Abstract
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has recently emerged in entomology as a technique to identify arthropods and their blood meal source. In this study, female Anopheles gambiae were fed on five host blood sources: ocelot (Leopardus pardalis), binturong (Arctictis binturong), springbok (Antidorcas marsupialis), jaguar (Panthera onca) and Hamadryas baboon (Papio hamadryas), while Anopheles coluzzii were fed on three hosts: dromedary (Camelus dromedarius), Barbary sheep (Ammotragus lervia) and pig (Sus scrofa). We obtained the MS spectra from 240 engorged mosquito abdomens and selected high quality ones from 72 mosquito abdomens to upgrade our home-made database. We excluded from the analysis any spectra of low quality (n = 80), and the remaining 88 specimens were subjected to a blind test analysis against the home-made database. We obtained 100% correct identification of the blood meal source for the specimens collected, 1, 12 and 24 h post-feeding, whereas for the specimens collected 36 h post-feeding, the correct identification rate decreased dramatically. We confirm here that MALDI-TOF MS can be used to identify the blood meal origin of freshly engorged mosquitoes, which opens new perspectives for further studies, including the impact of the mosquito species on blood meal identification.
Keywords: Blood meal identification, MALDI-TOF MS, Anopheles coluzzii, Anopheles gambiae
Abstract
L’identification par spectrométrie de masse à temps de vol par désorption/ionisation assistée par matrice (MALDI-TOF MS) a récemment émergé en entomologie pour l’identification des arthropodes et de leur source de sang. Des femelles d’Anopheles gambiae ont été nourries de sang de cinq hôtes, ocelot (Leopardus pardalis), binturong (Arctictis binturong), springbok (Antidorcas marsupialis), jaguar (Panthera onca) et babouin Hamadryas (Papio hamadryas), et des femelles d’Anopheles coluzzii ont été nourries sur trois hôtes, dromadaire (Camelus dromedarius), mouflon à manchettes (Ammotragus lervia) et porc (Sus scrofa). Nous avons obtenu les spectres MS à partir de 240 abdomens de moustiques engorgés et avons sélectionné ceux de 72 abdomens de moustiques de haute qualité pour améliorer notre base de données maison. Nous avons exclu de l’analyse les spectres de faible qualité (n = 80) et les 88 échantillons restants ont été soumis à une analyse de test en aveugle contre la base de données maison. Nous avons obtenu 100 % d’identification correcte de la source de sang pour les échantillons collectés, 1, 12 et 24 heures après l’alimentation, mais le taux d’identification correct a diminué de façon spectaculaire pour les échantillons collectés 36 heures après l’alimentation. Nous confirmons ici que la MALDI-TOF MS peut être utilisée pour identifier l’origine des repas sanguins des moustiques fraîchement engorgés et ouvre de nouvelles perspectives pour d’autres études, y compris l’impact des espèces de moustiques sur l’identification des repas sanguins.
Introduction
The analysis and identification of mosquito blood meals is essential to the study of vector bite behavior (anthropophilic or zoophilic). Several methods have been developed to identify the vertebrate host of mosquito blood meals, including serological tools such as precipitin and ELISA tests [3, 4]. Although these methods provide valuable information, they present several drawbacks, including the availability of specific antisera against antibodies, and their cross-reactivity [3, 4]. A molecular biology approach has been adopted as an effective strategy to identify mosquito blood sources [3, 4]. Nevertheless, DNA sequencing can be costly and time consuming [3, 4].
The matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) process is based on acidic extraction of proteins from an organism of interest, which are co-crystallized with a matrix. The molecules are ionized and propelled in a flight tube according to their mass-to-charge ratio. The detection of each molecule will generate an individual peak, therefore providing an overall spectrum which will be highly specific to the sample [22]. In recent years, this process has revolutionized clinical microbiology [21]. Recently, MALDI-TOF MS has emerged as an innovative tool to identify arthropods [8, 24, 25]. Also, in preliminary reports, MALDI-TOF MS has appeared as a promising tool to identify mosquito blood meals, using mosquitoes experimentally engorged in the lab, as well as using engorged mosquito abdomens crushed on Whatman filter papers (WFPs) collected during entomological field surveys [16–18, 23]. In these studies, a home-made MALDI-TOF MS database was established with abdomen MS spectra from mosquitoes freshly engorged on several vertebrate hosts [17, 18]. Other recent studies have used a tandem mass spectrometry approach to identify tick and triatomine blood meals [6, 7, 10, 19] by the trypsin digestion of the samples [6, 7, 10, 11, 19].
The goal of this study was to confirm the discriminative power of the MALDI-TOF MS tool for mosquito blood meal identification using a large panel of host blood vertebrates. For this purpose, Anopheles coluzzii and Anopheles gambiae Giles mosquitoes, two cryptic species among the main vectors of malaria in Africa [1, 5], were artificially fed with different host species that were not previously included in the home-made database.
Materials and methods
Ethical statement
The Sus scrofa blood sample was collected by a veterinary specialist, with the agreement of the owners, in accordance with standards relating to animal welfare. All other animals were living in zoos and were first anesthetized by a veterinarian responsible for monitoring their health. This protocol was approved by the Animal Ethics Committee of Marseille (C2EA-14), and by the French authorities. The blood samples were processed and stored in accordance with the World Health Organization’s Good Clinical Laboratory Practice guidelines and documents on blood sample handling procedures. (Good Clinical Laboratory Practice, 2009). Mosquitoes were reared using the International Conference on Harmonization/Good Laboratory Practices (ICH/GLP) procedures.
Laboratory rearing of Anopheles gambiae Giles and Anopheles coluzzii
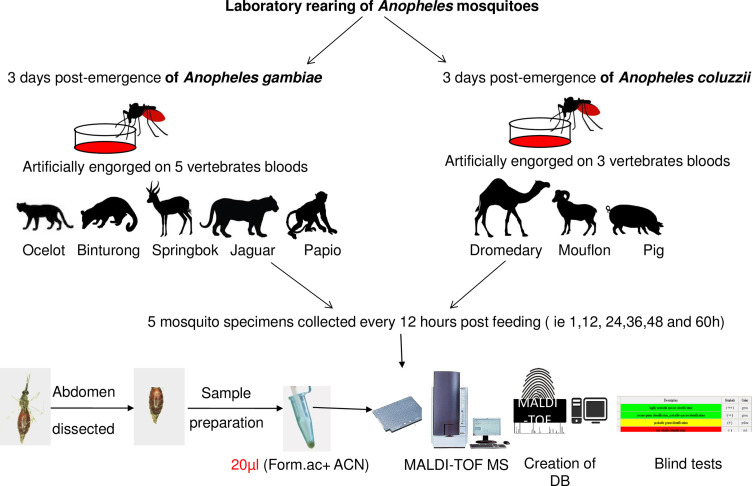
Both mosquito species were reared using standard methods at a temperature of 26 ± 1 °C, relative humidity of 70–90%, and over a photoperiod of 12 h (light/dark). Anopheles coluzzii adult females were artificially fed through a Parafilm-membrane on three blood vertebrates, namely Camelus dromedarius, Ammotragus lervia and Sus scrofa. The An. gambiae Giles females were artificially engorged on five vertebrate blood sources, namely Leopardus pardalis, Arctictis binturong, Antidorcas marsupialus, Panthera onca and Papio hamadryas for 2 h, as previously described [17, 18]. Engorged females were transferred to a new cage and fed with 10% glucose solution. Five engorged females were harvested between 1 and 60 h post-blood feeding, every 12 h (i.e. 1, 12, 24, 36, 48, and 60 h). The mosquito abdomens were placed in individually labeled vials. A flowchart illustrating the main steps is presented in Figure 1.
Figure 1.
Experimental workflow for blood meal identification using MALDI-TOF MS (Form.ac: Formic acid ; ACN: acetonitrile ; DB: database).
Spectral analysis
The individual mosquito abdomens were manually crushed and prepared as described previously [17]. MALDI-TOF MS spectra from engorged mosquito specimens were evaluated by analyzing the average spectra obtained from the four spectra of each sample tested using Flex Analysis 3.3 and ClinPro-Tools 2.2 software. These four spectra correspond to the four spots of the same sample on the MALDI-TOF target plate. High quality spectra (n = 40) were selected to create a dendrogram. Dendrograms are based on the results of a Composite Correlation Index (CCI) matrix. The CCIs are calculated by dividing spectra into intervals and comparing these intervals across a dataset. The composition of correlations of all intervals provides the CCI, which is used as a parameter that defines the distance between spectra [9].
Database upgrading and blind tests
In order to upgrade the MS arthropod home-made database (Additional file: Table S1), the four high quality spectra from at least three specimens per species, that were fed on the same blood and harvested at the same time point, were combined by the automated function of MALDI-Biotyper software v.3.3 to create 72 reference spectra. Spectra of low quality were excluded from the analysis. Subsequently, a blind test against the updated database was performed with all remaining MS spectra from the abdominals of clogged mosquitoes on eight separate host bloods. The results of the database queries are presented as Log Score Values (LSVs) for each spectrum given corresponding to a matched degree of signal intensities of mass spectra of the query and the reference spectra. LSVs ranged from 0 to 3 [9]. LSVs greater than 1.8 were considered as the threshold value for relevant identification, as previously published [17].
Results
A total of 240 MS spectra from 150 Anopheles gambiae Giles and 90 Anopheles coluzzii engorged abdomens, collected from 1 to 60 h post-blood feeding and spotted in quadruplicate, were obtained.
Comparison of these MS spectra revealed high reproducibility of protein profiles between the same vertebrate blood samples from mosquito abdomen protein extracts using Flex Analysis software (Fig. 2). Among the 240 spectra, 72 spectra of the highest quality were used to upgrade the home-made database. This database previously contained reference spectra for several arthropods, including reference spectra derived from the legs of 50 mosquito species, as well as reference spectra of Anopheles gambiae abdomens engorged on 17 different hosts (Table S1).
Figure 2.
MALDI-TOF MS spectra of Anopheles gambiae Giles and Anopheles coluzzii abdomen protein extracts engorged on host blood vertebrates. All mosquitoes were collected only at times 1 and 24 h post-feeding. Alignment of MS spectra from: H1_An_coluzzii_Camelus dromedarius (A), H24_An_coluzzii_ Camelus dromedarius (B), H1_ An_coluzzii Ammotragus lervia (C), H24_ An_coluzzii Ammotragus lervia(D), H1_ An_coluzzii _Sus scrofa (E), H24_ An_coluzzii _Sus scrofa (F), H1_ An_gambiae _Papio hamadryas (G), H24_ An_gambiae _Papio hamadryas (H), H1_An_gambiae_ Antidorcas marsupialus (I), H24_An_gambiae_ Antidorcas marsupialus (J), H1_An_gambiae_Arctictis binturong (K), H24_An_gambiae_ Arctictis binturong (L), H1_ An_gambiae_ _Panthera onca (M), H24_ An_gambiae_ _Panthera onca (N), H1_An_gambiae_Leopardus pardalis (O), and H24_An_gambiae_ Leopardus pardalis (P). a.u. arbitrary units; m/z mass-to-charge ratio.
We excluded from the analysis the spectra of low quality (n = 80) collected from 48 to 60 h post-blood feeding and the remaining MS spectra from 88 mosquito abdomens collected from 1, 12, 24 and 36 h, engorged on eight blood vertebrates, were tested against the home-made database. The specimens (n = 48) collected 1, 12 and 24 h post-feeding, revealed 100% correct identification of the host blood origin. The log score values (LSVs) for these freshly engorged specimens ranged from 1.912 to 2.918 (Table 1). Regarding the specimens collected at 36 h post-feeding (n = 40), the percentage of correct identification (with LSVs ranging from 1.396 to 2.207) dropped to less than 10%. Among them, 4/40 Anopheles coluzzii abdomens engorged on Sus scrofa blood (n = 2) and Ammotragus lervia blood (n = 2) were correctly identified with an LSV ranging from 1.800 to 2.207. For the remaining specimens collected at 36 h (n = 36, 90%), an incorrect blood source was identified (Table 1). As for the specimens collected 48 and 60 h post-feeding (n = 80), all MS spectra were of low quality.
Table 1.
Mosquito engorged abdomen spectra used for the home-made database and identification according to post-feeding period.
| Mosquito species | Blood meal source | Time of collection (hours) | Number of specimens used to upgrade the DB | Number of specimens used for blind tests | LSVs obtained from blind tests against DB | Mean of range | Blood meal identified by MS |
|---|---|---|---|---|---|---|---|
| Anopheles coluzzii | Ammotragus lervia | 1–24 | 9 | 6 | [2.518–2.918] | 2.756 | Ammotragus lervia |
| Anopheles coluzzii | Ammotragus lervia | 36 | 0 | 5 | [1.789–2.011] | 1.873 | / |
| Anopheles coluzzii | Camelus dromedarius | 1–24 | 9 | 6 | [2.418–2.830] | 2.706 | Camelus dromedarius |
| Anopheles coluzzii | Camelus dromedarius | 36 | 0 | 5 | [1. 597–2.029] | 1.764 | / |
| Anopheles coluzzii | Sus scrofa | 1–24 | 9 | 6 | [2.112–2.711] | 2.615 | Sus scrofa |
| Anopheles coluzzii | Sus scrofa | 36 | 0 | 5 | [1. 397–2.207] | 1.905 | / |
| Anopheles gambiae Giles | Antidorcas marsupialus | 1–24 | 9 | 6 | [2.530–2.807] | 2.613 | Antidorcas marsupialus |
| Anopheles gambiae Giles | Antidorcas marsupialus | 36 | 0 | 5 | [1. 562–1.941] | 1.764 | / |
| Anopheles gambiae Giles | Arctictis binturong | 1–24 | 9 | 6 | [2.585–2.892] | 2.754 | Arctictis binturong |
| Anopheles gambiae Giles | Arctictis binturong | 36 | 0 | 5 | [1.746 –1.892] | 1.818 | / |
| Anopheles gambiae Giles | Leopardus pardalis | 1–24 | 9 | 6 | [2.703–2.860] | 2.773 | Leopardus pardalis |
| Anopheles gambiae Giles | Leopardus pardalis | 36 | 0 | 5 | [1.747 –1.891] | 1.818 | / |
| Anopheles gambiae Giles | Papio hamadryas | 1–24 | 9 | 6 | [2.640–2.853] | 2.756 | Papio hamadryas |
| Anopheles gambiae Giles | Papio hamadryas | 36 | 0 | 5 | [1.652 –2.278] | 1.998 | / |
| Anopheles gambiae Giles | Panthera onca | 1–24 | 9 | 6 | [2.316–2.602] | 2.521 | Panthera onca |
| Anopheles gambiae Giles | Panthera onca | 36 | 0 | 5 | [1. 268–1.395] | 1.342 | / |
| Total | 72 | 88 |
Abbreviation: DB, home-made database.
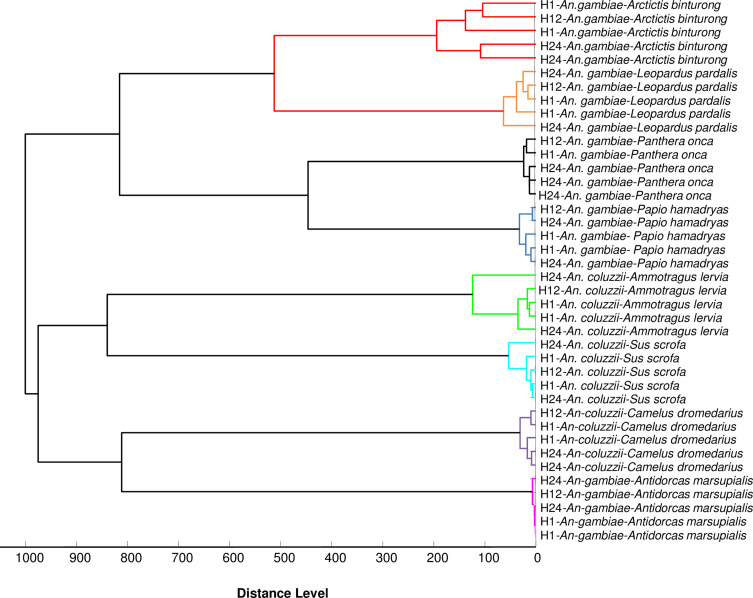
Cluster analysis revealed grouping on separate mosquito branches according to blood origin (Fig. 3). The MSP dendrogram showed that the spectra obtained from the abdomen of An. gambiae Giles, engorged on different felids (Leopardus pardalis and Panthera onca), were close (Fig. 3). The MSP dendrogram also revealed that the spectra obtained from the abdomen of Anopheles gambiae Giles, fed on different carnivore bloods, were close (Fig. 3).
Figure 3.
MSP dendrogram of MALDI-TOF MS spectra from Anopheles gambiae Giles and Anopheles coluzzii abdomens collected 1, 12 and 24 h post-feeding. MS spectra from two specimens from 1 to 24 h post-feeding and one specimen from 12 h post-feeding are represented. Blood meal host origins are indicated in the graph. Cluster analysis was performed by MALDI-Biotyper software v.3.3.
Discussion and conclusions
Our results confirm that MALDI-TOF MS is a promising tool for mosquito blood meal identification. The MS spectra generated from the abdomen of Anopheles gambiae Giles and Anopheles coluzzii revealed specific and reproducible blood results up to 24 h after feeding. In contrast, specimens collected 36 hours post-feeding revealed only 10% correct identification of the host blood origin, as suggested by preliminary studies [17, 18].
Cluster analysis revealed that all specimens fed with the same blood were clustered in the same branch (Fig. 3). Moreover, MALDI-TOF MS generated distinct MS profiles from the abdomens of Anopheles gambiae Giles freshly engorged with bloods of closely related animals, highlighting the specificity of this tool.
On the basis of this study and previous reports [17, 18], the field application of MALDI-TOF MS blood meal identification requires freshly engorged specimens. However, this is not problematic since field mosquitoes are caught in houses in the morning and traps are collected early, less than 24 h after the last blood meal [22]. In addition, Sella’s score visually determines the time and stage of digestion of mosquito blood meals and may be used to choose the mosquitoes that would give spectra of quality [14].
The MALDI-TOF approach may appear limited by the cost of the device and database comprehensiveness. Indeed, the correct identification of blood meals depends strictly on the presence of reference spectra in the database. Specimens without corresponding species reference spectra in the database matched with low log score values (<1.8). This underlines the need to continue database expansion with MS spectra from new vertebrate hosts [18]. However, reference spectra can be shared, avoiding the difficulties of creating a database. Nevertheless, when the MALDI-TOF device is purchased for clinical microbiology purposes, it can also be used for medical entomology at no additional cost [20].
This study opens new perspectives and further research is needed to better determine the usefulness of MALDI-TOF MS in medical entomology, and in particular for blood meal identification. In this study, the two included mosquito species, Anopheles gambiae Giles and Anopheles coluzzii were fed on different vertebrate bloods. However, An. gambiae and An. coluzzi are cryptic species, and it is not known at this stage whether similar results would be obtained by feeding mosquitoes of different genera. Although not noticeable for closely related species, the mosquito proteome might influence the obtained spectra and affect the resulting blood meal identification. If this were the case, the consequence would be the need to build an in-home-made database with a larger number of mosquitoes.
The MALDI-TOF MS tool has proven its effectiveness in identifying individual blood meals. However, in the field it is important to be able to identify multiple blood meals, as some studies have shown that mixed blood meals can be found in up to 10% of mosquitoes collected [13, 15]. Multiple host feedings (i.e. single mosquitoes taking blood from different types of hosts) have been observed and described by other authors in the field of entomological surveys [2, 12]. The effectiveness of the MALDI-TOF MS method opens other perspectives such as the identification of interrupted blood meals.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by the French Government under the “Investissements d’avenir” program managed by the French Agence Nationale de la Recherche (ANR), (Méditerranée Infection 10-IAHU-03). Our thanks go to Baptiste Mulot, veterinary surgeon at the Beauval zoological park, who provided animal blood samples. We also thank Sirama Niaré for his help with experimentation.
Author contributions
PP designed the study. PP and FT designed and developed the protocol. FT performed the experiments. PP, FT, and ML analyzed the data. BD and OD contributed reagents, materials, and analysis tools. FT wrote the paper. PP, ML, and OD reviewed and contributed to editing the paper. All authors agreed to publication.
Cite this article as: Tandina F, Laroche M, Davoust B, K Doumbo O & Parola P. 2018. Blood meal identification in the cryptic species Anopheles gambiae and Anopheles coluzzii using MALDI-TOF MS. Parasite 25, 40.
References
- 1. Coetzee M, Hunt RH, Wilkerson R, Della TA, Coulibaly MB, Besansky NJ. 2013. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa, 36(19), 246–274. [PubMed] [Google Scholar]
- 2. Degefa T, Yewhalaw D, Zhou G, Lee MC, Atieli H, Githeko AK, Yan G. 2017. Indoor and outdoor malaria vector surveillance in western Kenya: implications for better understanding of residual transmission. Malaria Journal, 16, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fyodorova MV, Savage HM, Lopatina JV, Bulgakova TA, Ivanitsky AV, Platonova OV, Platonov AE. 2006. Evaluation of potential West Nile virus vectors in Volgograd region, Russia, 2003 (Diptera: Culicidae): species composition, bloodmeal host utilization, and virus infection rates of mosquitoes. Journal of Medical Entomology, 43, 552–563. [DOI] [PubMed] [Google Scholar]
- 4. Gomes B, Sousa CA, Vicente JL, Pinho L, Calderon I, Arez E, Almeida AP, Donnelly MJ, Pinto J. 2013. Feeding patterns of molestus and pipiens forms of Culex pipiens (Diptera: Culicidae) in a region of high hybridization. Parasites & Vectors, 6, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hay SI, Sinka ME, Okara RM, Kabaria CW, Mbithi PM, Tago CC, Benz D, Gething PW, Howes RE, Patil AP, Temperley WH, Bangs MJ, Chareonviriyaphap T, Elyazar IR, Harbach RE, Hemingway J, Manguin S, Mbogo CM, Rubio-Palis Y, Godfray HC. 2010. Developing global maps of the dominant Anopheles vectors of human malaria. PLoS Medecine, 7, e1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Honig V, Carolan HE, Vavruskova Z, Massire C, Mosel MR, Crowder CD, Rounds MA, Ecker DJ, Ruzek D, Grubhoffer L, Luft BJ, Eshoo MW. 2017. Broad-range survey of vector-borne pathogens and tick host identification of Ixodes ricinus from Southern Czech Republic. FEMS Microbiology Ecology, 93(11), 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keller JI, Ballif BA, St Clair RM, Vincent JJ, Monroy MC, Stevens L. 2017. Chagas disease vector blood meal sources identified by protein mass spectrometry. PLoS One, 12, e0189647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laroche M, Almeras L, Pecchi E, Bechah Y, Raoult D, Viola A, Parola P. 2017. MALDI-TOF MS as an innovative tool for detection of Plasmodium parasites in Anopheles mosquitoes. Malaria Journal, 16, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laroche M, Berenger JM, Gazelle G, Blanchet D, Raoult D, Parola P. 2017. MALDI-TOF MS protein profiling for the rapid identification of Chagas disease triatomine vectors and application to the triatomine fauna of French Guiana. Parasitology, 145(5), 665–675. [DOI] [PubMed] [Google Scholar]
- 10. Laskay UA, Breci L, Vilcins IM, Dietrich G, Barbour AG, Piesman J, Wysocki VH. 2013. Survival of host blood proteins in Ixodes scapularis (Acari: Ixodidae) ticks: a time course study. Journal of Medical Entomology, 50, 1282–1290. [DOI] [PubMed] [Google Scholar]
- 11. Laskay UA, Burg J, Kaleta EJ, Vilcins IM, Telford Iii SR, Barbour AG, Wysocki VH. 2012. Development of a host blood meal database: de novo sequencing of hemoglobin from nine small mammals using mass spectrometry. Biology Chemistry, 393, 195–201. [DOI] [PubMed] [Google Scholar]
- 12. Lekweiry KM, Salem MS, Cotteaux-Lautard C, Jarjaval F, Marin-Jauffre A, Bogreau H, Basco L, Briolant S, Boukhary AO, Brahim KO, Pages F. 2016. Circumsporozoite protein rates, blood-feeding pattern and frequency of knockdown resistance mutations in Anopheles spp. in two ecological zones of Mauritania. Parasites & Vectors, 9, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Logue K, Keven JB, Cannon MV, Reimer L, Siba P, Walker ED, Zimmerman PA, Serre D. 2016. Unbiased characterization of Anopheles mosquito blood meals by targeted high-throughput sequencing. PLoS Neglected and Tropical Diseases, 10, e0004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martinez-de la Puente J, Ruiz S, Soriguer R, Figuerola J. 2013. Effect of blood meal digestion and DNA extraction protocol on the success of blood meal source determination in the malaria vector Anopheles atroparvus. Malaria Journal, 12, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moreno M, Saavedra MP, Bickersmith SA, Prussing C, Michalski A, Tong RC, Vinetz JM, Conn JE. 2017. Intensive trapping of blood-fed Anopheles darlingi in Amazonian Peru reveals unexpectedly high proportions of avian blood-meals. PLoS Neglected and Tropical Diseases, 11, e0005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niaré S, Almeras L, Tandina F, Yssouf A, Bacar A, Toilibou A, Doumbo O, Raoult D, Parola P. 2017. MALDI-TOF MS identification of Anopheles gambiae Giles blood meal crushed on Whatman filter papers. PLoS One, 12, e0183238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Niaré S, Berenger JM, Dieme C, Doumbo O, Raoult D, Parola P, Almeras L. 2016. Identification of blood meal sources in the main African malaria mosquito vector by MALDI-TOF MS. Malaria Journal, 15, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niaré S, Tandina F, Davoust B, Doumbo O, Raoult D, Parola P, Almeras L. 2017. Accurate identification of Anopheles gambiae Giles trophic preferences by MALDI-TOF MS. Infection, Genetics and Evolution. pii: S1567-1348(17)30315-5. [DOI] [PubMed] [Google Scholar]
- 19. Onder O, Shao W, Kemps BD, Lam H, Brisson D. 2013. Identifying sources of tick blood meals using unidentified tandem mass spectral libraries. Nature Communication, 4, 1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sambou M, Aubadie-Ladrix M, Fenollar F, Fall B, Bassene H, Almeras L, Sambe-Ba B, Perrot N, Chatellier S, Faye N, Parola P, Wade B, Raoult D, Mediannikov O. 2015. Comparison of matrix-assisted laser desorption ionization-time of flight mass spectrometry and molecular biology techniques for identification of Culicoides (Diptera: ceratopogonidae) biting midges in Senegal. Journal of Clinical Microbiology, 53, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiology, 5, 1733–1754. [DOI] [PubMed] [Google Scholar]
- 22. Sougoufara S, Sokhna C, Diagne N, Doucoure S, Sembene PM, Harry M. 2017. The implementation of long-lasting insecticidal bed nets has differential effects on the genetic structure of the African malaria vectors in the Anopheles gambiae complex in Dielmo. Senegal. Malaria Journal, 16, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tandina F, Niaré S, Laroche M, Koné AK, Diarra AZ, Ongoiba A, Berenger JM, Doumbo OK, Raoult D, Parola P. 2018. Using MALDI-TOF MS to identify mosquitoes collected in Mali and their blood meals. Parasitology, 7, 1–13. [DOI] [PubMed] [Google Scholar]
- 24. Yssouf A, Almeras L, Berenger JM, Laroche M, Raoult D, Parola P. 2015. Identification of tick species and disseminate pathogen using hemolymph by MALDI-TOF MS. Ticks and Tick Borne Diseases, 6, 579–586. [DOI] [PubMed] [Google Scholar]
- 25. Yssouf A, Almeras L, Raoult D, Parola P. 2016. Emerging tools for identification of arthropod vectors. Future Microbiology, 11, 549–566. [DOI] [PubMed] [Google Scholar]