Abstract
Nontuberculous mycobacterial lung disease (NTM), including Mycobacterium avium complex (MAC), is a growing health problem in North America and worldwide. Little is known about the molecular alterations occurring in the tissue microenvironment during NTM pathogenesis. Utilizing next generation sequencing, we sequenced sputum and matched lymphocyte DNA in 15 MAC patients for a panel of 19 genes known to harbor cancer susceptibility associated mutations. Thirteen of 15 NTM subjects had a diagnosis of breast cancer (BCa) before or after NTM infection. Thirty three percent (4/12) of these NTM-BCa cases exhibited at least 3 somatic mutations in sputa compared to matched lymphocytes. Twenty four somatic mutations were detected with at least one mutation in ATM, ERBB2, BARD1, BRCA1, BRCA2, AR, TP53, PALB2, CASP8, BRIP1, NBN and TGFB1 genes. All four NTM-BCa patients harboring somatic mutations also exhibited 15 germ line BRCA1 and BRCA2 mutations. The two NTM subjects without BCa exhibited twenty somatic mutations spanning BRCA1, BRCA1, BARD1, BRIP1, CHEK2, ERBB2, TP53, ATM, PALB2, TGFB1 and 3 germ line mutations in BRCA1 and BRCA2 genes. A single copy loss of STK11 and AR gene was noted in NTM-BCa subjects. Periodic screening of sputa may aid to develop risk assessment biomarkers for neoplastic diseases in NTM patients.
Introduction
The incidence and prevalence of nontuberculous mycobacterial lung disease (NTM) is increasing in USA and worldwide1–9. In the USA, a prevalence of 20 NTM cases out of 100,000 in 1997 and 47 cases out of 100,000 in 2007 was observed with an 8.2% annual increase8. NTM lung disease in the U.S.A. is primarily caused by Mycobacterium avium complex (MAC) and poses considerable challenges in diagnosis and treatment1–4. NTM may occur in patients with or without a predisposing condition1–9. Postmenopausal women and individuals with various conditions such as bronchiectasis and cystic fibrosis are at risk of developing NTM disease1–10. Notably, the frequency of NTM patients with nodular bronchiectasis has increased over the past few years8. However, the underlying cellular and physiologic abnormalities causing NTM lung disease are not well understood9. In our recent study8, we observed that some women with stable NTM disease developed breast cancer (BCa) later in their lifetime, suggesting that NTM infection could be a potential risk factor for chronic inflammation and cellular transformation similar to Helicobacter pylori associated gastrointestinal transformation11. We rationalize that assessment of molecular abnormalities at infection sites and the surrounding environment would be important for continuous monitoring of the NTM infected patients. This will aid to develop suitable biomarkers for disease surveillance, treatment guidance and risk assessment.
Next generation sequencing is a powerful tool for detecting molecular abnormalities in tissues and body fluids and guiding biomarker and therapeutic development in various diseases including cancer. In the present study, on a next generation sequencing platform, we sequenced sputum and matched lymphocyte DNA in 15 women with NTM lung disease (MAC) for a panel 19 cancer predisposing genes12–34. Thirteen out of 15 NTM subjects had a diagnosis of BCa before or after NTM lung infection. Thirty three percent (4/12) of these NTM-BCa cases has been detected with 24 somatic mutations with at least one mutation in ATM, ERBB2, BARD1, BRCA1, BRCA2, AR, TP53, PALB2, CASP8, BRIP1, NBN and TGFB1 genes. Numerous (N = 15) germ line BRCA1 and BRCA2 mutations were also detected in these four subjects. The two NTM subjects with no history of BCa exhibited 20 somatic mutations spanning BRCA1, BRCA1, BARD1, BRIP1, CHEK2, ERBB2, TP53, ATM, PALB2, and TGFB1 and 3 germ line mutations in BRCA1 and BRCA2 genes. In addition to genomic mutations, copy number loss in STK11, AR gene was evident in one NTM subject with BCa.
Results
Clinicopathological characteristics of the NTM affected women
Based on the available clinical history, thirteen out of the fifteen NTM patients we sequenced had also been diagnosed with BCa (Table 1). Notably, four out of these thirteen subjects were detected with NTM disease first and then BCa within a period of 4–6 years (Table 1). All the NTM subjects in our study cohort were positive for Bronchiectasis. Hormonal contraceptives were used by 67% (10/15) of the women with both NTM and BCa. In addition, 64% (9/14, no information available for 1 case) of these patients had undergone postmenopausal hormone therapy as well.
Table 1.
Demographic information of the NTM-BCa and NTM subjects with the mutation spectrum and CNV.
| NTM-BCa/NTM casesa | Age at NTM diagnosis | Age at BCa Diagnosis | BCa Stage | bSmoking/Drinking | Weight (lb) | Height (inches) | cBMI | dSomatic Mutation | eGermline Mutation | fCNV |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 75 | 38 | UK | UK | 120 | 63 | 21.3 | N | N | Y |
| 2 | 60 | 65 | 0 | N/N | 132 | 65 | 22.0 | N | N | N |
| 3 | 66 | 45 | I | N/N | 143 | 65 | 23.8 | Y | Y | N |
| 4 | 57 | 61 | I | N/N | 180 | 69 | 25.0 | N | N | Y |
| 5 | 71 | 75 | 0 | N/N | 120 | 64 | 20.6 | N | N | N |
| 6 | 73 | 68 | II | N/Y | 150 | 63 | 26.6 | N | N | N |
| 7 | 54 | 52 | II | N/N | 132 | 66 | 21.3 | N | N | N |
| 8 | 63 | 58 | I | FS/N | 120 | 67 | 18.8 | N | N | Y |
| 9 | 61 | 45 | 0 | FS/Y | 145 | 66 | 23.4 | N | N | Y |
| 10 | 55 | 51 | III | N/N | 98 | 61 | 18.5 | N | N | Y |
| 11 | 68 | 68 | 0 | FS/N | 156 | 54 | 37.6 | Y | Y | Y |
| 12 | 55 | 61 | IB | N/Y | 157 | 67 | 24.6 | Y | Y | N |
| 13 | 66 | 56 | II | N/N | 91 | 65 | 15.1 | Y | Y | N |
| NTM01 | 49 | NA | NA | N/Y | 109 | 65 | 18.1 | Y | Y | N |
| NTM03 | 58 | NA | NA | Y/Y | 114 | 66 | 18.4 | Y | Y | N |
aRows 1–13: Women with both NTM and breast cancer; Rows 14–15 (NTM01/NTM03): Women with NTM disease only.
bTobacco and alcohol usage history. UK: Unknown; FS: Former smoker; Y: Yes; N: No. cBMI: Body mass index determined following NHLBI criteria: Underweight = <18.5; Normal weight = 18.5–24.9; Overweight = 25–29.9; Obesity = BMI of 30 or greater. dMutation in sputum DNA compared to matched normal lymphocyte DNA; eMutation in both sputum and matched normal lymphocytes. fCopy number variation in sputa compared to the matched normal lymphocytes.
Pattern of the somatic genomic variants in the sputum of NTM subjects with breast cancer
In this study, we have undertaken next generation sequencing (NGS) analysis of a 19-gene signature panel associated with cancer susceptibility and predisposition12–34 (Table 2) in women with NTM lung disease with (13) or without (two) a diagnosis of BCa. Matched lymphocytes and sputum DNA samples from thirteen NTM subjects with a history of BCa (NTM-BCa) and two subjects with NTM disease without BCa (NTM) were sequenced utilizing this high-throughput sequencing platform. Stringent data analysis and validation criteria were employed to determine both somatic and germ line mutations35. One subject did not pass quality control (NTM-BCa02) and was excluded from further analysis. Overall, we have detected numerous non-synonymous (Fig. 1A) and synonymous somatic mutations in these subjects (#PRJNA431897). Many unique somatic mutations were also identified in these samples (Fig. 1B). Thirty three percent (4/12) of the NTM subjects with a previous history of breast cancer (NTM-BCa) exhibited at least 3 somatic mutations in the sputum when compared to the matched lymphocytes (Table 2, Fig. 1). A total of 24 somatic mutations were detected in the sputum samples of these subjects with at least one mutation in ATM, ERBB2, BARD1, BRCA1, BRCA2, TP53, PALB2, CASP8, BRIP1, NBN and TGFB1 gene. All the mutations were missense in nature (Table 3, Figs 1 and 2). We detected a novel ERBB2 sequence variant (C-A, Ala > Glu; Table 3) in one NTM-BCa subject (NTM-BCa11), not reported previously. The genes most frequently harbored somatic mutations include ERBB2 (N = 5), BARD1 (N = 3), and BRCA2 (N = 3) (Fig. 2). Notably, somatic mutations in BRCA1 and BRCA2 were detected in 75% (3/4) NTM-BCa subjects exhibiting mutations in the sputum (Table 3, Fig. 3).
Table 2.
The panel of 19 cancer predisposing genes sequenced in the sputa of NTM affected women.
| Gene | GenBank Reference | OMIM referencea | Description | CDDS (bp)b | Number of exonsc |
|---|---|---|---|---|---|
| AR | NM_000044.3 | 313700 | Androgen receptor | 10661 | 8 |
| ATM | NM_000051.3 | 607585 | Serine-protein kinase ataxia telangiectasia mutated | 13147 | 68 |
| BARD1 | NM_000465.3 | 601593 | BRCA associated RING domain 1 | 5523 | 11 |
| BRCA1 | NM_007298.3 | 113705 | Breast cancer 1, early onset | 3699 | 22 |
| BRCA2 | NM_000059.3 | 600185 | Breast cancer 2, early onset | 11386 | 27 |
| BRIP1 | NM_032043.2 | 605882 | BRCA1-interacting protein | 8166 | 20 |
| CASP8 | NM_001080124.1 | 601763 | Apoptosis-related cysteine protease 8 | 2750 | 9 |
| CDH1 | NM_004360.3 | 192090 | Cadherin 1 | 4815 | 16 |
| CHEK2 | NM_001005735.1 | 604373 | Serine/threonine checkpoint kinase 2 | 1991 | 16 |
| DIRAS3 | NM_004675.2 | 605193 | GTP-binding Ras-like protein 3 | 1642 | 2 |
| ERBB2 | NM_001005862.1 | 164870 | Avian erythroblastic leukemia viral concogene homolog 2 | 4816 | 30 |
| NBN | NM_002485.4 | 602667 | Nibrin | 4639 | 16 |
| PALB2 | NM_024675.3 | 601355 | Partner and localizer of BRCA2 | 4069 | 13 |
| PTEN | NM_000314.4 | 601728 | Phosphatase and tensin | 5572 | 9 |
| RAD50 | NM_005732.3 | 604040 | DNA repair protein AD50 homolog | 6597 | 25 |
| RAD51 | NM_001164269.1 | 179617 | DNA repair protein RAD51A homolog | 2147 | 10 |
| STK11 | NM_000455.4 | 602216 | Serine/threonine protein kinase 11 | 3286 | 10 |
| TGFB1 | NM_000660.4 | 190180 | Transforming growth factor B1 | 2217 | 7 |
| P53 | NM_000546.5 | 191170 | Tumor protein p53 | 2591 | 11 |
aOnline Mendelian Inheritance in Man; bCDDS: coding region of the genes in base pairs; cNumber of coding exons in each gene.
Figure 1.
Nature of synonymous and nonsynonymous somatic mutations in women with pulmonary NTM disease. (A) Total somatic mutations tallied across all samples. Red bar represents missense mutations, blue bar represents synonymous mutations. (B) Unique somatic mutations in genes of interest from all samples (same data as (B), but mutations shared across samples are only counted once). Red bar represents missense mutations, blue bar represents synonymous mutations.
Table 3.
Somatic mutations of the predisposing gene panel in NTM affected women with a diagnosis of breast cancer.
| Samplea | Chromosomeb | Genec | Positiond | Nature of Mutation | Referencee | LYMf | Sputumg | Amino acid Changeh |
|---|---|---|---|---|---|---|---|---|
| NTM-BCa03 | Chr11 | ATM | 108175463 | Missense | A | A | T | Asp > Val |
| Chr2 | BARD1 | 215632255 | Missense | C | C | T | Val > Met | |
| Chr2 | BARD1 | 215674224 | Missense | G | G | A | Pro > Ser | |
| Chr13 | BRCA2 | 32906729 | Missense | A | A | C | Asn > His | |
| Chr17 | BRIP1 | 59763347 | Missense | A | A | G | Ser > Pro | |
| Chr2 | CASP8 | 202122995 | Missense | A | A | G | Lys > Arg | |
| Chr17 | ERBB2 | 37884037 | Missense | C | C | G | Pro > Ala | |
| Chr17 | TP53 | 7579472 | Missense | G | G | C | Pro > Arg | |
| NTM-BCa11 | Chr2 | BARD1 | 215674175 | Missense | G | G | A | Ala > Val |
| Chr17 | ERBB2 | 37856502 | Missense | C | C | A | Ala > Glu* | |
| Chr16 | PALB2 | 23646191 | Missense | T | T | C | Gln > Arg | |
| NTM-BCa12 | Chr17 | BRCA1 | 41244936 | Missense | G | G | A | Pro > Leu |
| Chr13 | BRCA2 | 32906729 | Missense | A | A | C | Asn > His | |
| Chr13 | BRCA2 | 32914592 | Missense | C | C | T | Arg > Cys | |
| Chr17 | BRIP1 | 59763347 | Missense | A | A | G | Ser > Pro | |
| Chr17 | ERBB2 | 37879588 | Missense | A | A | G | Ile > Val | |
| Chr17 | ERBB2 | 37884037 | Missense | C | C | G | Pro > Ala | |
| Chr16 | PALB2 | 23646191 | Missense | T | T | C | Gln > Arg | |
| NTM-BCa13 | Chr11 | ATM | 108175463 | Missense | A | A | T | Asp > Val |
| Chr2 | CASP8 | 202122995 | Missense | A | A | G | Lys > Arg | |
| Chr17 | ERBB2 | 37879588 | Missense | A | A | G | Ile > Val | |
| Chr8 | NBN | 90990479 | Missense | C | C | G | Glu > Gln | |
| Chr19 | TGFB1 | 41858921 | Missense | G | G | A | Pro > Leu | |
| Chr17 | TP53 | 7579472 | Missense | G | G | C | Pro > Arg |
aNTM-BCa: Women with both NTM and a diagnosis of breast cancer; bChromosomal location of each of the gene exhibiting mutation; cName of the gene affected; dChromosome positions with reference to GRCh37.p13. eReference sequence of the human genome; fSequence detected in the lymphocytes DNA (LYM); gSequence detected in the sputum DNA; hCorresponding change in the amino acid. An asterisk (*) indicates for a novel variant not reported previously.
Figure 2.
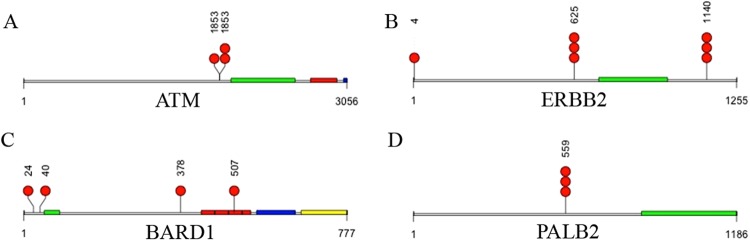
Nature of nonsynonymous (missense) somatic mutations in cancer associated genes ATM (A), ERBB2 (B), BRD1 (C) and PALB2 (D) in the NTM infected women. A red dot represents a mutation present in a single individual. The amino acid position affected by mutation was indicated above the red dot representing mutation. Multiple red dots indicate mutation present in more than one individual. Colored blocks indicate domains as described below for each gene. (A) ATM (Serine-protein kinase); green: FAT, red: Phosphatidylinositol 3- and 4-kinase, blue: FATC. Note: position 1853 possessed two different mutations. (B) BARD1 (BRCA associated RING domain 1); green: RING finger domain, red: ANK repeats, blue: BRCT1, yellow: BRCT2. (C) ERBB2 (Avian erythroblastic leukemia viral oncogene homolog 2); green: protein tyrosine kinase. (D) PALB2 (Partner and localizer of BRCA2); green: WD40 repeat domain. The total length of the amino acids for each gene was also indicated below the domains.
Figure 3.
Nonsynonymous (missense) mutations in cancer associated genes in the sputum of NTM subjects. Red dots represent somatic mutations, blue dots represent germ line mutations in specific genes present in a single individual. The amino acid positions affected by mutations were indicated above the red or blue dots representing sequence variants. Multiple dots indicate mutation present in more than one individual. Colored blocks indicate domains as described below for each gene. (A) BRCA1 (Breast cancer 1); green: RING finger domain, red: BRCT1 domain, blue: BRCT2 domain. (B) BRCA2 (Breast cancer 2); green: BRCA2 repeats. The total length of the amino acids for each gene was also indicated below the domains.
Pattern of somatic genomic variants in the sputum of women with NTM disease
Similar to the NTM patients with breast cancer history, both the NTM subjects (2/2, 100%) in the absence of breast cancer (Table 1) exhibited a number of somatic mutation in the sputum (Table 4, Figs 1 and 2). A total of 20 somatic mutations were detected in the sputum of these subjects with 11 mutations in one patient (NTM01) and 9 mutations in the other (NTM03) (Table 4). The majority of the somatic mutation were spanning BRCA1 (N = 9) and BRCA2 (N = 2) and ERBB2 (N = 2) genes along with a single mutation in BARD1, BRIP1, CHEK2, TP53, ATM, PALB2 and TGFB1 molecules (Table 4, Figs 2 and 3). Two ERBB2 gene mutations (chromosome position 37884037 and 37879588) that were detected in the NTM-BCa subjects (Table 3) were also present in the NTM subjects (Table 4). Similar was the case for mutations in TP53 (chromosome position 7579472) and PALB2 (chromosome position 23646191) for the NTM-BCa and NTM subjects (Tables 3 and 4).
Table 4.
Somatic mutations of the predisposing genes in women with NTM infection in the absence of breast cancer.
| Samplea | Chromosomeb | Genec | Positiond | Nature of mutation | Referencee | LYMf | Sputumg | Amino acid Changeh |
|---|---|---|---|---|---|---|---|---|
| NTM01 | chr2 | BARD1 | 215645464 | missense | C | C | G | Arg > Ser |
| chr17 | BRCA1 | 41223094 | missense | T | T | C | Ser > Gly | |
| chr17 | BRCA1 | 41244000 | missense | T | T | C | Lys > Arg | |
| chr17 | BRCA1 | 41244435 | missense | T | T | C | Glu > Gly | |
| chr17 | BRCA1 | 41244936 | missense | G | G | A | Pro > Leu | |
| chr13 | BRCA2 | 32906480 | missense | A | A | C | Asn > His | |
| chr13 | BRCA2 | 32911463 | missense | A | A | G | Asn > Asp | |
| chr17 | BRIP1 | 59763347 | missense | A | A | G | Ser > Pro | |
| chr22 | CHEK2 | 29121019 | missense | G | G | A | Arg > Cys | |
| chr17 | ERBB2 | 37884037 | missense | C | C | G | Pro > Ala | |
| chr17 | TP53 | 7579472 | missense | G | G | C | Pro > Arg | |
| NTM03 | chr11 | ATM | 108175462 | missense | G | G | A | Asp > Asn |
| chr17 | BRCA1 | 41223094 | missense | T | T | C | Ser > Gly | |
| chr17 | BRCA1 | 41243840 | missense | A | A | C | Asn > Lys | |
| chr17 | BRCA1 | 41244000 | missense | T | T | C | Lys > Arg | |
| chr17 | BRCA1 | 41244435 | missense | T | T | C | Glu > Gly | |
| chr17 | BRCA1 | 41244936 | missense | G | G | A | Pro > Leu | |
| chr17 | ERBB2 | 37879588 | missense | A | A | G | Ile > Val | |
| chr16 | PALB2 | 23646191 | missense | T | T | C | Gln > Arg | |
| chr19 | TGFB1 | 41858876 | missense | C | C | G | Arg > Pro |
aNTM: Women with NTM disease only; bChromosomal location of each of the gene exhibiting mutation; cName of the gene affected; dChromosome positions with reference to GRCh37.p13. eReference sequence of the human genome; fSequence detected in the lymphocytes DNA (LYM); gSequence detected in the sputum DNA; hCorresponding change in the amino acid.
The spectrum of BRCA1 and BRCA2 germ line variants in the sputum of women with NTM-BCa
Germ line mutation in BRCA1 and BRAC2 genes are known risk factors for BCa development in women harboring mutation in these genes25,28,32. Other than somatic mutation, all the above described NTM-BCa patients (4/12, 33%) have exhibited a number of germline mutations in BRCA1 and BRCA2 genes (Table 5, Fig. 3). A total of 15 germ line BRCA1 (N = 8) and BRCA2 (N = 7) gene mutations were detected in these subjects (Table 4, Fig. 3). All the mutations were missense in nature. Thus, both somatic as well as germ line mutations in BRCA1 and BRCA2 genes were evident in the NTM infected women with BCa (Tables 3 and 5; Fig. 3). Notably, subject NTM-BCa11, a past smoker (14 years) who harbored a novel ERBB2 variant (Table 3) and germ line BRCA2 mutation (Table 5), had a family history of BCa, and was diagnosed with BCa and NTM at the same age (Table 1).
Table 5.
Distribution of germline mutations in BRCA1 and BRCA2 in women both NTM-BCa and NTM disease.
| Samplea | Chromosomeb | Genec | Positiond | Nature of mutation | Referencee | LYMf | Sputumg | Amino acid Changeh |
|---|---|---|---|---|---|---|---|---|
| NTM-BCa03 | Chr17 | BRCA1 | 41223094 | missense | T | C | C | Ser > Gly |
| Chr17 | BRCA1 | 41244000 | missense | T | C | C | Lys > Arg | |
| Chr17 | BRCA1 | 41244435 | missense | T | C | C | Glu > Gly | |
| Chr17 | BRCA1 | 41244936 | missense | G | A | A | Pro > Leu | |
| Chr17 | BRCA1 | 41245471 | missense | C | T | T | Asp > Asn | |
| Chr19 | BRCA2 | 32906593 | missense | C | A | A | Ser > Arg | |
| NTM-BCa11 | Chr13 | BRCA2 | 32906480 | missense | A | C | C | Asn > His |
| Chr13 | BRCA2 | 32911463 | missense | A | G | G | Asn > Asp | |
| NTM-BCa12 | Chr17 | BRCA1 | 41223094 | missense | T | C | C | Ser > Gly |
| Chr17 | BRCA1 | 41244000 | missense | T | C | C | Lys > Arg | |
| Chr17 | BRCA1 | 41244435 | missense | T | C | C | Glu > Gly | |
| Chr13 | BRCA2 | 32914815 | missense | G | A | A | Arg > His | |
| Chr13 | BRCA2 | 32929309 | missense | A | G | G | His > Arg | |
| NTM-BCa13 | Chr13 | BRCA2 | 32906729 | missense | A | C | C | Asn > His |
| Chr13 | BRCA2 | 32914592 | missense | C | T | T | Arg > Cys | |
| NTM01 | Chr17 | BRCA1 | 41243840 | missense | A | C | C | Asn > Lys |
| Chr13 | BRCA2 | 32906729 | missense | A | C | C | Asn > His | |
| NTM03 | Chr17 | BRCA1 | 41245471 | missense | C | T | T | Asp > Asn |
aNTM-BCa: Women with NTM disease and breast cancer, NTM: Women with NTM disease; bChromosomal location of each of the gene exhibiting mutation; cName of the gene affected; dChromosome positions with reference to GRCh37.p13. eReference sequence of the human genome; fSequence detected in the lymphocytes DNA (LYM); gSequence detected in the sputum DNA; hCorresponding change in the amino acid.
Distribution of BRCA1 and BRCA2 germ line variants in the sputum of NTM affected women
We also detected 3 germline mutation spanning BRCA1 and BRCA2 genes in both the NTM subjects (2/2, 100%) (Table 5, Fig. 3). Among the 3 germ line mutation, 2 were similar to that observed in the NTM patients with breast cancer history (chromosome position: 41245471, BRCA1; 32906729, BRCA2). However, the germ line mutation in BRCA1 (position: 41243840) was only detected in the NTM subject in the absence of BCa (Table 4).
Copy number variation (CNV) in different genomic and chromosomal regions in the NTM affected women
The CNV analysis did not detect any breakpoints within genes for any samples (data not shown). No single sample contained more than one CNV in a target gene, although gains and losses in copy number periodically occurred throughout the rest of the genome. Lowering the CNV log2 call threshold to 0.2 (the default option in CNVkit) resulted in additional gains and losses of some chromosomal segments (data not shown), but only the high-confidence calls from a more stringent log2 threshold of 0.3 were presented. A single copy loss of a lung and breast cancer risk associated gene STK11 (a.k.a. LKB1) was noted in 25% (3/12) of the NTM subjects with breast cancer history (Fig. 4A). A single copy loss of the Androgen Receptor gene (AR) was also noted in one NTM subject with BCa history (Fig. 4B). A single copy loss in chromosome 5 and 16 was noted in BCa-NTM10 (Fig. 4C) and copy number loss in chromosome 17 was noted in subject BCa-NTM11 (Fig. 4D) who had a history of breast cancer and smoking as described above. These CNVs observed in chromosome 5, 16 and 17 were not associated with the 19 gene panels we have analyzed (Fig. 4C).
Figure 4.
Copy number variation in NTM affected patients. (A) Single copy loss of STK11 (named in gray box) as indicated by blue arrows in 3 subjects with NTM and breast cancer (NTM-BCa). (B) Copy number loss of androgen receptor (AR) gene (named in gray box), indicated by blue arrow in one subject with NTM and breast cancer (NTM-BCa). (C) Copy number loss in chromosomal region 5 and 16 (green arrows) in a woman with NTM and breast cancer (NTM-BCa). (D) Copy number loss in chromosome 17 (green arrow) in another woman with NTM and breast cancer (NTM-BCa).
Discussion
Pulmonary NTM disease (especially disease due to MAC) is a rising health concern in USA and throughout the world8. Many of these NTM patients develop therapeutic resistance posing significant challenges to disease management1,9. Infection with MAC particularly in the treatment resistant patients may lead to molecular changes associated with inflammation and tumorigenesis in surrounding epithelial tissues niche. In a recent study, infection of normal human lung airway epithelial cells with MAC triggered enhanced expression of CCL20, IL-32 and CXCL8 proteins36. These proteins are known to promote BCa growth, invasion and progression to metastasis37–43. These molecules were also demonstrated to promote lung cancer40,43–47. A recent study also uncovered functional involvement of intratumoral pathogenic bacteria in facilitating chemotherapeutic resistance in colon cancer patients48. Thus, NTM infected patients may remain at risk of developing neoplastic disease in their life time. Investigation of the molecular genetic alterations in the surrounding tissue microenvironment of the infected sites is important and could aid in developing disease monitoring and risk assessment strategies.
Next generation sequencing platform has revolutionized characterization of molecular genetic abnormalities resulting from infection or genotoxic damages in various affected cell types49–52. Free circulating DNA released from the infected or malignant cells often serves as monitoring/surveillance biomarkers and can also offer better therapeutic guidance53–55. Increased sputum production is one of the major symptoms due to NTM lung infection and sputum could be a valuable resource to identify free DNA not only associated with NTM pathogenesis but also inflammatory changes resulted from infection. Free circulating DNA associated with altered methylation, inflammation and cancer has been detected in sputum of COPD and lung cancer patients56–60. We identified cancer associated predisposing genetic mutations in sputum of women with NTM lung infection with or without a diagnosis of BCa. This novel finding confirms the presence of mutated DNA in sputum samples of NTM infected patients. Numerous genes exhibiting somatic mutations in these subjects such as EBBB2, PALB2, TP53, ATM, STK11 and TGFB1 are involved in various malignancies including BCa16,18,23,61. The majority of the women in our study cohort had been diagnosed with BCa before or after the diagnosis of NTM disease. Patient 11 (NTM-BCa11), a former smoker with a high BMI and family history of breast cancer was simultaneously diagnosed with NTM and early stage BCa (stage 0) at age 68. This patient was detected with a novel somatic ERBB2 mutation and germ line BRCA2 mutation accompanied by copy number loss in chr.17. Similarly, patient 12 (NTM-BCa-12) with a history of alcohol consumption who was diagnosed with NTM (MAC) at age 55 and stage-IB breast cancer at age 61 exhibited numerous somatic and germ line BRCA1 and BRCA2 mutations in the sputum. The two NTM subjects with low BMI and exhibiting somatic/germ line BRCA1 and BRCA2 mutations had presented with abnormal mammograms during their routine examination. Except for subject NTM01, they also had a history of alcohol consumption and tobacco smoking. Collectively, these findings suggest an association between NTM (MAC) lung disease and BCa development in these women and warrants breast examinations and routine screening of sputum for predisposing mutation detection.
STK11 (a.k.a. LKB1) is a critical regulator of mammary tumorigenesis62–64. Functional inactivation or loss of STK11 was shown to promote breast cancer initiation and progression to metastasis62–64. A functional coordination between STK11 with ERBB2 in mediating these effects in mammary tumorigenesis was also demonstrated62–64. The STK11 is also one of the most frequently inactivated genes in non-small cell lung cancer65. Loss of STK11 copy in multiple NTM-BCa subjects also suggests association with malignant transformation in these women. Notably, one of these women with STK11 alteration (NTM-BCa04) was diagnosed with NTM disease at age 57 and stage-I breast cancer at 61. These findings further suggest a functional correlation between NTM and malignant disease development in these women. Androgen receptor (AR) expression predicts better prognosis and survival of breast cancer patients34 and reduced AR expression promotes initiation of ERBB2 induced mammary tumorigenesis66. Thus, the loss of AR copy number observed in one NTM-BCa subject could also be associated with neoplastic transformation in this subject.
Germ line pathogenic variants in BRCA1 and BRCA2 are predisposing genetic factors associated with enhanced risk of BCa in the lifetime of an individual as demonstrated in numerous studies12,25,26,28,31–33. In this study, 40% of NTM affected women irrespective of their BCa diagnosis status exhibited germ line BRCA1 and BRCA2 mutations. Thus, NTM patients with long term infections and predisposing cancer associated genetic mutations may be at risk of developing malignant diseases in their life-time. Comparing the number of somatic mutations between the NTM-BCa (4/13) and the NTM (2/2) groups, there was no statistically significant relationship between breast cancer status and somatic mutation (p = 0.06) among NTM patients. Similarly, no statistically significant relationship was established for NTM-BCa with germ line (p = 0.06) mutation and copy number variations (p = 0.214).
To our knowledge, this is the first study, which revealed cancer associated gene mutations (both somatic and germ line) in sputum of NTM (MAC) infected subjects with or without a diagnosis of breast cancer. These findings suggest that chronic infection with NTM may trigger inflammation and cellular transformation surrounding the infection sites (immune and epithelial cells). Therefore, these subjects may potentially be at risk of acquiring transformational changes due to chronic NTM infection. Earlier, we have detected oncogenic ECM1 protein in the circulating exosomes of these subjects8. These findings collectively suggest for an increased risk of the NTM affected subjects towards oncogenic transformation. Helicobacter pylori infection is a relevant example facilitating gastrointestinal tumorigenesis11. This study suggests a novel avenue for study and may warrants monitoring of these subjects not only for NTM progression but also cellular transformation. In the clinical setting, molecular assessment of sputa by high throughput sequencing of NTM affected subjects may identify novel genetic alterations. A consensus panel of such molecular alterations could serve as biomarker for monitoring the risk of developing neoplastic disease in these patients as do BRCA1, BRCA2 and ER/PR/HER2 biomarkers for BCa25–27,52.
Methods
Human samples and ethical statement
Matched normal lymphocytes and sputa with relevant clinical information such as age, grade, stage, diagnosis etc. were collected from 13 NTM-BCa and 2 NTM subjects (de-identified, Table 1). The Institutional Review Board of The University of Texas Health Science Center at Tyler approved this study (#974). All subjects had sputum cultures which were culture positive for MAC infection by acid fast bacilli (AFB) sputum analysis8. Informed consent was obtained from all the patients. All methods were performed in accordance with the relevant guidelines and regulations.
Sputum collection and quality control
Routine expectorated sputa were collected and cultured as necessary for detection of AFB67–69. Samples were processed using standard decontamination procedures, fluorochrome microscopy and cultured on solid and liquid media as recommended by the Clinical and Laboratory Standards Insititute (CLSI) guidelines for mycobacteria detection and culture67,69. MAC isolates were identified using AccuProbe (Hologic Gen-Probe Inc)68. For decontamination, the N-acetyl-L-cysteine-sodium hydroxide method alone or in combination with oxalic acid was used70. All methods were performed in accordance with relevant guidelines and regulations.
DNA extraction and quantification
The lymphocytes were isolated from whole blood as described71. Genomic DNA was extracted from lymphocytes and sputa by digesting samples with 1% sodium dodecyl sulfate/proteinase K mixture overnight at 55 °C. The DNA was then extracted by phenol-chloroform, and ethanol precipitation and suspended in Tris-EDTA buffer and concentration was measured using the Nanodrop System. For sequencing analysis, 1 µg of DNA was used.
Next generation sequencing of the predisposing human gene panel
Utilizing next generation sequencing (NGS) platform35, we sequenced sputum and matched lymphocyte DNA of 15 NTM subjects for a panel 19 genes known to harbor mutations associated with cancer susceptibility and neoplastic transformation (Table 2)12–34. A total of 313 exons (coding regions) covering 63619 base pair regions of these 19 genes were mapped. We utilized a custom oligonucleotide-based capture with sequencing of regions within these 19 genes on Illumina HiSeq platform.
Data analysis and validation
The analysis pipeline utilizes genome analysis tool kit (GATK) standards, includes quality assessment with FASTQC72 followed by mapping reads to human reference genome GRCh37.p13 (hg19). SNVs and indels with depth of coverage >10 were called using Burrows-Wheeler Aligner (BWA)73 and Sequence Alignment/Map (SAM) tools74, with annotation from the Human Genetic Mutation Database using SnpEff75. When multiple annotations for gene location were available, the most severe was reported (e.g., missense variant scored instead of non-coding exon variant). Following Winter et al.32, variants were classified as somatic if they were present only in the sputa; and germ line if they were present in both the lymphocyte and sputa compared to the reference sequence. Variants present only in the lymphocytes were excluded from subsequent analysis. Sequence data have been submitted to the NCBI Sequence Read Archive and can be found under accession (BioProject #PRJNA431897). Scripts used for filtering and visualizing results can be found at https://github.com/k8hertweck/breastCancerNTM.
Copy number variation analysis
CNVs were assessed using CNVkit76. This software uses both target (e.g., from cancer associated genes) and off-target reads to call copy number across the genome, and is most accurate in detecting CNVs larger than 1 mega base pair (Mbp) spanning multiple exons (or captured regions). Log2 values for segment calls were summarized for visualization purposes. Given the uncertainty in assessing levels of copy number heterogeneity associated with these samples, the log2 threshold of 0.3 (as recommended by the CNV kit manual) was applied to call gains or losses in copy number of target genes.
Statistical analysis
We employed Binomial test for proportion to compare mutation outcome among various groups.
Acknowledgements
The work was supported by the UT Health Science Center at Tyler (S.D.). We thank all the patients for providing their precious samples for this study.
Author Contributions
S.D., J.V.P. and D.E.G. conceived and designed the study. S.D., A.K. and K.L.H. performed experiments and data analysis. Specimens and clinical data provided by J.V.P., D.E.G., B.E. and R.J.W. S.D., J.V.P., B.E. and R.J.W. and E.J.M., H.N., K.P.S. contributed reagents, materials and statistical analyses. S.D., J.V.P., D.E.G., E.J.M. wrote the manuscript. All authors reviewed and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Griffith DE, Aksamit TR. Understanding nontuberculous mycobacterial lung disease: it’s been a long time coming. F1000Res. 2016;5:2797. doi: 10.12688/f1000research.9272.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lake MA, Ambrose LR, Lipman MC, Lowe DM. "Why me, why now?" Using clinical immunology and epidemiology to explain who gets nontuberculous mycobacterial infection. BMC Med. 2016;14:54. doi: 10.1186/s12916-016-0606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith DE, Aksamit TR. Bronchiectasis and nontuberculous mycobacterial disease. Clin Chest Med. 2012;33:283–295. doi: 10.1016/j.ccm.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Honda JR, Knight V, Chan ED. Pathogenesis and risk factors for nontuberculous mycobacterial lung disease. Clin Chest Med. 2015;36:1–11. doi: 10.1016/j.ccm.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Feld R, Bodey GP, Gröschel D. Mycobacteriosis in patients with malignant disease. Arch Intern Med. 1976;136:67–70. doi: 10.1001/archinte.1976.03630010051009. [DOI] [PubMed] [Google Scholar]
- 6.Sood G, Parrish N. Outbreaks of nontuberculous mycobacteria. Curr Opin Infect Dis. 2017;2017(30):404–409. doi: 10.1097/QCO.0000000000000386. [DOI] [PubMed] [Google Scholar]
- 7.Aksamit TR, Philley JV, Griffith DE. Nontuberculous mycobacterial (NTM) lung disease: the top ten essentials. Respir Med. 2014;108:417–425. doi: 10.1016/j.rmed.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Philley JV, et al. Exosome secretome and mediated signaling in breast cancer patients with nontuberculous mycobacterial disease. Oncotarget. 2017;8:18070–18081. doi: 10.18632/oncotarget.14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsson LO, et al. Pulmonary disease by non-tuberculous mycobacteria - clinical management, unmet needs and future perspectives. Expert Rev Respir Med. 2017;11:977–989. doi: 10.1080/17476348.2017.1386563. [DOI] [PubMed] [Google Scholar]
- 10.Diel R, et al. Microbiological and Clinical Outcomes of Treating Non-Mycobacterium Avium Complex Nontuberculous Mycobacterial Pulmonary Disease: A Systematic Review and Meta-Analysis. Chest. 2017;152:120–142. doi: 10.1016/j.chest.2017.04.166. [DOI] [PubMed] [Google Scholar]
- 11.Abreu MT, Peek RM., Jr. Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146:1534–1546. doi: 10.1053/j.gastro.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couch FJ, et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Li M, Chen P, Ba Q. High expression of PALB2 predicts poor prognosis in patients with advanced breast cancer. FEBS Open Bio. 2017;8:56–63. doi: 10.1002/2211-5463.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nepomuceno, T.C. et al. The Role of PALB2 in the DNA Damage Response and Cancer Predisposition. Int J Mol Sci18(9) (2017). [DOI] [PMC free article] [PubMed]
- 15.Xu Y, Gao P, Lv X, Zhang L, Zhang J. The role of the ataxia telangiectasia mutated gene in lung cancer: recent advances in research. Ther Adv Respir Dis. 2017;11:375–380. doi: 10.1177/1753465817725716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi M, Kipps T, Kurzrock R. ATM Mutations in Cancer: Therapeutic Implications. Mol Cancer Ther. 2016;15:1781–91. doi: 10.1158/1535-7163.MCT-15-0945. [DOI] [PubMed] [Google Scholar]
- 17.DeLeonardis K, et al. Challenges in Interpreting Germline Mutations in BARD1 and ATM in Breast and Ovarian Cancer Patients. Breast J. 2017;23:461–464. doi: 10.1111/tbj.12764. [DOI] [PubMed] [Google Scholar]
- 18.Busch EL, et al. Somatic mutations in CDH1 and CTNNB1 in primary carcinomas at 13 anatomic sites. Oncotarget. 2017;8:85680–85691. doi: 10.18632/oncotarget.21115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ping Z, et al. ERBB2 mutation is associated with a worse prognosis in patients with CDH1 altered invasive lobular cancer of the breast. Oncotarget. 2016;7:80655–80663. doi: 10.18632/oncotarget.13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, et al. Frequency of pathogenic germline mutation in CHEK2, PALB2, MRE11, and RAD50 in patients at high risk for hereditary breast cancer. Breast Cancer Res Treat. 2017;161:95–102. doi: 10.1007/s10549-016-4034-2. [DOI] [PubMed] [Google Scholar]
- 21.Wiegmans AP, Miranda M, Wen SW, Al-Ejeh F, Möller A. RAD51 inhibition in triple negative breast cancer cells is challenged by compensatory survival signaling and requires rational combination therapy. Oncotarget. 2016;7:60087–60100. doi: 10.18632/oncotarget.11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarzynska JM. Two faces of TGF-beta1 in breast cancer. Mediators Inflamm. 2014;2014(():141747. doi: 10.1155/2014/141747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cimino-Mathews A, et al. Androgen receptor expression is usually maintained in initial surgically resected breast cancer metastases but is often lost in end-stage metastases found at autopsy. Hum Pathol. 2012;43:1003–1011. doi: 10.1016/j.humpath.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tung N, et al. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. J Clin Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tedaldi G, et al. Multiple-gene panel analysis in a case series of 255 women with hereditary breast and ovarian cancer. Oncotarget. 2017;8:47064–47075. doi: 10.18632/oncotarget.16791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aloraifi F, et al. Detection of novel germline mutations for breast cancer in non-BRCA1/2 families. FEBS J. 2015;282:3424–3437. doi: 10.1111/febs.13352. [DOI] [PubMed] [Google Scholar]
- 27.Walsh T, et al. Genetic Predisposition to Breast Cancer Due to Mutations Other Than BRCA1 and BRCA2 Founder Alleles Among Ashkenazi Jewish Women. JAMA Oncol. 2017;3:1647–1653. doi: 10.1001/jamaoncol.2017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheikh A, et al. The spectrum of genetic mutations in breast cancer. Asian Pac J Cancer Prev. 2015;16:2177–2185. doi: 10.7314/APJCP.2015.16.6.2177. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda N, et al. Identification of frequent somatic mutations in inflammatory breast cancer. Breast Cancer Res Treat. 2017;163:263–272. doi: 10.1007/s10549-017-4165-0. [DOI] [PubMed] [Google Scholar]
- 30.Graffeo. R, et al. Time to incorporate germline multigene panel testing into breast and ovarian cancer patient care. Breast Cancer Res Treat. 2016;160:393–410. doi: 10.1007/s10549-016-4003-9. [DOI] [PubMed] [Google Scholar]
- 31.Afghahi A, Kurian AW. The Changing Landscape of Genetic Testing for Inherited Breast Cancer Predisposition. Curr Treat Options Oncol. 2017;18:27. doi: 10.1007/s11864-017-0468-y. [DOI] [PubMed] [Google Scholar]
- 32.Winter C, et al. Targeted sequencing of BRCA1 and BRCA2 across a large unselected breast cancer cohort suggests that one-third of mutations are somatic. Ann Oncol. 2016;27:1532–1538. doi: 10.1093/annonc/mdw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraus C, et al. Gene panel sequencing in familial breast/ovarian cancer patients identifies multiple novel mutations also in genes others than BRCA1/2. Int J Cancer. 2017;140:95–102. doi: 10.1002/ijc.30428. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, He X, Yu Q, Eng C. Androgen receptor-induced tumor suppressor, KLLN, inhibits breast cancer growth and transcriptionally activates p53/p73-mediated apoptosis in breast carcinomas. Hum Mol Genet. 2013;22:2263–2272. doi: 10.1093/hmg/ddt077. [DOI] [PubMed] [Google Scholar]
- 35.Kannan A, Hertweck KL, Philley JV, Wells RB, Dasgupta S. Genetic Mutation and Exosome Signature of Human Papilloma Virus Associated Oropharyngeal Cancer. Sci Rep. 2017;7:46102. doi: 10.1038/srep46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuyama, M. et al. Transcriptional Response of Respiratory Epithelium to Nontuberculous Mycobacteria. Am J Respir Cell Mol Biol, 10.1165/rcmb.2017-0218OC (2017). [DOI] [PMC free article] [PubMed]
- 37.Muscella A, Vetrugno C, Marsigliante S. CCL20 promotes migration and invasiveness of human cancerous breast epithelial cells in primary culture. Mol Carcinogenesis. 2017;56:2461–2473. doi: 10.1002/mc.22693. [DOI] [PubMed] [Google Scholar]
- 38.Lee SK, et al. Human antigen R-regulated CCL20 contributes to osteolytic breast cancer bone metastasis. Sci Rep. 2017;7:9610. doi: 10.1038/s41598-017-09040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsigliante S, Vetrugno C, Muscella A. Paracrine CCL20 loop induces epithelial-mesenchymal transition in breast epithelial cells. Mol Carcinogenesis. 2016;55:1175–1186. doi: 10.1002/mc.22360. [DOI] [PubMed] [Google Scholar]
- 40.Ha H, Debnath B, Neamati N. Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases. Theranostics. 2017;7:1543–1588. doi: 10.7150/thno.15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu. Q, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016;31:61–71. doi: 10.1016/j.cytogfr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Chen F, Tang L. IL-32 promotes breast cancer cell growth and invasiveness. Oncol Lett. 2015;9:305–307. doi: 10.3892/ol.2014.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong JT, et al. Interleukin 32, inflammation and cancer. Pharmacol Ther. 2017;174:127–137. doi: 10.1016/j.pharmthera.2017.02.025. [DOI] [PubMed] [Google Scholar]
- 44.Wang B, et al. Production of CCL20 from lung cancer cells induces the cell migration and proliferation through PI3K pathway. J Cell Mol Med. 2016;20:920–929. doi: 10.1111/jcmm.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang GZ, et al. Tobacco smoke induces production of chemokine CCL20 to promote lung cancer. Cancer Lett. 2015;363:60–70. doi: 10.1016/j.canlet.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Zhang. XP, et al. Role of CCL20/CCR6 and the ERK signaling pathway in lung adenocarcinoma. Oncol Lett. 2017;14:8183–8189. doi: 10.3892/ol.2017.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng. Q, et al. Interleukin-32 contributes to invasion and metastasis of primary lung adenocarcinoma via NF-kappaB induced matrix metalloproteinases 2 and 9 expression. Cytokine. 2014;65:24–32. doi: 10.1016/j.cyto.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 48.Geller LT, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adalsteinsson VA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rheinbay E, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lesurf R, et al. Genomic characterization of HER2-positive breast cancer and response to neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041 (Alliance) trial. Ann Oncol. 2017;28:1070–1077. doi: 10.1093/annonc/mdx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng CKY, et al. The Landscape of Somatic Genetic Alterations in Metaplastic Breast Carcinomas. Clin Cancer Res. 2017;23:3859–3870. doi: 10.1158/1078-0432.CCR-16-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guibert, N. et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol Jan 9 (2018). [DOI] [PMC free article] [PubMed]
- 54.Diamandis EP, Fiala C. Can circulating tumor DNA be used for direct and early stage cancer detection? F1000Res. 2017;6:2129. doi: 10.12688/f1000research.13440.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Widschwendter M, et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 2017;9:116. doi: 10.1186/s13073-017-0500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Drift MA, et al. Can free DNA be detected in sputum of lung cancer patients? Lung Cancer. 2008;61:385–390. doi: 10.1016/j.lungcan.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 57.Guzmán L, et al. Analysis of aberrant methylation on promoter sequences of tumor suppressor genes and total DNA in sputum samples: a promising tool for early detection of COPD and lung cancer in smokers. Diagn Pathol. 2012;7:87. doi: 10.1186/1746-1596-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao L, Hruban RH, Boyle JO, Tockman M, Sidransky D. Detection of oncogene mutations in sputum precedes diagnosis of lung cancer. Cancer Res. 1994;54:1634–1637. [PubMed] [Google Scholar]
- 59.Keohavong P, et al. Detection of K-ras and p53 mutations in sputum samples of lung cancer patients using laser capture microdissection microscope and mutation analysis. Anal Biochem. 2004;324:92–99. doi: 10.1016/j.ab.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 60.Hubers AJ, Prinsen CFM, Sozzi G, Witte BI, Thunnissen E. Molecular sputum analysis for the diagnosis of lung cancer. Br J Cancer. 2013;109:530–537. doi: 10.1038/bjc.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polyak K, Metzger, Filho O. SnapShot: breast cancer. Cancer Cell. 2012;22:562–562.e1. doi: 10.1016/j.ccr.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 62.Li J, et al. Loss of LKB1 disrupts breast epithelial cell polarity and promotes breast cancer metastasis and invasion. J Exp Clin Cancer Res. 2014;33:70. doi: 10.1186/s13046-014-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrade-Vieira R, Xu Z, Colp P, Marignani PA. Loss of LKB1 expression reduces the latency of ErbB2-mediated mammary gland tumorigenesis, promoting changes in metabolic pathways. PLoS One. 2013;8:e56567. doi: 10.1371/journal.pone.0056567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dupuy F, et al. LKB1 is a central regulator of tumor initiation and pro-growth metabolism in ErbB2-mediated breast cancer. Cancer Metab. 2013;1:18. doi: 10.1186/2049-3002-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Facchinetti F, et al. LKB1/STK11 mutations in non-small cell lung cancer patients: Descriptive analysis and prognostic value. Lung Cancer. 2017;112:62–68. doi: 10.1016/j.lungcan.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Hodgson MC, et al. Reduced androgen receptor expression accelerates the onset of ERBB2 induced breast tumors in female mice. PLoS One. 2013;8:e60455. doi: 10.1371/journal.pone.0060455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallace RJ, Jr., et al. Initial clarithromycin monotherapy for Mycobacterium avium-intracellulare complex lung disease. Am J Respir Cri Care Med. 1994;149:1335–1341. doi: 10.1164/ajrccm.149.5.8173775. [DOI] [PubMed] [Google Scholar]
- 68.Wallace RJ, Jr., Brown BA, Griffith DE, Girard WM, Murphy DT. Clarithromycin regimens for pulmonary Mycobacterium avium complex. The first 50 patients. Am J Respir Crit Care Med. 1996;153:1766–1772. doi: 10.1164/ajrccm.153.6.8665032. [DOI] [PubMed] [Google Scholar]
- 69.Brown-Elliott BA, Nash KA, Wallace RJ., Jr. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev. 2012;25:545–582. doi: 10.1128/CMR.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wallace RJ, Jr., et al. Macrolide/Azalide therapy for nodular/bronchiectatic mycobacterium avium complex lung disease. Chest. 2014;146:276–282. doi: 10.1378/chest.13-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bandaru A, et al. Phosphorylated STAT3 and PD-1 regulate IL-17 production and IL-23 receptor expression in Mycobacterium tuberculosis infection. Eur J Immunol. 2014;44:2013–24. doi: 10.1002/eji.201343680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andrews, S. FastQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (2016).
- 73.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li. H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-wide copy number detection and visualization from targeted sequencing. PLOS Computational Biology. 2014;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]