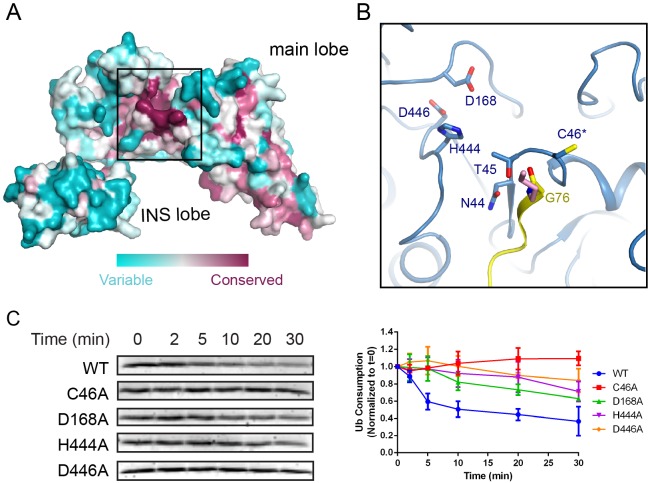
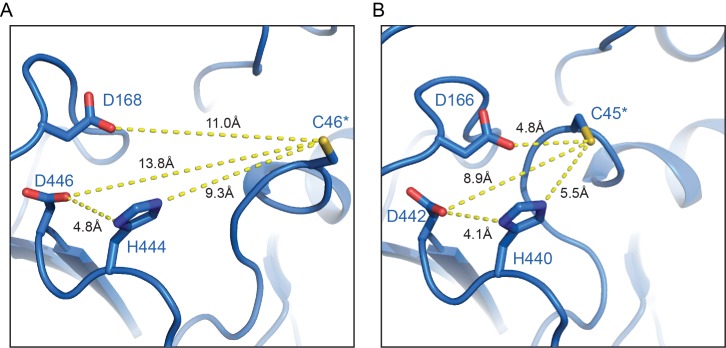
Figure 7. Conserved acidic residues near the SNL domain catalytic site.
(A) Surface conservation analysis of the SNL domain. The conservation was calculated from all SidC homologous sequences from all Legionella species with available genomic sequences using the ConSurf server, with the most conserved residues colored in purple and the least conserved residues in cyan. Note that the catalytic site of the SNL domain is concentrated with the most conserved residues. (B) A zoomed-in view of the most conserved residues at the catalytic site, including H444 and two acidic residues, D168 and D446. (C) Multiple turnover ubiquitination activity assays of SidC mutants of the conserved residues near the SidC active site monitored by the consumption of Ub. Left panel: representative SDS-gel of Ub remaining by the ubiquitination reaction at the indicated time points. Right panel: Quantified intensity of the remaining Ub at the indicated time points. The error bar represents the standard deviation of three independent experiments.