Abstract
Risk variants for schizophrenia affect more than 100 genomic loci, yet cell- and tissue-specific roles underlying disease liability remain poorly characterized. We have generated for two cortical areas implicated in psychosis, dorsolateral prefrontal cortex and anterior cingulate cortex, 157 reference maps from neuronal, neuronal-depleted and bulk tissue chromatin for two histone marks associated with active promoters and enhancers, H3-trimethyl-lysine 4 (H3K4me3) and H3-acetyl-lysine 27 (H3K27ac). Differences between neuronal and neuronal-depleted chromatin states were the major axis of variation in histone modification profiles followed by substantial variability across subjects and cortical areas. Thousands of significant histone quantitative trait loci (hQTLs) were identified in neuronal and neuronal-depleted samples. Risk variants for schizophrenia, depressive symptoms and neuroticism were significantly overrepresented in neuronal H3K4me3 and H3K27ac landscapes. Our PsychENCODE and CommonMind sponsored resource highlights the critical role of cell-type specific signatures at regulatory and disease-associated non-coding sequences in the human frontal lobe.
INTRODUCTION
Recent progress in understanding the genetic basis of many psychiatric diseases has identified both rare and common variants responsible for genetic risk1. Integrating epigenomics data from disease-relevant cell types and tissues promises to enhance interpretation of these risk variants and the mechanisms by which they confer disease liability2. This includes the exploration of non-coding regulatory DNA, and its epigenetic variation in mediating the effects of genetic risk variants2–4. Thus, the long-term goal of the PsychENCODE5, 6 and CommonMind7 consortia is to generate a large-scale epigenomics resource for the human brain to serve as a foundation for integrative genomics in psychiatric research6. To this end, nucleosomal histone modifications contribute to genome organization and function, with various histone methylation and acetylation markings—including H3-trimethyl-lysine 4 (H3K4me3) and H3-acetyl-lysine 27 (H3K27ac)—considered key regulators for active promoters and enhancers and other cis-regulatory non-coding sequences8. Importantly, molecular regulators for such types of open chromatin-associated histone modifications rank as top scoring biological pathways by genome-wide association in schizophrenia and bipolar disorder9, further underscoring the importance of fine mapping histone landscapes in brain. However, to date, only a few publically available histone datasets and resources exist for the human brain5, 10, 11, all of which were created from bulk tissue homogenate. These tissue homogenate-based resources have clearly contributed to a deeper understanding of the genetic risk architecture of common psychiatric disease. However, there is evidence that even in the context of normal cortical development and aging, vast portions of the neuronal genome show a very different histone modification landscape in comparison to the surrounding glia and other non-neuronal cells12, 13. Unfortunately, the degree to which cell type- and region-specific epigenomic signatures mediate the influence of genetic risk factors for psychiatric disease remains largely unexplored.
Here we present the largest dataset to date of open chromatin-associated histone modifications mapped separately in neurons versus the remaining neuron-depleted cell fraction from two higher order brain areas implicated in schizophrenia and other psychiatric diseases14: dorsolateral prefrontal cortex (PFC) and anterior cingulate cortex (ACC). Our publicly accessible resource, available at psychencode.org and https://www.synapse.org/#!Synapse:syn4566010 includes data, results and UCSC browser visualizations for cell-type specific maps from N=129 samples, complemented with another N=28 maps from tissue homogenate from adult control subjects without known neurological or psychiatric disease (Table S1). This epigenomics resource reveals hitherto unexplored insights into cell- and region-specific histone methylation and acetylation landscapes, including sites with extraordinarily high inter-individual variability. We elucidate the role of genetic regulation in influencing chromatin state and identify thousands of significant histone quantitative trait loci (hQTLs). We report striking enrichments of risk variants for schizophrenia, educational attainment, neuroticism and depressive symptoms highly specific to neuronal chromatin, thereby critically confirming cell type as a key variable in the neurogenomic architecture of psychiatric disease.
RESULTS
Samples and Sequencing
Nuclei were extracted from frozen-thawed gray matter collected from two frontal lobe areas implicated in higher order processing serving cognition and emotion: the dorsolateral prefrontal cortex (PFC) at the superior frontal gyrus, and the anterior cingulate cortex (ACC) positioned immediately dorso-anterior to the corpus callosum (Figure 1A, left panel). ChIP-Seq from chromatin immunoprecipitates with anti-H3K4me3 and anti-H3K27ac antibodies followed by 100 base pair paired end sequencing was performed for neuronal and non-neuronal nuclei separately after NeuN neuronal marker immunotagging and fluorescence-activated sorting (Figure 1A, right panel). NeuN, broadly expressed in the vast majority of cortical excitatory and inhibitory neurons15 is a prototype neuronal marker in adult human cortex16. We herein refer to the NeuN+ fraction as neuronal and the NeuN− fraction as ‘neuron-depleted’, while acknowledging that each of these two cell types is comprised of many different subpopulations17. Performing cell type-specific ChIP-Seq on 17 subjects (14 males and 3 females) × 2 brain regions, we generated N=129 cell-type specific (N=63 H3K4me3; N=66 H3K27ac), and N=28 tissue homogenate-based libraries from N=19 additional controls (N=11 H3K4me3, 4 female, 7 male; N=17 H3K27ac, 8 female, 9 male) passing ENCODE quality controls (>10 million uniquely mapped reads, normalized strand coefficient (NSC) > 1 and PCR bottleneck coefficient > 0.8, Figure S1 and Table S2).
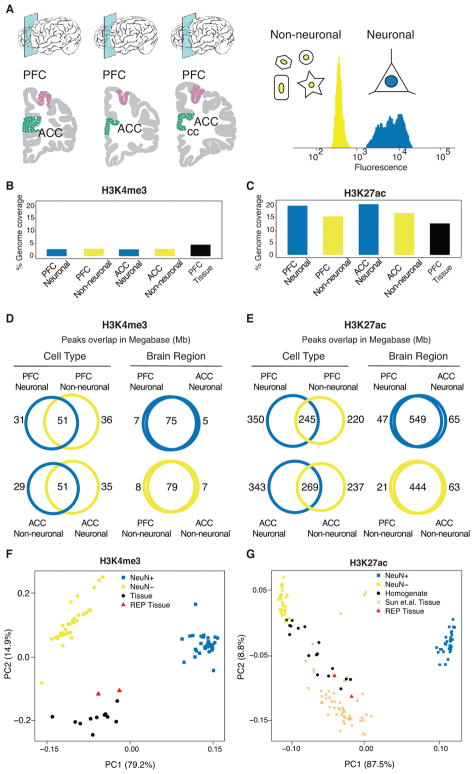
Figure 1. Cell- and region-specific histone modification profiling in the human frontal lobe.
(A) (left) Region-of-interest, dorsolateral prefrontal cortex (PFC) and anterior cingulate cortex (ACC) positioned dorsal and anterior from rostral genu of corpus callosum (cc). (right) representative FACS nuclei sorting showing fluorescence of NeuN antibody binding separating nuclei into neuronal (NeuN+) and neuron-depleted (NeuN−) fraction. (B,C) The genome wide coverage of ChIP-Seq peaks for each consolidated data set: PFC neuronal, PFC neuron-depleted, ACC neuronal and ACC neuron-depleted, separately for (B) H3K4me3, N (brains) = 17 PFC NeuN+, 14 ACC NeuN+, 17 PFC NeuN−, 15 ACC NeuN−, and (C) H3K27ac, N (brains) = 17 PFC NeuN+, 17 ACC NeuN+, 17 PFC NeuN−, 15 ACC NeuN−, (D,E) Venn diagrams by histone mark (panel D: H3K4me3, panel E: H3K27ac) cell type (blue: neuronal, gold: neuron-depleted), and brain region, summarizing the overlap (expressed in Mb) of called peaks (MACS2, P value<0.01). (F,G) PCA of pairwise correlations between each pair of ChIP-Seq log2 counts per million of samples from each mark. Visualization of first two principal components where each sample/data point shown in the figure are from 1) our cell-specific and homogenate dataset: PFC neurons, ACC neurons in blue squares, PFC non-neurons, ACC-non neurons are in golden squares (N (samples) = 63-H3K4me3, 66-H3K27ac) and PFC homogenate (N(samples) = 11-H3K4me3, 17-H3K27ac) in black circles 2) REP Roadmap Epigenomic project5: ACC and PFC homogenate are in red triangles and 3) Sun et.al.10: PFC homogenate in orange cross markers (N (brains) 53-H3K27ac).
For downstream analysis, we consolidated multiple ChIP-Seq datasets by cell type for each brain region and histone mark as 1) H3K4me3-PFC neuronal 2) H3K4me3-PFC neuron-depleted 3) H3K4me3-ACC neuronal 4) H3K4me3-ACC neuron-depleted and 5) H3K27ac-PFC neuronal 6) H3K27ac-PFC neuron-depleted 7) H3K27ac-ACC neuronal 8) H3K27ac-ACC neuron-depleted. Tissue homogenate samples for each histone mark were consolidated as 9) H3K4me3-PFC HBCC homogenate and 10) H3K27ac-PFC HBCC homogenate (although all our samples were acquired through the HBCC brain bank, the “HBCC” prefix was only used for the homogenate samples in order to distinguish them from the Roadmap Epigenomics Project tissue homogenates in our subsequent analysis.) Table S3 shows the list of samples in each of the 10 consolidated datasets (see Methods for a detailed description of the consolidation steps). The average number of uniquely mapped and non-redundant reads for the consolidated datasets by cell-type and brain region ranged from 13–41 million for H3K4me3 and 23–125 million for H3K27ac, reflecting that H3K27ac samples were sequenced at twice the coverage depth due to their larger width (Figure S1A). The subsequent steps of peak calling, read quantification of each peak, exploration of technical and biological covariates, differential modification analysis and functional annotation of peak sets (see Figure S2 for workflow diagram) were performed on each consolidated dataset. Across all individuals and both histone marks, ~50–70% of consolidated peaks in the cell type-specific data and ~20–40% in the tissue homogenate data had read coverage of at least 1 count per million (CPM) (Figure S3).
In order to evaluate the specificity of our histone-modification maps, we compared the peak coordinates to published H3K4me3 and H3K27ac maps from the Roadmap Epigenomics Project (REP) covering 111 tissues5. The maximum similarity (estimated based on Jaccard’s J) was found when our consolidated subset was compared to the REP brain tissues, while overlap with non-neural and peripheral REP tissues was lower (Figure S4 and Table S4). For both brain regions and epigenetic marks, our neuron-depleted samples which overwhelmingly are comprised of non-neuronal cells, had a higher similarity with REP brain samples than neuronal samples. Likewise, our NeuN− H3K27ac landscapes displayed a higher similarity with H3K27ac and also H3-acetyl-histone lysine 9 (H3K9ac) landscapes collected from bulk cortex tissue (homogenate) from independent brain cohorts10, 11. These observations, taken together, likely reflect the fact that the majority of cells residing in cortical gray matter are indeed non-neuronal18.
Genome-wide analysis of H3K4me3 and H3K27ac peaks reveal cell type specificity
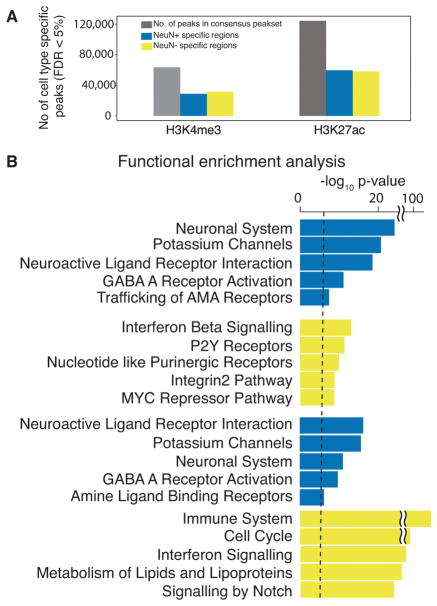
The cell type-specific peaksets (peaks called on consolidated datasets 1–8, above) varied by the fraction of the genome covered by peak regions, as well as by the degree of overlap with other subsets. As expected, the 61,000 – 95,000 narrow H3K4me3 peaks (range reflecting different cell types and cortical areas) covered a much smaller fraction of the genome than the 91,000 – 116,000 broader H3K27ac peaks (Table S5). For example, in PFC neurons, H3K4me3 peaks covered 82Mb (2.8%) of the genome, while H3K27ac covered 595Mb (19.8%) in the same subset (Figure 1B–E). Only minimal differences in the percentage of genomic coverage by H3K4me3 peaks (2.7–2.9%) was observed across cell types, whereas H3K27ac showed much higher genomic coverage for neuronal (19.8–20.4%) than neuron-depleted (15.5–16.9%) chromatin (Figure 1C).
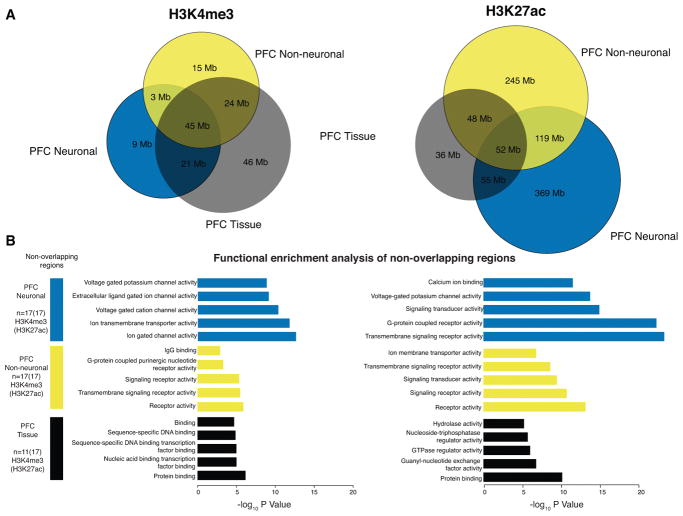
Principal component analysis (PCA) revealed distinct clusters of neuronal, neuron-depleted/non-neuronal and homogenate samples for both histone marks (Figure 1F,G and Figure S5A), however, samples from the PFC and ACC clustered together (see Figure S5B). This indicates a relatively high degree of epigenetic difference between neuronal and non-neuronal chromatin compared to a minimal difference between cortical areas. In contrast, chromatin from our PFC tissue homogenate samples and additional homogenate brain tissue from other sources5, 10, fall in between the FACS sorted cells along the first principal component (Figure 1F,G). Notably, the HBCC homogenate PFC samples are much more similar to the non-neuronal component and the fraction of NeuN− nuclei in our tissue homogenates comprised, on average, 60–70% of the total population of (Figure S6, Table S6), which is consistent with the fact that non-neuronal cells out number neurons by 1.6–2:1 in the human frontal lobe18. To further explore this similarity of PFC homogenate with non-neuronal cells, we quantified and analyzed the non-overlapping regions of PFC neuronal, PFC neuron-depleted and PFC HBCC homogenate peaksets. PFC neuronal chromatin included vast amounts of H3K27ac (369Mb) and H3K4me3 (46Mb) peak sequences not shared with either neuron-depleted or tissue homogenate, while only 245Mb (H3K27ac) and 15Mb (H3K4me3) of peak sequences were unique to non-neuronal chromatin not shared with tissue chromatin extracts or neurons (Figure 2A). Taken together, these characteristics illustrate a crucial advantage of cell-specific data over homogenate data. Functional enrichment of genes in close proximity to these non-overlapping modified peak regions using GREAT19 indicated distinct biological functions by cell type (Figure 2B, Table S7A–F). Neuron-specific H3K4me3 and H3K27ac peaks were enriched for ion channels, neurotransmitter signaling and synaptic genes, while genome regions marked in neuron-depleted and tissue homogenate peak sets showed enrichment for broader, less defined categories (Figure 2B).
Figure 2. Functional enrichment of non-overlapping cell- and tissue-specific histone peaks.
(A) Venn diagrams showing overlap in Mb of peak regions between neuronal (blue), non-neuronal (gold) and homogenate (black) for H3K4me3 (left panel) and H3K27ac (right panel). (B) Functional enrichments evaluated using GREAT 19 of peak regions that are unique to each of the 3 sets. Bar plots in blue, gold and black correspond to –log10 p-value from hypergeometric test of pathway enrichment results of peaks that are unique to neuronal, non-neuronal and homogenate respectively. H3K4me3, N (brains) = 17 PFC NeuN+, 17 PFC NeuN−, 11 PFC tissue homogenate, and H3K27ac N (brains) = 17 PFC NeuN+, 17 PFC NeuN−, 17 PFC tissue homogenate.
While this analysis has described large-scale trends, cell specificity of histone modification is readily visualized at the gene level. As representative examples, we consider CAMK2A and OLIG1, which are neuronal and non-neuronal specific genes, respectively (see Figure S7).
Collectively, our findings affirm that the neuronal epigenomic landscape is distinct from both non-neuronal and tissue homogenate landscapes. Although our findings indicate that chromatin maps from homogenate may omit critical neuron-specific epigenomic signatures, they do however provide a better representation of non-neuronal chromatin. Indeed, analysis of published brain histone QTLs from H3K27ac profiles in cortical homogenate10 showed modest enrichment for overlap with our NeuN− H3K27ac peaks, but a depletion for overlap with our NeuN+ H3K27ac peaks (Figure S8). This enrichment was highly specific as these hQTLs where depleted for overlap with H3K4me4 peaks. Moreover, analysis of another type of H3-acetyl mark (H3K9ac) from brain tissue homogenate11 showed only depletion for overlap with the two marks from neuronal and neuron-depleted chromatin in this study (Figure S8).
Neuronal histone modification landscapes show strong enrichment of schizophrenia GWAS loci
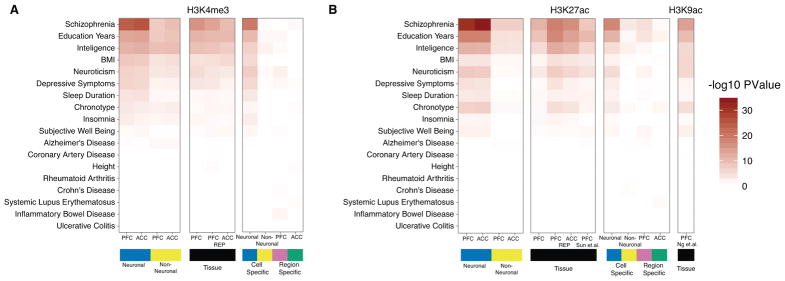
Due to the distinct histone modification landscapes between neuronal and non-neuronal cells in the frontal lobe, we wanted to better understand the role of cell- and region-specific epigenomic regulation associated with various psychiatric and non-psychiatric traits. To this end, we used the LD-score partitioned heritability method20 to examine the enrichment of common genetic variants identified by genome wide association studies (GWAS) within genomic regions with cell type-specific histone modifications. Altogether, 18 different types of brain and non-brain related diseases and conditions were included in these analyses (Figure 3).
Figure 3. Enrichment of heritability for brain and non-brain related phenotypes within cell- and tissue-specific histone peaks.
Using LD-score regression to partitioned heritability, we tested if the genetic variants contributing to 18 brain and non-brain-related phenotypes were enriched for (A) H3K4me3 and (B) H3K27ac. In (A), heritability enrichment analysis was preformed on multiple sets of genome regions that are visualized in 3 blocks. (1) Regions marked in blue and golden show enrichment values from PFC neuronal: 63,642 (105,075) peaks, ACC neuronal: 61,043 (116,714) and PFC neuronal-depleted: 95,501 (91,037) peaks, ACC-neuronal-depleted: 87,292 (101,885) from consolidated H3K4me3 (H3K27ac) datasets. (2) Regions marked in black show enrichment value of PFC tissue from our dataset: 158,345(183,885) peaks and PFC tissue: 75,912(317,582) peaks, ACC tissue: 79,844(260,288) peaks from the Roadmap Epigenomics project for H3K4me3(H3K27ac) marks. (3) Regions showing statistically significant differential histone modification either between the two cell types or two brain regions. Results for peaks that show increased histone modification in neurons: 28,838(59,588) peaks or non-neurons: 31,790(58,120) peaks from H3K4me3(H3K27ac) marks are indicated in blue and gold, respectively. Results for peaks that show increased histone modification in PFC: 696(10,665) peaks or ACC: 508(10,797) peaks from H3K4me3(H3K27ac) are indicated in purple and green, respectively. (B) Layout is the same as in (A) except that enrichment of 56,503 peaks from PFC tissue from Sun, et al.10 and 26,384 peaks from Ng et al. 11 were added. We note that regions marked in neurons consistently show the most significant enrichment for sequences associated with genetic risk for schizophrenia. H3K4me3, N (brains) = 17 PFC NeuN+, 14 ACC NeuN+, 17 PFC NeuN−, 15 ACC NeuN−, and 11 PFC tissue homogenate; H3K27ac. N (brains) = 17 PFC NeuN+, 17 ACC NeuN+, 17 PFC NeuN−, 15 ACC NeuN−, 17 PFC tissue homogenate (HBCC).
The strongest enrichment was found for schizophrenia-associated loci; weaker (nonetheless significant) enrichments were found for the genetic architectures associated with education years, intelligence, neuroticism, depressive symptoms, body mass index (BMI), chronotype and sleep duration (Table S8 A,B). Strikingly, each of these enrichments were almost exclusively limited to the neuronal histone modification landscapes of the PFC and ACC, suggesting that the aforementioned GWAS datasets link disease-associated vulnerabilities specifically to neurons. Indeed, the neuron-specific enrichment for sequences implicated in schizophrenia risk were consistently more significant than the comparatively weaker enrichment for these risk sequences in the histone modification maps from brain tissue homogenate herein, as well as in published H3K27ac10 and H3K9ac11 maps from brain tissue homogenate (Figure 3, Figure S9). For non-brain related traits such as height, coronary artery disease, Crohn’s disease and ulcerative colitis, we observed little enrichment for peaks from either neuronal and neuron-depleted chromatin. Furthermore, the strongest enrichment of brain related traits was identified in the non-overlapping peak regions of PFC neurons compared with the PFC HBCC homogenate peaksets, which further corroborates the association of GWAS loci of neuropsychiatric diseases with neuronal chromatin regions (Figure S9, Table S8 C,D). Finally, LD-score regression coefficients 21 from the enrichment analysis of schizophrenia were significantly larger in neuronal as compared to neuron-depleted chromatin and this effect was consistently observed for both histone marks in the two cortical regions, ACC and PFC (Figure S10). However, neither neuronal nor neuron-depleted PFC and ACC chromatin showed any significant overlap with Alzheimer’s disease associated variants, consistent with the hypothesis that Alzheimer’s disease risk variants are enriched for regulatory sequences within cells of myeloid origin3, 22–24
Decomposing quantitative variation in histone modification into multiple components
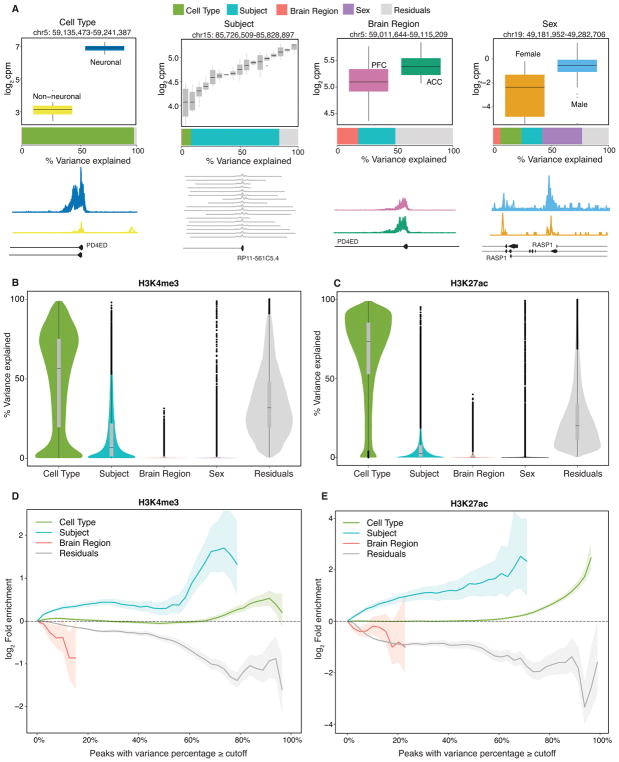
Quantitative epigenetic variation could be attributed to biological variation across cell types, subjects, brain regions and sexes. In order to quantify the percentage of variation in histone modification in each peak region that is attributable to each of these four variables plus residual variation, we fit a linear mixed model using variancePartition25 (Figure 4). Since variance percentages sum to 100%, these values can be easily compared across variables, peak regions and histone marks. The variance percentages are easily interpretable visually: a peak region with high variation across cell types shows distinct levels of histone modification in neuronal versus neuron-depleted chromatin (Figure 4A). The genome-wide trend across all peak regions for each mark indicates that cell type is the strongest source of variation in histone modification, followed by subject (Figure 4B,C). In contrast, variation across brain regions is very limited. Finally, as expected, variation across sexes was minimal genome-wide while exerting a strong effect on chrX and chrY linked genes. Thus, to further clarify the extent by which epigenomic differences between male and female frontal cortex are driven by histone peaks located on the sex chromosomes, we conducted principal component analysis of our 83 H3K27ac samples (11 female, 72 male) including cell-type specific and tissue homogenate datasets. Indeed, inclusion of regions on chrX and chrY resulted in strong sex-specific clustering on the 4th principal component while male and female brains completely intermixed when the analyses with repeated under exclusion of histone-tagged sequences specific to the X and Y chromosomes (Figure S11).
Figure 4. Decomposing multiples sources of epigenetic variation.
The contribution of epigenetic variation across 2 cell types, 17 subjects, 2 brain regions and 2 sexes, plus residual variation, were quantified using a linear mixed model implemented in variancePartition. (A) Representative examples of H3K4me3 consensus peaks (128,467 peaks from n=63 samples) where one source explains a large fraction of the epigenetic variation. Box plots indicate the log2 counts per million stratified by cell type, subject, brain region and sex. Box plot black horizontal line indicates median, box demarcates log2cpm in IQR (inter quartile range) for a given peak region, vertical line above and below the box plot show the 1.5IQR of upper (lower) quartile. Barplot below the box plot indicates the fraction of epigenetic variation in the peak explained by each variable. Genome coverage plot (bottom row) of ChIP-Seq signal (n=17 individuals) from data subset PFC neuronal for each peak region shown in the boxplot. (B–C) Violin plots indicate the genome-wide distribution of epigenetic variation across 4 variables, plus the residual variation for (B) H3K4me3 (128,467 peaks from n=63 samples) and (C) H3K27ac (147,539 peaks from n=66 samples). Each point represents a peak, and the width of the violin plot represents the number of peaks. Bar plot indicate the median and 25% and 75% quantiles. (D,E) Fold enrichment of histone QTL’s identified in lymphoblastoid cell lines26 and post mortem PFC10 for peaks with variance explained by each variable exceeding the cutoff indicated on the x-axis for (D) H3K4me3 (n=63 samples) and (E) H3K27ac (n=66 samples), respectively. Results for sex are not shown because enrichment for only autosomal genes was considered. Shaded regions indicate 90% confidence interval from 10,000 permutations.
Finally, in order to interpret the peak regions with the highest variation across subjects, we computed the overlap of peak regions from the current dataset with regions that have genome-wide significant histone QTLs identified in lymphoblastoid cell lines (LCL) and human brains for the H3K4me326 and H3K27ac10 epigenetic marks, respectively. Indeed, peak regions with high variation across subjects were enriched for regions that are histone QTLs in LCLs and human brains for their respective histone mark (Figure 4D,E). This is consistent with variation in histone modification across subjects being driven at least in part by genetic regulatory variation10, 26, 27. Importantly, this enrichment is limited to loci subject to epigenomic regulation that is common between neuronal and neuron-depleted chromatin (this study) and tissue extract and lymphoblast lines from previous studies10, 26.
Genetic regulation of histone modification in neuronal and neuronal depleted fractions
To examine whether there is cell type specific genetic regulation of histone modifications (as has been observed for gene expression6, 28 previously) we applied RASQUAL (Robust Allele-specific Quantitation and Quality Control), a QTL approach integrating allele-specific and between-subject differences29. Indeed, each of the 4 neuronal, neuron-depleted and bulk tissue chromatin preparations harbored thousands of histone quantitative trait loci (hQTLs), ranging from 6695 to 8042 for H3K27ac and 1565 to 3517 for H3K4me3 at FDR < 0.05, depending on chromatin fraction (Figure S12, Table S9). Of note, H3K27ac-tagged chromatin showing unexpectedly strong enrichment for Gene Ontology GREAT biological processes such as neurofilament organization, regulation of synaptic plasticity, associative learning, catecholamine-dependent signaling and various other pathways highly relevant to the neurobiology of schizophrenia and other common psychiatric disease (Figure S13).
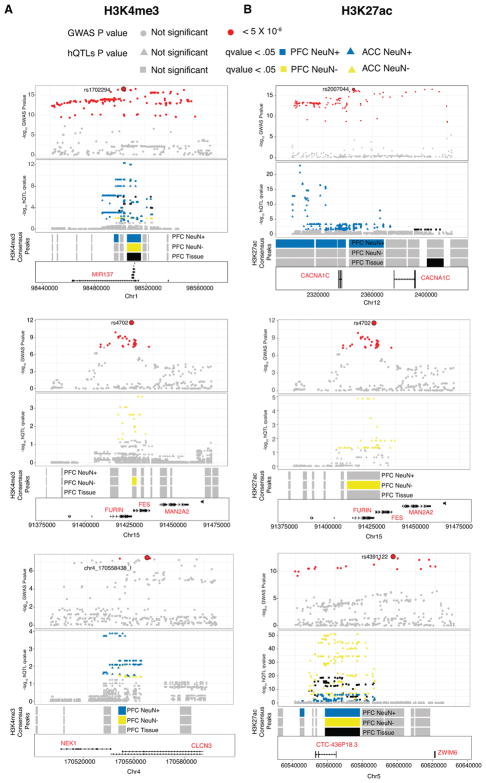
Since hQTLs calling is slightly underpowered due to the small sample size (n=36), we took a simple approach of comparing our cell specific and bulk tissue hQTLs with data from genome wide association studies (GWAS) for schizophrenia 30. We took all associations with p < 5 ×10−8 that are in high LD (r2 > 0.8) with the lead SNP and evaluated their overlap with our hQTLs. Comparisons across bulk tissue, neuronal and neuronal-depleted chromatin revealed strong cell type-specific effects for many of these risk-associated loci. For example, H3K4me3 peaks near MIR137 showed stronger hQTLs in neuronal samples than neuronal depleted and bulk tissue samples with localization of lead SNP rs1702294. H3K27ac peaks showed even stronger cell specific hQTL signal near the voltage-gated calcium channel CACNA1C in neuronal samples, whereas peaks for both histone marks near FURIN showed an hQTL single only in neuronal depleted samples (Figure 5, S12, Table S10A–J).
Figure 5. Overlap of cell specific and homogenate hQTLs with genome wide significant loci in schizophrenia.
A few representative genome wide significant loci in schizophrenia that overlap with cell specific and homogenate hQTLs are shown. All significant SNPs (p-value<5×10−8) are colored in red with lead SNP as a big red circle and rest are shown in gray. Corresponding to these representative GWAS loci, overlapping cell specific and homogenate hQTLs are shown in gray with significant hQTLs (RASQUAL q value < .05) colored in blue (neurons), gold (non-neurons) and black (homogenate). Consensus peak regions for which hQTLs were called are shown in gray separately for neurons, non-neurons and homogenates. These tracks are colored as blue, gold and black if they have any significant hQTL (RASQUAL q value < .05) that overlaps with the representative genome wide significant loci (p-value<5×10−8). From top to bottom, loci shown are MIR137, FURIN, CLCN3 for H3K4me3 (A) and CACNA1C, FURIN and ZWIM6 for H3K427ac (B). H3K4me3, N (brains) = 17 PFC NeuN+, 14 ACC NeuN+, 17 PFC NeuN−, 15 ACC NeuN−, and 11 PFC tissue homogenate; H3K27ac. N (brains) = 17 PFC NeuN+, 17 ACC NeuN+, 17 PFC NeuN−, 15 ACC NeuN−, 17 PFC tissue homogenate (HBCC).
Epigenomic variation between ACC and PFC
Cell type is the major source of quantitative variation in histone modification, with 55,628 H3K4me3 and 117,708 H3K27ac peak regions epigenetically different across cell types at FDR 5% (Figure 6A, Figure S14A). Unsurprisingly, for each histone mark, functional enrichment by gene categories was highly specific for cell type (Figure 6B). However, differences in histone modification between ACC and PFC were much smaller due to the similarity between the brain regions. Differential histone modification analysis between brain regions in neuronal cells identified 508 H3K4me3 and 10,797 H3K27ac peaks with increased modification in ACC, as well as 696 H3K4me3 and 10,665 H3K27ac peaks in PFC (Figure S14B, S15A–C, Table S11, S12). Interestingly, there was minimal region-specific signal in neuron-depleted chromatin with only 27 H3K4me3 and 18 H3K27ac peaks with increased modification in ACC were identified, and none in PFC (Figure S15D). These results indicate dramatically higher regional specificity for the population of neurons compared to their surrounding non-neuronal cells in the frontal lobe. It remains to be determined whether the differential histone acetylation landscapes in PFC vs. ACC neurons are reflective of differences in neurocognitive function between these cortical areas. For example, there is robust ACC activation with regard to reward processing, pain, affect and emotion31. In contrast, dorsolateral PFC is frequently implicated in the regulation of goal directed behavior including working memory and executive functions32. We note that multiple peak regions that are differentially modified between PFC and ACC neurons are proximal to neuropsychiatric risk genes (Table S13, S14). These include the forkhead transcription factor FOXP1 which functions synergistically with a related molecule, FOXP2, to regulate cognition and speech33, the exocytosis-regulator CADPS2 which is essential for axonal release of brain-derived nerve growth factor (BDNF)34, and the GRIK4 kainate receptor relevant for a broader range of disorders on the autism, mood and psychosis spectrum 35, 36.
Figure 6. Regions differentially modified in neuronal and non-neuronal cell types.
(A) Bar plot of counts of differentially modified peaks for each neuronal (blue) and neuron-depleted (gold) chromatin for H3K4me3 (top) and H3K27ac (bottom) at FDR < 5%. Differential modification analysis was performed on the normalized read counts matrix with columns as genomic regions 128,467 (147,539) and 4 types of samples PFC neuronal, ACC neuronal, PFC neuron-depleted and ACC neuron-depleted from 17 individuals (brains) as rows N (samples) = 63 (66) after QC for H3K4me3 (H3K27ac) (B) Functional enrichments for genes near differentially modified peaks computed with GREAT. Bar plots in corresponding to significant peaks (neuronal: 28,838(59,588) and non-neuronal 31,790(58,120) from H3K4me3(H3K27ac) marks identified in datasets from (A) N (samples) = 63 (66) H3K4me3 (H3K27ac) show top 5 pathways from REACTOME, Pathway Interaction Database and KEGG databases with –log10 p-value from hypergeometric test for neuronal and non-neuronal peaks respectively. Dashed line shows the Bonferroni cutoff at 2×10−5.
Transcriptional signatures of promoter-bound histone methylation and acetylation
While histone peaks from neuronal and neuron-depleted chromatin were bound to promoters, introns and intergenic elements (Figure S16), annotation of the H3K4me3 and H3K27ac peak sequences in the consolidated subsets revealed a large majority of sequences (70–79% of H3K4me3-tagged and 57–68% of H3K27ac-tagged) were bound to promoters within 5Kb of annotated transcription start sites (TSS). We therefore examined whether levels of H3K4me3 and H3K27ac modification are associated with gene expression magnitude in an independent set of post mortem brain samples from dorsolateral PFC from the CommonMind Consortium7. To this end, we calculated the number of ChIP-Seq reads aligned within 15kb of the annotated TSS of genes in 5 gene sets grouped by expression magnitude. As expected from findings in peripheral cells and tissues8, both H3K4me3 and H3K27ac ChIP-Seq reads were enriched around the TSS of genes with high levels of expression compared to genes with low levels of expression (Figure S17).
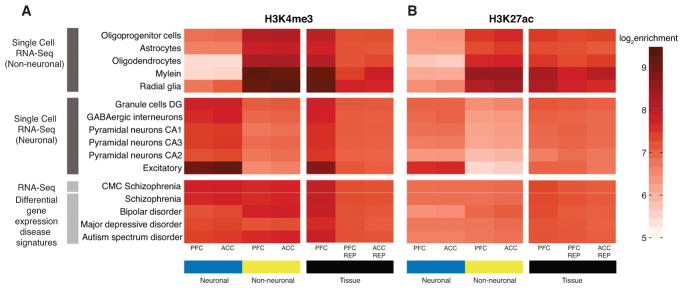
In our final analyses, we examined the association of neuronal, neuron-depleted and homogenate chromatin landscape with gene expression magnitudes in multiple subtypes of neurons and glia recently identified by massively parallel profiling of single brain nuclei17. Indeed, there were strong cell-type specific chromatin effects, with neuron-depleted chromatin showing strong enrichment for oligodendrocyte- and astrocyte-specific transcripts while, conversely, neuronal chromatin profiles were stronger associated with transcripts of the various types of neurons as compared to glia (Figure 7). In contrast, these enrichments showed no cell type specificity for chromatin fractions prepared from tissue homogenate (Figure 7). Not limiting our analyses to cell types, we examined the association of neuronal, neuron-depleted and homogenate chromatin with differentially expressed transcripts in multiple cohorts of subjects diagnosed with autism, bipolar disorder or schizophrenia37. Both cell-type and homogenate chromatin fractions showed moderate levels of enrichment with these disease-related gene sets (Figure 7).
Figure 7. Cell-type specific histone acetylation and methylation profiles are associated with differential enrichment for neuronal and glial transcripts.
(A,B) The colored tiles illustrates the log2 magnitude of enrichment of ChIP-seq counts (A, H3K4me3; B, H3K27ac) within 15 Kb downstream and upstream of transcription start site (TSS) of gene sets that are identified as neuronal and non-neuronal cell types from (top) scRNA-Seq 17 defining various neuronal and glial subtypes as indicated and (bottom) disease-associated gene expression profiles 37. Enrichments were quantified for cell-specific datasets (blue) PFC neuronal, ACC neuronal, (golden) PFC neuron-depleted, ACC neuron-depleted and PFC tissue (homogenate) including Human Brain Collection Core (HBCC), and the REP Roadmap Epigenomics Project5.
DISCUSSION
Interpreting the functional consequences of recently identified genetic variants contributing to the risk of neuropsychiatric disease requires a deeper understanding of the epigenomic context of these variants in brain and other tissues2–4, 6, 10, 11. We built the largest dataset of cell type specific reference maps for NeuN+ neuronal and NeuN− (overwhelmingly non-neuronal) histone modification landscapes for H3K4me3 and H3K27ac, which are typically associated with active promoter and enhancer regions, respectively. Importantly, non-neuronal chromatin showed a high degree of concordance with epigenomic landscapes of cortical homogenates from multiple sources. In contrast, histone methylation and acetylation landscapes form ACC and PFC neurons showed considerable ‘epigenomic distance’ to neuron-depleted and tissue homogenate samples (Figure 1F,G, S5A) suggesting they are likely a poor surrogate for neuron-specific alterations in the context of cognitive function and neurological disease. Given that the differences between neuronal and non-neuronal H3K4me3 and H3K27ac landscapes are the major axis of epigenomic variation (Figure 4), it will be essential for future studies to pursue additional sample fractionation by cell type, in order to capture the estimated 16 neuronal populations defined by single cell RNA sequencing in human cerebral cortex38 and potentially similar degrees of heterogeneity in glia as recently reported for mouse brain39. Such a higher resolution approach is expected to reveal vast numbers of genomic loci with an epigenomic signature unique to a specific type of neuron or glia, and provide deeper insight into the interrelation of transcriptome and histone modification landscapes. We also note the unexpectedly large quantitative H3K27ac differences between cell types, with a much larger genome coverage (20%) in neuronal chromatin decorated by histone acetylation versus only 15–16% genome coverage in neuron-depleted chromatin. The extended H3K27ac coverage broadly included intronic and intergenic sequences in addition to many promoter-bound peaks (Figure S12). While the functional implications of the extended H3K27ac peak coverage in the neuronal genome remains to be explored, we note that drugs interfering with the regulation of histone acetylation, including histone deacetylase inhibitors (HDACi) and suppressors of histone-acetyl-reader proteins, show a surprisingly broad therapeutic profile, improving cognition and neuronal function in a wide range of neuropsychiatric disease models40, 41,42. Furthermore, consistent with previous gene expression profiles in adult frontal cortex43, the transcriptional histone marks of the present study, H3K4me3 and H3K27ac, showed few sex-specific histone methylation and acetylation differences in the autosomal genome (Figure S11). However, previous DNA methylation profiling in cortical tissue homogenate from elderly brains revealed sex-specific effects for approximately 10% of age-sensitive methyl-CpG marks44. Presently, it is not known whether sex-specific regulation of histone modifications is increased in aged brain.
One primary goal of the PsychENCODE Consortium is to explore regulatory non-coding DNA associated within the genetic risk architectures of common neuropsychiatric disorders6. Using linkage disequilibrium-score regression to partition heritability20, we found strong, specific enrichments for schizophrenia, and somewhat weaker association with depression, neuroticism and education attainment in both H3K4me3 and H3K27ac peaks (Figure 3). This effect is primarily if not exclusively driven by neuronal chromatin (Figure 3, S8), with minimal or no contribution from neuron-depleted chromatin. Intriguingly, the strongest association with brain region specific peaks identifies risk variants for schizophrenia and educational attainment specifically in PFC neurons, consistent with the key role of the PFC in executive function Taken together, these findings underscore the importance of ‘epigenomic fine mapping’ with maximal region- and cell-type specific resolution for the human brain, in order to link the genetic risk architectures of neuropsychiatric disorders to selected cell populations or neural circuits.
Our cell type specific reference maps, accessible through the PsychENCODE Knowledge Portal and UCSC browser on Synapse (https://www.synapse.org/#!Synapse:syn4566010) is a valuable resource that will empower future studies exploring the epigenetic foundations of cell type specific genome organization and function in human brain, with important implications for the neurobiology of common psychiatric disease.
METHODS
Brains
All tissue donors of the present study were from the Human Brain Collection Core (HBCC) at the National Institutes of Health. None of the brains had known neurological or psychiatric disease. All brains had undergone a detailed neuropathological exam (incl. Bielschowsky stain) and were considered normal by histopathology. Demographics of the brain cohort, and toxicology and neuropathology reports are summarized Table S1. Sample size: No statistical methods were used to pre-determine sample sizes but our sample sizes exceeded those reported in previous publications focused on cell-type specific histone profiling in human brain (references 12, 13, 45) by several-fold.
Antibodies, ChIP-Seq library preparation and sequencing
Nuclei were extracted from approximately 300mg aliquots of frozen frontal (dorsolateral prefrontal and anterior cingulate gyrus) cortex tissue, immunotagged with Anti-NeuN-Alexa488 (Cat# MAB377X, EMD Millipore) antibody which robustly stains human cortical neuron nuclei45,46 for subsequent fluorescence-activated nuclei sorting. Next, chromatin of sorted nuclei was digested with micrococcal nuclease and subsequently pulled down with anti-histone antibodies, followed by library preparation and sequencing. Two histone antibodies, anti-H3K4me3 (Cat# 9751BC, lot 7; Cell Signaling, Danvers, MA) and anti-H3K27ac (Cat# 39133, Lot# 01613007; Active Motif, Carlsbad, CA) were used for immunoprecipitation. Antibody specificity was tested using peptide binding assays and immunoblotting of nuclear extracts from human postmortem cortical tissue. A commercially available histone H3 peptide array (Cat# 16–667; Millipore) containing 46 peptides representing 46 different histone H3 posttranslational modifications was used as previously described45. All procedures were performed as described in the recent PsychENCODE methods paper, providing a detailed description of the protocol45. For each cell-type specific ChIP-assay, a minimum of 400,000 sorted neuronal (NeuN+) or neuron-depleted/non-neuronal (NeuN−) nuclei was required as starting material. For selected gene promoters ChIP-PCR was conducted to validate cell-type specific peak profiles (Figure S18 and ref.45,46). Furthermore, quality controls for nuclei post-FACS included visual inspection under the microscope as described45. Of note, due to our stringent FACS gating criteria with maximized specificity (not sensitivity)(Figure S19), 100% of sorted nuclei in the neuronal fraction showed green fluorescence confirming NeuN+ status, while 100% of sorted nuclei in the non-neuronal fraction only showed blue DAPI stain, confirming NeuN− status. For the prefrontal cortex samples, we collected (mean±SD) NeuN+(PFC) 667,675±196,847 and NeuN-(PFC) 611,025±203,172 nuclei. For the anterior cingulate cortex samples, we collected NeuN+(ACC) 490,585±184,358 and NeuN-(ACC) 653,743±389,284 nuclei.
Additional ChIP-seq studies were conducted with homogenized dorsolateral prefrontal cortex as input. To this end, frozen human postmortem brain tissue (approximately 20–200mg) was homogenized in lysis buffer and the total nuclei were purified. The nuclei solution was resuspended in 300ul of douncing buffer, treated with 2uL of micrococcal nuclease (0.2U/uL) for 5 minutes at 28 degrees Celsius, followed by 30uL of 500mM of EDTA to stop the reaction. After this initial procedure for nuclei preparation and digestion, the sample was processed in the same manner as described for the FACS sorted nuclei samples.
Randomization and blinding
To avoid batch effects and other confounds, samples underwent repeated rounds of randomization, including (i) chromatin immunoprecipitation procedures and (ii) library preparation. Blinding was not relevant to this study, analysts were aware of data generation, processing and donor metadata.
ChIP-Seq Alignment
Sequenced cell-specific and homogenate ChIP-Seq FASTQ files were aligned to Hg19 (Feb 2009, GRCh37) human genome using the Burrows-Wheeler Aligner (BWA-0.7.8-r455) method with default settings47.. The output files were exported as BAM files.
Filtering and quality control
PCR duplicates in aligned BAM files were removed using picard 2.2.4 tool48. After filtering out duplicates, all BAM files were preprocessed to remove unmapped reads and any inter-chromosomal read pairs of length >10 kb. The mapped reads were subsampled to the median number of paired-end reads of each dataset; H3K4me3=13M and H3K27ac=23M (Figure S1). Any samples after removing duplicates with sequencing depth < 10M (from ENCODE49) were flagged in this study. These uniformly subsampled files were used for further downstream analysis.
Experimental design and statistical analyses
For a general overview of the bioinformatical analyses see Figure S2. To determine the best Peak calling method, we used p-value from irreproducible discovery rate (IDR) analysis where the input was peaks called using MACS2, PeakSeq and SPP methods. To identify differentially modified histone peaks across cell types and brain regions, we applied quasi-likelihood negative binomial generalized log-linear model on normalized CPM matrix. For multiple testing correction of identified differential peaks, we used Benjamini-Hochberg method on the p-values to control false discovery rate. For pathway enrichment analysis of differentially modified peaks, we used p-values from hypergeometric test computed by GREAT and did multiple testing correction using Bonferroni correction method. To test overlap of identified peaks with disease and trait-associated genetic variants, we used LDSR method which take p-values of peak regions as an input. See methods paragraphs below for additional details on statistical methods.
Variant Calling
Variants were called from BAM files using GATK 3.5–050 to produce gVCF files. Variant concordance analysis was performed to identify any mislabeling issues. Variants on chr22 were merged using GATK’s CombineVCFs functionality. Variant concordance between all pairs of samples was evaluated with bcftools v1.351. Two mislabeled samples were identified and were relabeled appropriately for all downstream analysis.
Comparison of peak calling methods
For each histone mark, we consolidated BAMs across all individuals for PFC neuronal set and subsampled 3 files. Our approach to determine best peak calling method was to derive irreproducible discovery rate (IDR)52 after calling peaks using MACS (v.2.1.0)53, SPP (v.1.13)54 and PeakSeq55 methods. Afterwards, the method which gave maximum number of overlapping regions between subsamples at 5% IDR was used for peak calling on full dataset. We find that MACS2 is the best peak caller method with maximum number of peak regions at 5% IDR for both marks (Figure S1D). ENCODE uses IDR on technical replicates of samples to determine the reproducibility of peaks52 while we have used it globally on our dataset. We applied following parameters in MACS2: SE, SE no model, PE, PE no model and p-values = 0.01, 0.1, 0.5, SPP: FDR 0.01, 0.05, 0.99, background model=simulated, minimum interpeak distance=150 and PeakSeq: target FDR=0.01, 0.05, 0.99.
Consolidating Datasets
For cell-specific data set, we consolidated uniformly processed BAMs by cell type for each brain region. For example H3K4me3 modified Chip-seq BAMs from neuronal cells from PFC brain region for all individuals (n=17) were consolidated as H3K4me3-PFC neuronal data set. Consolidating the BAMs by cell type for each brain region produces 8 large BAM files for both marks: 1) H3K4me3-PFC neuronal 2) H3K4me3-PFC neuron-depleted 3) H3K4me3-ACC neuronal 4) H3K4me3-ACC neuron-depleted and 5) H3K27ac-PFC neuronal 6) H3K27ac-PFC neuron-depleted 7) H3K27ac-ACC neuronal 8) H3K27ac-ACC neuron-depleted.
Chip-seq BAMs for homogenate were generated from one brain region, therefore, all individuals BAMs were consolidated into 2 large BAM files for both marks as 1) H3K4me3-PFC HBCC homogenate 2) H3K27ac-PFC HBCC homogenate. Similarly input samples were consolidated separately for cell-specific and homogenate datasets. For details of set of individual files contributing to consolidated dataset (Table S3). We use these cell-specific (n=8) and homogenate (n=2) consolidated BAMs for further downstream analysis.
Peak Calling
Narrow peak regions were called for H3K4me3 histone mark datasets on each of the consolidated cell-specific and homogenate BAMs: 1) H3K4me3-PFC neuronal 2) H3K4me3-PFC neuron-depleted 3) H3K4me3-ACC neuronal 4) H3K4me3-ACC neuron-depleted 5) H3K4me3-PFC HBCC homogenate with Poisson p-value = 0.01 with --keep-dup all --nomodel --extsize = fragment length. Broad peak regions were called for H3K27ac histone mark datasets on each of the consolidated BAMs 6) H3K27ac-PFC neuronal 7) H3K27ac-PFC neuron-depleted 8) H3K27ac-ACC neuronal 9) H3K27ac-ACC neuron-depleted and 10) H3K27ac-PFC HBCC homogenate using latter parameters. The consolidated cell-type and homogenate input control samples were used as control inputs for peak calling on cell-specific and homogenate datasets respectively.
All called peaks were filtered from blacklisted49 region peaks and p-values > 3.05 (p-value obtained from IDR analysis) for downstream analysis. For each mark, the coordinates for peaks for each set PFC neuronal, PFC neuron-depleted, ACC neuronal, ACC neuron-depleted used in this study are given for cell specifc-H3K4me3=syn11306591, homogenate-H3K4me3=syn11306589, cell specific-H3K27ac= syn9998643 and homogenate-H3K27ac= syn11485660. Before calling peaks using MACS2, we first ran SPP to find the fragment length using maximum strand cross correlation (Figure S1B, S2). For QC parameters (NSC, RSC, PBC and number of mapped reads) of uniformly reprocessed and consolidated ChIP-Seq sets we used phantompeakqualtools54. The NSC of all samples used in this study were above threshold of 1.1 (Figure S1C). We provide summarized QC parameters of individual files (Table S2) and consolidated (Table S5) data sets respectively.
Functional enrichment of non-overlapping cell- and tissue-specific histone peaks
In order to interpret the specificity of cell-type and homogenate data, we identified their respective unique or non-overlapping peak regions. Non-overlapping regions in a dataset are defined as all genomic regions except the ones that have at least 50% overlap with the dataset they are compared with. We have examined the biological function of nearby genes of these non-overlapping peak regions using Genomic Regions Enrichment of Annotations Tool (GREAT)19. The settings for genomic regions used are (proximal: 5.0 kb upstream, 5.0kb upstream and distal to 100 kb.)
Gene set enrichment analysis based on single cell RNA-seq
We next examined the difference between the Chip-seq signal in cell specific and homogenate datasets by measuring the enrichment of gene sets identified in neuronal and neuron-depleted chromatin subtypes by massive parallel profiling of single brain nuclei17. For neuronal subtypes we used identified gene sets for excitatory neurons (n=24), pyramidal neurons CA1 (n=132), pyramidal neurons CA2 (n=111), pyramidal neurons CA3 (n=50), GABAergic interneurons (n=145) and granule cells DG (n=163) and for non-neuronal subtypes we used identified genes sets for radial glia (n=10), myelin (n=16), oligodendrocytes (n=120), astrocytes (n=155) and oligoprogenitor cells (n=42).
nsgplot v2.6156 was used to quantify ChIP-Seq reads enrichment of 7 datasets for both marks: 1) PFC neuronal 2) PFC neuron-depleted, 3) ACC neuronal 4) ACC neuron-depleted, 5) PFC HBCC homogenate, 6) PFC REP Homogenate 7) ACC REP homogenate for abovementioned neuronal and non-neuronal gene sets as a function of 15kb distance upstream, downstream around TSS for both marks. We calculated the magnitude of area under these ChIP-Seq reads enrichment curve to examine the difference between the enrichment of cell-specific and homogenate datasets for neuronal and non-neuronal subtypes.
In addition to these genesets we measured the enrichment of ChIP-Seq reads for neuropsychiatric disease signatures as well. We curated these genesets for 1) CMC schizophrenia (n=693) based on RNA-seq differential gene expression between cases and controls from PFC region from 690 individuals (p-value <= 0.05) and 2) schizophrenia (n=884), 3) bipolar disorder (n=179), 4) major depressive disorder (n=25) 5) autism spectrum disorder (n=933) based on differential gene expression cerebral cortex region from microarray studied done on 715 individuals (p-value <= 0.05 and log2FC >= 2.)
Quantification of ChIP-seq signal in each peak
To determine the reads coverage across the whole genoßme for a BAM file, we used featureCounts from subread 1.5.257. The data input to featureCounts consists of a) uniformly processed BAM files and b) a consensus peak file in Simplified Automation Format (SAF). The consensus peak signals for H3K4me3 and H3K27ac were generated by taking the union of MACS2 narrowPeak files of cell-specific and homogenate consolidated datasets which are 1) H3K4me3-PFC neuronal, 2) H3K4me3-PFC non-neuronal, 3) H3K4me3-ACC neuronal, 4) H3K4me3-ACC neuron-depleted and 5) H3K4me3-PFC HBCC homogenate and union of MACS2 broadPeaks files of 5) H3K27ac-PFC neuronal, 6) H3K27ac-PFC neuron-depleted, 7) H3K27ac-ACC neuronal, 8) H3K27ac-ACC neuron-depleted respectively. featureCounts quantifies number of reads for each sample in every peak region of consensus signal. The counts were put together in a matrix separately for H3K4me3 and H3K27ac marks with 74 (cell-specifc=63, homogenate=11) samples from 28 individuals (cell-specific=17, homogenate=11) as rows for H3K4me3 and 83 (cell-specific=66, homogenate=17) from 34 individuals (cell-specific=17, homogenate=17) as rows for H3K27ac and 107,480 (152,590) peak regions as columns for H3K4me3 (H3K27ac). This matrix was converted into log2 counts per million (CPM) using TMM normalization58 to correct for the total number of reads. The log2 CPM matrix was used for downstream analysis.
Decomposing variation into multiple components with variancePartition
For cell-specific dataset for each histone mark, the epigenetic variance of each peak was decomposed into variation attributable to cell type, subject, brain region, sex, plus the residual variation:
These 4 variables are categorical and so were modeled as random effects. The analysis was performed by modeling the log2 CPM with a linear mixed model implemented in variancePartition v1.4.125 and treating each variable as a random effect. Each peak was considered separately and the results for all peaks were aggregated afterwards. Results were summarized in terms of the fraction of total variation explained by each variable for each peak.
A variancePartition analysis was also performed on additional metadata variables such as QC statistics (i.e. NSC, RSC, PCR PBC and NRF), and sample processing batches (library preparation date of chip, chip DNA volume, chip DNA amount (ng), total chip DNA in a library, library preparation operator, library AMpure beads lot, library PCR cycles number, library volume, library sequencing batch, library sequencing submission date, library preparation library batch.) Continuous variables were modeled as fixed effects and categorical variables were modeled as random effects. The percentage variation explained by technical variables such as experimental batches or QC statistics was either relatively small or better explained by the 4 major variables described above.
Principal components analysis
As a QC step, performed PCA on the log2 CPM matrix in order to identify outliers. 8 samples were identified as outliers (Table S2) and these corresponded to samples that barely passed our previous QC cutoffs. These samples were excluded from further analysis.
Differential histone modification
For cell-specific dataset for each mark, we performed differential analysis to identify peak regions with significant differences 1) across the cell types (neuronal and non-neuronal) 2) across brain regions (ACC neuronal and PFC neuronal) and (ACC neuron-depleted and PFC neuron-depleted) using edgeR v.3.14.059. The CPM matrix was prefiltered to regions with CPM >1 in at least 5 samples for both histone marks and normalized using calcNormFactors function which uses trimmed mean of M-values (TMM)58. The edgeR software modeled the reads counts matrix as negative binomial distribution using cell types and brain regions as covariates. We fit the normalized CPM matrix to quasi-likelihood negative binomial generalized log-linear model using glmQLFit function with robust = TRUE option. The Quasi-Likelihood F-test was then applied to test to identify peak regions that are significantly different across cell types and brain region (for both neuronal and non-neuronal cell types) using glmQLFTest (glmQLFit object, contrast=cell type or brain region). Multiple testing was done by applying the Benjamini-Hochberg method on the p-values to control false discovery rate60. The total number of differential peaks was determined at a FDR of 5%. The coordinates of neuronal, non-neuronal, ACC neuronal, PFC neuronal, PFC neuronal, PFC non-neuronal genomic regions are given in Table S8, S9.
Comparison with Roadmap Epigenomics Project
For each mark, we measured the similarity of genomic regions of cell specific (4) and homogenate (1) consolidated data sets: ACC neuronal, PFC neuronal, PFC neuronal, PFC neuron-depleted/non-neuronal and PFC HBCC homogenate with REP data from 111 tissues. We used bedtools jaccard –a sample bed file –b REP bed file61. This command outputs Jaccard index parameter (see Table S4) which is evaluated as
Cell composition analysis
Cell type proportions were quantified using R library CellMix62. Using our neuronal and non-neuronal ChIP-seq datasets, we generated cell type signatures to run deconvolution on homogenate samples to quantify the proportion of each cell type for every sample. We first created the basis set for neuronal and non-neuronal cell types by taking the mean of RPKM values for each peak across neuronal samples and neuron-depleted samples respectively for both marks. We defined as our input matrix the HBCC homogenate samples’ RPKM matrix. We then used the “lsfit” method from CellMix library for decomposition of RPKM matrix to calculate the coefficients of neuronal and non-neuronal cell types.
Genic annotation
We used CHIPSeeker v.1.8.963 to annotate peaks to seven distinct categories: promoter, 5′UTR, exons, introns, 3′UTR, downstream(<=3 kb) and distal intergenic regions within 5Kb of downstream and upstream of the transcription start site. The transcript database used for the annotation is ENSEMBL v75 for GRCh37.70.
Correlation of ChIP-Seq reads counts with RNA-Seq expression
We next examined the enrichment of ChIP-Seq reads counts around transcription start site (TSS) region of protein coding genes with RNA-Seq expressions from 537 individuals for 20,330 genes from PFC brain regions. We use nsgplot v2.6156 to plot ChIP-Seq read enrichment of combined PFC neuronal data sets as a function of 15kb distance upstream, downstream around TSS for both marks. Enrichments plots were made for all protein coding genes grouped into 5 categories sorted by the RPKM mean values across 537 subjects from the CommonMind RNA-Seq dataset 64.
Histone QTL enrichment analysis
The overlap between peak regions with a) histone QTLs detected in lymphoblastoid cell lines (LCL)26 and peak regions exceeding a variance percentage cutoff for a particular variable for both marks is computed. This overlap is then compared to the overlap computed from randomly permutated variance percentages. Each peak region is assigned a value based on the percentage of variance explained by a particular variable in the variancePartition analysis. At each of 40 cutoff values, the overlap between peak regions with values exceeding this cutoff and the peak regions with a histone QTL for the same histone mark in LCLs/PFC is evaluated using the Jaccard index.
The overlap was computed for the observed data and 10,000 datasets with the variance percentages randomly permutated. At each cutoff, the enrichment is computed as
The mean enrichment value and the 90% confidence interval are shown in the plot. Only regions on autosomes are considered leaving 9,575 H3K4me3 hQTLs in LCLs and 1,912 H3K27ac hQTLs in post mortem PCF. Permutation and overlap calculations were performed using regioneR65.
We ran similar analysis to test the overlap between the peak regions with a) hQTLs in H3K27ac modified peak regions from PFC homogenate from Sun et.al. b) hQTLs in H3K9ac modified peak regions from PFC homogenate from Ng et.al. with peak regions from 1) H3K27ac-PFC neuronal 2) H3K27ac-PFC non-neuronal, 3) H3K27ac-ACC neuronal 4) H3K27ac-ACC non-neuronal datasets and 5) differentially modified neuronal and 6) non-neuronal H3K27ac modified peak regions.
The overlapobserved is the jaccard index between a,b hQTL regions and 1–6 datasets. The overlappermuted is the jaccard index between abovementioned 1–6 datsets and 1000 datsets obtained by randomly permuting x no. of peak regions from a) PFC homogenate from Sun et.al b) PFC homogenate from Ng et.al. (x = length of hQTLs)
Pathway Enrichment analysis
Genomic Regions Enrichment of Annotations Tool (GREAT)19 was used to interpret differentially modified peaks in terms of the biological function of nearby genes. We took the sets of peaks that showed significant (< 5% FDR) differences across cell types (neurons and non-neurons) from edgeR analysis and tested for functional enrichment using the consensus peaks for each mark as a background (see Table S7, S9). The settings for genomic regions used are (proximal: 5.0 kb upstream, 5.0kb upstream and distal to 100 kb.) Since many of the gene set from different databases are redundant, we only considered REACTOME, KEGG and PID for a total of gene sets. Significance testing for the enrichment analysis was based on the binomial test compute by GREAT and using a Bonferroni cutoff 4.7 X 10−5 based on these tests.
Overlap of identified peaks with disease and trait-associated genetic variants
To assess if the genomic regions carrying the two assayed histone marks in the different brain regions and cell types play a role in the various traits and diseases, we examined the overlap with common genetic variants identified by genome-wide association studies (GWAS). For this, we employed LD-score partitioned heritability21, which estimates if common genetic variants in the genomic regions of interest explain more of the heritability of a given trait than genetic variants not overlapping the genomic regions of interest normalized by the number of variants in either category. The algorithm allows for correction of the general genetic context of the annotation using a baseline model of broad genomic annotations (like coding, intronic, and conserved). By using this baseline model, the algorithm focuses on enrichments above those expected from the genetic context of the interrogated regions. We applied the method to a range of GWAS traits with presumed involvement of the brain66–71 and well powered studies of traits not believed to involve the brain72–74. For the Alzheimer’s GWAS, see Materials and Methods for the Alzheimer’s GWAS. We used the European only versions of the summary statistics when available. This led to only coronary artery disease having a somewhat mixed ancestry (77% Europeans). We excluded the broad MHC-region (chr6:25–35MB) and otherwise used default parameters.
Allele specific QTL analysis
We used RASQUAL29 to call cell- and tissue- specific cis-hQTLs (histone QTLs) in our dataset PFC neuronal, ACC neuronal, PFC non-neuronal, ACC non-neuronal and PFC tissue homogenate for each of the two histone marks, H3K27ac and H3K4me3. RASQUAL uses allele specific reads counts at heterozygous sites to increase power to detect cis-hQTLs correlated with quantitative variation in histone modification.
With RASQUAL a feature in our ChIP-seq dataset is defined by a set of start and end coordinates of identified peaks for calling a cis-hQTLs. RASQUAL requires few data preprocessing steps before calling cis-hQTLs. 1) ChIP-seq read counts and offset matrices as text and bin files for each dataset and mark. We used bedtools –nuc option to obtain GC content for each identified peak region and use that as an input for offset calculation from counts matrix in their custom makeOffset.R script file. All text files were converted to bin file using text2bin.R script file. 2) Covariates text and bin file for each dataset and mark. The confounding factors in ChIP-seq reads counts are obtained by applying PCA onto log FPKMs with and without permutation and outputs the first N components whose variances are greater than those from permutation results. We used the makeCovariates.R script file and found 5 components as covariates for PFC neuronal, ACC neuronal, PFC non-neuronal, ACC non-neuronal dataset for H3K4me3 and H3K27ac marks whereas PFC homogenate samples have 3 (4) components for H3K4me3 (H3K27ac). 3) Allele specific counts VCF file. createASVF.sh script file was used to count allele specific reads for every individual for a given SNP within a feature. We used whole genome sequenced data of 17 individuals to generate allele specific counts. WGS paired-end 150bp reads were aligned to the GRCh37 human reference using the Burrows-Wheeler Aligner (BWA-MEM v0.78) and processed using the best-practices pipeline that includes marking of duplicate reads by the use of Picard tools (v1.83, http://picard.sourceforge.net), realignment around indels, and base recalibration via Genome Analysis Toolkit (GATK v3.2.2). All individuals WGS data was merged into a single vcf file and was used as one of the inputs to createASVF.sh.
We ran RASQUAL feature on 90767 H3K4me3 peaks of PFC neuronal (n=17), ACC neuronal (n=14), PFC non-neuronal (n=17), ACC non-neuronal (n=15) and PFC homogenate (n=11) and 127773 H3K27ac peaks from PFC neuronal (n=17), ACC neuronal (n=17), PFC non-neuronal (n=17), ACC non-neuronal (n=15) and PFC homogenate( n=17). SNPs were tested within 10 kb of cis region from peak start and end points. We used Benjamini-Hochberg q-value <0.05 as a threshold to determine the significant cis-hQTLs.
hQTL-GWAS overlap
In order to test the overlap of significant hQTLs (RASQUAL qvalue > 0.05) with GWAS identified SCZ loci, we took the list of lead SNPs and SNPs in LD (R^2 > 0.8) with the latter. The list was downloaded from https://www.med.unc.edu/pgc/results-and-downloads/downloads. We report the overlapping loci separately with cell specific hQTLs:- PFC NeuN+, PFC NeuN−, ACC NeuN+, ACC NeuN− ) and PFC HBCC tissue homogenate. hQTLs for both marks are listed in Table S1075.
Materials and Methods for the Alzheimer’s GWAS
Summary statistics for Alzheimer’s disease were provided by the International Genomics of Alzheimer’s Project (IGAP). IGAP is a large two-stage study based upon genome-wide association studies (GWAS) on individuals of European ancestry. In stage 1, IGAP used genotyped and imputed data on 7,055,881 single nucleotide polymorphisms (SNPs) to meta-analyse four previously-published GWAS datasets consisting of 17,008 Alzheimer’s disease cases and 37,154 controls (The European Alzheimer’s disease Initiative – EADI the Alzheimer Disease Genetics Consortium – ADGC The Cohorts for Heart and Aging Research in Genomic Epidemiology consortium – CHARGE The Genetic and Environmental Risk in AD consortium – GERAD). In stage 2,11,632 SNPs were genotyped and tested for association in an independent set of 8,572 Alzheimer’s disease cases and 11,312 controls. Finally, a meta-analysis was performed combining results from stages 1 & 2.
Data Availability Statement for the current study
The data analyzed for this article is available through the psychENCODE Knowledge Portal (psychencode.org). Access to the data is controlled by the NIMH Repository and Genomics Resources (NRGR) https://www.nimhgenetics.org. See instructions for in the PsychENCODE Knowledge Portal: https://www.synapse.org/#!Synapse:syn4921369. Data and results are at https://www.synapse.org/#!Synapse:syn4566010. The site includes link to UCSC browser visualizations.
URLs
Data access instructions (for ChIP-seq data presented in our paper): https://www.synapse.org/#!Synapse:syn4921369/wiki/235539
Data, results and visualizations (for ChIP-seq data presented in our paper): https://www.synapse.org/#!Synapse:syn4566010
Psychiatric Genomics Consortium: med.unc.edu/pgc
International Genomics of Alzheimer’s Project: web.pasteur-lille.fr/en/recherche/u744
The Social Science Genetic Association Consortium: ssgac.org
Sleep phenotypes: www.t2diabetesgenes.org/data
Genetic Investigation of ANthropometric Traits: portals.broadinstitute.org/collaboration/giant
Coronary Artery Disease: cardiogramplusc4d.org
International Inflammatory Bowel Disease Genetics Consortium: ibdgenetics.org
CommonMind Consortium: commonmind.org
Roadmap Epigenomics Project: roadmapepigenomics.org
Grubert, et al. (Cell 2015) histone QTLs: http://chromovar3d.stanford.edu/
Supplementary Material
Acknowledgments
Data were generated as part of the PsychENCODE Consortium, supported by: U01MH103339, U01MH103365, U01MH103392, U01MH103340, U01MH103346, R01MH105472, R01MH094714, R01MH105898, R21MH102791, R21MH105881, R21MH103877, and P50MH106934 awarded to: Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (USC), Mark Gerstein (Yale), Daniel Geschwind (UCLA), Thomas M. Hyde (LIBD), Andrew Jaffe (LIBD), James A. Knowles (USC), Chunyu Liu (UIC), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Nenad Sestan (Yale), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (UCSF), Patrick Sullivan (UNC), Flora Vaccarino (Yale), Sherman Weissman (Yale), Kevin White (UChicago) and Peter Zandi (JHU).
Data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138 and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories and the NIMH Human Brain Collection Core. CMC Leadership: Pamela Sklar, Joseph Buxbaum (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), Raquel Gur, Chang-Gyu Hahn (University of Pennsylvania), Keisuke Hirai, Hiroyoshi Toyoshiba (Takeda Pharmaceuticals Company Limited), Enrico Domenici, Laurent Essioux (F. Hoffman-La Roche Ltd), Lara Mangravite, Mette Peters (Sage Bionetworks), Thomas Lehner, Barbara Lipska (NIMH).
Data on coronary artery disease/myocardial infarction have been contributed by CARDIoGRAMplusC4D investigator. We additionally thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant no. 503480), Alzheimer’s Research UK (Grant no. 503176), the Wellcome Trust (Grant no. 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant no. 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728.
This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. We are extremely grateful to Dr. Jordi Ochando and Christopher Bare and other personnel of the Icahn School of Medicine at Mount Sinai’s Flow Cytometry Core for providing and teaching cell sorting expertise.
We thank Menachem Fromer, Eli Stahl, Laura Huckins, Li Shen, Geetha Senthil and Thomas Lehner for helpful discussion. This paper is dedicated to the memory of Pamela Sklar.
Footnotes
Author contributions: Wet lab work including tissue processing, nuclei sorting and ChIP-seq library generation: Y.J., L.B., M.K., E.Z., R.J., J.R.W., R.P., B.S.K.. Data processing and coordination: Y.J., M.K., D.H.K., J.S.J., L.S., S.K.S., M.A.P., Y.W., H.S.. Bioinformatics and computational genomics: K.G., G.E.H., M.E.H., N.J.F., E.M., Z.W.. Provided brain tissue and resources: B.T.H., B.K.L. Conceived study design (including wet lab and/or bioinformatical analyses pipelines): Y.J., K.G., G.E.H., P.R., P.S., S.A.. Wrote the paper: K.G., G.E.H., P.S., P.R., S.A.
Competing interests: The authors declare no competing interests.
References
- 1.Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science (New York, NY) 2015;349:1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nature neuroscience. 2016;19:1397–1407. doi: 10.1038/nn.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotech. 2010;28 doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farh KKH, et al. Genetic and Epigenetic Fine-Mapping of Causal Autoimmune Disease Variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roadmap Epigenomics, C., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akbarian S, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–1712. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 9.Network & Pathway Analysis Subgroup of Psychiatric Genomics, C. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun W, et al. Histone Acetylome-wide Association Study of Autism Spectrum Disorder. Cell. 2016;167:1385–1397e1311. doi: 10.1016/j.cell.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 11.Ng B, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung I, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. Coordinated cell type-specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet. 2013;9:e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charney DS, Sklar PB, Buxbaum JD, Nestler EJ. Charney & Nestler’s neurobiology of mental illness. Oxford University Press; New York, NY: 2018. [Google Scholar]
- 15.Mancarci BO, et al. Cross-laboratory analysis of brain cell type transcriptomes with applications to interpretation of bulk tissue data. 2017 doi: 10.1523/ENEURO.0212-17.2017. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huttner HB, et al. The age and genomic integrity of neurons after cortical stroke in humans. Nat Neurosci. 2014;17:801–803. doi: 10.1038/nn.3706. [DOI] [PubMed] [Google Scholar]
- 17.Habib N, et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherwood CC, et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc Natl Acad Sci U S A. 2006;103:13606–13611. doi: 10.1073/pnas.0605843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nature biotechnology. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature genetics. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang K-l, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci. 2017 doi: 10.1038/nn.4587. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gjoneska E, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer/’s disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj T, et al. Polarization of the Effects of Autoimmune and Neurodegenerative Risk Alleles in Leukocytes. Science (New York NY) 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman GE, Schadt EE. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics. 2016;17:483. doi: 10.1186/s12859-016-1323-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grubert F, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1051–1065. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waszak Sebastian M, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell. 2015;162:1039–1050. doi: 10.1016/j.cell.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Consortium, G.T. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science (New York, NY) 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumasaka N, Knights AJ, Gaffney DJ. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schizophrenia Working Group of the Psychiatric Genomics, C., et al. Biological Insights From 108 Schizophrenia-Associated Genetic Loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posner MI, Rothbart MK, Sheese BE, Tang Y. The anterior cingulate gyrus and the mechanism of self-regulation. Cogn Affect Behav Neurosci. 2007;7:391–395. doi: 10.3758/cabn.7.4.391. [DOI] [PubMed] [Google Scholar]
- 32.Homayoun H, Moghaddam B. NMDA receptor hypofunction produces opposite effects on prefrontal cortex interneurons and pyramidal neurons. J Neurosci. 2007;27:11496–11500. doi: 10.1523/JNEUROSCI.2213-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Fevre AK, et al. FOXP1 mutations cause intellectual disability and a recognizable phenotype. Am J Med Genet A. 2013;161A:3166–3175. doi: 10.1002/ajmg.a.36174. [DOI] [PubMed] [Google Scholar]
- 34.Sadakata T, et al. Reduced axonal localization of a Caps2 splice variant impairs axonal release of BDNF and causes autistic-like behavior in mice. Proc Natl Acad Sci U S A. 2012;109:21104–21109. doi: 10.1073/pnas.1210055109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griswold AJ, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet. 2012;21:3513–3523. doi: 10.1093/hmg/dds164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaguchi DM, Glatt SJ. GRIK4 polymorphism and its association with antidepressant response in depressed patients: a meta-analysis. Pharmacogenomics. 2014;15:1451–1459. doi: 10.2217/pgs.14.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandal MJ, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. 2016 doi: 10.1176/appi.focus.17103. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeisel A, et al. Molecular architecture of the mouse nervous system. 2018 doi: 10.1016/j.cell.2018.06.021. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan JM, et al. Autism-like syndrome is induced by pharmacological suppression of BET proteins in young mice. J Exp Med. 2015;212:1771–1781. doi: 10.1084/jem.20151271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penney J, Tsai LH. Histone deacetylases in memory and cognition. Sci Signal. 2014;7 doi: 10.1126/scisignal.aaa0069. [DOI] [PubMed] [Google Scholar]
- 42.Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, et al. Association of DNA methylation in the brain with age in older persons is confounded by common neuropathologies. Int J Biochem Cell Biol. 2015;67:58–64. doi: 10.1016/j.biocel.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kundakovic M, et al. Practical Guidelines for High-Resolution Epigenomic Profiling of Nucleosomal Histones in Postmortem Human Brain Tissue. Biological Psychiatry. 2017;81:162–170. doi: 10.1016/j.biopsych.2016.03.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Y, et al. Isolation of neuronal chromatin from brain tissue. BMC Neuroscience. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fennell AWKTT. Picard tools version 1.90. 2013. [Google Scholar]
- 49.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. 2011:1752–1779. [Google Scholar]
- 53.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotech. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotech. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 58.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 61.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaujoux R, Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013;29:2211–2212. doi: 10.1093/bioinformatics/btt351. [DOI] [PubMed] [Google Scholar]
- 63.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 64.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gel B, et al. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics. 2016;32:289–291. doi: 10.1093/bioinformatics/btv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature genetics. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okbay A, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okbay A, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nature genetics. 2016 doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones SE, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS genetics. 2016;12:e1006125. doi: 10.1371/journal.pgen.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature genetics. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Consortium, C.D. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nature genetics. 2015 doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nature genetics. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wetterstrand K. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) Available at: http://www.genome.gov/sequencingcosts. Accessed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed for this article is available through the psychENCODE Knowledge Portal (psychencode.org). Access to the data is controlled by the NIMH Repository and Genomics Resources (NRGR) https://www.nimhgenetics.org. See instructions for in the PsychENCODE Knowledge Portal: https://www.synapse.org/#!Synapse:syn4921369. Data and results are at https://www.synapse.org/#!Synapse:syn4566010. The site includes link to UCSC browser visualizations.