Abstract
Francisella tularensis, a highly infectious, intracellular bacterium possesses an atypical type VI secretion system (T6SS), which is essential for the virulence of the bacterium. Recent data suggest that the HSP100 family member, ClpB, is involved in T6SS disassembly in the subspecies Francisella novicida. Here, we investigated the role of ClpB for the function of the T6SS and for phenotypic characteristics of the human pathogenic subspecies holarctica and tularensis. The ∆clpB mutants of the human live vaccine strain, LVS, belonging to subspecies holarctica, and the highly virulent SCHU S4 strain, belonging to subspecies tularensis, both showed extreme susceptibility to heat shock and low pH, severely impaired type VI secretion (T6S), and significant, but impaired intracellular replication compared to the wild-type strains. Moreover, they showed essentially intact phagosomal escape. Infection of mice demonstrated that both ΔclpB mutants were highly attenuated, but the SCHU S4 mutant showed more effective replication than the LVS strain. Collectively, our data demonstrate that ClpB performs multiple functions in the F. tularensis subspecies holarctica and tularensis and its function is important for T6S, intracellular replication, and virulence.
Introduction
The zoonotic disease tularemia is caused by the virulent, intracellular, Gram-negative coccobacillus designated Francisella tularensis1. Infection can occur through a variety of entry sites, including ingestion, direct contact with non-intact skin or mucous membranes, inhalation, or via the bite of a tick or fly vector2. Strains virulent for humans belong to the subspecies holarctica and tularensis, both of which are highly contagious and the latter subspecies may cause fatal human infection, whereas subspecies holarctica cause potentially serious, but not fatal disease. The pathogenicity of Francisella is intimately dependent on the Francisella Pathogenicity Island (FPI), a gene cluster encoding a functional, but atypical type VI secretion system (T6SS)3,4.
The T6SSs are transenvelope complexes specialized in the delivery of effector proteins directly to the target cell membranes of both bacterial and eukaryotic cells, thereby modulating the host-bacterial or bacterial-bacterial interactions5,6. The highly dynamic assembly of T6SS starts by formation of a membrane complex7 that recruits a baseplate complex8–11. At the assembled baseplate, the polymerization of a long tube is initiated, which then becomes wrapped by a sheath12–15. The spike and tube then combine with effector molecules, which are exerted upon sheath contraction15–18.
The FPI gene cluster of Francisella encodes 17 proteins, most of which are required for phagosomal escape and survival inside the host and 8 of them have low sequence similarity to canonical T6SS proteins19–22. Recently, a mesh-like structure was observed in F. novicida and, despite low sequence similarity, its sheath is similar to the contractile sheath of canonical T6SS, indicating the presence of a functional T6SS14,23,24. Techniques have been implemented to detect secretion of FPI proteins and several secreted proteins have been identified, although much remains to be understood about their functions, e.g., no effector function has been assigned to any FPI protein25–27. Despite the presence of many functional T6SS homologues, Francisella lacks the two ATPases, IcmF/TssM and ClpV, both of which may provide the energy required for secretion in prototypical T6SS28,29. An IcmF homologue (termed PdpB) is present, but lacks the Walker A motif necessary for the ATPase activity22. Campylobacter jejuni, Helicobacter hepaticus, and Salmonella choleraesuis also lack the ClpV homologue, but demonstrate a functional T6SS30–33, indicating that ClpV is not essential for T6S of all the species. Instead, a related member of the ClpV family, the ClpB ATPase, may substitute. The hexameric ClpB molecular chaperone belongs to the ring-forming Clp/Hsp100 proteins34, which form two distinct subfamilies; class I proteins, ClpA, ClpB and ClpV, and class II proteins, ClpX and HslU29,34. ClpB confers thermotolerance to a range of species via its unfoldase activity35, a role executed jointly with the co-chaperones DnaK, DnaJ, and GrpE36. ClpV, although being a class I Clp/Hsp100 protein, is not involved in thermotolerance, however, it has been identified in vitro37 and in vivo18 as crucial for the disassembly of the contractile sheath in prototypical T6SS37. This disassembly activity contributes to the reassembly of an extended sheath28,38. Recently, colocalization of ClpB with the contracted sheath of Francisella novicida T6SS has been demonstrated and it has been suggested to play an essential role for sheath disassembly18,23. Although experimental evidence is lacking, ClpB may also provide energy for the translocation of the T6SS substrate molecules. However, in the absence of ClpB, the assembly is still partially active, demonstrating that its role for T6S is not essential28,38.
The ΔclpB mutant of the highly virulent strain SCHU S4 of F. tularensis subspecies tularensis has been extensively studied since it is highly attenuated and confers very effective protection in the mouse against challenge with virulent F. tularensis strains39–41. There are several characterized ΔclpB mutant of subspecies holarctica described. One was derived from a Swedish patient isolate and found to be more attenuated, yet, confer superior protection compared to the human live vaccine strain of the same subspecies42. The latter strain was empirically derived from a Russian patient isolate and subsequently passaged in the US and designated the live vaccine strain, LVS43. It was tested extensively in human volunteers during the 1960s44 and also utilized to protect laboratory staff. It led to an almost 90% reduction of laboratory-acquired tularemia45. A ΔclpB mutant of the LVS strain has been characterized and observed to induce a more robust proinflammatory response than did the parental strain46.
In the present study, we demonstrate that the ∆clpB mutants of the LVS strain, subspecies holarctica, and the highly virulent SCHU S4 strain, subspecies tularensis, both are exquisitely susceptible to heat shock and low pH, show defective intracellular growth, concomitantly with impaired T6S.
Results
ΔclpB mutants are highly susceptible to heat shock and low pH
Survival of ΔclpB mutants of any bacterial species is severely compromised during heat stress, since resolubilization of protein aggregates that result from the stress is predominantly dependent on ClpB. To determine the role of ClpB protein of F. tularensis in stress tolerance, we monitored the survival of LVS and the SCHU S4 ΔclpB mutants under various stress conditions.
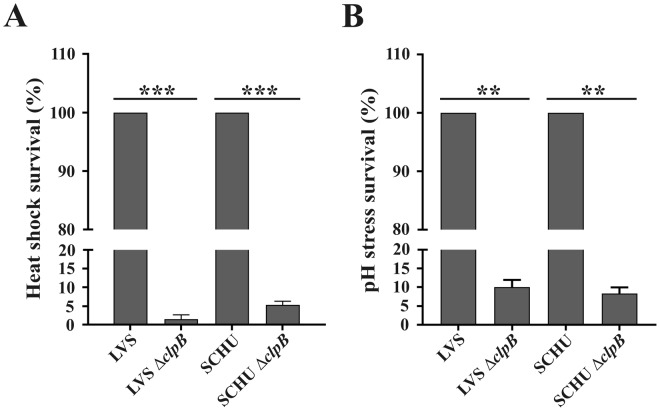
When subjected to high temperature (50 °C), as expected, the LVS and SCHU S4 ΔclpB mutants showed compromised survival and their numbers decreased to 1.5% and 5.3%, respectively, of the numbers of the wild-type strains after 30 min (Fig. 1A) while the CFU of their wild-type strains, 8.4 and 8.9 log10, respectively, did not drop significantly.
Figure 1.
Survival of indicated F. tularensis strains during heat and pH stress. Overnight grown bacteria were diluted in fresh growth media and subjected to stress at 50 °C for 30 min (A) or pH 4.5 for 1 h (B). Survival of each strain was monitored as viable bacteria and plotted as percentage of survival, setting wild-type strains survival as 100%. There was no significant killing of any wild-type strain during the treatment. Data are the average survival percentage for at least three independent experiments for each condition. Values are mean ± SD. **P < 0.01; *** indicates P < 0.001 vs. the corresponding wild-type strain.
We next evaluated the susceptibility of the ΔclpB mutant to low pH by exposing bacteria to a pH of 4.5 for 1 h. As in the case of the heat stress response, each of the two wild-type strains was relatively unaffected, while each of the ΔclpB mutants was highly susceptible as their survival was less than 10% of the corresponding wild-type strains (P < 0.001; Fig. 1B).
The responses of the ΔclpB mutants to other forms of stress were also investigated. O2− is continuously generated as a by-product of the respiratory chain during bacterial growth. To investigate the resistance to ROS, bacteria were exposed to either paraquat in a disc diffusion assay, or to SIN-1. The former catalyzes the formation of O2−, whereas the latter is a surrogate for ONOO− susceptibility, which is bactericidal against F. tularensis in activated macrophages47. Both the LVS and SCHU S4 ΔclpB mutants and wild-type strains showed the same susceptibility to 40 mM of paraquat or 0.40 mM of SIN-1 (data not shown).
The F. tularensis ΔclpB mutants show compromised intracellular replication
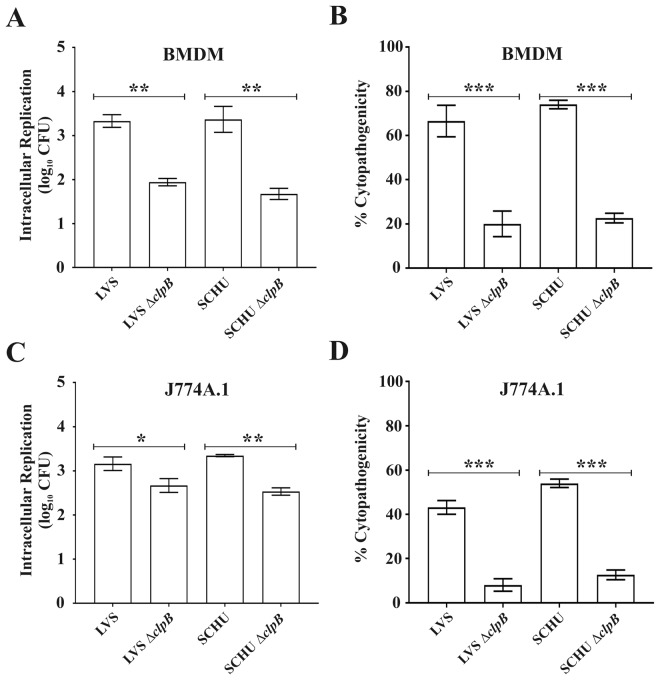
To determine whether the clpB mutation conferred compromised ability to replicate intracellularly, we investigated the ability of the LVS, or SCHU S4 ΔclpB mutants to multiply within murine BMDM or the macrophage-like cell line J774 for 24 h. All mutants showed significant replication in BMDM, approximately 100-fold, however, compared to each of the wild-type strains, the replication was significantly compromised (P < 0.01 vs. the corresponding wild-type strain; Fig. 2A). The absolute differences between mutants and wild-type strains were less marked in J774 cells; however, they were still significant (Fig. 2C). The cytopathogenic effects were very similar between the ΔclpB mutants, but much lower (P < 0.001) for the mutants than the effects seen upon infection with LVS or SCHU S4 (Fig. 2B,D). This suggests that the deletion of clpB is important, although not essential, for the intracellular replication and cytopathogenic effects.
Figure 2.
Intracellular growth and LDH release from F. tularensis-infected BMDM and J774A.1 cells. Growth of the LVS or SCHU S4 ΔclpB mutants and the corresponding wild type strains were analyzed by lysis of infected BMDM (A) and J774A.1 (C) cells at 0 h and 18 h and the number of CFU determined. The net growth mean values ± SEM of at least three independent experiments are shown. Supernatants of cultures of BMDM (B) and J774A.1 (D) cells infected with the indicated strains were harvested at 18 h. The activity was expressed as a percentage of the level of uninfected lysed cells (positive lysis control). The mean values ± SEM of triplicate wells from one representative experiment of two are shown. The asterisk indicates that the values of the ∆clpB mutants differed significantly compared to wild type strains (*P < 0.05; **P < 0.01; ***P < 0.001). Unpaired t-test with Welch’s correction was used to compare values.
The LVS and SCHU S4 ΔclpB mutants show intact phagosomal escape
In view of the compromised ability of the ΔclpB mutants to replicate intracellularly, we investigated whether the mutants also showed compromised phagosomal escape, since the two phenotypes are intimately linked for FPI mutants, i.e., those that are defective for the escape from the phagosomal compartment also lack intracellular replication3. The most common marker used to investigate the intraphagosomal localization by immunofluorescence is LAMP-1, which is a late endosomal and lysosomal marker acquired within 30 min by the Francisella-containing phagosome48,49. Thus, we compared the degree of co-localization in BMDM between LAMP and the SCHU S4 and LVS strains, or the corresponding ΔclpB mutants, each expressing the green fluorescent protein (GFP), at 3 h and 6 h. As control, the ΔiglC mutant of LVS, which lacks phagosomal escape, was used. At both the 3 and 6 h time points, there were no significant differences between the LVS and the ΔclpB mutant strains, demonstrating a normal phagosomal escape of the two mutants (Fig. 3).
Figure 3.
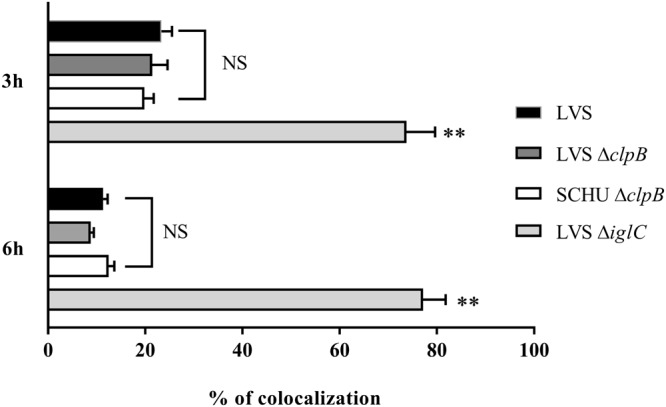
Co-localization of GFP-expressing strains of F. tularensis with LAMP-1. BMDM cells were infected with the indicated strains expressing GFP for 2 h at an MOI of 30. After 3 and 6 h, specimens were labeled for the late endosomal and lysosomal marker LAMP-1. At least 100 infected cells from multiple cover slips were examined in each experiment. Co-localization was analyzed using a fluorescence microscope (Nikon Eclipse 90i, Nikon, Japan). Results are representative of two experiments and presented as % of co-localization of GFP-expressing bacteria with LAMP-1. The asterisks indicate that the values of the LVS ∆iglC mutant differed significantly compared to LVS at 3 h and 6 h (**P < 0.01); however, the differences between LVS and LVS ∆clpB or SCHU ∆clpB were non-significant (NS P > 0.05). Unpaired t-test with Welch’s correction was used to compare values.
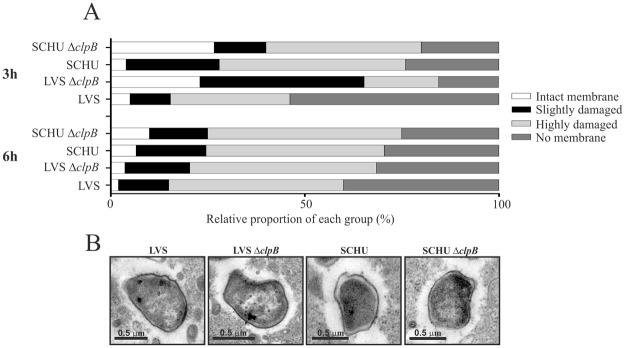
To further corroborate the results regarding the phagosomal escape, we also performed transmission electron microscopy. BMDM were infected with SCHU S4, LVS, or the respective ΔclpB mutants. The phagosomal membranes were more intact in cells infected with the ΔclpB mutants at the 3 h time point, but there were no significant differences between the SCHU S4 and LVS strains and the ΔclpB mutants at the 6 h time point (Fig. 4A). Representative images are shown in Fig. 4B).
Figure 4.
Assessment of the phagosomal membrane integrity of BMDM cells infected with indicated F. tularensis strains by the TEM assay. BMDM cells were infected for 2 h at an MOI of 1,000, washed, further incubated for 3 and 6 h, and then processed for TEM. To examine the membrane integrity of phagosome, at least 100 bacteria from different sections were analyzed for each time point and categorized as follows: Intact, >90% of the phagosomal membrane intact; Slightly damaged, 50–90% of the membrane intact; Highly damaged, 10–50% of the membrane intact; No membrane, <10% of the membrane intact.
Collectively, the findings using the two assays to determine phagosomal escape demonstrate similar results and the SCHU S4 and LVS ΔclpB mutant exhibited essentially intact phagosomal escape.
The F. tularensis ΔclpB mutants show compromised T6S
We and others have previously reported that the ΔclpB null mutants of F. tularensis SCHU S4 LVS, and a clinical isolate of holarctica are highly attenuated in vivo39,40,42,50 and we hypothesized that a possible mechanism may be impairment of the T6S. Therefore, the secretion of FPI proteins was investigated using the TEM-1 β-lactamase secretion assay, which is a sensitive method to assess intracellular protein secretion in general and FPI secretion specifically25,51. Therefore, TEM-1-fusions of IglC, IglE and VgrG, all FPI proteins known to be secreted, were expressed in LVS, SCHU ∆clpB and LVS ∆clpB and, in addition, IglE-FS1, a frame-shift mutant of IglE (first 2 to 6 amino acids), since it is secreted at much higher levels compared to the wild-type IglE52. The SCHU S4 strain could not be used because of biosafety restrictions; instead, a mutant, Δggt was used. SCHU Δggt is highly attenuated and deficient in the gamma-glutamyltranspeptidase (ggt), essential for the utilization of glutathione; however, the presence of cysteine allows it to replicate intracellularly53.
J774 macrophages cells were infected with the aforementioned strains expressing the β-lactamase fusions and translocation of the β-lactamase chimeras was assessed at 18 h after start of infection. The proportion of cells infected with LVS expressing fusions to IglC, IglE, VgrG, or IglE-FS1 demonstrating blue fluorescence ranged from 3.0 to 38.3% for LVS, depending on the fusion analyzed, similar to previously reported values54, and from 12.3 to 48.0% for SCHU S4, whereas both ∆clpB mutants demonstrated very low levels, from 0.3 to 2.9% for the LVS mutant and from 2.5 to 9.0% for the SCHU S4 mutant (Table 1). The lower values for each ∆clpB mutant were highly significant for each of the constructs (P < 0.001), demonstrating that ClpB of both subspecies plays a very important role for efficient T6S during infection. Representative images of the IglE fusion are shown (Fig. 5)
Table 1.
Secretion of FPI-TEM fusions upon infection of F. tularensis to J774A.1 cells.
| Strain | Secretion of FPI-β-lactamase fusions | |||
|---|---|---|---|---|
| LVS | LVS ∆clpB | SCHU ∆ggt | SCHU ∆clpB | |
| IglE-TEM | 10.6 ± 1.0a | 0.4 ± 0.2*** | 14.9 ± 1.9 | 5.4 ± 0.8*** |
| FS1-TEM | 38.3 ± 2.8 | 2.9 ± 0.3*** | 48.0 ± 3.9 | 9.0 ± 0.1*** |
| IglC-TEM | 8.7 ± 1.1 | 0.3 ± 0.1*** | 14.0 ± 2.5 | 2.5 ± 0.2*** |
| VgrG-TEM | 3.0 ± 0.5 | 0.7 ± 0.2*** | 12.3 ± 1.3 | 4.3 ± 0.4*** |
aThe percentages of blue fluorescent cells (indicating secretion of the β-lactamase fused protein) after 18 h of infection was determined by live-cell microscopy.
*** indicates that P < 0.001 when each value was compared to the corresponding value for the parental strain.
Figure 5.
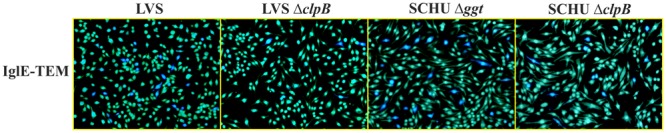
Representative examples of secretion of β-lactamase-tagged IglE protein in J774A.1 macrophages. Macrophages were infected either with LVS, LVS ∆clpB, SCHU ∆ggt or SCHU ∆clpB expressing IglE-TEM fusions for 18 h and the fluorescence was determined. TEM β-lactamase activity was identified by the blue fluorescence emitted by the cleaved CCF2 product, whereas uncleaved CCF2 substrate emitted a green fluorescence. The experiments were repeated at least 4 times using duplicate samples. Representative images of each of 4 samples are shown.
To confirm that the effect of the clpB mutation was specifically related to T6S and not to a general defect in FPI protein expression, Western blot analysis was performed using antibodies against IglA, IglB, IglC, IglD, IglE, IglH, PdpA, PdpB, PdpC, and VgrG. Lysates of LVS and SCHU and their corresponding ∆clpB mutants were analyzed and no differences were observed between the wild-type strain and the respective ∆clpB mutant (Fig. S1), demonstrating that the T6S defect observed in the mutants was due to a specific secretion defect and not due to impaired FPI expression.
ClpB markedly affects the virulence of F. tularensis
The attenuated phenotypes observed for the ΔclpB mutants with respect to phagosomal escape, LDH release, intracellular replication and T6S suggested that they would also show attenuated phenotypes in vivo. This has been confirmed previously with regard to the ΔclpB mutant of SCHU S4, which has been studied in much detail39,42. The SCHU S4 strain is highly virulent in mice with a LD50 below 10 CFU55.
To perform a comparative analysis of the in vivo phenotype of the ΔclpB mutants, mice were subcutaneously infected with the LVS strain or the ΔclpB mutants of SCHU S4 or LVS followed by enumeration of bacteria in spleen and liver, the main target organs of tularemia, after an inoculum of approximately 1 × 104 CFU. The bacterial numbers were highest on day 5 and thereafter diminished and were barely detectable on days 15 and 21 (Fig. 6). Regardless of time point, numbers of the LVS ΔclpB mutant were low or non-detectable. In contrast, the SCHU S4 ΔclpB mutant reached high numbers, significantly higher than the LVS strain, on days 5 and 9. For example, on day 5, the bacterial numbers were 2 log10 higher in spleen of the former strain. In separate experiments, the LD50 values were estimated and, despite the much higher bacterial numbers of the SCHU S4 ΔclpB mutant, its LD50 value was in fact higher than that of LVS, approximately 1 × 107 vs. 1 × 106 CFU (Fig. S2). These values are in agreement with previously reported ones42,56.
Figure 6.
Replication of F. tularensis strains in liver and spleen following infection of mice. After intradermal inoculation with 1 × 104 CFU of the indicated strain, mice were sacrificed on day 5, 9, 15 and 21, and bacterial burdens in spleen and liver were determined. The mean ± SEM for six mice per group and time point are shown. A significant difference in the bacterial numbers of mutant strains vs. LVS is indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; NS P > 0.05.
Taken together, these results clearly demonstrate that ClpB plays a critical role for the virulence in subspecies holarctica and, notably, that the SCHU S4 ΔclpB mutant demonstrates more rapid replication that does the LVS strain, despite that the LD50 value of the former is higher.
Discussion
The virulence of F. tularensis is intimately linked to its intracellular replication and a prerequisite for this is an intact T6SS, encoded by the Francisella pathogenicity island3,19. There has been much progress regarding the structure of this atypical T6SS. Recent studies have revealed that F. novicida T6SS assembles on the bacterial cell poles in vitro as well as during macrophage infection and constitutes a sheath nearly as long as the bacterial length with a mesh-like architecture, consisting of IglA/IglB14,23. ClpB was found to be specifically co-localized with the contracted sheath and was required for the disassembly, in the same way as described for ClpV in other T6SS-containing bacteria18,23,37. It should be noted, that there are other bacteria besides Francisella that lack the ClpV homologue, but still have functional T6S, e.g., Campylobacter, and Salmonella30–33. In view of this information and the present findings on Francisella, it is likely that ClpB, besides being critical for heat shock survival, also serves a critical role for T6S of those bacteria lacking ClpV. T6SS of several enterobacteria rely on the ATPases ClpV and IcmF for the energy required for the function of the T6SS28,29, but since F. tularensis lacks both ClpV and the Walker A motif in IcmF, which is critical for ATPase activity22, it is, therefore, possible that in the absence of ClpV, ClpB, besides contributing to sheath disassembly, generates the energy required for T6S. However, in view of the fact that a low level of secretion was observed in each ΔclpB mutant, the contribution, if any, of ClpB to generate energy is dispensable.
In the present study, we demonstrate that, as expected, and in agreement with previous data23,50, ClpB displays an essential role for the heat shock survival of subspecies holarctica and tularensis. There is precedence for the involvement of the ClpB-DnaK disaggregation complex in the assembly of the T6SS, since the insertion of a transmembrane segment within the essential T6SS component TssL of E. coli, is modulated by DnaK57. The findings regarding the LVS ΔclpB mutant concur with previously published data50.
To further characterize the intracellular phenotypes of the ΔclpB mutants, we also investigated their phagosomal escape. Normally, there is close correlation between the intracellular replication and phagosomal escape, as evidenced by numerous findings on various FPI mutants that all lack phagosomal escape and do not replicate intracellularly3. We observed that the ΔclpB mutants of SCHU S4 and LVS escaped as quickly as did the wild-type strains and, therefore, this was not a cause for their impaired replication. The escape of the LVS mutant has been studied previously and although the degree of escape was not quantified, it was concluded that it displayed a cytoplasmic localization50. We have previously demonstrated that FPI mutants lacking T6S, e.g., ΔiglA and ΔiglC, replicated at least as rapidly as LVS upon microinjection, regardless of cell type58. Therefore, it was concluded that the critical function of the T6SS is to execute phagosomal escape, but it is not essential for the cytoplasmic replication. Based on this hypothesis, the much compromised T6S of the ΔclpB mutants may not result in defective intracellular replication and an explanation for the compromised intracellular replication may be the exquisite susceptibility to low pH of both ΔclpB mutants, also previously demonstrated for the LVS mutant50. In further support of the hypothesis, it was observed in the latter study and also in the present study that the importance of ClpB varied depending on the host cell type. It is likely that the stress encountered, and thereby the relative contribution of ClpB, is not as severe in certain cell lines, such as the J774 macrophage-like cells, as in ex vivo-derived cells, such as BMDM.
The two ΔclpB mutants showed very low secretion of IglC and other FPI proteins. Previous studies have consistently demonstrated the need for a functional T6S for phagosomal escape, however, the minimal T6S of the ΔclpB mutants of F. tularensis SCHU S4 and LVS demonstrated that even very low levels of T6S are sufficient for the bacteria to escape. In fact, the two mutants showed rather distinct T6S, since the levels observed using the β-lactamase secretion assay were very low for the LVS mutant, whereas the SCHU S4 mutant demonstrated levels distinctly lower than the wild-type strains, but still much higher than the LVS mutant. Despite these differences, there were no obvious differences in their phagosomal escape.
Although both ΔclpB mutants demonstrated similar, defective intracellular replication, they showed very distinct phenotypes in vivo, since the LVS ΔclpB mutant was very compromised and disseminated only marginally to liver and spleen. In contrast, the SCHU S4 ΔclpB mutant disseminated and replicated more effectively than did the LVS strain. Paradoxically, the LD50 value of the ΔclpB mutant was still 10-fold higher. One possible explanation could be that the lethality of infection is partly dependent on T6S effectors and, therefore, despite the more effective replication of the SCHU S4 ΔclpB mutant, it is not as lethal as the LVS strain due to its compromised T6S.
Collectively, our data demonstrate a critical role of ClpB in both of the human-pathogenic subspecies holarctica and tularensis for the normal T6S. The ΔclpB mutants also demonstrate defective intracellular replication and virulence and also impaired handling of stress stimuli, such as heat shock and low pH. The exact contribution of ClpB and the T6SS to all of the observed phenotypes is still unclear and it will be important in future studies to elucidate this, since it will provide essential information about the control and regulation of the T6SS of Francisella and possibly other T6SS that lack ClpV.
Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S1. F. tularensis strains were cultured either in Tryptic soy broth (TSB) supplemented with 0.1% cysteine (w/v) and 0.1% glucose (w/v) or in chamberlain defined medium (CDM). Kanamycin (10 μg/ml) was added to the medium when needed. All bacteriological work related to the SCHU S4 strain was carried out in a biosafety level 3 facility certified by the Swedish Work Environment Authority. As decided by the Swedish Work Environment Authority, The SCHU S4 ΔclpB and Δggt mutants are classified as BSL2 agents.
Cultivation and infection of macrophages
The J774A.1 mouse macrophage-like cell line (ATCC TIB-67) or bone marrow-derived macrophages (BMDMs) were used in the cell infection assays. J774A.1 macrophages were cultured and maintained in DMEM (GIBCO BRL, Grand Island, NY, USA) with 10% heat-inactivated FBS (GIBCO). BMDMs were isolated by flushing bone marrow cells from the femurs and tibias of C57BL/6 mice as described previously59. These cells were cultured for 4 days in DMEM containing 10% FBS, 5 μg/ml gentamicin and 20% conditioned media (CM) from L929 cells (ATCC no CCL-1) overexpressing M-CSF, after which they were grown in medium lacking gentamicin. Macrophages were seeded in tissue culture plates in DMEM with 10% FBS and incubation overnight. A multiplicity of infection (MOI) of 200 was used in all infection experiments, with the exception of the TEM study, where an MOI of 1,000 was used. After 24 h post infection, the macrophage monolayers were lysed in 0.1% deoxycholate, serially diluted plated on agar plates for determination of viable counts.
Lactate dehydrogenase (LDH) release assay
At indicated time points, culture supernatant were collected from infected wells and the release of LDH, indicative of cytopathogenicity, determined using the LDH assay kit according to the manufacturer’s instructions (Promega, Madison, WI). Uninfected J774A.1 cells lysed in 0.1% deoxycholate served as a positive control, and the value for this control was arbitrarily considered as 100% cell lysis. Samples absorbance was expressed as a percentage of the positive control value. The assay was performed with triplicate samples and was repeated at least three times.
Temperature and pH susceptibility test
Bacteria were grown overnight on modified GC-agar, resuspended in Chamberlain to give an OD600 of 0.5, and 0.25 ml of the culture was placed in a 2 ml Eppendorf tube and placed statically in a water bath at 50 °C for 30 min. The tubes were then placed on ice, serially diluted in PBS and plated on agar plates to determine the number of viable bacteria. To determine the sensitivity to pH stress, overnight agar-grown bacteria were resuspended in TSB, pH 4.5 and incubated at 37 °C for 1 h. At indicated time points, aliquots were sampled, serially diluted in PBS and plated to determine the number of viable bacteria.
Paraquat and peroxynitrite susceptibility assay
Susceptibility of F. tularensis strains to O2− was determined by use of the O2− generating compound paraquat dichloride hydrate (Sigma-Aldrich, St. Louis, USA) in a disc diffusion assay as previously described47. Susceptibility to peroxynitrite was tested as previously described after incubation for 3 h in the presence of 0.40 mM SIN-147.
β-lactamase secretion assay
The TEM1 β-lactamase secretion assay was performed essentially following the same protocol as described before25. In brief, 1.5 × 105 J774 cells/well were seeded onto BD Falcon 8-wells glass chambers slides (BD Biosciences, Bedford, MA, USA) and incubated overnight before infected with the indicated bacterial strain. After 2 h of infection, cells were washed and incubated further with 5 µg/ml of gentamicin (defined as time point zero). After 30 min, cells were washed and after an additional 18 h, washed and loaded with CCF2/AM (Invitrogen, CA, USA) and Probenicid (Sigma). Translocation of β-lactamase fusions was determined with a live-cell imaging microscope (Nikon Eclipse Ti-E) equipped with a Nikon DS-U2/L2 camera, using a Chroma beta-lactamase double filter. For statistical analysis of blue vs, green fluorescent cells, an average of 3,000–6,000 cells that included pictures from three experiments for a given strain was counted.
Intracellular immunofluorescence assay
To assess phagosomal escape of GFP-expressing ∆clpB mutants of F. tularensis, the ∆clpB mutants (all expressing pKK289Km-GFP) were used in the cell infections as described previously21. As negative controls, LVS ∆iglC60 was used. At indicated time points, cells were fixed, and stained using an antibody recognizing the LAMP-1 glycoprotein (BD Pharmingen, San Jose, CA). Colocalization of GFP-labeled F. tularensis and LAMP-1 was analyzed with an epifluorescence microscope (Nikon Eclipse 90i, Nikon Instruments Europe BV, Netherlands). Two biological replicates were used and a minimum of 100 bacteria per strain and time point were scored.
Transmission electron microscopy
The analysis was carried out as previously described54. BMDM cells were infected for 2 h, washed, and incubated for 3 h or 6 h. Monolayers were washed and fixed with 2.5% glutaraldehyde, washed in 0.1 M sodium cacodylate buffer, scraped, and treated with 1% osmium tetroxide. Embedded cell pellets were cut into 70 nm sections and stained with uranyl acetate and lead citrate before analyzed with a JEM 1230 transmission electron microscope (Jeol Ltd., Tokyo, Japan). At least 100 bacteria from different sections were analyzed for each time point and categorized as having (i) an intact phagosomal membrane, (ii) a slightly damaged phagosomal membrane (<50% of membrane integrity affected), (iii) a highly damaged phagosomal membrane (>50% of membrane integrity affected), or (iv) no residual membrane.
Western blot analysis
The samples were dissolved in sample buffer, boiled before analyzed applied on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane. Blots were probed by either rabbit polyclonal antibodies against IglA, PdpC (BEI Resources), IglH and VgrG, (Inbiolabs, Tallinn, Estonia) or mouse monoclonal antibodies against IglB, IglC, IglD, PdpA and pdpB (BEI Resources). To probe against IglE52, rat polyclonal antibodies was used. Polyclonal IgY chicken antibodies, specific to FupA was used (Agrisera).
Mouse infection
For determination of the killing capacity of each strain, C57BL/6 female mice (n = 6) were infected intradermally with approximately 1 × 104 CFU for each F. tularensis strain. Mice were examined twice daily and euthanized if they displayed signs of severe disease. At days 5, 9, 15 and 21, infected mice were killed and serial dilutions of the homogenized organs were plated. The animal experiments were approved by the local Ethical Committee on Laboratory Animals, Umeå, Sweden (approval no. A67-14) and all experiments were performed in accordance to the national guidelines and regulations.
Statistical analysis
For the statistical analysis, GraphPad Prism 7 (GraphPad Software Inc., CA, USA) was used.
Electronic supplementary material
Acknowledgements
This work was supported by grants 2013-4581 and 2013-8621 from the Swedish Research Council and a grant from the Medical Faculty, Umeå University, Umeå, Sweden, and the JC Kempe Memorial Foundation (JCK-1624). We thank Jeanette Bröms for providing constructs and helpful comments.
Author Contributions
A.A. wrote the main manuscript text and performed much of the experimental work. E.J. and I.G. performed some of the experimental work. A.S. planned the experimental work and wrote the main manuscript text. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29745-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sjöstedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 2.Oyston PC, Sjöstedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 3.Bröms JE, Sjöstedt A, Lavander M. The role of the Francisella tularensis pathogenicity island in Type VI secretion, intracellular survival, and modulation of host cell signaling. Front Microbiol. 2010;1:136. doi: 10.3389/fmicb.2010.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celli J, Zahrt TC. Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb Perspect Med. 2013;3:a010314. doi: 10.1101/cshperspect.a010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hachani A, Wood TE, Filloux A. Type VI secretion and anti-host effectors. Curr Opin Microbiol. 2016;29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Alcoforado Diniz J, Liu YC, Coulthurst SJ. Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell Microbiol. 2015;17:1742–1751. doi: 10.1111/cmi.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand E, et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 8.Rigard M, et al. Francisella tularensis IglG belongs to a novel family of PAAR-like T6SS proteins and harbors a unique N-terminal extension required for virulence. PLoS Pathog. 2016;12:e1005821. doi: 10.1371/journal.ppat.1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Planamente S, et al. TssA forms a gp6-like ring attached to the type VI secretion sheath. EMBO J. 2016;35:1613–1627. doi: 10.15252/embj.201694024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shneider MM, et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nazarov, S. et al. Cryo-EM reconstruction of Type VI secretion system baseplate and sheath distal end. EMBO J 37, 10.15252/embj.201797103 (2018). [DOI] [PMC free article] [PubMed]
- 12.Brunet YR, Henin J, Celia H, Cascales E. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 2014;15:315–321. doi: 10.1002/embr.201337936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudryashev M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell. 2015;160:940–951. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat Microbiol. 2017;2:1507–1512. doi: 10.1038/s41564-017-0020-7. [DOI] [PubMed] [Google Scholar]
- 16.Flaugnatti N, et al. A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol. 2016;99:1099–1118. doi: 10.1111/mmi.13292. [DOI] [PubMed] [Google Scholar]
- 17.Silverman JM, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nano FE, et al. A Francisella tularensis Pathogenicity Island required for intramacrophage growth. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindgren H, et al. Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol. 2004;53:953–958. doi: 10.1099/jmm.0.45685-0. [DOI] [PubMed] [Google Scholar]
- 21.Bönquist L, Lindgren H, Golovliov I, Guina T, Sjöstedt A. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect Immun. 2008;76:3502–3510. doi: 10.1128/IAI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barker JR, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brodmann M, Dreier RF, Broz P, Basler M. Francisella requires dynamic type VI secretion system and ClpB to deliver effectors for phagosomal escape. Nat Commun. 2017;8:15853. doi: 10.1038/ncomms15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemens DL, Lee BY, Horwitz MA. The Francisella Type VI Secretion System. Front Cell Infect Microbiol. 2018;8:121. doi: 10.3389/fcimb.2018.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bröms JE, Meyer L, Sun K, Lavander M, Sjöstedt A. Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS One. 2012;7:e50473. doi: 10.1371/journal.pone.0050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hare RF, Hueffer K. Francisella novicida pathogenicity island encoded proteins were secreted during infection of macrophage-like cells. PLoS One. 2014;9:e105773. doi: 10.1371/journal.pone.0105773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eshraghi A, et al. Secreted effectors encoded within and outside of the Francisella Pathogenicity Island promote intramacrophage growth. Cell Host Microbe. 2016;20:573–583. doi: 10.1016/j.chom.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forster A, et al. Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes. J Biol Chem. 2014;289:33032–33043. doi: 10.1074/jbc.M114.600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlieker C, Zentgraf H, Dersch P, Mogk A. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem. 2005;386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- 30.Lertpiriyapong K, et al. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One. 2012;7:e42842. doi: 10.1371/journal.pone.0042842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrivastava S, Mande SS. Identification and functional characterization of gene components of Type VI Secretion system in bacterial genomes. PLoS One. 2008;3:e2955. doi: 10.1371/journal.pone.0002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bleumink-Pluym NM, van Alphen LB, Bouwman LI, Wosten MM, van Putten JP. Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog. 2013;9:e1003393. doi: 10.1371/journal.ppat.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA + chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 35.Rosenzweig R, et al. ClpB N-terminal domain plays a regulatory role in protein disaggregation. Proc Natl Acad Sci USA. 2015;112:E6872–6881. doi: 10.1073/pnas.1512783112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlee, S., Beinker, P., Akhrymuk, A. & Reinstein, J. A chaperone network for the resolubilization of protein aggregates: direct interaction of ClpB and DnaK. J Mol Biol336, 275–285, S0022283603014815 (2004). [DOI] [PubMed]
- 37.Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietrosiuk A, et al. Molecular basis for the unique role of the AAA + chaperone ClpV in type VI protein secretion. J Biol Chem. 2011;286:30010–30021. doi: 10.1074/jbc.M111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conlan JW, et al. Differential ability of novel attenuated targeted deletion mutants of Francisella tularensis subspecies tularensis strain SCHU S4 to protect mice against aerosol challenge with virulent bacteria: Effects of host background and route of immunization. Vaccine. 2010;28:1824–1831. doi: 10.1016/j.vaccine.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen H, et al. Molecular immune responses to aerosol challenge with Francisella tularensis in mice inoculated with live vaccine candidates of varying efficacy. PLoS One. 2010;5:e13349. doi: 10.1371/journal.pone.0013349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryden P, et al. Correlates of protection following vaccination of mice with gene deletion mutants of Francisella tularensis subspecies tularensis strain, SCHU S4 that elicit varying degrees of immunity to systemic and respiratory challenge with wild-type bacteria. Mol Immunol. 2012;54:58–67. doi: 10.1016/j.molimm.2012.10.043. [DOI] [PubMed] [Google Scholar]
- 42.Golovliov I, Twine SM, Shen H, Sjöstedt A, Conlan W. A DeltaclpB mutant of Francisella tularensis subspecies holarctica strain, FSC200, is a more effective live vaccine than F. tularensis LVS in a mouse respiratory challenge model of tularemia. PLoS One. 2013;8:e78671. doi: 10.1371/journal.pone.0078671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conlan JW. Tularemia vaccines: recent developments and remaining hurdles. Future Microbiol. 2011;6:391–405. doi: 10.2217/fmb.11.22. [DOI] [PubMed] [Google Scholar]
- 44.Hornick RB, Eigelsbach HT. Aerogenic immunization of man with live Tularemia vaccine. Bacteriol Rev. 1966;30:532–538. doi: 10.1128/br.30.3.532-538.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burke DS. Immunization against tularemia: analysis of the effectiveness of live Francisella tularensis vaccine in prevention of laboratory-acquired tularemia. J Infect Dis. 1977;135:55–60. doi: 10.1093/infdis/135.1.55. [DOI] [PubMed] [Google Scholar]
- 46.Barrigan LM, et al. Infection with Francisella tularensis LVS clpB leads to an altered yet protective immune response. Infect Immun. 2013;81:2028–2042. doi: 10.1128/IAI.00207-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindgren H, Stenman L, Tärnvik A, Sjöstedt A. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect. 2005;7:467–475. doi: 10.1016/j.micinf.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 48.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjöstedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003;71:5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meibom KL, et al. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol Microbiol. 2008;67:1384–1401. doi: 10.1111/j.1365-2958.2008.06139.x. [DOI] [PubMed] [Google Scholar]
- 51.Charpentier X, et al. Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoS Pathog. 2009;5:e1000501. doi: 10.1371/journal.ppat.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bröms, J. E., Meyer, L. & Sjöstedt, A. A mutagenesis-based approach identifies amino acids in the N-terminal part of Francisella tularensis IglE that critically control Type VI system-mediated secretion. Virulence, 1–27, 10.1080/21505594.2016.1258507 (2016). [DOI] [PMC free article] [PubMed]
- 53.Alkhuder K, Meibom KL, Dubail I, Dupuis M, Charbit A. Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog. 2009;5:e1000284. doi: 10.1371/journal.ppat.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bröms JE, Meyer L, Lavander M, Larsson P, Sjöstedt A. DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity. PLoS One. 2012;7:e34639. doi: 10.1371/journal.pone.0034639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conlan JW, Chen W, Shen H, Webb A, KuoLee R. Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb Pathog. 2003;34:239–248. doi: 10.1016/S0882-4010(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 56.Salomonsson E, et al. Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect Immun. 2009;77:3424–3431. doi: 10.1128/IAI.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aschtgen MS, Zoued A, Lloubes R, Journet L, Cascales E. The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 Type VI secretion system, is inserted by YidC. Microbiology open. 2012;1:71–82. doi: 10.1002/mbo3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyer L, Bröms JE, Liu X, Rottenberg ME, Sjöstedt A. Microinjection of Francisella tularensis and Listeria monocytogenes reveals the importance of bacterial and host factors for successful replication. Infect Immun. 2015;83:3233–3242. doi: 10.1128/IAI.00416-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bröms JE, Lavander M, Meyer L, Sjöstedt A. IglG and IglI of the Francisella pathogenicity island are important virulence determinants of Francisella tularensis LVS. Infect Immun. 2011;79:3683–3696. doi: 10.1128/IAI.01344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gallagher LA, et al. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci USA. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.