Abstract
Feline interferon beta is a cytokine that belongs to the type I IFN family, with antitumor, antiviral and immunomodulatory functions. In this work, recombinant feline interferon beta (rFeIFNβ) was expressed in insect larvae that constitute important agronomic plagues. rFeIFNβ accumulated in the hemolymph of Spodoptera frugiperda larvae infected with recombinant baculovirus and was purified by Blue-Sepharose chromatography directly from larval homogenates on day 4 post-infection. rFeIFNβ was recovered after purification with a specific activity of 1 × 106 IU mg−1. By this method, we obtained 8.9 × 104 IU of purified rFeIFNβ per larva. The product was biologically active in vitro, with an antiviral activity of 9.5 × 104 IU mL−1, as well as a potent antitumor activity comparable to that of the commercial FeIFNω. The glycosylation of rFeIFNβ was confirmed by peptide-N-glycosidase F digestion. Our findings provide a cost-effective platform for large-scale rFeIFNβ production in laboratory research or veterinary medicine applications.
Keywords: Feline interferon beta, Baculovirus, Insect larvae, Veterinary medicine
Introduction
Interferons (IFNs) are cytokines that induce resistance to virus infection, an attribute discovered by Isaacs and Lindenmann (1957). Since then, IFNs have been shown to regulate not only antiviral, but also antitumor, apoptotic, and cellular immune responses, which makes them of major interest in human and veterinary medicine. Type I IFNs include α, β, κ, ω, ε and ζ subtypes. Transient synthesis of IFNα and IFNβ can be induced by live or inactivated viruses, bacterial oligosaccharides, foreign genetic material, IL-l, IL-2, and TNF-α. Secreted IFNs bind to the dimeric IFN α/β receptor (IFNAR), consisting of IFNAR1 and IFNAR2, in nearby uninfected cells. This binding triggers intracellular cascades, generally via the JAK–STAT pathway, which induce the expression of nearly 30 IFN-stimulated genes (Kruth 1998). IFNs also activate phagocytic cells, and increase the expression of class I and II MHC molecules in antigen-presenting cells. With regard to their antiproliferative activity, IFNs have several effects on neoplastic cells, including modulation of oncogene expression, down-regulation of c-myc, c-fos, and c-Ha-ras and antiangiogenic activity (Lindner 2002).
In veterinary medicine, the commercial Virbagen Omega® (Sakurai et al. 1992), a feline IFN-ω-like compound that belongs to the IFNα cluster (Nagai et al. 2004; Yang et al. 2007), is a good treatment choice for dogs and cats infected with oncogenic viruses. This compound is active against feline leukemia virus, feline immunodeficiency virus, feline calicivirus and canine parvovirosis, reducing the mortality rate and clinical signs over time, and thus improving the quality of life of the animal. Besides, recombinant feline IFNα (rFeIFNα) has been shown to have antiviral activity against rotavirus, feline panleukopenia virus and feline infectious peritonitis virus in vitro (Mochizuki et al. 1994). Although it has been found that the incubation of cells with feline IFNβ (FeIFNβ) before viral challenge could also provide antiviral protection (Weiss and Toivio-Kinnucan 1988), the main difference with IFNα is that IFNβ is more effective at activating anti-proliferative responses and pro-apoptotic pathways in tumor cells (Damdinsuren et al. 2007). IFNβ also has a vital role in the control of macrophage differentiation and osteoclastogenesis (Coelho et al. 2005), lymphoid development, myelopoiesis and septic shock toxicity (Chawla-Sarkar et al. 2001). It has been recently reported that these unique functional properties awarded to IFNβ are due to an exclusive way of ligation to IFNAR1, independently of IFNAR2, forming an IFNAR1-IFNβ complex that modulates the expression of a different set of genes not related to JAK–STAT signaling (de Weerd et al. 2013).
The coding region for FeIFNβ encodes a predicted protein of 186 amino acids, consisting of a signal sequence of 21 amino acids and a mature IFNβ of 165 amino acids. Its amino acid sequence has four potential N-glycosylation sites, two cysteine residues and one phosphorylatable tyrosine residue. FeIFNβ shares 73 and 60% amino acid sequence homology with canine and human IFNβ, respectively, whereas only 32% with FeIFNα.
Previous to the development of recombinant DNA technology, type I IFNs were extracted from virus-infected white blood cells, representing less than 0.1% of the total protein (Pestka 2007). Thus, recombinant production of IFN represents an attractive technology to obtain large quantities of this cytokine.
Insect-derived baculoviruses are broadly applied in biotechnology as vectors for recombinant protein production. Some attractive properties of baculoviruses such as Autographa californica Multiple Nucleopolyhedrovirus (AcMNPV) are that (1) they can be manipulated in biosafety level 1 facilities since, due to their narrow host range (insects), they do not cause disease in vertebrates, plants or microorganisms (Burges et al. 1980; Kost and Condreay 2002), (2) they can provide high levels of recombinant proteins, and (3) they show appropriate post-translational modifications, in contrast to prokaryotic expression systems (van Oers 2011). The use of insect larvae instead of insect cell cultures has the advantage of allowing higher protein expression (Romero et al. 2011; Targovnik et al. 2016). Spodoptera frugiperda larvae (Lepidoptera: Noctuidae) constitute an important crop pest widely distributed in North and South America (Sparks 1979). These larvae can be reared under laboratory conditions with automated rearing equipment and without allergenic reactions in human handlers. These advantages, together with the advantage of not needing the large volumes of cell culture needed in insect cell culture expression, simplify the scale-up and reduce the costs of recombinant protein mass production. Despite several cytokines have been expressed using the baculovirus-insect system (Stifter et al. 2014; Maeda et al. 1985; Okano et al. 2000; Iwata et al. 1996; Argyle et al. 1998) the biotechnological production of FeIFNβ has never been achieved.
The aim of this work was to express and purify rFeIFNβ using the baculovirus-insect larvae system as a platform and to evaluate its in vitro biological activity.
Materials and methods
Materials
The EcoRI and BamHI restriction endonucleases were provided by Promega (Madison, WI, USA). PageRuler™ Prestained Protein Ladder from Thermo Scientific (Thermo Fisher Scientific, Waltham, MA, USA) and Blue Plus Protein Marker from TransGen Biotech (Beijing, China) were used as molecular weight (MW) standards for SDS-PAGE and Western blot. The 100-bp DNA ladder was from Promega. The insect cell line IPBL-Sf9 from S. frugiperda (Sf9) was provided by the Asociación Banco Argentino de Células (ABAC, Pergamino, Buenos Aires, Argentina). Sf900 II insect culture media and the antibiotic–antimycotic solution were from Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA). S. frugiperda larvae were from AgIdea S.A. (Pergamino, Buenos Aires, Argentina).
Gene construction
The full-length FeIFNβ sequence was obtained from the National Center for Biotechnology Information data bank (GenBank accession no. AB021707.1). The cDNA containing two restriction site sequences for EcoRI and BamHI was synthesized by GenScript (Piscataway, NJ, USA), and provided cloned in the pUC18 plasmid (pUC18-FeIFNβ). First, pUC18-FeIFNβ was amplified in Escherichia coli DH5α under ampicillin selection and purified using the AxyPrep Miniprep kit (Axygen Biosciences, CA, USA). Then, pUC18-FeIFNβ was subjected to a PCR to remove the natural FeIFNβ signal peptide sequence to use the baculoviral glycoprotein 67 (gp67) signal peptide sequence afterwards. For this reaction, two primers designed in our laboratory were used: forward (5ʹ–3ʹ): CGGATCCGTGTCCTACAAGTTGCTGGG (the underlined sequence corresponds to the BamHI site); reverse (5ʹ–3ʹ): GGAATTCTTAGTTTTGCAGGTAGTCAGTCAA (the underlined sequence corresponds to the EcoRI site). The PCR conditions (25 µL final volume) were: 0.2 µM each primer, 1× Pfu buffer, 0.3 mM each dNTP and 2.5 U Pfu DNA polymerase (Promega). The PCR program was: a first step of 3 min at 95 °C, a second step of 95 °C for 30 s, 54 °C for 30 s, and 72 °C for 1.30 min (30 cycles). An additional extension step of 5 min at 72 °C was then applied. Free primers from the PCR product were removed using the PCR Wizard™ SV gel and PCR Clean-up System (Promega).
Second, FeIFNβ gene was cloned using BamHI and EcoRI sites into the pAcGP67-B vector (BD Biosciences Pharmingen, San Diego, CA, USA), which contains a sequence for the GP67 signal peptide that targets the recombinant protein for secretion (pAcFeIFNβ).
Recombinant baculovirus production
A monolayer of 1 × 106 Sf9 cells was cotransfected with pAcFeIFNβ (1 µg) and 250 ng of Baculogold Bright AcMNPV DNA (BD Biosciences Pharmingen) using Cellfectin™ reagent (Invitrogen), in a six-well plate. After 4 days of incubation at 27 °C, the cell culture medium was collected and centrifuged (3000×g for 10 min). Cotransfection and homologous recombination efficiency were determined by monitoring green fluorescent protein expression under fluorescence microscopy, since this reporter protein is encoded in the baculoviral genome. It is worth noting that, after homologous recombination, purification of the virus from the supernatant was not necessary, because the viral genome has a lethal deletion that is only reversed through homologous recombination with pAcFeIFNβ. Thus, viable viruses present in the cotransfection supernatant are all recombinant (AcMNPV–FeIFNβ).
Then, a round of amplifications was performed as follows: the fourth-day cotransfection supernatant was used to infect 2 × 106 cells seeded in T-25 flasks at a low multiplicity of infection (MOI) of 0.02. Cells were then incubated for 4 days at 27 °C, and 100 µL of the supernatant of the first amplification step was used to initiate a second amplification round, and so on, up to four amplifications.
The amplified AcMNPV–FeIFNβ was titrated by plaque assay (O’Reilly et al. 1994). This high-titer AcMNPV–FeIFNβ was the viral stock used for protein production.
Expression in insect cell cultures
Suspension cultures of Sf9 cells were grown in Sf900II medium, which was supplemented with 1% (V/V) antibiotic–antimycotic solution (containing Penicillin 10,000 units mL−1, Streptomycin 10,000 µg mL−1 and Amphotericin B 25 µg mL−1) and 1% (V/V) Fetal Bovine Serum (FBS, NATOCOR, Córdoba, Argentina). The sterile Erlenmeyer flasks were orbitally shaken at 100 rpm at 27 °C. Sf9 cells were subcultured before reaching a density of 6 × 106 cells mL−1. Besides, the suspension volume did not exceed 10% of the total volume of the Erlenmeyer flask.
rFeIFNβ was produced in suspension cultures in log-phase (95–99% viability). The Erlenmeyer flasks contained 10 mL of suspension at a cell density of 1 × 106 cells mL−1. One flask was used for each MOI with recombinant baculovirus (MOI of 0.05, 2 and 5). Infected cultures were incubated in an orbital shaker at 27 °C for 5 days. rFeIFN expression in the supernatants was determined every day post-infection (DPI). The cells were removed by centrifugation (3000×g for 10 min) and biological activity was measured in the supernatants. The results are expressed as the mean ± standard deviation of at least three determinations from independent experiments. Supernatants of Sf9 cells infected with a baculovirus that contains horseradish peroxidase (HRP) gene instead of IFN (non-related baculovirus) were included as negative control.
Expression in insect larvae
Spodoptera frugiperda larvae were reared in 12-well plates at 23–25 °C in a 70% humidified chamber, with a 16:8 light:dark photoperiod, and fed on a high-wheat germ diet until they reached their fifth instar (20 days of age).
For all the experiments, fifth-instar larvae were injected with 50 µL of the recombinant baculovirus stock (diluted to 1 × 107 PFU mL−1) near the third prolegs. To characterize and quantify the recombinant protein produced, larvae that were alive and fluorescent under UV light were harvested, which is between 3 and 6 DPI, and frozen immediately at − 20 °C until they were processed for analysis. Larvae infected with a non-related recombinant baculovirus were included as control.
Generation of S. frugiperda extract
Infected larvae were homogenized using a marble mortar and pestle in groups of three (n = 6). One mL of the extraction buffer [20 mM phosphate buffer, pH 7.2, with 10 mg glutathione crystals and 1/100 (V/V) protease inhibitor cocktail (Sigma-Aldrich, Saint Louis, MO, USA)] was used for each larva, 300 mg average weight. Then, the larval extract was centrifuged at 10,000×g for 10 min at 4 °C and the pellet was discarded. The supernatant was filtered through Whatman paper using a glass Buchner funnel to remove the lipid fraction remaining at the top. This crude extract contained the total soluble proteins and was used to measure the biological activity of rFeIFNβ. Results are expressed as the mean ± standard deviation from independent experiments.
Purification
The S. frugiperda crude extract was pre-conditioned by a SEC PD10 column (GE Healthcare, Chicago, IL, USA) equilibrated with 20 mM phosphate buffer, pH 7.2 (buffer A). Then, the sample (3.5 mL) was diluted to 10.5 mL to facilitate the interaction with the matrix and 10 mL were loaded on the 1 mL Blue-Sepharose column (HiTrap Blue HP, GE Healthcare), also equilibrated with buffer A. We used a flow rate of 0.5 mL min−1. Ten column volumes of buffer A were passed through the column, and then washing was performed using ten column volumes of buffer A with 0.5 M NaCl. Finally, the elution took place by increasing the ionic strength to 1M NaCl. All the fractions were collected and subjected to SDS-PAGE and Western blot analysis.
Determination of total protein concentration
Total protein concentration was determined by following the Bradford microassay protocol (Bradford 1976), using the Quick Start™ Bradford reagent (BioRad, Hercules, CA, USA). The samples used were crude larval extract and the purified rFeIFNβ fraction.
Western blot analysis
Sf9 cell culture supernatants and larval extracts were resolved by SDS-PAGE (15% polyacrylamide gels). Before loading the samples into the wells, they were heated for 5 min at 100 °C in sample buffer [125 mM Tris–HCl, pH 6.8, 4% (w/v) SDS, 20% (w/v) glycerol, 0.01% (w/v) bromophenol blue, 10% (v/v) 2-mercaptoethanol]. One of the lanes was reserved for the protein marker, which allowed determining the MW of the protein bands. The resulting gels were either stained with Coomassie Blue R-250 or transferred onto nitrocellulose membranes (GE Healthcare). Membranes were then incubated at 4 °C in blocking solution [0.05% phosphate-buffered saline (PBS)-Tween—3% skim milk] for 16 h. After one 10-min wash, mouse anti-FeIFNβ polyclonal antibody (1:2500 in 0.05% PBS-Tween—1% skim milk) was added (synthesized by GenScript, from the immunogenic peptide EVPEEIKKSQRFQKC). Polyclonal rabbit anti-Mouse immunoglobulin conjugated with HRP (1:2000 in 0.05% PBS-Tween—1% skim milk) was used as the secondary antibody (DakoCytomation, Denmark). Development was carried out with an enhanced chemiluminescent substrate and high performance chemiluminescence films (CL-X Posure™, Thermo Fisher Scientific).
In vitro antiviral activity
The antiviral activity of rFeIFNβ was evaluated by means of the cytopathic effect (CPE) inhibition assay (Rubinstein et al. 1981), with minor modifications. Briefly, three components were used: (1) vesicular stomatitis virus (VSV, ATCC VR-158, Indiana Strain, Manassas, VA, USA) at a titer of 1 × 107 PFU mL−1, (2) Crandell Feline Kidney Cells (CRFK, ATCC CCL-94, Manassas), and (3) rFeIFNβ, in Sf9 cell culture supernatants, crude larval extract or purified by Blue-Sepharose chromatography.
CRFK cells were grown in an atmosphere of 95% air, 5% CO2 at 37 °C. The medium used was Modified Eagle’s Medium (MEM, Gibco™, Thermo Fisher Scientific) supplemented with 1% antibiotic–antimycotic solution and 10% FBS. On day one of the experiment, monolayers of CRFK cells were prepared in 96-well tissue culture plates at a density of 5 × 104 cells/well (100 µL). Then, they were incubated in triplicate with twofold serial dilutions of the sample containing rFeIFNβ, which was previously filtered through a 0.22-µm membrane. On day two, after a 16-h incubation for rFeIFNβ incorporation and signaling pathway development, CRFK cells were challenged with VSV at a MOI of 0.25. Two controls were included: cells treated without rFeIFNβ or VSV (cell viability control), and cells not treated with rFeIFNβ but infected with VSV (viral control). On day three or four, according to the appearance of 100% CPE on the viral control, CRFK cells were fixed in 1% (V/V) formaldehyde, washed with PBS and stained with 0.1% (W/V) crystal violet (Biopack, Buenos Aires, Argentina). After a final wash with deionized water, the absorbance was measured in an ELISA reader at 595 nm. The results are expressed as IU mL−1, based on a standard curve made with the antiviral activity of the commercial Virbagen Omega® (Virbac, France), which was assayed in parallel in the same tissue culture plate. Sf9 cell culture supernatants, as well as crude larval extracts, infected with a non-related baculovirus were assayed in independent experiments. These negative controls were tested using the same dilutions as the rFeIFNβ samples, so as to discard that baculovirus particles contributed to the antiviral effect (Gronowski et al. 1999; Hervas-Stubbs et al. 2007).
In vitro antitumor activity
Feline mammary carcinoma cells (AlRB) (Villaverde et al. 2016) were cultured in Dulbecco’s MEM and Ham’s nutrient mixture F12 (Thermo Fisher Scientific) supplemented with 10% FBS, 10 mM HEPES (pH 7.4) and 1% antibiotic–antimycotic solution in a 5% CO2 humidified chamber at 37 °C. The cell line was maintained in monolayer and sub-cultured by trypsinization (trypsin 0.25%, EDTA 0.02% in PBS) before confluence was reached.
The cytotoxic activities of the infected larval extract and purified product were evaluated on AlRB monolayers seeded on 96-well plates (4 × 104 cells in 100 µL per well). To exclude the possibility of a cytotoxic effect due to baculovirus infection, a control larval extract from larvae infected with a non-related baculovirus was assayed at the same time. The AlRB monolayers were treated with each sample: control larval extract, rFeIFNβ larval extract, 5000 IU mL−1 of purified rFeIFNβ or 5000 IU mL−1 of commercial rFeIFNω, and cultured under the above-mentioned conditions. Each sample was previously filtered through a 0.22 µM membrane for sterilization. After 5 days of treatment, cell viability was quantified with the acidic phosphatase assay (Friedrich et al. 2009). The percentage of cell survival was calculated from the ratio of the absorbance between treated and untreated control cells. The results are expressed as the mean ± standard deviation of at least three determinations. The results were statistically analyzed by one-way ANOVA using GraphPad Prism 6 software (GraphPad Software Inc., USA) and considered significant only when p < 0.05.
Analysis of the presence of viral DNA
The Viral Nucleic Acid Extraction Kit (Real Biotech Corporation, Banqiao, Taiwan) was used to purify the viral DNA from three samples: (1) S. frugiperda crude extract, (2) S. frugiperda crude extract passed through a PD10 column, and (3) Blue-Sepharose eluate containing rFeIFNβ. The DNA obtained was resuspended in 50 µL of DNAse-free water. Then, a PCR reaction was performed using a pair of primers: forward (5ʹ–3ʹ) TCCGGATTATTCATACCGTCCCACCATC; reverse (5ʹ–3ʹ): GCTTCATCGTGTCGGGTTTAACATTACGG that hybridize to the recombinant baculovirus DNA, 9 bp upstream the ATG initiation codon and 51 bp downstream the stop codon, respectively. The PCR conditions (50 µL final volume) were: 0.2 µM each primer, 1× hot FIREPol reaction buffer, 0.2 mM dNTPs, 2.5 mM MgCl2 and 2.5 U FIREPol DNA polymerase (Solis BioDyne, Tartu, Estonia), with 1 µL DNA. The PCR program was: a first step of 95 °C for 15 min, a second step of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min (35 cycles). An additional extension step of 5 min at 72 °C was then applied. The PCR products were seeded on a 1% agarose (Sigma-Aldrich) gel to reveal the presence of AcMNPV–FeIFNβ in each of the three samples and the positive control (bacterial positive clone), as a band of around 800 bp.
Glycosylation assay
Purified rFeIFNβ was subjected to peptide-N-glycosidase F (Roche, Mannheim, Germany) digestion as follows: 72 µL of eluate was mixed with denaturing buffer (SDS 2.5%, DTT 0.4 M) to 80 µL. After heating at 100 °C for 10 min, reaction buffer was added (Na+ phosphate buffer 0.5 M pH 7.5, NP40 10%). The mixture was divided in two Eppendorf tubes, one of which was added with 3 µL glycosidase and the other used as a negative control. The two tubes were incubated at 37 °C overnight, after addition of protease inhibitor.
Results and discussion
Expression of rFeIFNβ in insect cell lines and larvae
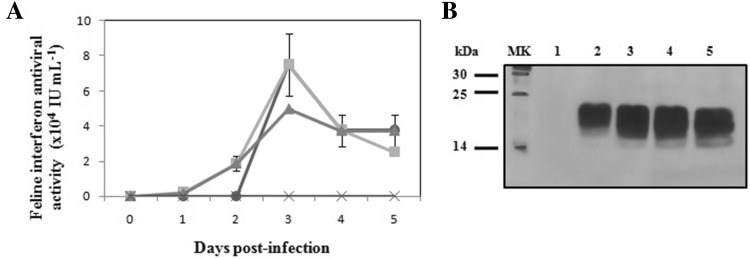
FeIFNβ was cloned under control of the polyhedrin promoter and fused in-frame to the viral secretion signal GP67. First, to determine rFeIFNβ expression and localization, we infected Sf9 cells with AcMNPV–FeIFNβ at MOIs of 0.05, 2 and 5 and analyzed the culture supernatants on different DPI. Although the Sf9 cell line used was adapted to serum-free medium which would facilitate further purification of product, we added 1% FBS, because it not only led to a higher cell growth rate, but also generated a tenfold increase in IFN expression (Targovnik et al. 2014). The expression kinetics curve showed that the rFeIFNβ antiviral activity increased gradually, achieving a maximum at 3 DPI (7.5 ± 1.8 × 104 IU mL−1) at MOIs 0.05 and 2, higher than at MOI 5 (5 × 104 IU mL−1). However, an important decrease was observed at 4 and 5 DPI (Fig. 1a). No activity was detected in negative control experiments using supernatants of Sf9 cell cultures infected with a non-related baculovirus. rFeIFNβ was effectively secreted into the supernatant of the expression cultures. After 4 DPI, the decrease in biological activity could be attributed to instability inherent to cytokines, like molecular aggregation previously reported (Rodriguez et al. 2005), because there is no evident decrease in rFeIFNβ quantity due to protease activity as judged by the Western blot (Fig. 1b). The MW of the protein, around 24 kDa, corresponds to a post-translational modification of FeIFNβ, whose sequence-deduced MW is 19.9 kDa. This protein has four potential N-glycosylation sites.
Fig. 1.
Time-course analysis of feline interferon β (FeIFNβ) expression in Sf9 cells. a Antiviral activity of FeIFNβ in supernatants of Sf9 cell cultures infected at different MOI. (Circle) MOI 0.05; (square) MOI 2; (triangle) MOI 5; (cross) negative control supernatants (Sf9 cell cultures infected with a non-related baculovirus). b Western blot analysis of the expression kinetics of FeIFNβ at MOI 2. MK: molecular weight marker, 1–5: 1–5 days post-infection
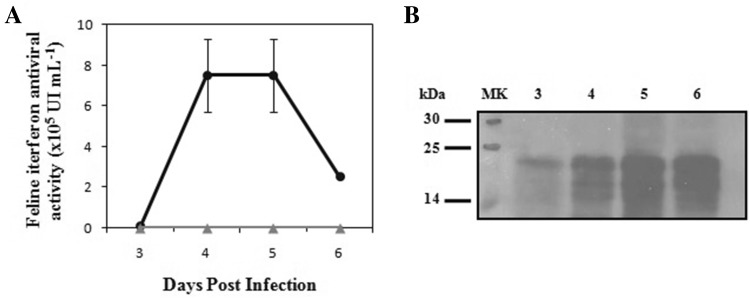
Second, we achieved the expression of rFeIFNβ directly in S. frugiperda larvae. The inoculum of AcMNPV–FeIFNβ used for infection was 5 × 105 PFU per larva, because we have previously shown that 1 × 106 PFU result lethal for the larvae, while 1 × 105 PFU result in poor protein expression levels (Targovnik et al. 2014). Larvae started showing signs of infection, such as motility and appetite loss, at 1 DPI. However, green fluorescence became evident under UV light only after 3 DPI, so larvae were harvested. This was also done at 4, 5 and 6 DPI, but mortality of larvae after 6 DPI prevented further harvesting. Then, frozen larvae were separated into groups of three (n = 6 per DPI), and the larval extracts were tested for antiviral activity. The antiviral activity of rFeIFNβ was evident at 3 DPI [(9.4 ± 2.2) × 103 IU mL−1], with a maximum at 4 and 5 DPI [(7.5 ± 1.7) × 105 IU mL−1] and finally decreased at 6 DPI [2.5 × 105 IU mL−1] (Fig. 2a). No antiviral activity was observed in negative controls consisting of crude extracts from larvae infected with a non-related baculovirus. Given these results, day 4 post-infection was selected as the optimal day for larval harvesting. As judged by Western blot analysis, rFeIFNβ expression was efficient in S. frugiperda larvae, with a MW that corresponded to the glycosylated form of the protein (Fig. 2b). Glycosylation is essential for cytokines because it confers greater stability, interaction with specific receptors and pharmacokinetic profile (Martina et al. 1998; Chamorey et al. 2002). The decrease in antiviral activity at 6 DPI was not due to proteolytic activity but rather to cytokine instability, since no decrease in quantity was observed in the Western blot (Fig. 2b). This is supported by the same behavior observed in Sf9 cell culture supernatants (Fig. 1b). The fact that samples infected with a non-related baculovirus did not provide protection against VSV infection proved that although contaminating baculovirus particles may potentiate IFN antiviral activity on mammalian cells (Gronowski et al. 1999; Hervas-Hubbs et al. 2007), this did not occur with the sample dilutions we used.
Fig. 2.
Time-course analysis of FeIFNβ expression in crude S. frugiperda larval extracts. a Antiviral activity against vesicular stomatitis virus. Circle: FeIFNβ larval extracts. Triangle: negative control larval extracts (larvae infected with a non-related baculovirus). b Western blot analysis of the expression kinetics of FeIFNβ. MK: molecular weight marker, 3–6 days: 3–6 post-infection
Purification
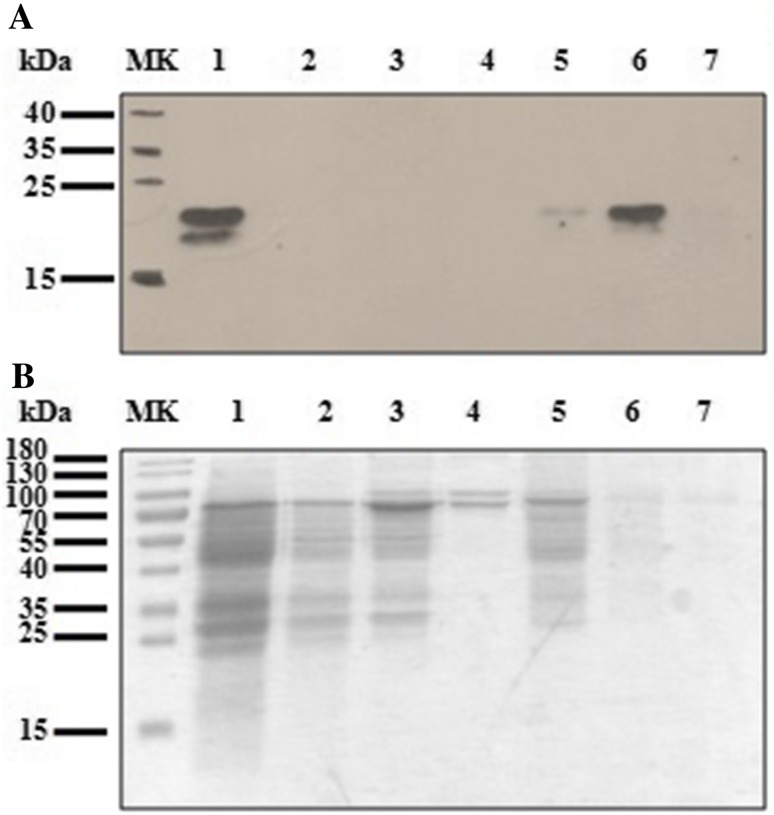
No purification tag was included in rFeIFNβ to facilitate this step because some loss of biological activity has been previously reported (Schmeisser et al. 2006). In this work, Blue-Sepharose chromatography was used to purify the rFeIFNβ from the crude larval extract. rFeIFNβ bound to the matrix with high affinity, since no recombinant protein was detected in the passthrough (Fig. 3a). After washing steps with 0.5 M NaCl, rFeIFNβ was eluted with 1 M NaCl (Fig. 3a). Further addition of ethylene glycol did not improve the elution (data not shown). Most contaminating proteins appeared in the passthrough fraction (no adsorption to Blue-Sepharose) or eluted in the 0.5 M NaCl fractions, as judged by SDS-PAGE (Fig. 3b). The sensibility of SDS-PAGE was not enough to visualize the band corresponding to rFeIFNβ in the assayed conditions. A mixture of hydrophobic and ionic interactions took place between IFN (theoretical pI 6.1) and several amino and sulfonic acid ionizable groups of the Cibacron Blue dye, which allowed the purification of rFeIFNβ in only one step. This supposes an advantage over the two-step purification process used for the commercial Virbagen Omega®. rFeIFNβ was recovered in the elution fraction, with a yield of 75%, a purification factor of 30 and a specific activity of 1 × 106 IU mg−1. We obtained 8.9 × 104 rFeIFNβ IU per larva and 3.63 × 105 rFeIFNβ IU per gram of larvae.
Fig. 3.
Blue-Sepharose purification. a Western blot. b SDS-PAGE with Coomassie Blue staining. MK: molecular weight marker; 1: original larval extract; 2: passthrough; 3: wash1; 4: wash2; 5: NaCl 1 M (fraction 1); 6: NaCl 1 M (fraction 2); 7: NaCl 1 M (fraction 3)
In comparison with the process in S. frugiperda larvae previously reported by Targovnik et al. (2014) some differences could be remarked: using the same inoculum of recombinant baculovirus for the larval infection, the optimal day of harvest for rFeIFNβ was 4 DPI, while for rFeIFNα it was 5 DPI. Each cytokine presented different levels of expression in crude S. frugiperda extracts (7.5 × 105 and 1.1 × 106 UI mL−1), demonstrating that the yield of the platform varies even among two cytokines belonging to type I IFN family and both with feline origin.
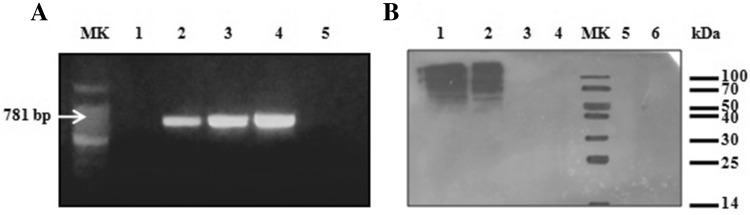
The absence of remaining viral DNA was proven through agarose 1% gel electrophoresis (Fig. 4a). Recombinant baculovirus DNA was found in crude larval extracts, but not in the purified product fraction after Blue-Sepharose chromatography. Therefore, this one-step purification method allows obtaining FeIFN without baculovirus contamination, which makes baculovirus inactivation unnecessary.
Fig. 4.
Analyses of contaminants on purified FeIFNβ fraction. a Agarose 1% gel: viral DNA presence analysis. MK: molecular weight marker (100 pb); 1: negative control (H2O); 2: positive control (AcMNPV DNA); 3: original larval extract; 4: original larval extract post-PD10; 5: purified FeIFNβ fraction. b Western blot: S. frugiperda proteins analysis. 1: original larval extract; 2: passthrough; 3: wash1; 4: wash2; MK: protein marker; 5: wash3; 6: purified FeIFNβ fraction. Western blot was developed with a specific antiserum raised against total S. frugiperda extract
We also evaluated the presence of S. frugiperda proteins in the purified rFeIFNβ fraction by a Western blot that used a polyclonal antiserum directed against the total S. frugiperda homogenate. Although immunogenic larval proteins appeared as several high MW bands in the crude extract and in the passthrough fraction, none were found in the eluate (Fig. 4b). This result implies that although the recovered rFeIFNβ is not 100% pure, the contaminating proteins might not cause an immune response after administration to animals.
Characterization of purified rFeIFNβ
The crude extract of S. frugiperda expressing rFeIFNβ (non-purified) demonstrated both antiviral and antitumor activities in in vitro bioassays.
On the one hand, the ability of purified rFeIFNβ to inhibit the cytotoxic effect of VSV was evaluated on CRFK cells. The antiviral activity value was 9.5 × 104 IU mL−1. Both purified and non-purified rFeIFNβ (Fig. 2a) generated protection against VSV infection.
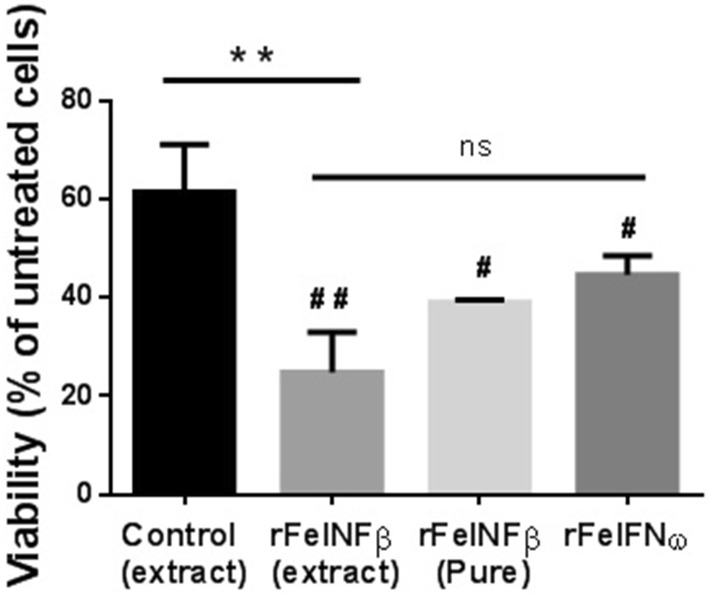
On the other hand, the antitumor activity was evaluated on AlRB, a feline mammary carcinoma cell line that has been previously reported as being sensitive to interferon omega (Villaverdeet al. 2016). AlRB cells were incubated with larval extracts (control or rFeIFNβ) or with purified products (rFeIFNβ or commercial rFeIFNω), the latter used as standard (Virbagen Omega®). As it is shown in Fig. 5, rFeIFNβ larval extract drastically decreased AlRB cells viability from 60% (control larval extract) to less than 25% (p < 0.01). Because of the complexity of the larval extract, the control showed some inhibition of the cell viability, but it was significantly lower than that of rFeIFNβ larval extract. In addition, purified rFeIFNβ decreased AlRB cells viability from 100% (untreated cells) to less than 40% (p < 0.05). This effect was comparable to that observed with 5000 IU mL−1 of commercial rFeIFNω (Fig. 5) and with previously reported effects of FeIFNω gene therapy (Villaverde et al. 2016).
Fig. 5.
In vitro antitumor activity. Black: negative control larval extract (larvae infected with a non-related baculovirus); gray: rFeIFNβ larval extract; pale gray: rFeFNβ purified product (5000 IU mL−1); dark gray: standard rFeIFNω (commercial Virbagen Omega®, 5000 IU mL−1) **p < 0.01 vs. control larval extract; #p < 0.05 and ##p < 0.01 vs. untreated AlRB cells
Deglycosylation
To further characterize the rFeIFNβ, it was subjected to the action of peptide-N-glycosidase F. The cleavage of asparagine-bound N-glycans induced a shift in the MW of the protein to a lower one, as evidenced through Western blot (Fig. 6). In this case, 24 kDa became 20 kDa, nearer its sequence-deduced MW (19.9 kDa).
Fig. 6.
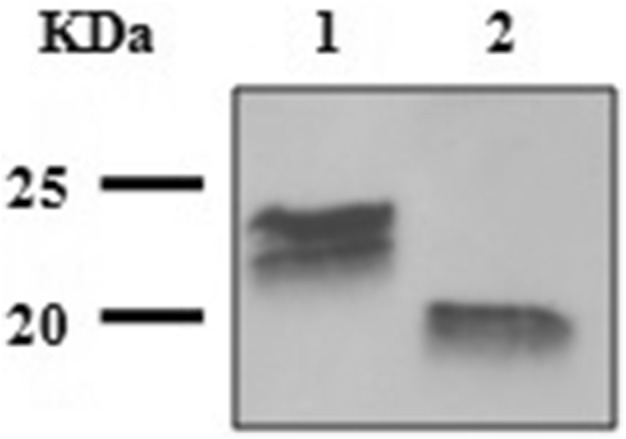
FeIFNβ deglycosylation. Lane 1: purified FeIFNβ fraction; lane 2: purified FeIFNβ fraction treated with peptide-N-glycosidase F
Thus, the eukaryotic environment allowed the N-glycosylation of the biologically active rFeIFNβ. With regards to the double-band more evident in Fig. 3a, it has been sustained that, similar to their native counterparts, glycoprotein glycans are unlikely to be homogenous, giving rise to a profile of partially trimmed, high mannose structures and hybrids (Shi and Jarvis 2007). The predominant MW of 24 kDa corresponds to paucimannose structures, according to bioinformatic analysis and previous works (Kulakosky et al. 1998). Multiple bands due to glycosylation have previously described by other authors with the expression of recombinant IFN in insect cells and Bombyx mori larvae (Na et al. 2008; Usami et al. 2011). In this case, the biological activity of the rFeIFNβ was not altered by this glycosylation heterogenicity.
Conclusions
Spodoptera frugiperda larvae, a plague with no economic value, appear as an interesting platform to express biologically active rFeIFNβ. Purification of rFeIFNβ from the larval extract through Blue-Sepharose chromatography is a low-cost strategy, easy for scaling-up. In a typical experiment, with a small lot-to-lot variation, the amount of rFeIFNβ recovered from a single larva was 8.9 × 104 IU.
As supported by our results, rFeIFNβ becomes an interesting cytokine for future applications in veterinary medicine due to its proven antiviral and antitumor activities. The process herein described may be used for future assessment of rFeIFNβ biological activity in vivo, as well as scaling-up rFeIFNβ production with a low cost.
Acknowledgements
This work was supported by Grants from Agencia Nacional de Promoción Científica y Tecnológica de Argentina and from Universidad de Buenos Aires (PICT 2014-3350; UBACyT 2016-20020150100145BA). AMT, MSV, FJW and MVM are career researchers of the Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina (CONICET). MBA is a research fellow of CONICET. GJM is a research fellow of Agencia de Promoción Científica y Tecnológica (ANPCyT).
Abbreviations
- AcMNPV
Autographa californica multiple nucleopolyhedrovirus
- FeIFN
Feline interferon
- rFeIFN
Recombinant feline interferon
- MW
Molecular weight
- FBS
Fetal bovine serum
- VSV
Vesicular stomatitis virus
- MEM
Modified Eagle’s medium
- MOI
Multiplicity of infection
- DPI
Day(s) post-infection
Author contributions
MBA worked out almost all of the technical details and performed the numerical calculations; GJM and IS contributed with analytical methods; MSV performed antitumor activity assays; FJW contributed in downstream processing; AMT contributed in discussion; MVM was involved in planning and supervised the work. All authors discussed the results and commented on the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Contributor Information
Mariana Bernadett Arregui, Email: arreguimb@gmail.com.
Gregorio Juan Mc Callum, Email: mccallumgj@gmail.com.
Ignacio Smith, Email: nachosmith@live.com.ar.
Marcela Solange Villaverde, Email: marcelavillaverde@hotmail.com.
Federico Javier Wolman, Email: fwolman@ffyb.uba.ar.
Alexandra Marisa Targovnik, Email: atargovnik@yahoo.com.
María Victoria Miranda, Phone: +54-11-5287-4679, Email: mvic@ffyb.uba.ar.
References
- Argyle DJ, Harris M, Lawrence C, McBride K, Barron R, McGillivray C, Onions DE. Expression of feline recombinant interferon-gamma in baculovirus and demonstration of biological activity. Vet Immunol Immunopathol. 1998;64(2):97–105. doi: 10.1016/S0165-2427(98)00127-5. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burges HD, Croizier G, Huber J. A review of safety tests on baculoviruses. Entomophaga. 1980;25(4):329–340. doi: 10.1007/BF02374693. [DOI] [Google Scholar]
- Chamorey AL, Magne N, Pivot X, Milano G. Impact of glycosylation on the effect of cytokines. A special focus on oncology. Eur Cytok Netw. 2002;13(2):154–160. [PubMed] [Google Scholar]
- Chawla-Sarkar M, Leaman DW, Borden EC. Preferential induction of apoptosis by interferon (IFN)-beta compared with IFN-alpha2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin Cancer Res. 2001;7(6):1821–1831. [PubMed] [Google Scholar]
- Coelho LF, Magno de Freitas Almeida G, Mennechet FJ, Blangy A, Uze G. Interferon-alpha and -beta differentially regulate osteoclastogenesis: role of differential induction of chemokine CXCL11 expression. Proc Natl Acad Sci USA. 2005;102(33):11917–11922. doi: 10.1073/pnas.0502188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damdinsuren B, Nagano H, Wada H, Kondo M, Ota H, Nakamura M, Noda T, Natsag J, Yamamoto H, Doki Y, Umeshita K, Dono K, Nakamori S, Sakon M, Monden M. Stronger growth-inhibitory effect of interferon (IFN)-beta compared to IFN-alpha is mediated by IFN signaling pathway in hepatocellular carcinoma cells. Int J Oncol. 2007;30(1):201–208. [PubMed] [Google Scholar]
- de Weerd NA, Vivian JP, Nguyen TK, Mangan NE, Gould JA, Braniff SJ, Zaker-Tabrizi L, Fung KY, Forster SC, Beddoe T, Reid HH, Rossjohn J, Hertzog PJ. Structural basis of a unique interferon-beta signaling axis mediated via the receptor IFNAR1. Nat Immunol. 2013;14(9):901–907. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat Protoc. 2009;4(3):309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- Gronowski AM, Hilbert DM, Sheehan KC, Garotta G, Schreiber RD. Baculovirus stimulates antiviral effects in mammalian cells. J Virol. 1999;73(12):9944–9951. doi: 10.1128/jvi.73.12.9944-9951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas-Stubbs S, Rueda P, Lopez L, Lecrerc C. Insect baculoviruses strongly potentiate adaptative immune responses by inducing Type I IFN. J Immunol. 2007;178(4):2361–2369. doi: 10.4049/jimmunol.178.4.2361. [DOI] [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond Ser B Biol Sci. 1957;147(927):258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- Iwata A, Saito T, Mizukoshi-Iwata N, Fujino M, Katsumata A, Hamada K, Sokawa Y, Ueda S. Cloning and expression of the canine interferon-beta gene. J Interferon Cytok Res. 1996;16(10):765–770. doi: 10.1089/jir.1996.16.765. [DOI] [PubMed] [Google Scholar]
- Kost TA, Condreay JP. Innovations—biotechnology: baculovirus vectors as gene transfer vectors for mammalian cells: biosafety considerations. Appl Biosaf. 2002;7(3):167–169. doi: 10.1177/153567600200700312. [DOI] [Google Scholar]
- Kruth SA. Biological response modifiers: interferons, interleukins, recombinant products, liposomal products. Vet Clin N Am Small Anim Pract. 1998;28(2):269–295. doi: 10.1016/S0195-5616(98)82005-6. [DOI] [PubMed] [Google Scholar]
- Kulakosky PC, Hughes PR, Wood HA. N-Linked glycosylation of a baculovirus-expressed recombinant glycoprotein in insect larvae and tissue culture cells. Glycobiology. 1998;8(7):741–745. doi: 10.1093/glycob/8.7.741. [DOI] [PubMed] [Google Scholar]
- Lindner DJ. Interferons as antiangiogenic agents. Curr Oncol Rep. 2002;4(6):510–514. doi: 10.1007/s11912-002-0065-4. [DOI] [PubMed] [Google Scholar]
- Maeda S, Kawai T, Obinata M, Fujiwara H, Horiuchi T, Saeki Y, Sato Y, Furusawa M. Production of human alpha-interferon in silkworm using a baculovirus vector. Nature. 1985;315(6020):592–594. doi: 10.1038/315592a0. [DOI] [PubMed] [Google Scholar]
- Martina JA, Daniotti JL, Maccioni HJ. Influence of N-glycosylation and N-glycan trimming on the activity and intracellular traffic of GD3 synthase. J Biol Chem. 1998;273(6):3725–3731. doi: 10.1074/jbc.273.6.3725. [DOI] [PubMed] [Google Scholar]
- Mochizuki M, Nakatani H, Yoshida M. Inhibitory effects of recombinant feline interferon on the replication of feline enteropathogenic viruses in vitro. Vet Microbiol. 1994;39(1–2):145–152. doi: 10.1016/0378-1135(94)90095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Z, Huipeng Y, Lipan L, Cuiping C, Umashankar ML, Xingmeng L, Xiaofeng W, Bing W, Weizheng C, Cenis JL. Efficient production of canine interferon-alpha in silkworm Bombyx mori by use of a BmNPV/Bac-to-Bac expression system. Appl Microbiol Biotechnol. 2008;78(2):221–226. doi: 10.1007/s00253-007-1296-y. [DOI] [PubMed] [Google Scholar]
- Nagai A, Taira O, Ishikawa M, Hiramatsu K, Hohdatsu T, Koyama H, Arai S, Sato H, Nakano K, Maehara N. Cloning of cDNAs encoding multiple subtypes of feline interferon-alpha from the feline epitherial cell line. J Vet Med Sci. 2004;66(6):725–728. doi: 10.1292/jvms.66.725. [DOI] [PubMed] [Google Scholar]
- O’Reilly DR, Miller LK, Luckow VA. Baculovirus expression vectors: a laboratory manual. Oxford: Oxford University Press; 1994. [Google Scholar]
- Okano F, Satoh M, Ido T, Okamoto N, Yamada K. Production of canine IFN-gamma in silkworm by recombinant baculovirus and characterization of the product. J Interferon Cytok Res. 2000;20(11):1015–1022. doi: 10.1089/10799900050198462. [DOI] [PubMed] [Google Scholar]
- Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem. 2007;282(28):20047–20051. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Spearman M, Huzel N, Butler M. Enhanced production of monomeric interferon-beta by CHO cells through the control of culture conditions. Biotechnol Progress. 2005;21(1):22–30. doi: 10.1021/bp049807b. [DOI] [PubMed] [Google Scholar]
- Romero LV, Targovnik AM, Wolman FJ, Cascone O, Miranda MV. Rachiplusia nu larva as a biofactory to achieve high level expression of horseradish peroxidase. Biotechnol Lett. 2011;33(5):947–956. doi: 10.1007/s10529-011-0540-9. [DOI] [PubMed] [Google Scholar]
- Rubinstein S, Familletti PC, Pestka S. Convenient assay for interferons. J Virol. 1981;37(2):755–758. doi: 10.1128/jvi.37.2.755-758.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Ueda Y, Sato M, Yanai A. Feline interferon production in silkworm by recombinant baculovirus. J Vet Med Sci. 1992;54(3):563–565. doi: 10.1292/jvms.54.563. [DOI] [PubMed] [Google Scholar]
- Schmeisser H, Kontsek P, Esposito D, Gillette W, Schreiber G, Zoon KC. Binding characteristics of IFN-alpha subvariants to IFNAR2-EC and influence of the 6-histidine tag. J Interferon Cytok Res. 2006;26(12):866–876. doi: 10.1089/jir.2006.26.866. [DOI] [PubMed] [Google Scholar]
- Shi X, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell system. Curr Drug Targets. 2007;8(10):1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AN. A review of the biology of the fall armyworm. Fla Entomol. 1979;62(2):82–87. doi: 10.2307/3494083. [DOI] [Google Scholar]
- Stifter SA, Gould JA, Mangan NE, Reid HH, Rossjohn J, Hertzog PJ, de Weerd NA. Purification and biological characterization of soluble, recombinant mouse IFNbeta expressed in insect cells. Prot Express Purif. 2014;94:7–14. doi: 10.1016/j.pep.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Targovnik AM, Villaverde MS, Arregui MB, Fogar M, Taboga O, Glikin GC, Finocchiaro LME, Cascone O, Miranda MV. Expression and purification of recombinant feline interferon in the baculovirus-insect larvae system. Process Biochem. 2014;49(6):917–926. doi: 10.1016/j.procbio.2014.03.013. [DOI] [Google Scholar]
- Targovnik AM, Arregui MB, Bracco LF, Urtasun N, Baieli MF, Segura MM, Simonella MA, Fogar M, Wolman FJ, Cascone O, Miranda MV. Insect larvae: a new platform to produce commercial recombinant proteins. Curr Pharm Biotechnol. 2016;17(5):431–438. doi: 10.2174/138920101705160303163947. [DOI] [PubMed] [Google Scholar]
- Usami A, Ishiyama S, Enomoto C, Okazaki H, Higuchi K, Ikeda M, Yamamoto T, Sugai M, Ishikawa Y, Hosaka Y, Koyama T, Tobita Y, Ebihara S, Mochizuki T, Asano Y, Nagaya H. Comparison of recombinant protein expression in a baculovirus system in insect cells (Sf9) and silkworm. J Biochem. 2011;149(2):219–227. doi: 10.1093/jb/mvq138. [DOI] [PubMed] [Google Scholar]
- van Oers MM. Opportunities and challenges for the baculovirus expression system. J Invertebr Pathol. 2011;107 Suppl:S3–S15. doi: 10.1016/j.jip.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Villaverde MS, Targovnik AM, Miranda MV, Finocchiaro LME, Glikin GC. Cytotoxic effects induced by interferon-ω gene lipofection through ROS generation and mitochondrial membrane potential disruption in feline mammary carcinoma cells. Cytokine. 2016;84(Supplement C):47–55. doi: 10.1016/j.cyto.2016.05.018. [DOI] [PubMed] [Google Scholar]
- Weiss RC, Toivio-Kinnucan M. Inhibition of feline infectious peritonitis virus replication by recombinant human leukocyte (alpha) interferon and feline fibroblastic (beta) interferon. Am J Vet Res. 1988;49(8):1329–1335. [PubMed] [Google Scholar]
- Yang LM, Xue QH, Sun L, Zhu YP, Liu WJ. Cloning and characterization of a novel feline IFN-omega. J Interferon Cytok Res. 2007;27(2):119–127. doi: 10.1089/jir.2006.0094. [DOI] [PubMed] [Google Scholar]