Chem Soc. 2015;137:16012–16015. doi: 10.1021/jacs.5b10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. Ribosomally Synthesized Thiopeptide Antibiotics Targeting Elongation Factor Tu. J Am Chem Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 125.Anderson B, Hodgkin DC, Viswamitra MA. The Structure of Thiostrepton. Nature. 1970;225:233–235. doi: 10.1038/225233a0. [DOI] [PubMed] [Google Scholar]
- 126.Ebata M, Miyazaki K, Otsuka H. Studies on Siomycin. I. Physicochemical Properties of Siomycins A, B, and C. J Antibiot. 1969;22:364–368. [PubMed] [Google Scholar]
- 127.Tori K, Tokura K, Yoshimura Y, Terui Y, Okabe K, Otsuka H, Matsushita K, Inagaki F, Miyazawa T. Structures of Siomycin-B and -C and Thiostrepton-B Determined by NMR Spectroscopy and Carbon-13 Signal Assignments of Siomycins, Thiostreptons, and Thiopeptin-B. J Antibiot. 1981;34:124–129. doi: 10.7164/antibiotics.34.124. [DOI] [PubMed] [Google Scholar]
- 128.Prange T, Ducruix A, Pascard C, Lunel J. Structure of Nosiheptide, a Polythiazole-Containing Antibiotic. Nature. 1977;265:189–190. doi: 10.1038/265189a0. [DOI] [PubMed] [Google Scholar]
- 129.Kirst HA. The Spinosyn Family of Insecticides: Realizing the Potential of Natural Products research. J Antibiotics. 2010;63:101–111. doi: 10.1038/ja.2010.5. [DOI] [PubMed] [Google Scholar]
- 130.Evans DA, Black WC. Total Synthesis of (+)-A83543A [(+)-Lepicidin A] J Am Chem Soc. 1993;115:4497–4513. [Google Scholar]
- 131.Mergott DJ, Frank SA, Roush WR. Total Synthesis of (−)-Spinosyn A. Proc Natl Acad Sci USA. 2004;101:11955–11959. doi: 10.1073/pnas.0401247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Paquette LA, Gao Z, Ni Z, Smith GF. Total Synthesis of Spinosyn A. 1. Enantioselective Construction of a Key Tricyclic Intermediate by a Multiple Configurational Inversion Scheme. J Am Chem Soc. 1998;120:2543–2552. [Google Scholar]
- 133.Paquette LA, Collado I, Purdie M. Total Synthesis of Spinosyn A. 2. Degradation Studies Involving the Pure Factor and Its Complete Reconstitution. J Am Chem Soc. 1998;120:2553–2562. [Google Scholar]
- 134.Waldron C, Matsushima P, Rosteck PR, Broughton MC, Turner J, Madduri K, Crawford KP, Merlo DJ, Baltz RH. Cloning and Analysis of the Spinosad Biosynthetic Gene Cluster ofSaccaropolyspora spinosa. ChemBiol. 2001;8:487–499. doi: 10.1016/s1074-5521(01)00029-1. [DOI] [PubMed] [Google Scholar]
- 135.Kim HJ, Ruszczycky MW, Choi S-h, Liu Y-n, Liu H-w. Enzyme-catalysed [4+2] Cycloaddition is a Key Step in the Biosynthesis of Spinosyn A. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Townsend CA. A “Diels-Alderase” at Last. ChemBioChem. 2011;12:2267–2269. doi: 10.1002/cbic.201100431. [DOI] [PubMed] [Google Scholar]
- 137.Martin CJ, Timoney MC, Sheridan RM, Kendrew SG, Wilkinson B, Staunton J, Leadlay PF. Heterologous Expression in Saccharopolyspora erythaea of a Pentaketide Synthase Derived from the Spinosyn Polyketide Synthase. Org Biomol Chem. 2003;1:4144–4147. doi: 10.1039/b310740j. [DOI] [PubMed] [Google Scholar]
- 138.Kim HJ, Pongdee R, Wu Q, Hong L, Liu H-w. The Biosynthesis of Spinosyn in Saccharopolyspora spinosa: Synthesis of the Cross-Bridging Precursor and Identification of the Function of SpnJ. J Am Chem Soc. 2007;129:14582–14584. doi: 10.1021/ja076580i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Isiorho EA, Jeon B-s, Kim NH, Liu H-w, Keatinge-Clay AT. Structural Studies of the Spinosyn Forosaminyltransferase, SpnP. Biochemistry. 2014;53:4292–4301. doi: 10.1021/bi5003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim HJ, Choi S-h, Jeon B-s, Kim N, Pongdee R, Wu Q, Liu H-w. Chemoenzymatic Synthesis of Spinosyn A. Angew Chem Int Ed. 2014;53:13553–13557. doi: 10.1002/anie.201407806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen YL, Chen YH, Lin YC, Tsai KC, Chiu HT. Functional Characterization and Substrate Specificity of Spinosyn Rhamnosyltransferase by in Vitro Reconstitution of Spinosyn Biosynthetic Enzymes. J Biol Chem. 2009;284:7352–7363. doi: 10.1074/jbc.M808441200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fage CD, Isiorho EA, Liu Y, Wagner DT, Liu H-w, Keatinge-Clay AT. The Structure of SpnF, a Standalone Enzyme that Catalyzes [4 + 2] Cycloaddition. Nat Chem Biol. 2015;11:256–258. doi: 10.1038/nchembio.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Byrne MJ, Lees NR, Han LC, van der Kamp M, Mulholland AJ, Stach JEM, Willis CL, Race PR. The Catalytic Mechanism of a Natural Diels-Alderase Revealed in Molecular Detail. J Am Chem Soc. 2016;138:6095–6098. doi: 10.1021/jacs.6b00232. [DOI] [PubMed] [Google Scholar]
- 144.Zheng Q, Guo Y, Yang L, Zhao Z, Wu Z, Zhang H, Liu J, Cheng X, Wu J, Yang H, Jiang H, Pan L, Liu W. Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4. Cell Chem Biol. 2016;23:352–360. doi: 10.1016/j.chembiol.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 145.Hess BA, Jr, Smentek L. Concerted Highly Asynchronous Enzyme-catalyzed [4 + 2] Cycloaddition in the Biosynthesis of Spinosyn A; Computational Evidence. Org Biomol Chem. 2012;10:7503–7509. doi: 10.1039/c2ob25827g. [DOI] [PubMed] [Google Scholar]
- 146.Gordeev EG, Ananikov VP. Computational Study of a Model System of Enzyme-Mediated [4 + 2] Cycloaddition Reaction. PLoS ONE. 2015;10:e0119984. doi: 10.1371/journal.pone.0119984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pham HV, Houk KN. Diels-Alder Reactions of Allene with Benzene and Butadiene: Concerted, Stepwise, and Ambimodal Transition States. J Org Chem. 2014;79:8968–8976. doi: 10.1021/jo502041f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Quadrelli P, Romano S, Toma L, Caramella P. A Bispericyclic Transition Structure Allows for Efficient Relief of Antiaromaticity Enhancing Reactivity and Endo Stereochemistry in the Dimerization of the Fleeting Cyclopentadienone. J Org Chem. 2003;68:6035–6038. doi: 10.1021/jo034401j. [DOI] [PubMed] [Google Scholar]
- 149.Thomas JB, Waas JR, Harmata M, Singleton DA. Control Elements in Dynamically Determined Selectivity on a Bifurcating Surface. J Am Chem Soc. 2008;130:14544–14555. doi: 10.1021/ja802577v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang Z, Hirschi JS, Singleton DA. Recrossing and Dynamic Matching Effects on Selectivity in a Diels-Alder Reaction. Angew Chem Int Ed. 2009;48:9156–9159. doi: 10.1002/anie.200903293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Caramella P, Quadrelli P, Toma L. An Unexpected Bispericyclic Transition Structure Leading to 4 + 2 and 2 + 4 Cycloadducts in the Endo Dimerization of Cyclopentadiene. J Am Chem Soc. 2002;124:1130–1131. doi: 10.1021/ja016622h. [DOI] [PubMed] [Google Scholar]
- 152.Hong YJ, Tantillo DJ. Biosynthetic Consequences of Multiple Sequential Post-Transition-State Bifurcations. Nat Chem. 2014;6:104–111. doi: 10.1038/nchem.1843. [DOI] [PubMed] [Google Scholar]
- 153.Truhlar DG, Garrett BC. Variational Transition-State Theory. Acc Chem Res. 1980;13:440–448. [Google Scholar]
- 154.Truhlar DG. Accuracy of Trajectory Calculations and Transition State Theory for Thermal Rate Constants of Atom Transfer Reactions. J Phys Chem. 1979;83:188–199. [Google Scholar]
- 155.Schobert R, Schlenk A. Tetramic and Tetronic Acids: An Update on New Derivatives and Biological Aspects. Bioorg Med Chem. 2008;16:4203–4221. doi: 10.1016/j.bmc.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 156.Tao W, Zhu M, Deng Z, Sun Y. Biosynthesis of Tetronate Antibiotics: A Growing Family of Natural Products with Broad Biological Activities. Sci China Chem. 2013;56:1364–1371. [Google Scholar]
- 157.Vieweg L, Reichau S, Schobert R, Leadlay PF, Sussmuth RD. Recent Advances in the Field of Bioactive Tetronates. Nat Prod Rep. 2014;31:1554–1584. doi: 10.1039/c4np00015c. [DOI] [PubMed] [Google Scholar]
- 158.Park HR, Furihata K, Hayakawa Y, Shinya K. Versipelostatin, Novel GRP78/Bip Molecular Chaperone Down-regulator of Microbial Origin. Tetrahedron Lett. 2002;43:6941–6945. [Google Scholar]
- 159.Park HR, Chijiwa S, Furihata K, Hayakawa Y, Shinya K. Relative and Absolute Configuration of Versipelostatin, a Down-Regulator of Molecular Chaperone GRP78 Expression. Org Lett. 2007;9:1457–1460. doi: 10.1021/ol070042t. [DOI] [PubMed] [Google Scholar]
- 160.Ueda J-y, Chijiwa S, Takagi M, Shinya K. A Novel Versipelostatin Analogue, Versipelostatin F Isolated from Streptomyces versipellis 4083-SVS6. J Antibiot. 2008;61:752–755. doi: 10.1038/ja.2008.89. [DOI] [PubMed] [Google Scholar]
- 161.Zhao P, Ueda J-y, Kozone I, Chijiwa S, Takagi M, Kudo F, Nishiyama M, Shinya K, Kuzuyama T. New Glycosylated Deriviatives of Versipelostatin, the GRP78/Bip Molecular Chaperone Down-regulator, from Streptomyces versipellis 4083-SVS6. Org Biomol Chem. 2009;7:1454–1460. doi: 10.1039/b817312e. [DOI] [PubMed] [Google Scholar]
- 162.Ding W, Willams DR, Northcote P, Siegel MM, Tsao R, Ashcroft J, Morton GO, Alluri M, Abbanat D, Maiese WM, Ellestad GA, Pyrroindomycins N. Antibiotics Produced by Streptomyces rugosporus sp. LL-42D005 I. Isolation and Structure Determination. J Antibiot. 1994;47:1250–1257. doi: 10.7164/antibiotics.47.1250. [DOI] [PubMed] [Google Scholar]
- 163.Singh MP, Peterson PJ, Jacobus NV, Mroczenski-Wildey MJ, Maiese WM, Greenstein M, Steinberg DA. Pyrroindomycins, Novel Antibiotics Produced by Streptomyces rugosporus sp. LL-42D005 II. Biological Activities. J Antibiot. 1994;47:1258–1265. doi: 10.7164/antibiotics.47.1258. [DOI] [PubMed] [Google Scholar]
- 164.Abbanat D, Maiese W, Greenstein M. Biosynthesis of the Pyrroindomycins by Streptomyces rugosporus LL-42D005; Characterization of Nutrient Requirements. J Antibiot. 1999;52:117–126. doi: 10.7164/antibiotics.52.117. [DOI] [PubMed] [Google Scholar]
- 165.Zehner S, Kotzsch A, Bister B, Sussmuth RD, Mendez C, Salas JA, van Pee KH. A Regioselective Tryptophan 5-Halogenase Is Involved in Pyrroindomycin Biosynthesis in Streptomyces rugosporus LL-42D005. Chem Biol. 2005;12:445–452. doi: 10.1016/j.chembiol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 166.Wu Q, Wu Z, Qu X, Liu W. Insights into Pyrroindomycin Biosynthesis Reveal a Uniform Paradigm for Tetramate/Tetronate Formation. J Am Chem Soc. 2012;134:17342–17345. doi: 10.1021/ja304829g. [DOI] [PubMed] [Google Scholar]
- 167.Keller-Schierlein W, Muntwyler R, Pache W, Zahner H. Stoffwechselprodukte von Mikroorganismen 73. Mitteilung [1] Chlorothricin und Deschlorothoricin. Helv Chim Acta. 1969;52:127–142. [Google Scholar]
- 168.Muntwyler R, Keller-Schierlein W. Stoffwechselprodukte von Mikroorganismen. 107. Mitteilung [1]. Die Struktur des Chlorothricins, eines neuartigen Makrolid-Antibioticums. Helv Chim Acta. 1972;55:2071–2094. doi: 10.1002/hlca.19720550627. [DOI] [PubMed] [Google Scholar]
- 169.Brufani M, Cerrini S, Fedeli W, Mazza F, Muntwyler, Helm R. Chim Acta. 1972;55:2094–2102. doi: 10.1002/hlca.19720550628. [DOI] [PubMed] [Google Scholar]
- 170.Holzbach R, Pape H, Hook D, Kreutzer EF, Chang CJ, Floss HG. Biosynthesis of the Macrolide Antibiotic Chlorothricin: Basic Building Blocks. Biochemistry. 1978;17:556–560. doi: 10.1021/bi00596a029. [DOI] [PubMed] [Google Scholar]
- 171.Mascaretti OA, Chang CJ, Floss HG. Biosynthsis of the Antibiotic Chlorothricin: Assignment of the Carbon-13 Magnetic Resonance Spectrum. J Nat Prod. 1979;42:455–462. [Google Scholar]
- 172.Mascaretti OA, Chang CJ, Hook D, Otsuka H, Kreutzer EF, Floss HG. Biosynthesis of the Macrolide Antibiotic Chlorothricin. Biochemistry. 1981;20:919–924. doi: 10.1021/bi00507a042. [DOI] [PubMed] [Google Scholar]
- 173.Lee JJ, Lee JP, Keller PJ, Cottrell CE, Chang CJ, Zahner H, Floss HG. Further Studies on the Biosynthesis of Chlorothricin. J Antibiot. 1986;39:1123–1134. doi: 10.7164/antibiotics.39.1123. [DOI] [PubMed] [Google Scholar]
- 174.Jia XY, Tian ZH, Shao L, Qu XD, Zhao QF, Tang J, Tang GL, Liu W. Genetic Characterization of Chlorothricin Gene Cluster as a Model for Spirotetronate Antibiotic Biosynthesis. Chem Biol. 2006;13:575–585. doi: 10.1016/j.chembiol.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 175.Li Y, Li J, Tian Z, Xu Y, Zhang J, Liu W, Tan H. Coordinative Modulation of Chlorothricin Biosynthesis by Binding of the Glycosylated Intermediates and End Product to a Responsive Regulator ChlF1. J Biol Chem. 2016;291:5406–5417. doi: 10.1074/jbc.M115.695874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Waitz JA, Horan AC, Kalyanpur M, Lee BK, Loebenberg D, Marquez JA, Miller G, Patel MG. Kijanimicin (Sch 25663), A Novel Antibiotic Produced by Actinomadura kijaniata SCC1256 Fermentation, Isolation, Characterization and Biological Properties. J Antibiot. 1981;34:1101–1106. doi: 10.7164/antibiotics.34.1101. [DOI] [PubMed] [Google Scholar]
- 177.Mallams AK, Puar MS, Rossman RR. Kijanimicin. 1. Structures of the Individual Sugar Components. J Am Chem Soc. 1981;103:3938–3940. [Google Scholar]
- 178.Mallams AK, Puar MS, Rossman RR. Kijanimicin. 2. Structure and Absolute Stereochemistry of Kijanimicin. J Am Chem Soc. 1981;103:3940–3943. [Google Scholar]
- 179.Horan AC, Brodsky BC. A Novel Antibiotic-Producing Actinomadura, Actinomadura kijaniata sp. nov. Int J Syst Bacteriol. 1982;32:195–200. [Google Scholar]
- 180.Mallams AK, Puar MS, Rossman RR, McPhail AT, Macfarlane RD, Stephens RL. Kijanimicin. Part 3. Structure and Absolute Stereochemistry of Kijanimicin. J Chem Soc, Perkin Trans 1. 1983:1497–1534. [Google Scholar]
- 181.Zhang H, White-Phillip JA, Melanon CEI, Kwon H-j, Yu W-l, Liu H-w. Elucidation of the Kijanimicin Gene Cluster: Insights into the Biosynthesis of Spirotetronate Antibiotics and Nitrosugars. J Am Chem Soc. 2007;129:14670–14683. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kusumi T, Ichikawa A, Kakisawa H, Tsunakawa M, Konishi M, Oki T. The Structures of Quartromicins A1: A2 and A3: Novel Macrocyclic Antiviral Antibiotics Possessing Four Tetronic Acid Moieties. J Am Chem Soc. 1991;113:8947–8948. [Google Scholar]
- 183.Tsunakawa M, Tenmyo O, Tomita K, Naruse N, Kotake C, Miyaki T, Konishi M, Oki T. Quartromicin, a Complex of Novel Antiviral Antibiotics I. Production, Isolation, Physico-Chemical Properties and Antiviral Activity. J Antibiot. 1992;45:180–188. doi: 10.7164/antibiotics.45.180. [DOI] [PubMed] [Google Scholar]
- 184.Roush WR, Barda DA. Partial Stereochemical Assignment of Quartromicins A3 and D3. Org Lett. 2002;4:1539–1542. doi: 10.1021/ol025771h. [DOI] [PubMed] [Google Scholar]
- 185.He HY, Pan HX, Wu LF, Zhang BB, Chai HB, Liu W, Tang GL. Quartromicin Biosynthesis: Two Alternative Polyketide Chains Produced by One Polyketide Synthase Assembly Line. Chem Biol. 2012;19:1313–1323. doi: 10.1016/j.chembiol.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 186.Wu LF, He HY, Pan HX, Han L, Wang R, Tang GL. Characterization of QmnD3/QmnD4 for Double Bond Formation in Quartromicin Biosynthesis. Org Lett. 2014;16:1578–1581. doi: 10.1021/ol500111n. [DOI] [PubMed] [Google Scholar]
- 187.Riedlinger J, Reicke A, Zahner H, Krismer B, Bull AT, Maldonado LA, Ward AC, Goodfellow M, Bister B, Bischoff D, Sussmuth RD, Fiedler HP. Abyssomicins, Inhibitors of the para-Aminobenzoic Acid Pathway Produced by the Marine Verrucosispora Strain AB-18-032. J Antibiot. 2004;57:271–279. doi: 10.7164/antibiotics.57.271. [DOI] [PubMed] [Google Scholar]
- 188.Bister B, Bischoff D, Strobele M, Riedlinger J, Reicke A, Wolter F, Bull A, TH, Zahner, Fiedler HP, Sussmuth RD. Abyssomicin C—A polycyclic Antibiotic from a Marine Verrucosispora Strain as an Inhibitor of the p-Aminobenzoic Acid/Tetrahydrofolate Biosynthesis Pathway. Angew Chem Int Ed. 2004;43:2574–2576. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- 189.Keller S, Nicholson G, Drahl C, Sorensen E, Fiedler HP, Sussmuth RD. Abyssomicins G and H and atrop-Abyssomicin C from the Marine Verrucosispora Strain AB-18-032. J Antibiot. 2007;60:391–394. doi: 10.1038/ja.2007.54. [DOI] [PubMed] [Google Scholar]
- 190.Roh H, Uguru GC, Ko HJ, Kim S, Kim BY, Goodfellow M, Bull AT, Kim KH, Bibb MJ, Choi IG, Stach JEM. Genome Sequence of the Abyssomicin- and Proximicin-Producing Marine Actinomycete Verrucosispora maris AB-18-032. J Bacteriol. 2011;193:3391–3392. doi: 10.1128/JB.05041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gottardi EM, Krawczyk JM, von Suchodoletz H, Schadt S, Muhlenweg A, Uguru G, Pelzer S, Fiedler HP, Bibb MJ, Stach JEM, Sussmuth RD. Abyssomicin Biosynthesis: Formation of an Unusual Polyketide, Antibiotic-Feeding Studies and Genetic Analysis. Chem Bio Chem. 2011;12:1401–1410. doi: 10.1002/cbic.201100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang Q, et al. Abyssomicins from the South China Sea Deep-Sea Sediment Verrucosispora sp.: Natural Thioether Michael Addition Adducts as Antitubercular Prodrugs. Angew Chem Int Ed. 2013;52:1231–1234. doi: 10.1002/anie.201208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tomita F, Tamaoki T, Shirahata K, Kasai M, Morimoto M, Ohkubo S, Mineura K, Ishii S. Novel Antitumor Antibiotics, Tetrocarcins. J Antibiot. 1980;33:668–670. doi: 10.7164/antibiotics.33.668. [DOI] [PubMed] [Google Scholar]
- 194.Tomita F, Tamaoki T. Tetrocarcins, Novel Antitumor Antibiotics I. Producing Organism, Fermentation and Antimicrobial Activity. J Antibiot. 1980;33:940–945. doi: 10.7164/antibiotics.33.940. [DOI] [PubMed] [Google Scholar]
- 195.Tamaoki T, Kasai M, Shirahata K, Ohkubo S, Morimoto M, Mineura K, Ishii S, Tomita F. Tetrocarcins, Novel Antitumor Antibiotics II. Isolation, Characterization and Antitumor Activity. J Antibiot. 1980;33:946–950. doi: 10.7164/antibiotics.33.946. [DOI] [PubMed] [Google Scholar]
- 196.Hirayama N, Kasai M, Shirahata K, Ohashi Y, Sasada Y. The Structure of Tetronolide, the Aglycone of Antitumor Antibiotic Tetrocarcin. Tetrahedron Lett. 1980;21:2559–2560. [Google Scholar]
- 197.Tamaoki T, Tomita F. Biosynthesis and Production of Tetrocarcin A, a New Antitumor Antibiotic in Chemically Defined Medium. Agric Biol Chem. 1982;46:1021–1026. [Google Scholar]
- 198.Fang J, Zhang Y, Huang L, Jia X, Zhang Q, Zhang X, Tang G, Liu W. Cloning and Characterization of the Tetrocarcin A Gene Cluster from Micromonospora chalcea NRRL 11289 Reveals a Highly Conserved Strategy for Tetronate Biosynthesis in Spirotetronate Antibiotics. J Bacteriol. 2008;190:6014–6025. doi: 10.1128/JB.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jiang ZD, Jensen PR, Fenical W. Lobophorins A and B, New Antiinflammatory Macrolides Produced by a Tropical Marine Bacterium. Bioorg Med Chem Lett. 1999;9:2003–2006. doi: 10.1016/s0960-894x(99)00337-6. [DOI] [PubMed] [Google Scholar]
- 200.Niu S, Li S, Chen Y, Tian X, Zhang H, Zhang G, Zhang W, Yang X, Zhang S, Ju J, Zhang C. Lobophorins E and F, New Spirotetronate Antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J Antibiot. 2011;64:711–716. doi: 10.1038/ja.2011.78. [DOI] [PubMed] [Google Scholar]
- 201.Wei RB, Xi T, Li J, Wang P, Li FC, Lin YC, Qin S. Lobophorin C and D, New Kijanimicin Derivatives from a Marine Sponge-Associated Actinomycetal Strain AZS17. Mar Drugs. 2011;9:359–368. doi: 10.3390/md9030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Li S, Xiao J, Zhu Y, Zhang G, Yang C, Zhang H, Ma L, Zhang C. Dissecting Glycosylation Steps in Lobophorin Biosynthesis Implies an Iterative Glycosyltransferase. Org Lett. 2013;15:1374–1377. doi: 10.1021/ol400342e. [DOI] [PubMed] [Google Scholar]
- 203.Xiao J, Zhang Q, Zhu Y, Li S, Zhang G, Zhang H, Saurav K, Zhang C. Characterization of the Sugar-O-methyltransferase LobS1 in Lobophorin Biosynthesis. Appl Microbiol Biotechnol. 2013;97:9043–9053. doi: 10.1007/s00253-013-5083-7. [DOI] [PubMed] [Google Scholar]
- 204.Lin Z, Koch M, Pond CD, Mabeza G, Seronay RA, Concepcion GP, Barrows LR, Olivera BM, Schmidt EW. Structure and Activity of Lobophorins from a Turrid Mollusk-associated Streptomyces sp. J Antibiot. 2014;67:121–126. doi: 10.1038/ja.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yue C, Niu J, Liu N, LY, Liu M, Li Y, Wang M, Shao M, Qian S, Bao Y, Huang Y. Cloning and Identification of the Lobophorin Biosynthetic Gene Cluster from Marine Streptomyces olivaceus Strain FXJ7.023. Pak J Pharm Sci. 2016;29:287–293. [PubMed] [Google Scholar]
- 206.Igarashi Y, Iida T, Oku N, Watanabe H, Furihata K, Miyanouchi K. Nomimicin, a New Spirotetronate-class Polyketide from an Actinomycete of the GenusActinomadura. JAntibiot. 2012;65:355–359. doi: 10.1038/ja.2012.30. [DOI] [PubMed] [Google Scholar]
- 207.Igarashi Y, Ogura H, Furihata K, Oku N, Indananda C. Thamchaipenet Maklamicin, an Antibacterial Polyketide from Endophytic Micromonospora sp. J Nat Prod. 2011;74:670–674. doi: 10.1021/np100727h. [DOI] [PubMed] [Google Scholar]
- 208.Daduang R, Kitani S, Sudoh Y, Pait IGU, Thamchaipenet A, Ikeda H, Igarashi Y, Nihira T. 29-Deoxymaklamicin, a New Maklamicin Analogue Produced by a Genetically Engineered Strain of Micromonospora sp. NBRC 110955. J Biosci Bioeng. 2015;120:608–613. doi: 10.1016/j.jbiosc.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 209.Daduang R, Kitani S, Hashimoto J, Thamchaipenet A, Igarashi Y, Shinya K, Ikeda H, Nihira T. Characterization of the Biosynthetic Gene Cluster for Maklam-icin, a Spirotetronate-class Antibiotic of the Endophytic Micromonospora sp. NBRC 110955. Microbiol Res. 2015;180:30–39. doi: 10.1016/j.micres.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 210.Ohtsuka T, Kudoh T, Shimma N, Kotaki H, Nakayama N, Itezono Y, Fujisaki N, Watanabe J, Yokose K, Seto H. Tetronothiodin, a Novel Cholecystokinin Type-B Receptor Antagonist Produced by Streptomyces sp. J Antibiot. 1992;45:140–143. doi: 10.7164/antibiotics.45.140. [DOI] [PubMed] [Google Scholar]
- 211.Kuwahara T, Kudoh T, Nagase H, Takamiya M, Nakano A, Ohtsuka T, Yoshizaki H, Arisawa M. Tetronothiodin, a Novel CCKB Receptor Ligand, Antagonizes Cholecystokinin-induced Ca2+ Mobilization in a Pituitary Cell Line. Eur J Pharmacol. 1992;221:99–105. doi: 10.1016/0014-2999(92)90777-2. [DOI] [PubMed] [Google Scholar]
- 212.Watanabe J, Fujisaki N, Fujimori K, Anzai Y, Oshima S, Sano T, Ohtsuka T, Watanabe K, Okuda T. Tetronothiodin, a Novel Cholecystokinin Type-B Receptor Antagonist Produced by Streptomyces sp. NR0489. I. Taxonomy, Yield Improvement and Fermentation. J Antibiot. 1993;46:1–10. doi: 10.7164/antibiotics.46.1. [DOI] [PubMed] [Google Scholar]
- 213.Ohtsuka T, Kotaki H, Nakayama N, Itezono Y, Shimma N, Kudoh T, Kuwahara T, Arisawa M, Yokose K, Seto H. Tetronothiodin, a Novel Cholecystokinin Type-B Receptor Antagonist Produced by Streptomyces sp. NR0489 II. Isolation, Characterization and Biological Activities. J Antibiot. 1993;46:11–17. doi: 10.7164/antibiotics.46.11. [DOI] [PubMed] [Google Scholar]
- 214.Ohtsuka T, Nakayama N, Itezono Y, Shimma N, Kuwahara T, Yokose K, Seto H. Tetronothiodin, a Novel Cholecystokinin Type-B Receptor Antagonist Produced by Streptomyces sp. NR0489 III. Structural Elucidation. J Antibiot. 1993;46:18–24. doi: 10.7164/antibiotics.46.18. [DOI] [PubMed] [Google Scholar]
- 215.Sun Y, Hahn F, Demydchuk Y, Chettle J, Tosin M, Osada H, Leadlay P. In vitro Reconstitution of Tetronate RK-682 Biosynthesis. Nat Chem Biol. 2010;6:99–101. doi: 10.1038/nchembio.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kanchanabanca C, Tao WX, Hong H, Liu YJ, Hahn F, Samborskyy M, Deng ZX, Sun YH, Leadlay PF. Unusual Acetylation-Elimination in the Formation of Tetronate Antibiotics. Angew Chem Int Ed. 2013;52:5785–5788. doi: 10.1002/anie.201301680. [DOI] [PubMed] [Google Scholar]
- 217.Zapf CW, Harrison BA, Drahl C, Sorensen EJ. A Diels-Alder Macrocyclization Enables an Efficient Asymmetric Synthesis of the Antibacterial Natural Product Abyssomicin C. Angew Chem Int Ed. 2005;44:6533–6537. doi: 10.1002/anie.200502119. [DOI] [PubMed] [Google Scholar]
- 218.Snider B, Zou Y. Synthesis of the Carbocyclic Skeleton of Abyssomicins C and D. Org Lett. 2005;7:4939–4941. doi: 10.1021/ol0518941. [DOI] [PubMed] [Google Scholar]
- 219.Butt NA, Moody CJ. Synthesis of Spirotetramates via a Diels-Alder Approach. Org Lett. 2011;13:2224–2227. doi: 10.1021/ol200477s. [DOI] [PubMed] [Google Scholar]
- 220.Hashimoto T, Hashimoto J, Teruya K, Hirano T, Shinya K, Ikeda H, Liu H-w, Nishiyama M, Kuzuyama T. Biosynthesis of Versipelostatin: Identification of an Enzyme-Catalyzed [4 + 2]-Cycloaddition Required for Macrocyclization of Spirotetronate-Containing Polyketides. J Am Chem Soc. 2015;137:572–575. doi: 10.1021/ja510711x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tian Z, Sun P, Yan Y, Wu Z, Zhang Q, Zhou S, Zhang H, Yu F, Jia X, Chen D, Mandi A, Kurtan T, Liu W. An Enzymatic [4 + 2] Cyclization Cascade Creates the Pentacyclic Core of Pyrroindomycins. Nat Chem Biol. 2015;11:259–265. doi: 10.1038/nchembio.1769. [DOI] [PubMed] [Google Scholar]
- 222.Koskiniemi H, Metsa-Ketela M, Dobritzsch D, Kallio P, Korhonen H, Mantsala P, Schneider G, Niemi J. Crystal Structures of Two Aromatic Hydroxylases Involved in the Early Tailoring Steps of Angucycline Biosynthesis. J Mol Biol. 2007;372:633–648. doi: 10.1016/j.jmb.2007.06.087. [DOI] [PubMed] [Google Scholar]
- 223.Kallio P, Patrikainen P, Belogurov GA, Mantsala P, Yang K, Niemi J, Metsa-Ketela M. Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE. Biochemistry. 2013;52:4507–4516. doi: 10.1021/bi400381s. [DOI] [PubMed] [Google Scholar]
- 224.Wang P, Bashiri G, Gao X, Sawaya M, Tang Y. Uncovering the Enzymes that Catalyze the Final Steps in Oxytetracycline Biosynthesis. J Am Chem Soc. 2013;135:7138–7141. doi: 10.1021/ja403516u. [DOI] [PubMed] [Google Scholar]
- 225.Wang T, Hoye TR. Diels-Alderase-Free, bis-Pericyclic [4 + 2] Dimerization in the Biosynthesis of (±)-Paracaseolide A. Nat Chem. 2015;7:641–645. doi: 10.1038/nchem.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Breslow R. Hydrophobic Effects on Simple Organic Reactions in Water. Acc Chem Res. 1991;24:159–164. [Google Scholar]
- 227.Blokzijl W, Blandamer MJ, Enberts JBFN. Diels-Alder Reactions in Aqueous Solutions. Enforced Hydrophobic Interactions between Diene and Dienophile. J Am Chem Soc. 1991;113:4241–4246. [Google Scholar]
- 228.Xu J, Deng Q, Chen J, Houk KN, Bartek J, Hilvert D, Wilson IA. Evolution of Shape Complementarity and Catalytic Efficiency from a Primordial Antibody Template. Science. 1999;286:2345–2348. doi: 10.1126/science.286.5448.2345. [DOI] [PubMed] [Google Scholar]
- 229.Sato M, Yagishita F, Mino T, Uchiyama N, Patel A, Chooi YH, Goda Y, Xu W, Noguchi H, Yamamoto T, Hotta K, Houk KN, Tang Y, Watanabe K. Involvement of Lipocalin-like CghA in Decalin-Forming Stereoselective Intramolecular [4 + 2] Cycloaddition. ChemBioChem. 2015;16:2294–2298. doi: 10.1002/cbic.201500386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kato N, Nogawa T, Hirota H, Jang JH, Takahashi S, Ahn JS, Osada H. A New Enzyme Involved in the Control of the Stereochemistry in the Decalin Formation during Equisetin Biosynthesis. Biochem Biophys Res Comm. 2015;460:210–215. doi: 10.1016/j.bbrc.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 231.Fujii R, Minami A, Gomi K, Oikawa H. Biosynthetic Assembly of Cytochalasin Backbone. Tetrahedron Lett. 2013;54:2999–3002. [Google Scholar]
- 232.Schumann J, Hertwerk C. Molecular Basis of Cytochalasan Biosynthesis in Fungi: Gene Cluster Analysis and Evidence for the Involvement of a PKS-NRPS Hybrid Synthase by RNA Silencing. J Am Chem Soc. 2007;129:9564–9565. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- 233.Li S, Srinivasan K, Tran H, Yu F, Finefield JM, Sunderhaus JD, McAfoos TJ, Tsukamoto S, Williams RM, Sherman DH. Comparative Analysis of the Biosynthetic Systems for Fungal Bicyclo[2.2.2]diazaoctane Indole Alkaloids: The (+)/(−)-Notoamide, Paraherquamide and Malbrancheamide Pathways. Med Chem Commun. 2012;3:987–996. doi: 10.1039/C2MD20029E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.He W, Liu M, Li X, Zhang X, Abdel-Mageed WM, Li L, Wang W, Zhang J, Han J, Dai H, Quinn RJ, Liu H-w, Luo H, Zhang L, Liu X. Fungal Biotransformation of Tanshinone Results in [4 + 2] Cycloaddition with Sorbicillinol: Evidence for Enzyme Catalysis and Increased Antibacterial Activity. Appl Microbiol Biotechnol. 2016;2016:1–9. doi: 10.1007/s00253-016-7488-6. [DOI] [PubMed] [Google Scholar]