Abstract
Over the past decade, unusual mortality outbreaks have decimated echinoderm populations over broad geographic regions, raising awareness globally of the importance of investigating such events. Echinoderms are key components of marine benthos for top-down and bottom-up regulations of plants and animals; population declines of these individuals can have significant ecosystem-wide effects. Here we describe the first case study of an outbreak affecting Antarctic echinoderms and consisting of an ulcerative epidermal disease affecting ~10% of the population of the keystone asteroid predator Odontaster validus at Deception Island, Antarctica. This event was first detected in the Austral summer 2012–2013, coinciding with unprecedented high seawater temperatures and increased seismicity. Histological analyses revealed epidermal ulceration, inflammation, and necrosis in diseased animals. Bacterial and fungal alpha diversity was consistently lower and of different composition in lesioned versus unaffected tissues (32.87% and 16.94% shared bacterial and fungal operational taxonomic units OTUs respectively). The microbiome of healthy stars was more consistent across individuals than in diseased specimens suggesting microbial dysbiosis, especially in the lesion fronts. Because these microbes were not associated with tissue damage at the microscopic level, their contribution to the development of epidermal lesions remains unclear. Our study reveals that disease events are reaching echinoderms as far as the polar regions thereby highlighting the need to develop a greater understanding of the microbiology and physiology of marine diseases and ecosystems health, especially in the era of global warming.
Introduction
Emerging diseases in marine invertebrates seem to be increasing in prevalence, complexity, and sometimes virulence, concomitant with global climate change1–5. In addition to their direct impact on affected species, epizootics in marine invertebrates can have important ecological ramifications. For example, a mass die off of sea urchins in the Caribbean led to profound ecosystem shifts manifested by overgrowth of algae and subsequent secular declines in hermatypic corals6. The impact of marine diseases, especially in isolated regions, is difficult to investigate because of limited opportunities for long-term observations, especially if the immediate effects are sublethal or can be confounded by other ecological interactions (e.g., competition, predation)5. In other cases, epizootics occur too rapidly to be investigated or are not investigated at all7,8. Conceptually, diseases are the outcomes of susceptible hosts interacting with causative agents and the environment9. Some infectious diseases are polymicrobial, requiring the cooperation of several pathogens such as in black band disease in corals that involves primary sulfate-oxidizing cyanobacteria Roseofilum reptotaenium and secondary sulfate-reducing Desulfovibrio bacteria10. Confirming causation in marine invertebrate diseases can be challenging because of limited knowledge of host physiology and anatomy and difficulty in laboratory manipulation of agents associated with disease. For instance, many marine microbes cannot be cultured3. Against these backdrops, integrating multiple diagnostic methods such as molecular techniques paired with classic approaches (e.g., field survey, microscopy, transmission experiments) seems the best practice for disease exploration in wild marine organisms5.
Echinoderms are fundamental constituents structuring benthic systems, because they are ubiquitous predators/grazers, as well as prey for top carnivores affecting top-down and bottom-up regulation of communities11–13. A number of diseases sometimes associated with bacteria, fungi, protozoans, algae, metazoans, viruses, or often multifactorial unknown etiologies have been reported to affect this phylum sometimes leading to mass mortalities14–20. In some cases, diseases of echinoderms have influenced ecosystem scale processes. Examples include coral declines and algal overgrowth subsequent to urchin die offs in the Caribbean6 and reforestation of kelp subsequent to protozoan induced die offs of urchins in Nova Scotia21. The most recent echinoderm epizootic was the devastating 2013–2014 Sea Star Wasting Disease (SSWD) that killed millions of sea stars of +20 different species over an extensive geographic range of the North Pacific American Coast. Affected animals were seen often after warm seawater episodes manifesting behavioural changes, twisted arms, deflated appearance, and white lesions on the aboral dermis that rapidly progressed to tissue degradation, loss of turgor, arm loss, and death by eventual disintegration – “melting”19,22,23. Experimental evidence (sensu24) pointed a Parvoviridae virus, named SSaDV – sea star-associated densovirus– as the best candidate causative agent, yet this was only consistent for one asteroid species19. Indeed, SSWD etiology encompasses diverse cocktails of potential pathogens, and is heterogeneous across environmental conditions, geographies and species25–27. As a result, SSWD was recently re-named as Asteroid Idiopathic Wasting Syndrome (AIWS)26. The grotesque manner in which sea stars melted, together with the virulence and magnitude of this syndrome, has led to scientific concern, in addition to seizing public attention23. Historically though, most disease outbreaks affecting sea stars have been brief and localized and not as extensively investigated as the AIWS8.
Deception Island (South Shetland Archipelago) is a horseshoe-shaped island near the Antarctic Peninsula that encloses an active flooded volcano (Port Foster’s Bay; Fig. S1), subject to intense temperature fluctuations28–30. After the last eruption in 1970, the local benthos experienced remarkable recolonization of primarily algae and echinoderms, together with detritivore communities31,32. Currently, three species of echinoderms predominate: the echinoid Sterechinus neumayeri, the ophiuroid Ophionotus victoriae, and among the three most common asteroids stands out Odontaster validus33,34. The latter species is a keystone predator that has a circum-Antarctic and eurybathic range (down to 1000 m depth)35,36. During routine surveys in the austral summer of 2012–2013 and 2016, O. validus specimens were observed manifesting focal epidermal depigmentation, anorexia, and arm loss, suggesting an incipient epizootic5,19,37. Interestingly, the outbreak concurred with unprecedented geothermal anomalies, sudden episodic elevations in seawater temperature, and seismic reactivation of the volcano29. This paper describes the gross, and microscopic pathology of this disease and the role of the associated microbiota.
Material and Methods
Ethical statement
Odontaster validus, Koehler 1906, were collected and handled in agreement with all applicable international and national guidelines and regulations for the care and use of animals in accordance with the current laws of Spain and the Comité Polar Español (CPE) through the Antarctic Treaty and the Madrid Protocol on Environmental Protection to the Antarctic Treaty (‘Act on Antarctic Activities and Protection of Antarctic Environment’). The target species is not endangered or protected, and those that survived after the study were released.
Study site and disease incidence
Transect surveys were conducted to assess the prevalence of epidermal lesions in sea stars around Port Foster’s bay (Deception Island; Fig. S1) during the Antarctic expedition ACTIQUIM-4 (January–February 2013; Figs 1 and 2). Five haphazardly chosen replicate 50-m linear transects were surveyed at 5 m and 15 m depth at eight sites around the bay (80 transects in total; Fig. 1). Apparently healthy O. validus and specimens with lesions were recorded within 2 m of each transect line. The census was repeated in 2016.
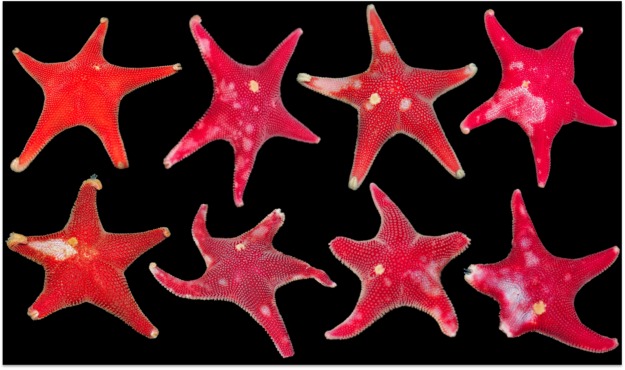
Figure 1.
Antarctic sea star Odontaster validus: Normal (upper left) and diseased (remainder) specimens. Note epidermal lesions characterized by multifocal to locally extensive punctate to amorphous distinct to indistinct areas of discoloration on the dorsum. Stars on bottom row manifest partial loss of arms.
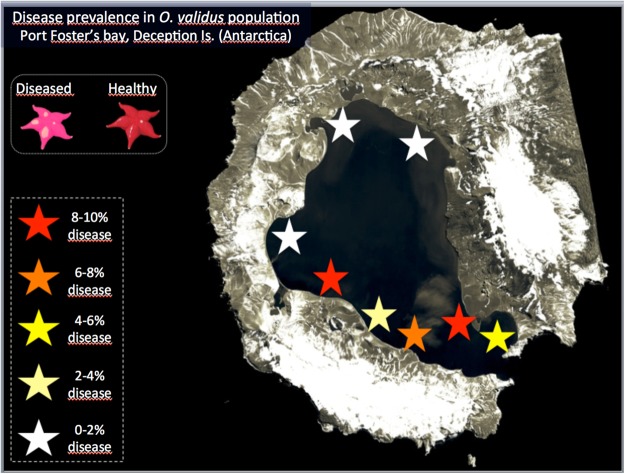
Figure 2.
Prevalence of Odontaster validus sea stars affected by visually evident lesions in populations within Port Foster’s bay, Deception Island, South Shetland Archipelago (Antarctica).
Sampling collections
Twenty-five healthy and diseased O validus (n = 50) were collected by SCUBA diving on February 2013. Tissue biopsies (1 cm3) were removed with sterile scalpel from 30 individuals (15 healthy and 15 diseased) as follows: 15 sections from healthy specimens (hereafter called “Healthy_#”), 15 affected lesion fronts from diseased sea stars (“Affected_#”), and 15 tissue areas several cm away from the lesions of the same diseased specimens (“NON_Affected_#”). Samples were divided in two subsamples, one preserved in 100% ethanol at −20 °C for DNA extraction and microbial characterization, and the other fixed in 2.5% paraformaldehyde in filtered sea water at 4 °C for histopathology studies.
Transmission trials
To assess potential disease transmission, ten 2L-aquaria, each with one apparently healthy and one diseased sea star, were maintained in the wet laboratory for five weeks under natural sunlight. Seawater in the aquaria was pumped directly from the bay nearby the BAE (Spanish Antarctic Base) and kept at −1 °C. The seawater was completely (100%) renewed twice daily. Progression of lesions in diseased specimens and appearance of lesions in healthy individuals was monitored daily with a stereomicroscope.
Histopathology examinations
Samples in 2.5% glutaraldehyde were embedded in paraffin at 60 °C overnight, sectioned at 5 µm, stained with hematoxylin and eosin, and examined under light microscopy.
DNA extraction and amplification of bacterial and fungal rRNA gene
DNA from healthy and lesioned tissues of healthy and diseased stars were extracted using a modified C-TAB organic extraction protocol for amplicon deep sequencing of ribosomal gene target markers on MiSeq (Illumina), for bacterial/archaeal and fungal community composition, which were performed with a two-PCR protocol and two dual-index strategy38,39. In the first PCR, we used bacterial specific primers to amplify the V3–V4 region (Escherichia coli position: 341–785) of the small-subunit ribosomal RNA (16S) gene (341 F and 785 R)39,40; and fungi-specific primers ITS1F41 and ITS2R42 targeting the internal transcribed spacer 1 (ITS1) region of fungi. Amplifications were performed in 25 µl reactions with NEBNext® Q5® Hot Start HiFi PCR Master Mix (New England Biolabs, Inc.), 0.8 µl BSA (Bovine Serum Albumin; 20 mg/ml), 1 µl of each 5 µM primer, and 1.5 µl of template. Reactions were under the thermocycling profile: 98 °C for 2 min, then 28 cycles of 98 °C for 15 s, 53 °C for 30 s, 72 °C for 30 s, final extension at 72 °C for 2 min. The second Index PCR to attach dual indexes and Illumina sequencing adapters used forward primers with the 5′-3′ Illumina i5 adapter (AATGATACGGCGACCACCGAGATCTACAC), an 8–10 bp barcode and a primer pad; and reverse fusion primers with 5′-3′ Illumina i7 adapter (CAAGCAGAAGACGGCATACGAGAT), an 8–10 bp barcode, a primer pad. Reactions were made in 25 μl with 0.5 µl of each 5 µM primer, and 1 µl of corresponding products from first amplicon PCR reactions diluted (1:30), and with a temperature regime of: 98 °C for 2 min, then 28 cycles of 98 °C for 15 s, 55 °C for 30 s, 72 °C for 30 s, final extension at 72 °C for 2 min. The PCR products were purified and pooled equimolar on Just-a-Plate™ 96 PCR Purification and Normalization Kit plates following manufacturer’s instructions (Charm Biotec). Then, paired-end sequencing was performed on an Illumina MiSeq sequencer 2 × 300 flow cell at 10 pM at Core Lab, Hawai’i Institute of Marine Biology (Hawai’i, USA).
Sequence processing and statistical analyses
Sequences demultiplexed by sample-specific barcodes were clustered/parsed using Pear 0.9.8 and quality controlled and filtered with Fastx-Toolkit. Sequence reads were then processed using the quantitative insights into microbial ecology (QIIME) pipeline 1.9.143. Applying Edgar44–46 pipelines, low quality reads and chimeric sequences were discarded (USEARCH, UCHIME2), and OTUs (operational taxonomic units) were identified (method: UCLUST, threshold: 97%). Representative sequences were chosen by consensus and assigned a taxonomic identity (method: UCLUST) based on reference databases: Silva_123 (release version SSU/LSU 123) for Bacteria47 and UNITE for Fungi48. Prior to downstream analysis, sequences classified as “unknown” at the kingdom level, potential host co-amplification (chloroplast, mitochondrion), and contaminants were removed.
Alpha diversity (within sample) was estimated with several methods (e.g., Simpson, Fisher) and plotted using non-parametric Shannon H’ and Chao49 indices, whereas OTU tables were built in Microsoft Excel with output files from QIIME and R pipelines. Beta diversity (between sample) was assessed by permutational multivariate analysis of variance – PERMANOVA50 at 9999 permutations to test for statistical differences (P-values) in bacterial and fungal community assemblages among tissue health states (i.e., Healthy_#”, “Non_Affected_#”, and “Affected_#”). Post hoc pair-wise comparisons among health states were conducted following main effects results. Bray-Curtis distance metrics at the OTU level were used to construct unconstrained two-dimensional principal coordinate analysis (PCoA) plots to visualize differences among bacterial and fungi community assemblages51. The similarity percentages (SIMPER)52 analyses based on Bray-Curtis similarity at the OTU level was used to determine the contribution of individual bacterial and fungal taxa to the dissimilarity between groups. Analysis of variance (ANOVA) was conducted on read data at the class level and also on most contributing bacterial and fungal OTUs according to SIMPER analyses to test for differences between health states. Where ANOVA resulted in significant differences, post hoc Tukey’s honest significance texts (HSD) tests were applied to assess pairwise differences. All multivariate analyses were performed in R-Studio 1.0.136 (R version 3.3.3) with the corresponding R-packages (e.g., PhyloSeq, Vegan, Bioconductor).
Data availability
The sequence data set was deposited in the NCBI Sequence Read Archive (SRA) database (accession numbers: SRP131970). Other related datasets generated during and/or analysed during the current study are available from the corresponding author on request.
Results
Disease incidence
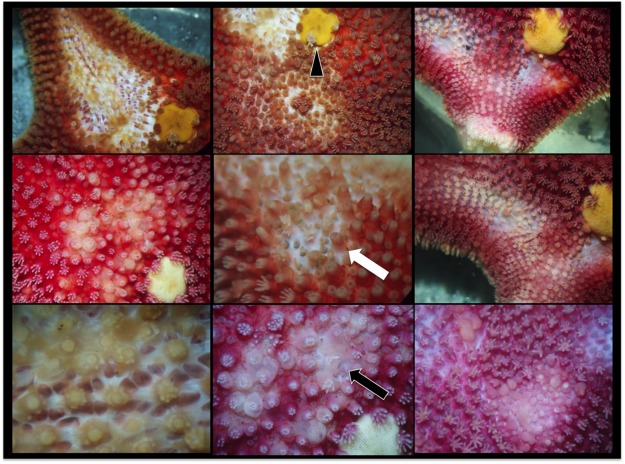
Lesions were limited to O. validus with a prevalence ranging from 0% at most inner areas of Port Foster’s caldera, up to 10% at sites closer to the entrance –Neptune’s Bellows (Fig. 2). Apparent early stages of disease comprised multifocal distinct to indistinct 1–5 mm pale to white spots that coalesced to larger ulcers leading to decay of spines, papulae and ablation of paxillae, with more severe cases revealing underlying ossicles or total amputation of arms (Figs 1 and 3).
Figure 3.
Higher magnification under stereoscope of epidermal lesions in Odontaster validus showing depigmentation (white arrow), and erosion of paxillae and epidermal ulceration (black arrow). Arrowhead pointing to the madreporite.
Transmission trial
In aquaria, epidermal lesions in affected stars progressed almost imperceptibly (1–2 mm increase in size over 5 weeks’ observation). Animals going from asymptomatic to the severe signs of disease were not observed in the time lapse during our campaigns. We saw no evidence of transmission between lesion and healthy stars.
Histopathology
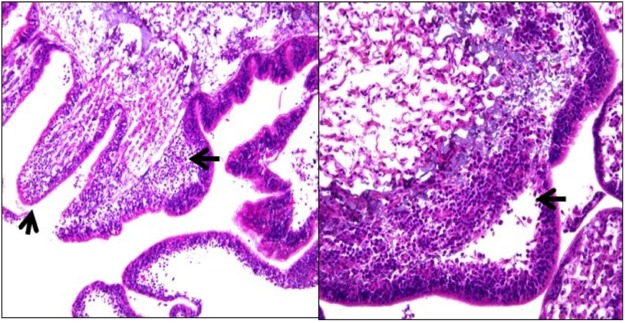
Histology of gross lesion fronts of diseased animals revealed cleft formation between epidermis and dermis with spaces infiltrated by hemocytes (inflammation). In more severe lesions, there was full thickness epidermal necrosis and ulceration with associated hemocyte infiltrates lifting off the epidermis from the underlying ossicles (ulceration). No characteristic accumulations of microorganisms were seen associated with the lesion. Five of 15 tissues with lesions had histologic evidence of inflammation/ulceration, whereas no histopathological signs were found in the 15 non-lesion tissues examined (Fig. 4).
Figure 4.
Histological sections of Odontaster validus epidermal lesions: (left) note inflammatory cell infiltrate lifting off epidermis from underlying ossicle (big black arrow) with diffuse ulceration (small black arrow); (right): note marked subepithelial infiltrates of inflammatory cells with cleft formation (arrow).
16S rRNA gene bacterial diversity and community composition
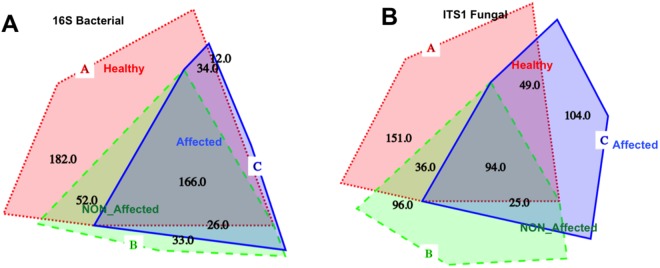
Sequences retrieved for the bacterial 16S hypervariable V3–V4 region yielded, after quality control and chimera filtering, 189,046 reads with an average length of ~427 bp. According to the number of observed OTUs, Shannon and Fisher indexes, and Chao1 estimation of species richness based on normalized reads (n = 1545), the bacterial diversity of tissues from healthy sea stars was remarkably higher than that from diseased samples, including apparently non-affected sections and lesion fronts (Table 1, Fig. S2). Simpson diversity index was similar for healthy tissues from healthy and affected sea stars, but yielded lower values in lesion areas. A total of 505 OTUs were detected based on a sequence similarity criterion of 97%, of which 434 were associated with Healthy tissues, whereas 277 and 238 were found in NON_Affected and Affected sections, respectively. In concordance with diversity statistics, the number of unique OTUs was the highest in healthy sea stars (182), with respect to apparently healthy (33), and lesion sections (12) from the diseased specimens (Table 1; Fig. 5).
Table 1.
Summary of 16S rRNA bacterial and ITS-1 fungal communities profiling from healthy and diseased Odontaster validus sea stars.
| Status | Healthy seastar | Diseased seastar | TOTAL | |||||
|---|---|---|---|---|---|---|---|---|
| Tissue | Healthy | NON_Affected | Affected | |||||
| Replicates (n) | 15 | 15 | 15 | 45 | ||||
| Genetic marker | 16S | ITS-1 | 16S | ITS1 | 16S | ITS-1 | 16S | ITS1 |
| Reads per sample | 1,545–6,271 | 3,129–3,4871 | 2,975–6,890 | 1,378–2,3487 | 2,269–5,917 | 1,380–1,5095 | 189,046 | 335,754 |
| Average length | 427 | 272 | ||||||
| Fisher index | 63.25436 | 39.62805 | 36.64054 | 31.09836 | 31.64202 | 35.7209 | 63.08028 | 64.90119 |
| Simpson index | 0.2812992 | 0.9484165 | 0.2953405 | 0.9442799 | 0.19308 | 0.9325904 | 0.2601939 | 0.9522523 |
| Observed OTUsa | 434 | 330 | 277 | 251 | 238 | 272 | 505 | 555 |
| Unique OTUsb | 182 | 151 | 33 | 96 | 12 | 104 | 227 | 351 |
| Number of Phyla | 13 | 3 | 9 | 3 | 10 | 4 | 14 | 4 |
| Number of Classes | 26 | 16 | 17 | 17 | 17 | 14 | 28 | 18 |
| Number of Order | 52 | 33 | 32 | 33 | 32 | 31 | 57 | 40 |
| Number of Families | 100 | 47 | 61 | 40 | 54 | 46 | 106 | 63 |
| Number of Genera | 182 | 90 | 95 | 79 | 79 | 82 | 207 | 136 |
Abbreviations: OTUs, operational taxonomic units; ITS, interspatial tanscripted spacer.
Alpha diversity statistics were calculated with phyloseq R-package on RStudio.
aOTUs can be shared among multiple samples and are based on 97% sequence similarity criteria according to Silva_123 database.
bNumber of unique OTUs characteristic for each tissue type after the sequences were referenced and combined by QIIME.
Figure 5.
Proportional Venn Diagrams depicting represented OTUs by tissue type for the bacterial (A) and fungal (B) communities associated with Odontaster validus sea stars. Healthy (in red): tissues from healthy sea stars; NON_Affected (in green): apparently healthy tissues from diseased specimens; and Affected (in blue): diseased tissues from diseased specimens.
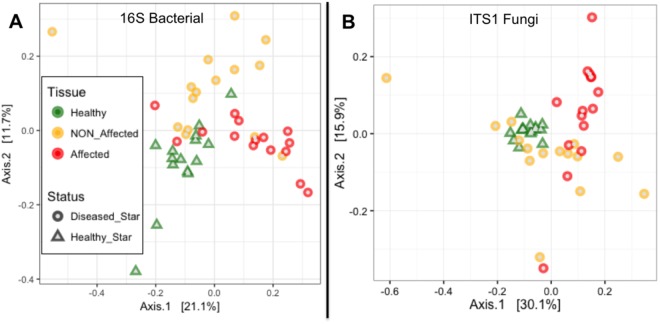
At the species level (97% sequence similarity), bacterial communities associated with O. validus were significantly different between health states (df = 1, pseudo-F = 1.996, Pperm < 0.05) and among tissue types (df = 1, pseudo-F = 2.923, Pperm < 0.001). For the same health status, specimen replicates displayed similar bacterial communities demonstrating homogeneity within tissue types (df = 28, pseudo-F = 1.318, Pperm > 0.05). Healthy and Affected tissues exhibited significant difference (df = 1, pseudo-F = 5.486, Pperm < 0.01), and in diseased sea stars the bacterial composition differed between Affected and NON_Affected tissues (df = 1, pseudo-F = 4.874, Pperm < 0.01). No significant differences were detected between healthy tissues sampled from healthy specimens versus healthy tissues sampled from diseased sea stars away from the lesion areas (i.e., Healthy and NON_Affected; df = 1, pseudo-F = 0.872, Pperm > 0.05; see Table S1). Segregation of bacterial community compositions associated with tissues of different health status can be visualized in PCoA ordination, where Affected tissues clustered loosely towards the positive end of the first axis. Along the negative end of this axis 1 there was a transition towards a loose clustering of Healthy and NON_Affected tissues, which shifted from one another along the centre of axis 2 (Fig. 6).
Figure 6.
Principal Components Analysis (PCoA) plots based on Bray-Curtis distances of Odontaster validus associated bacterial (A) and fungal (B) communities composition across three tissue types: healthy tissues from healthy sea stars (Healthy; green triangles), apparently healthy tissues from diseased sea stars (NON_Affected; yellow circles), and affected tissues from diseased sea stars (Affected; red circles). The proximity of samples on the PCoA illustrates the similarity of bacterial or fungal communities.
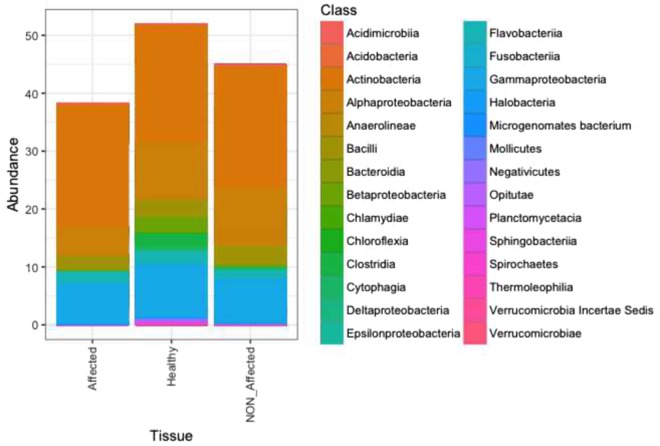
The most represented phylum in all tissue types was Actinobacteria (Fig. 7), which was dominated by the genus Rhodococcus (98%), and in particular by B_OTU240 (78.25% in Affected versus 63.16% in Healthy and 55.18% in NON_Affected, SIMPER, PANOVA < 0.05). According to SIMPER, Rhodococcus B_OTU240 along with B_OTU338 (Acinetobacter sp.) and B_OTU4 (Moritella sp.) are the most influencing OTUs associated with diseased sea stars (Table S2 and Fig. S3). Major differences among healthy states, though, were due to variations in the relative abundance of OTUs affiliated with members of phyla Proteobacteria, Firmicutes, and Bacteroidetes. These were consistently more highly represented in Healthy O. validus, followed by NON_Affected, and least abundant in lesion fronts. At the class level, graphing data and statistics (PANOVA < 0.05) showed that Alphaproteobacteria, Bacilli, and Cytophagia were particularly scant in lesion fronts. While Betaproteobacteria, Clostridia, Gammaproteobacteria, Sphingobacteriia, Negativicutes, and Bacteroidia were more abundant in Healthy sections, they were less abundant in both, NON_Affected and Affected tissues from diseased O. validus (Fig. 7). Twenty-one OTUs revealed >0.1% contribution to dissimilarity (SIMPER) and yielded loose clustering in PCoA plotting between health states (Fig. S3; see Table S2 and ESM1 for more detailed information).
Figure 7.
Class-level taxonomic diversity profiling of bacterial communities associated with tissues from healthy sea stars (Healthy), with apparently healthy (NON_Affected) and diseased (Affected) tissues from diseased specimens of Odontaster validus. “NA”: non-determined taxa.
ITS1 rRNA gene fungal diversity and community composition
Fungi associated with sea star tissues resulted in 335,754 classifiable, non-chimeric reads, averaging ca. 272 bp. The diversity of fungal communities from healthy sea stars was higher than those from Affected and NON_Affected tissues of diseased individuals, as indicated estimations of observed OTUs, Shannon and Fisher indexes, and Chao1 (normalized reads n = 1378; Table 1, Fig. S2). Simpson diversity index showed equivalent results for tissues from all health states. The total number of OTUs at a sequence similarity of 97% was 555. Healthy tissues yielded 151 unique OTUs, 104 were found for disease lesion fronts and 96 for apparently healthy sections from diseased specimens (Table 1; Fig. 5).
The PERMANOVA analysis did not reveal significant differences at the species level (97% sequence similarity) in the overall fungal composition associated with individual health states (df = 1, pseudo-F = 1.04, Pperm > 0.05), nor among different tissue types (df = 1, pseudo-F = 0.972, Pperm > 0.05). For the same health status, specimen replicates displayed homogeneity of fungal communities within tissue types (df = 28, pseudo-F = 1.005, Pperm > 0.05; Table S1). Although there was no statistical difference among health states at the community level, ordination plotting and SIMPER integrated with ANOVA analyses showed the existence of certain explanatory fungal taxa. In the PCoA ordination, Healthy-associated fungal communities appear tightly clustered and separated along axis 1 from a loose clustering corresponding to disease lesion fronts, which dispersed along the second PCoA axis. Fungi populations of NON_Affected tissues from diseased sea stars were the most scattered along both axes (Fig. 6).
No significant differences in fungal communities were recorded in samples coming from healthy versus diseased sea stars. Ascomycota followed by Basidiomycota were predominant in the three tissue types, along with a very small representation of Chytridiomycota. A notable percentage of sequences were unassigned –Fungi sp. Qualitative variability based on graphical and/or statistical (PANOVA < 0.05) data was observed for some class groups. Dothideomycetes, Tremellomycetes, and Microbotryomycetes were more abundant in Healthy, than in NON-Affected of Affeted tissues, whereas Agaricomycetes, Wallemiomycetes, Cystobasidiomycetes, and Eurotiomycetes were more associated with lesion fronts (Fig. 8). Qualitative changes became more evident at the OTU level. Indeed, from the SIMPER analysis, 57 OTUs contributed >0.3% in discriminating among health states. These provided relatively tight clusters (PCoA) according to tissue types (Fig. S3; see Table S3 and ESM1 for more detailed information).
Figure 8.
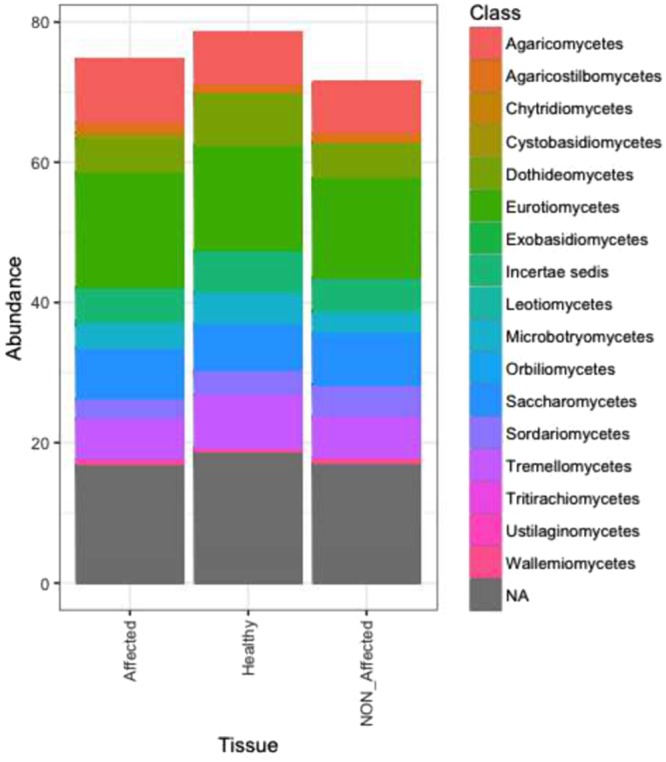
Class-level taxonomic diversity profiling of fungal (B) communities associated with tissues from healthy sea stars (Healthy), with apparently healthy (NON_Affected) and diseased (Affected) tissues from diseased specimens of Odontaster validus. “NA”: non-determined taxa.
Discussion
This is the first description of a novel emerging echinoderm disease occurring in the Antarctic Ocean based on a combination of microbial community profiling and pathology. Sea stars are abundant and comprise various species at Deception Island33,34; however, only one species, the most abundant – O. validus– manifested lesions. The outbreak was first detected in the austral summer of 2012–2013, coinciding with seawater and soil fluctuations characterized by spikes in elevated temperature, and increase in the frequency of seismic events. Similar geothermal anomalies were not recorded in the preceding or following years29. Prevalence of skin lesions in asteroids within Port Foster Bay ranged from barely over 0% in the innermost sites to 10% at locations adjacent to the bay’s opening, broadly following the gradient of species diversity in the region33,34. This trend was consistent over two surveys (2013,2016), and was correlated with O. validus distribution, which is largely driven by colonization, sedimentation, and sea current regimes34. In this context, what we saw in O. validus was similar to other disorders affecting echinoderms where disease was more prevalent after periods when temperature peaks, and in locations with higher agglomeration of counterparts, suggesting perhaps a density dependent process (e.g.,15,23,26,27). Cannibalism seems to be a rare behaviour in O. validus, hence density-dependent intraspecific aggression is an unlikely explanation for the lesions53. Physical or chemical traumas would neither explain the lesions; on the one hand because the closed shape of the bay reduces the entrance of strong currents or icebergs to cause abrasions; and on the other because human presence is too limited to generate pollution and chemical stress. In all these cases such insults would presumably be found at constant rates every year, yet, we only saw lesions in certain years.
The clinical signs described here differed from the AIWS of the NE Pacific, where multiple (>20) species were affected manifesting much more severe and rapidly progressing apparently transmissible clinical signs, i.e., wide spread arm loss and complete body disintegration19,23,54. Susceptibility of individuals to AIWS differed according to age (lower and slower in juveniles compared to adults), subhabitat, and sea star colour22,23. Moreover, subsequent to the AIWS epizootic, there was a sharp reduction in predation rate23. In contrast, in our study, only a single species was affected, and clinical signs were relatively mild (multifocal ulceration), did not appear to progress rapidly, and did not seem transmissible. This might be explained in part by the slower metabolism of Antarctic organisms in contrast to organisms in more temperate climates55,56, so it seems plausible that disease transmission and progression could be slow and, thence, imperceptible during the duration of a regular scientific polar campaign (~1.5 months). Another explanation could be that the etiology of lesions in O. validus is completely different than that affecting animals in AIWS. Colouration in O. validus can vary with age with juveniles being pink, purple, to orange and adults being bright red36. Most O. validus affected exhibiting lesions were red adults; however, juveniles of other colour-morphs were uncommon, so age comparisons were not possible. Additionally, behavioural consequences of the disease such as inactivity and appetite loss were seldom observed in predation experiments conducted in 2012–2015 (e.g.,57), This may be partially explained by stress, and the loss of respiratory surface and papulae.
Whilst microscopy revealed a moderate to marked inflammatory response with epidermal ulceration, there was no evidence of infectious agents visible on light microscopy, thus, it is unlikely that organisms visible at that level such as fungi, bacteria16 or parasites58 were responsible for the epidermal lesions seen here. Moreover, clinical signs (lack of progression, transmissibility) also argued against an infectious cause. However, light microscopy has low to no resolution when visualizing microorganisms of the size of mycoplasma and viruses, so these remain as possible infectious agents. Alternatively, non-infectious causes should also be considered, as skin ulceration is a common response to a variety of insults in echinoderms59,60, which may or may not have an infectious etiology.
Microbial community analyses failed to reveal bacterial or fungal putative pathogens associated with lesions. The consistently lower bacterial and fungal diversity found in the lesion fronts of affected asteroids contrasts with other studies, where an increase in microbial diversity followed a disease or stress response in, corals61,62, sponges63, and also echinoderms58, but agrees with other findings in marine invertebrates (e.g.,63). Some authors suggest that diseased tissues represent favourable niches for colonization of secondary invaders inflating microbial diversities due to host’s immune disruption caused by stress, together with a higher availability of nutrients from decaying tissues64. In fact, one of the major constraints when exploring the etiology of marine diseases during non-initial symptomatic stages (e.g., bleaching, spotting, or rotting) is to distinguish real pathogens from saprophytes growing on decaying hosts24,65. In this study, the relatively low microbial diversities recorded in the diseased fronts of O. validus may be explained by either a lower colonization rate of opportunistic microbes under Antarctic temperatures55 or to antimicrobial inhibitors (as suggested in63) elicited by immune response of the host, as described in other asteroid syndromes54. It is possible that the differences in microbiota could be due to microbiome shifts prior to the manifestation of lesions, leading to upsurge of opportunistic polymicrobial infections, and development of lesions without the implication of ‘legitimate’ pathogens (i.e. microbial dysbiosis sensu65). The empirical data to support this could be the higher consistency in microbial communities associated with healthy states across individuals, with respect to the lesion fronts and apparently healthy skin areas from specimens of diseased sea stars. All this illustrates that absent clear evidence of pathogens associated with lesions at the tissue and cellular level, conclusions gained from molecular studies are necessarily limited.
Actinobacteria were “core” members of the associated flora of O. validus, in agreement with other marine microbiomes66,67. The bulk of sequences within this phylum belonged to Rhodococcus, which harbours benign steroid producers and a few pathogens68. Proteobacteria, followed by Firmicutes and Bacteroidetes, displayed double the sequence abundances in Healthy tissues as compared to lesion fronts. Bacteroidetes has been associated with healthy states but also with disease in contrasting studies64,69; whereas, Proteobacteria comprises pathogens, opportunists, and nitrogen fixators70,71. Three Rhodobacteraceae OTUs and one Acinetobacter were particularly abundant in NON_Affected sections from diseased sea stars, where they could have proliferated as pre-invasive pathogens72,73. Accordingly, the family Rhodobacteraceae was correlated to coral disease outbreaks74. Instead, Clostridium (Firmicutes) strains, appearing exclusively in healthy sea stars, suggest symbiotic interactions75.
As in other marine invertebrates, Ascomycota and Basidiomycota were the most common fungi in O. validus, along with a small proportion of Chytridiomycota (mostly saprobic)76,77. Many sequences were not classified due to limitations in fungal databases76. Saccharomycetes yeasts contributed to the flora of all health states, where they could be forming symbiotic matrices77. Certain taxa with wide metabolic ranges in Dothideomycetes, Tremellomycetes, Microbotryomycetes, and Sordariomycetes were variably correlated to healthy or NON_Affected tissues, likely driving microbial shifts prior to lesion manifestation70,77–79. Eurotiomycetes, Agaricomycetes, Wallemiomycetes, and Cystobasidiomycetes were more common in diseased sea stars, and probably harboured opportunistic strains proliferating in distressed host tissues77,80. Besides the opportunistic Trichosporon guehoae81 (F_OTU556, Eurotiales), there were three exclusive strains (F_OTU520, F_OTU165, F_OTU450) that were closely associated with lesion fronts.
Temperature plays a critical role in pathogenesis of disease in marine invertebrates and poikilothermic vertebrates19,22,23,54,82–84. Antarctica is one of the most susceptible areas to climate change on Earth85, and Deception Island is one of the most temperature variable sites in the Southern Ocean28. Furthermore, marine ice has been proposed as an important abiotic reservoir for pathogens82. After 5-years of observation of this enigmatic disorder, the incidence of disease symptoms in O. validus populations seem to oscillate locally between over 0% and 10%. That said, and considering the coincidence of the first outbreak in 2012–2013 with an unprecedented local thermal anomaly29, we need to be vigilant as the disease could potentially boost with increasing temperatures. The isolated and unexpected nature of this disease illustrates gaps in our knowledge of demographics and health of marine invertebrates that play an inordinately important role in Antarctic benthic ecology. Most Antarctic species are characterized by a relatively slow growth and maturation times56. If this disease decreases sea stars’ populations in the coming years, recovery could be slow23,86–88, with direct consequences to the ecosystem35,36. Future studies might focus on clarifying the fitness and demographic impacts of epidermal lesions on O. validus and developing tools to understand pathology and pathogenesis, correlated with year-round temperature data. In the face of the alarming mortalities affecting sea star populations in the past years (e.g.,19,22,23), we believe this and upcoming research involving disorders in keystone Antarctic species are extremely relevant and urgent.
Electronic supplementary material
Acknowledgements
We are grateful to the Unidad de Tecnología Marina (UTM-CSIC), the BIO-Hespérides crew, and the “Gabriel de Castilla” Spanish Antarctic Base for providing logistic support during the Antarctic cruises. We greatly appreciate R. Rameyer (US NWHC) and A. Riesgo (NHM, London) support in the laboratory, and A. Eggers from the sequencing CORE Lab (HIMB – University of Hawai’i). Two anonymous reviewers are also acknowledged for their constructive suggestions. Amend Lab – UH and Gates Lab – HIMB provided infrastructure and supplies to perform the molecular analyses. Local, national and international regulations and conventions, and scientific ethical practices, have been respected. Project funding was obtained from the Spanish government through the ACTIQUIM and DISTANTCOM Projects (CGL2004-03356/ANT, CGL2007-65453/ANT, and CTM2010-17415/ANT; CTM2013-42667/ANT). L.N.-P. was supported by a postdoctoral fellowship from the Fundación Ramón Areces and Beatriu de Pinós Marie Curie CO-Fund Program (Catalonia). Mention of products or trade names does not imply endorsement by the US Government.
Author Contributions
Individual contributions are as follows: Conceived and designed the study: L.N.-P. Field surveys and collection of biological material: L.N.-P. C.A., J.M., C.A.; Microscopy analyses: T.W. Genetic barcoding work: L.N.-P., Data analyses: L.N.-P. Hosting/reagents/materials: C.A. Wrote the paper: L.N.P. T.W., J.M. Revised and contributed to the final version: all co-authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/21/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29684-0.
References
- 1.Harvell CD, et al. Emerging marine diseases: climate links and anthropogenic factors. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 2.Harvell CD, et al. Climate warming and disease risks for terrestrial and marine biota. Science. 2002;296:2158–2162. doi: 10.1126/science.1063699. [DOI] [PubMed] [Google Scholar]
- 3.Ritchie KB, Polson SW, Smith GW. Microbial disease causation in marine invertebrates: problems, practices and futures prospects. Hydrobiologia. 2001;460:131–139. doi: 10.1023/A:1013181718805. [DOI] [Google Scholar]
- 4.Hoegh-Guldberg O, Bruno JF. The impact of climate change on the world’s marine ecosystems. Science. 2010;328:1523–1528. doi: 10.1126/science.1189930. [DOI] [PubMed] [Google Scholar]
- 5.Burge CA, et al. Complementary approaches to diagnosing marine diseases: a union of the modern and the classic. Philos. Trans. Royal Soc. B. 2016;371:20150207. doi: 10.1098/rstb.2015.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lessios HA. The great Diadema antillarum die-off: 30 years later. Annu. Rev. Mar. Sci. 2016;8:267–283. doi: 10.1146/annurev-marine-122414-033857. [DOI] [PubMed] [Google Scholar]
- 7.Menge BA. Coexistence between the seastars Asterias vulgaris and A. forbesi in a heterogeneous environment: a non-equilibrium explanation. Oecologia. 1979;41:245–272. doi: 10.1007/BF00377430. [DOI] [PubMed] [Google Scholar]
- 8.Dungan ML, Miller TE, Thomson DA. Catastrophic decline of a top carnivore in the Gulf of California rocky intertidal zone. Science. 1982;216:989–991. doi: 10.1126/science.216.4549.989. [DOI] [PubMed] [Google Scholar]
- 9.Bruckner, A. W. Priorities for effective management of coral diseases. NOAA Technical Memorandum, NMFS-OPR-22 (Washington, DC, 2002).
- 10.Brownell AC, Richardson LL. Sulfate reducing bacteria as secondary and necessary pathogens in black band disease of corals. Rev. Bio.l Trop. 2014;62:249–257. [Google Scholar]
- 11.Hyman, L. H. The Invertebrates: Echinodermata, The coelomata Bilateria. In (McGraw-Hill New York, 1955).
- 12.Begon, M., Townsend, C. & Harper, J. Ecology: from individuals to ecosystems. In (Blackwell Publishing Oxford, 2006).
- 13.Lawrence, J. M. Starfish: biology and ecology of the Asteroidea. In (The Johns Hopkins University Press Baltimore, 2013).
- 14.Scheibling RE, Stephenson RL. Mass mortality of Strongylocentrotus droebachiensis (Echinodermata: Echinoidea) off Nova Scotia, Canada. Mar. Biol. 1984;78:153–164. doi: 10.1007/BF00394695. [DOI] [Google Scholar]
- 15.Lessios HA, et al. Mass mortality of Diadema antillarum on the Caribbean coast of Panama. Coral Reefs. 1984;3:173–182. doi: 10.1007/BF00288252. [DOI] [Google Scholar]
- 16.Jangoux M. Diseases of Echinodermata. I. Agents microorganisms and protistans. Dis. Aquat. Org. 1987;2:147–162. doi: 10.3354/dao002147. [DOI] [Google Scholar]
- 17.Jangoux M. Diseases of Echinodermata. 11. Agents metazoans (Mesozoa to Bryozoa) Dis. Aquat. Org. 1987;2:205–234. doi: 10.3354/dao002205. [DOI] [Google Scholar]
- 18.Bauer JC, Young CM. Epidermal lesions and mortality caused by vibriosis in deep-sea Bahamian echinoids: a laboratory study. Dis. Aquat. Org. 2000;39:193–199. doi: 10.3354/dao039193. [DOI] [PubMed] [Google Scholar]
- 19.Hewson I, et al. Densovirus associated with sea star wasting disease and mass mortality. Proc. Natl. Acad. Sci. USA. 2014;111:17278–17283. doi: 10.1073/pnas.1416625111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sweet M, Bulling M, Williamson JE. New disease outbreak affects two dominant sea urchin species associated with Australian temperate reefs. Mar. Ecol. Prog. Ser. 2016;551:171–183. doi: 10.3354/meps11750. [DOI] [Google Scholar]
- 21.Feehan CJ, Scheibling RE. Effects of sea urchin disease on coastal marine ecosystems. Mar. Biol. 2014;161:1467–1485. doi: 10.1007/s00227-014-2452-4. [DOI] [Google Scholar]
- 22.Eisenlord ME, et al. Ochre star mortality during the 2014 wasting disease epizootic: role of population size structure and temperature. Philos. Trans. Royal. Soc. B. 2016;371:20150212. doi: 10.1098/rstb.2015.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menge BA, et al. Sea star wasting disease in the keystone predator Pisaster ochraceus in Oregon: insights into differential population impacts recovery, predation rate, and temperature effects from long-term research. PloS ONE. 2016;11:e0153994. doi: 10.1371/journal.pone.0153994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivers TM. Viruses and Koch’s postulates. J. Bacteriol. 1937;33:1–12. doi: 10.1128/jb.33.1.1-12.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.FioRito R, Leander C, Leander B. Characterization of three novel species of Labyrinthulomycota isolated from ochre sea stars (Pisaster ochraceus) Mar. Biol. 2016;163:170. doi: 10.1007/s00227-016-2944-5. [DOI] [Google Scholar]
- 26.Hewson I, et al. Investigating the Complex Association Between Viral Ecology, Environment, and Northeast Pacific Sea Star Wasting. Front. Mar. Sci. 2018;5:77. doi: 10.3389/fmars.2018.00077. [DOI] [Google Scholar]
- 27.Miner CM, et al. Large-scale impacts of sea star wasting disease (SSWD) on intertidal sea stars and implications for recovery. PloS ONE. 2018;13(3):e0192870. doi: 10.1371/journal.pone.0192870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meredith, M. & King, J. Rapidclimate change in the ocean west of the Antarctic Peninsula during the second half of the 20th century. Geophys. Res. Lett. 32 (2005).
- 29. Berrocoso, M. et al. Caldera unrest detected with seawater temperature anomalies at Deception Island,Antarctic Peninsula. Bull. Volcan. 80(4), 10.1007/s00445-018-1216-2 (2018).
- 30.Smith K, Baldwin R. Ecosystem studies at Deception Island, Antarctica: an overview. Deep Sea Res. Part 2 Top Stud. Oceanogr. 2003;50:1595–1609. doi: 10.1016/S0967-0645(03)00081-X. [DOI] [Google Scholar]
- 31.Lovell LL, Trego KD. The epibenthic megafaunal and benthic infaunal invertebrates of Port Foster, Deception Island (South Shetland Islands, Antarctica). Deep Sea Res. Part 2 Top Stud. Oceanogr. 2003;50:1799–1819. doi: 10.1016/S0967-0645(03)00087-0. [DOI] [Google Scholar]
- 32.Angulo-Preckler C, Leiva C, Avila C, Taboada S. Macroinvertebrate communities from the shallow soft-bottoms of Deception Island (Southern Ocean): A paradise for opportunists. Mar. Environ. Res. 2017;127:62–74. doi: 10.1016/j.marenvres.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Barnes DKA, et al. Marine richness and gradients at Deception Island, Antarctica. Antarct. Sci. 2008;20:271–279. [Google Scholar]
- 34.Angulo-Preckler C, et al. Macrobenthic patterns at the shallow marine waters in the caldera of the active volcano of Deception Island, Antarctica. Cont. Shelf Res. 2018;157:20–31. doi: 10.1016/j.csr.2018.02.005. [DOI] [Google Scholar]
- 35.Dayton PK, Robilliard GA, Paine RT, Dayton LB. Biological accommodation in benthic community at McMurdo Sound Antarctica. Ecol. Monogr. 1974;44:105–128. doi: 10.2307/1942321. [DOI] [Google Scholar]
- 36.McClintock JB. Trophic biology of Antarctic echinoderms. Mar. Ecol. Prog. Ser. 1994;111:191–202. doi: 10.3354/meps111191. [DOI] [Google Scholar]
- 37.Lightner DV, Redman RM. Shrimp diseases and current diagnostic methods. Aquaculture. 1998;164:201–220. doi: 10.1016/S0044-8486(98)00187-2. [DOI] [Google Scholar]
- 38.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:1–11. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes—Application to identification of mycorhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 42.White, T. J., Bruns, T., Lee, S. & Taylor, J. W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In (eds Innis, M. A., Gelfand, D. H., Sninsky, J. J. & White, T. J.) PCR protocols: a guide to methods and applications. 315–322 (Academic Press Inc. New York, 1990).
- 43.Caporaso JG, et al. QIIME allows analysis of high throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 45.Edgar, R. C. UCHIME2: Improved chimera detection for amplicon sequences. Available at: 10.1101/074252 (2016).
- 46.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abarenkov K, et al. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol. 2010;186:281–285. doi: 10.1111/j.1469-8137.2009.03160.x. [DOI] [PubMed] [Google Scholar]
- 49.Chao A. Non-parametric estimation of the classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- 50.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Aust.Ecol. 2001;26:32–46. [Google Scholar]
- 51.Bray JR, Curtis JT. An ordination of upland forest communities of southern Wisconsin. Ecol. Monogr. 1957;27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 52.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 53.Kidawa A. Antarctic starfish, Odontaster validus, distinguish between fed and starved conspecifics. Polar Biol. 2001;24:408–410. doi: 10.1007/s003000100229. [DOI] [Google Scholar]
- 54.Fuess LE, et al. Up in arms: immune and nervous system response to sea star wasting disease. PLoS ONE. 2015;10:e0133053. doi: 10.1371/journal.pone.0133053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delille DE, Perret E. Influence of temperature on the growth potential of southern polar marine bacteria. Microb. Ecol. 1989;18:117–123. doi: 10.1007/BF02030120. [DOI] [PubMed] [Google Scholar]
- 56.Peck LS. Ecophysiology of Antarctic marine ectotherms: limits to life. Pol Biol. 2002;25:31–40. doi: 10.1007/s003000100308. [DOI] [Google Scholar]
- 57.Moles J, et al. Anti-predatory chemical defences in Antarctic benthic fauna. Mar. Biol. 2015;162:1813–1821. doi: 10.1007/s00227-015-2714-9. [DOI] [Google Scholar]
- 58.Bouland C, Jangoux M. Infestation of Asterias rubens (Echinodermata) by the ciliate Orchitophrya stellarum: effect on gonads and host reaction. Dis. Aquat. Organ. 1988;5:239–242. doi: 10.3354/dao005239. [DOI] [Google Scholar]
- 59.Becker P, et al. The skin ulceration disease in cultivated juveniles of Holothuria scabra (Holothuroidea, Echinodermata) Aquaculture. 2004;242:13–30. doi: 10.1016/j.aquaculture.2003.11.018. [DOI] [Google Scholar]
- 60.Christensen AM. Feeding biology of the sea-star Astropecten irregularis Pennant. Ophelia. 1970;8:1–134. doi: 10.1080/00785326.1970.10429554. [DOI] [Google Scholar]
- 61.Pantos O, et al. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ. Microbiol. 2003;5:370–382. doi: 10.1046/j.1462-2920.2003.00427.x. [DOI] [PubMed] [Google Scholar]
- 62.Pantos O, Bythell JC. Bacterial community structure associated with white band disease in the elkhorn coral Acropora palmata determined using culture independent 16S rRNA techniques. Dis. Aquat. Organ. 2006;69:79–88. doi: 10.3354/dao069079. [DOI] [PubMed] [Google Scholar]
- 63.Luter HM, Whalan S, Webster NS. Exploring the role of microorganisms in the disease-like syndrome affecting the sponge Ianthella basta. Appl. Environ. Microbiol. 2010;76:5736–5744. doi: 10.1128/AEM.00653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Webster NS, Xavier JR, Freckelton M, Motti CA, Cobb R. Shifts in microbial and chemical patterns within the marine sponge Aplysina aerophoba during a disease outbreak. Environ. Microbiol. 2008;10:3366–3376. doi: 10.1111/j.1462-2920.2008.01734.x. [DOI] [PubMed] [Google Scholar]
- 65.Egan S, Gardiner M. Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 2016;7:991. doi: 10.3389/fmicb.2016.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bull AT, Stach JEM. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 2007;15:491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 67.Ainsworth TD, et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015;9:2261–2274. doi: 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLeod MP, et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frías-López J, Zerkle AL, Bonheyo GT, Fouke BW. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 2002;68:2214–2228. doi: 10.1128/AEM.68.5.2214-2228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Madigan, M. & Martinko, J. Brock biology of microorganisms. 11th ed. (Prentice Hall, 2005).
- 71.Reinhold-Hurek B, Hurek T. Reassessment of the taxonomic structure of the diazotrophic genus Azoarcus sensu lato and description of three new genera and new species, Azovibrio restrictus gen. nov., sp. nov., Azospira oryzae gen. nov., sp. nov. and Azonexus fungiphilus gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2000;50:649–659. doi: 10.1099/00207713-50-2-649. [DOI] [PubMed] [Google Scholar]
- 72.Witt V, Wild C, Anthony KRN, Diaz-Pulido G, Uthicke S. Effects of ocean acidification on microbial community composition of, and oxygen fluxes through, biofilms from the Great Barrier Reef. Environ. Microbiol. 2011;13:2976–2989. doi: 10.1111/j.1462-2920.2011.02571.x. [DOI] [PubMed] [Google Scholar]
- 73.Elifantz H, Horn G, Ayon M, Cohen Y, Minz D. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 2013;85:348–357. doi: 10.1111/1574-6941.12122. [DOI] [PubMed] [Google Scholar]
- 74.Pollock FJ, Wada N, Torda G, Willis BL, Bourne DG. Repeated sampling of white syndrome-affected corals reveals distinct microbiome at disease lesion fronts. Appl. Environ. Microbiol. 83. 2016;02716:e02799. doi: 10.1128/AEM.02799-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maczulak, A. E. Clostridium. In (eds Maczulak, A. E. & Ruskin, R. H.) Encyclopedia of Microbiology. 168–173 (Facts on File Publisher New York, 2011).
- 76.Amend AS, Barshis DJ, Oliver TA. Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME J. 2012;6:1291–1301. doi: 10.1038/ismej.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blackwell M. The Fungi: 1, 2, 3 … million species? Am. J. Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 78.Richards TA, Jones MDM, Leonard G, Bass D. Marine fungi: their ecology and molecular diversity. Annu. Rev. Mar. Sci. 2012;4:495–522. doi: 10.1146/annurev-marine-120710-100802. [DOI] [PubMed] [Google Scholar]
- 79.Se-Kwon, K. Marine Microbiology: bioactive compounds and biotechnological applications. p. 535 (Wiley-VCH Verlag GmbH & Co Singapore, 2013).
- 80.Zalar P, de Hoog GS, Schroers HJ, Frank JM, Gunde-Cimerman N. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.) Anton. Leeuw. Int. J. G. 2005;87:311–28. doi: 10.1007/s10482-004-6783-x. [DOI] [PubMed] [Google Scholar]
- 81.Marty FM, Barouch DH, Coakley EP, Baden LR. Disseminated trichosporonosis caused by Trichosporon loubieri. J. Clin. Microbiol. 2003;41:5317–5320. doi: 10.1128/JCM.41.11.5317-5320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith AW, Skilling DE, Castello JD, Rogers SO. Ice as a reservoir for pathogenic human viruses: specifically, caliciviruses, influenza viruses, and enteroviruses. Med. Hypotheses. 2004;63:560–566. doi: 10.1016/j.mehy.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 83.Gudenkauf BM, Eaglesham JB, Aragundi WM, Hewson I. Discovery of urchin-associated densoviruses (family Parvoviridae) in coastal waters of the Big Island, Hawaii. J. Gen. Virol. 2014;95:652–658. doi: 10.1099/vir.0.060780-0. [DOI] [PubMed] [Google Scholar]
- 84.Burge CA, et al. Climate change influences on marine infectious diseases: implications for management and society. Annu. Rev. Mar. Sci. 2014;6:249–277. doi: 10.1146/annurev-marine-010213-135029. [DOI] [PubMed] [Google Scholar]
- 85.Peck LS. Prospects for surviving climate change in Antarctic aquatic species. Antarct. Sci. 2005;17:497–507. doi: 10.1017/S0954102005002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paine RT. Food web complexity and species diversity. Amer Nat. 1966;100:65–75. doi: 10.1086/282400. [DOI] [Google Scholar]
- 87.Paine RT. A note on trophic complexity and community stability. Amer. Nat. 1969;103:91–93. doi: 10.1086/282586. [DOI] [Google Scholar]
- 88.Paine RT. Intertidal community structure: experimental studies on the relationship between a dominant competitor and its principal predator. Oecologia. 1974;15:93–120. doi: 10.1007/BF00345739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data set was deposited in the NCBI Sequence Read Archive (SRA) database (accession numbers: SRP131970). Other related datasets generated during and/or analysed during the current study are available from the corresponding author on request.