Abstract
Post-translational modifications (PTMs) are chemical alterations that occur in proteins that play critical roles in various cellular functions. Lysine acetylation is an important PTM in eukaryotes, and it is catalyzed by lysine acetyltransferases (KATs). KATs transfer acetyl-coenzyme A to the internal lysine residue of substrate proteins. Arrest defective 1 (ARD1) is a member of the KAT family. Since the identification of its KAT activity 15 years ago, many studies have revealed that diverse cellular proteins are acetylated by ARD1. ARD1-mediated lysine acetylation is a key switch that regulates the enzymatic activities and biological functions of proteins and influences cell biology from development to pathology. In this review, we summarize protein lysine acetylation mediated by ARD1 and describe the biological meanings of this modification.
Subject terms: Protein folding, Protein folding
Protein modification: Effect on activity
Enzymes that modify proteins by adding an acetyl group have profound effects on metabolism and development, as well as disease. This process, known as acetylation, is carried out by KAT proteins, which are present throughout the body. Although acetyl groups are small, acetylation can change a protein’s electrical charge and shape, and even alter its function. Ji Hae Seo at Keimyung University School of Medicine in Daegu, South Korea, and co-workers reviewed the roles of KAT proteins in health and disease. They report that KAT proteins control gene expression, switch metabolic pathways on or off, and regulate development. Malfunction can lead to various disorders, including neurodegeneration and tumor growth. The researchers highlight several KAT proteins, in particular an enzyme that acetylates the amino acid lysine, that are promising targets for treatment of diseases, including cancer.
Introduction
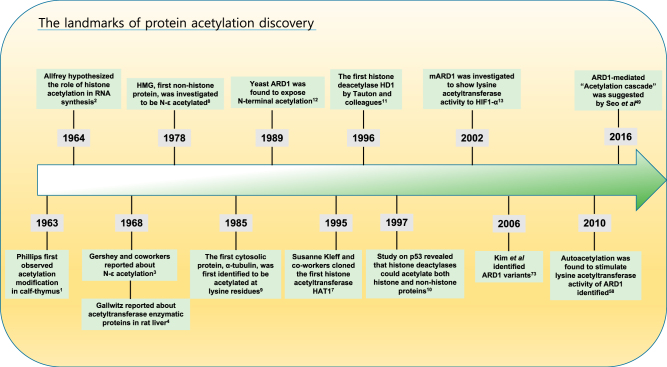
Post-translational modifications (PTMs) are an issue because they govern many biological processes during the development and disease by modifying the end products of expression. Acetylation is a major PTM in cells, along with phosphorylation, methylation, and ubiquitination. The first acetylation modification was identified in histone by Phillips in 1963 when he fractionated calf thymus nuclei to analyze histone components1. In 1964, Allfrey hypothesized that the acetylation of histone plays a critical role in the regulation of RNA synthesis2. Subsequently, many studies on histone modification, particularly histone acetylation, were performed. In 1968, in addition to the observation of Phillips that histone acetylation mainly occurred at the N-terminus of the polypeptide chain, Gershey et al. reported that histone was modified after synthesis by the attachment of acetyl groups to the ε-amino nitrogen of the lysine residues in the polypeptide chain3. In the same year, Gallwitz reported the occurrence of proteins that exhibited acetyltransferase activity in rat liver nuclei4. In the following years, many attempts to purify and characterize histone acetyltransferases (HATs) have been recorded in different organisms5,6. In 1995, Kleff et al. identified and cloned the first HAT, HAT17. A decade after the first description of histone acetylation, in 1978, the first non-histone protein, high-mobility group (HMG) was found to be acetylated at the ε-amino nitrogen of its lysine residues8. In 1985, cytosolic protein α-tubulin was first shown to be acetylated at a lysine residue9. However, it was not known that HATs could also acetylate non-histone proteins until the acetylation of p53 was identified in 199710. Thereafter, the broader term lysine acetyltransferases (KATs) was used instead of HATs. Although the deacetylation of lysine residues was believed to oppose N-ε-acetylation, and together with lysine acetylation, modulate cellular functions, the mechanism underlying deacetylation was unclear until Tauton et al. cloned and characterized the first histone deacetylase, HD1, in 199611. These groundbreaking findings have helped the study of protein acetylation for not only histone acetylation but also non-histone protein acetylation. Mullen et al. screened for yeast mutants with defective HAT activity and found that nat1, which is identical to ard1, was involved in N-terminal acetyltransferase (NAT) activity12. Thereafter, mouse ARD1 was reported to harbor KAT activity in addition to N-terminal acetyltransferase activity in a complex with NAT113. Herein, we provide an overview of protein acetylation. In particular, we focus on discussing the contributions of ARD1 to protein lysine acetylation (Fig. 1).
Fig. 1.
Timeline of breakthroughs that laid the foundation for the development of protein acetylation studies in developmental and pathological processes
Protein acetylation and deacetylation
Acetylation
Protein acetylation is a modification in which the acetyl group is transferred from the donor coenzyme A to either the Nα-termini of nascent polypeptide chains (termed N-terminal acetylation) or specific lysine residues on the polypeptide chains (termed lysine acetylation). N-terminal acetylation is a ubiquitous event in eukaryotes, as its rate of occurrence is more than 80% in humans and approximately 70% and 60% in Drosophila and yeast, respectively14,15. N-terminal acetylation has long been known to occur co-translationally and irreversibly. Nonetheless, previous studies have revealed that this modification also possibly occurs in a post-translational manner16,17.
In contrast to the irreversible reaction of N-terminal acetylation, post-translational lysine acetylation is catalyzed by the “writers” or KATs, which add acetyl groups to proteins, and is reversed by the “erasers” or lysine deacetylases (KDACs), which remove the acetyl groups from proteins18. Lysine acetylation is highly conserved across prokaryotes and eukaryotes19. The balance between lysine acetylation and deacetylation is tightly administered to ensure appropriate gene transcription, signaling machinery, and metabolism regulation20. Lysine acetylation neutralizes the positive charges that form on the amino group, and as a result, it has a significant impact on the electrostatic properties and conformation of the protein, thereby causing changes in the functions of proteins that regulate diverse biological events in the cytoplasm, nucleus, and mitochondria, such as the DNA-binding affinity of transcription factors, protein-protein interactions, and protein stability18. The acetylation of proteins at lysine residues is the signal to switch on or off signaling pathways or metabolic pathways, recognized by the “readers” that contain the recognizing domains, the so-called bromodomains21. Lysine acetylation also harmonically interplays with other modifications, such as phosphorylation, ubiquitination, and methylation, to regulate cellular events22. Therefore, malfunction of in vivo acetylation results in the improper regulation of biological processes, leading to disorders and diseases, namely, neurodegeneration, atherosclerosis, aneurysm, myocardial infarction, or tumorigenesis19.
Similar to the kinase proteins, KATs represent internal lysine acetylation and the acetylation of substrate proteins. Autoacetylation of KATs is essential for the acetyltransferase activity of KATs, as observed in the autoacetylation of ARD1, Tip60, MOF, p300, and p300/CBP-associated factor (PCAF). On the basis of sequence identity, KATs have been classified into three primary families, namely, the GNAT, MYST, and p300/CBP families (Table 1).
Table 1.
Classification and properties of lysine acetyltransferases and histone deacetylases
| KATs | |||||
|---|---|---|---|---|---|
| Class | Representatives | Autoacetylation ability | Diversity | ||
| GNAT | GCN5 | N/A | Prokaryotes and all eukaryotes | ||
| PCAF | Yes | ||||
| HAT1 | N/A | ||||
| ARD1 | Yes | ||||
| Nut1, Elp3, Hpa2/Hpa2, MCM3AP, and Eco1 | N/A | ||||
| MYST | HBO1 (MYST2 or KAT7) | N/A | Prokaryotes and all eukaryotes | ||
| MOF (MYST1 or KAT8) | Yes | ||||
| MORF (MYST4 or KAT6B) | Yes | ||||
| MOZ (MYST3 or KAT6) | N/A | ||||
| TIP60 (KAT5) | Yes | ||||
| p300/CBP | p300/CBP | Yes | Metazoan specific | ||
| Rtt109 | Yes | Yeast specific | |||
| HDACs | |||||
| Class | Members | Subcellular localization | Yeast homologous protein | Cofactor | Tissue expression |
| I | HDAC1 | Nucleus | Rpd3 | Zn2+ | Ubiquitous |
| HDAC2 | Nucleus | ||||
| HDAC3 | Nucleus/Cytoplasm | ||||
| HDAC8 | Nucleus | ||||
| IIA | HDAC4 | Nucleus/Cytoplasm | Hda1 | Zn2+ | Specific |
| HDAC5 | Nucleus/Cytoplasm | ||||
| HDAC7 | Nucleus/Cytoplasm | ||||
| HDAC9 | Nucleus/Cytoplasm | ||||
| IIB | HDAC6 | Cytoplasm | Hda1 | Zn2+ | Specific |
| HDAC10 | Nucleus | ||||
| IV | HDAC11 | Nucleus/Cytoplasm (depends on cell types and/or cellular environment) | Zn2+ | Ubiquitous | |
| SIRTUINS (Class III) | SIRT1 | Nucleus | Sir2 | NAD+ | Variable |
| SIRT2 | Cytoplasm | ||||
| SIRT3 | Mitochondria | ||||
| SIRT4 | Mitochondria | ||||
| SIRT5 | Mitochondria | ||||
| SIRT6 | Nucleus | ||||
| SIRT7 | Nucleus | ||||
The GNAT superfamily
GNAT, referred to as Gcn5-related-N-acetyltransferase, is the largest group among the KAT families. The members of the GNAT superfamily have conserved sequence motifs, especially the acetyl-CoA binding motif. The GNAT superfamily has several subfamilies. The HAT family is best studied in the midst of GNATs with GCN5 and PCAF as representatives. GCN5 was first identified in yeast as a transcriptional adaptor. Later, Tetrahymena GCN5 was reported as transcriptional-associated HAT type A23. Mammalian GCN5 and PCAF contain a HAT domain, a bromodomain, and PCAF-specific N-terminal domains24. For histone modifications, GCN5 shows enzymatic activity preference towards histone H3 at K9 and K14 and histone H4 at K8 and K1625. PCAF is one of the first acetyltransferases that was identified to have acetyltransferase activity towards non-histone proteins. A well-known substrate of PCAF is p5326. In addition to the GCN5/PCAF family, HAT1, Nut1, Elp3, Hpa2/Hpa3, MCM3AP, Eco1, and ARD1, belonging to the GNAT superfamily, exhibit acetyltransferase activity toward not only nuclear substrates but also cytosolic substrates. Because many ARD1 substrates have been investigated, the mechanistic regulation and biological functions of ARD1 will be discussed in detail in this review.
The MYST family
The acronym MYST was established on the basis of the first four members, namely: human MOZ, yeast Ybf2/Sas3, yeast Sas2, and mammalian TIP60. The MYST family of acetyltransferases has been found in all eukaryotes. In humans, five members of MYST have been identified, which are as follows: (1) MOF (MYST1 or KAT8), (2) HBO1 (MYST2 or KAT7), (3) MOZ (MYST3 or KAT6), (4) MORF (MYST4 or KAT6B), and (5) TIP60 (KAT5)24. The MYST family of proteins contains an evolutionarily conserved MYST domain composed of an acetyl-CoA-binding site and a C2HC zinc finger, except for the absence of C2HC zinc finger in yeast Esa1. Aside from the MYST domain, individual proteins in this family carry additional domains, such as the chromodomain, N-terminal part of Enok, plant homeodomain (PHD) zinc finger, and serine/methionine-rich domain, which play a unique role in the functions of each protein (reviewed in24). The MYST family has diverse functions in not only chromatin dynamics and gene regulation by acting on histone but also cell homeostasis and metabolism regulation by acting on non-chromatin substrates27.
The p300/CBP (CREB-binding protein) family
Unlike the other two acetyltransferase families, which have homologs in different evolutionary organisms, the p300/CBP family comprises p300/CBP, which is metazoan specific, and Rtt109, which is yeast specific28. Interestingly, Rtt109 displays neither sequence conservation nor functional similarity to other acetyltransferases but has a structure homologous to the p300/CBP HAT domain29. p300 and CBP have very well-preserved regions: a catalytic acetyltransferase domain, a nuclear receptor-interacting domain at the N-terminus, a bromodomain, three cysteine/histidine-rich (C/H) domains with PHD in C/H2, a KIX domain, and an interferon-binding domain at the C-terminus28.
Deacetylation
The balance of acetylation and deacetylation maintains cellular homeostasis. An imbalance of these modifications causes diseases and disorders. KDACs or so-called HDACs are different from KATs since this group of enzymes deacetylates not only acetylated histone proteins but also acetylated non-histone proteins. According to the homology of HDACs with yeast HDACs, 18 members of human HDACs are classified into two families. The conventional HDAC family, comprising 11 enzymes (HDAC1-11), is divided into four classes on the basis of their activity via a Zn2+-dependent mechanism, whereas the sirtuin family with seven enzymes (SIRT1-7) requires NAD+ as a cofactor for catalytic activity (Table 1)30.
Class I HDACs
Class I HDACs consist of HDAC1, HDAC2, HDAC3, and HDAC8, which are homologous to yeast Rpd3. This class of HDACs is predominantly localized to the nucleus and ubiquitously expressed in human cell lines and tissues31. HDAC1 and HDAC2 generally exist in multicomponent stable complexes, at least in three complexes—Sin3, NuRD, and CoREST—targeting different substrates. HDAC3 can form complexes with HDAC4, HDAC5, and HDAC7 in the presence of the coactivator SMRT or N-CoR32. The catalytic domain of HDAC8 has a nuclear localization signal, which is consistent with its main expression in the nucleus. HDAC8 can deacetylate its substrates without the presence of additional proteins33.
Class IIA HDACs
Class IIA is composed of four members: HDAC4, HDAC5, HDAC7, and HDAC9. Class IIA HDACs share sequence similarity to yeast Hda1. Similar to class I, class II HDACs share a homologous conserved catalytic domain. These proteins mainly act as transcriptional corepressors by deacetylating nucleosomal histones34. This class of deacetylases recruits class I proteins, particularly HDAC3, for repression activity. Class IIA HDACs show tissue-specific expression, indicating their differential roles in development.
Class IIB HDACs
HDAC6 and HDAC10 compose class IIB HDACs. HDAC6 and HDAC10 possess two deacetylase domains and a zinc finger domain at the C-terminus. HDAC6 mainly localizes to the cytoplasm and displays deacetylase activity towards cytoplasmic substrates, such as α-tubulin and cortactin35. The functions of HDAC10 are still unclear. Previous studies have implied that HDAC10 is predominantly localized to the nucleus in normal cells and to the cytoplasm in cancer cells36. HDAC10 potentially serves as a polyamine deacetylase37 and promotes cancer development, although its substrates are still unknown38.
Class IV HDAC
To date, HDAC11 is the only characterized enzyme of class IV HDAC. The subcellular localization of HDAC11 depends on cell types and/or the cellular environment. The expression of HDAC11 has been observed in the kidney, heart, brain, skeletal muscle, testis, and hematopoietic cells. HDAC11 has immune system-related and cell survival-related functions39,40. Similar to HDAC10, HDAC11 is poorly understood.
Sirtuin class
Class III HDACs include seven human sirtuins, which have two enzymatic activities: mono-ADP-ribosyltransferase and histone deactylase41. Sirtuins have sequence similarity to the yeast protein Sir2. SIRT1, SIRT2, and SIRT3 are considered to be true deacetylase enzymes. SIRT6 presents weak deacetylase activity and preferential ADP-ribosyltransferase activity. SIRT5 functions as a desuccinylase and demalonylase on carbamoyl phosphate synthetase and substrates other than deacetylase. SIRT7 specifically deacetylates histone H3 at lysine residue 18. Unlike other sirtuins, SIRT4 has ADP-ribosyltransferase and lipoamidase activities in addition to its deacetylase activity42. Sirtuins are expressed in various cell organelles. Each individual enzyme has a precise localization. SIRT1 is found primarily in the nucleus, but it shuttles to the cytoplasm under controlled conditions43. In contrast, SIRT2 is highly expressed in the cytosol, but it translocates to the nucleus during the G2/M transition44. SIRT6 and SIRT7 are nuclear proteins. In particular, SIRT3, SIRT4, and SIRT5 are expressed in the mitochondria, subsequently affecting metabolism regulation45.
ARD1-mediated protein lysine acetylation
ARD1 was originally identified as a NAT in yeast, and it acetylates the N-terminal amino acid of newly synthesized proteins from the ribosome12. However, mammalian ARD1 has been revealed to have KAT and NAT activities13. In the last decade, various proteins have been shown to be acetylated by ARD1 at their internal lysine residues46,47. In this review, we summarize the diverse cellular functions regulated by ARD1-mediated lysine acetylation and describe how the KAT function of ARD1 is regulated in cells.
KAT functions of ARD1
Cellular stress response
Heat shock protein (Hsp) 70 is a molecular chaperone that protects cellular proteins under stress conditions. It maintains cellular protein homeostasis by two opposing functions, protein repair and degradation48. Recently, Seo et al. found that in response to stress stimuli, such as reactive oxygen species (ROS), Hsp70 is rapidly acetylated at the K77 residue by ARD1 and then deacetylated again by HDAC449. Depending on the acetylation/deacetylation status, Hsp70 switches its function between protein repair and degradation. This switch is essential for maintaining protein homeostasis and cell survival under stress conditions. Therefore, ARD1-mediated acetylation of Hsp70 is a key regulatory mechanism that protects cells against stress conditions. On the basis of its anti-apoptotic properties, Hsp70 has been recognized as a promising target for cancer therapy; however, the development of a selective Hsp70 inhibitor is not easy because of the high similarities between members of the Hsp70 protein family50. It is worth noting that the K77 residue is specific for Hsp70 and not conserved in other members of the Hsp70 family. Indeed, the K77R mutation, which inhibits the acetylation of Hsp70, increased the susceptibility of cancer cells to anti-cancer drugs49. In addition to cancer treatment, Hsp70 acetylation also has a significant effect on neuroprotection. The K77Q mutation, which mimics the acetylation state of Hsp70, inhibited neuronal cell death due to neurotoxins in vitro and in vivo49. These results suggest that targeting ARD1-mediated Hsp70 acetylation could be helpful in the treatment of various diseases, such as cancer, neurodegeneration, and inflammatory diseases, in which Hsp70 function is dysregulated.
In addition, oxidative stress has a dual role in cell growth and death; thus, there is a question of whether ARD1 could have opposing functions in ROS-mediated cell proliferation and cell death. Indeed, Shin et al. reported that ARD1 negatively regulated the oxidative stress response51. ARD1-overexpressing cells were more susceptible to oxidative stress than control cells, and ARD1 transgenic mice showed more severe injuries in the kidney and liver under hyperoxic conditions than wild-type mice. As a molecular mechanism, Shin et al. suggested that ARD1 acetylates the K49 residue of methionine sulfoxide reductase A (MSRA), which is a thioredoxin-linked enzyme that converts methionine sulfoxide to methionine to protect amino acids from oxidative stress, resulting in the reduction of enzymatic activity of MSRA52. ARD1 could possibly have opposing roles between cell survival and cell death in response to oxidative stress, probably depending on the intensity or period of oxidative stress. These opposing functions might be mediated by acetylating different substrate proteins. While ARD1 protected cells against ROS through Hsp70 acetylation49, ARD1 worsened ROS-induced cellular damage by MSRA acetylation51. Therefore, although ARD1 plays a critical role in cellular redox balance, when we target ARD1 in oxidative stress-relative diseases, the opposing functions of ARD1 should be considered, and the downstream substrate proteins acetylated by ARD1 should also be carefully reviewed in relevant diseases.
Autophagy
Autophagy is a cellular defense system in response to cellular stress and plays a crucial role in maintaining cell homeostasis53. Recently, Qian et al. found that the KAT activity of ARD1 is needed to initiate autophagy and promote cell survival under harsh conditions54,55. Under glutamine deprivation and hypoxic conditions, ARD1 acetylates phosphoglycerate kinase 1 (PGK1) at the K388 residue, and then, acetylated PGK1 subsequently phosphorylates the S30 residue of Beclin1. Beclin1 is a core protein of the protein complex that contains vacuolar sorting protein 34 (VPS34) and class III phosphatidylinositol-3 kinase (PI3K). Phosphorylated Beclin1 enhances the activity of this protein complex by increasing the binding of phosphatidylinositol to VPS34 and thereby increases phosphatidylinositol-3-phosphate (PtdIns3P) generation, which is a required molecular event for autophagosome formation56. Therefore, ARD1-mediated PGK1 acetylation is essential for glutamine deprivation- and hypoxia-induced autophagy. PGK1 acetylation by ARD1 promotes brain tumor formation, and indeed, PGK1 K388 acetylation levels are correlated with poor prognosis in glioblastoma patients54. This report reveals the important roles of ARD1 in the regulation of cell metabolism and suggests that the K388 acetylation level of PGK1 could be useful as a prognosis marker in cancer patients.
Cell cycle
Yeast ARD1, as its name indicates, has an important role in the life cycle. Thus, human ARD1 may have similar biological functions related to cell cycle regulation. Lim et al. first investigated the function of human ARD1 in lung cancer cells and showed that the depletion of ARD1 resulted in a significant defect in cell cycle progression, especially at the G1-S phase57. Regarding the molecular mechanism, Lim et al. suggested that ARD1 acetylates β-catenin; however, the acetylated lysine site of β-catenin has not been identified yet. Later, Seo et al. also showed that ARD1-overexpressing cancer cells enhanced tumor growth in a mouse xenograft model and that the eradication of KAT activity of ARD1 significantly prevented cell proliferation in vitro and in vivo58. Wang et al. found that ARD1 is upregulated in human prostate cancer patients and promotes prostate cell proliferation in vitro and in vivo59,60. Mechanically, ARD1 acetylates the K618 residue of androgen receptor (AR). The K618R mutation, which prevents the ARD-mediated acetylation of AR, attenuated the proliferation of prostate cancer cells in vitro and in vivo. The regulation of AR signaling by ARD1 is reviewed more exhaustively in another article written by Liu’s group in this issue. Recently, Lee et al. also found that ARD1 controls cell cycle progression by regulating cellular dNTP levels61. Cellular dNTPs are the building blocks of DNA. Because balanced cellular dNTP levels are critical for proper DNA replication and repair, cellular dNTP levels are strictly controlled by two types of enzymes, dNTP synthetase and dNTPase. SAM domain and HD domain containing protein 1 (SAMHD1) is the only known dNTPase that cleaves dNTPs62. Since SAMHD1 was first identified in immune cells, where SAMHD1 inhibits retroviruses, such as human immunodeficiency virus type 1 (HIV-1), by the depletion of cellular dNTPs to block retroviral replication, most studies have focused on the anti-viral activity of SAMHD1 in immune cells63,64. However, SAMHD1 is ubiquitously expressed in human organs, suggesting that it has additional biological functions in non-immune cells. Lee et al. first reported the oncogenic characteristic of SMAHD1 in cancer cells and showed that the dNTPase activity of SAMHD1 is stimulated by ARD1-mediated acetylation61. SAMHD1 is acetylated at the K405 residue by ARD1, and acetylated SAMHD1 exhibits enhanced dNTPase activity compared with non-acetylated SAMHD1. During the cell cycle, SAMHD1 acetylation is the highest in the G1 phase, and a mutation of SAMHD1 that inhibits its acetylation induces a defect in the G1/S phase transition and inhibits cellular growth. These results suggest that regulation of the cellular dNTP pool could be useful for inhibiting cancer development and that modulation of the KAT activity of ARD1 or SMAHD1 acetylation level is a possible approach for controlling the cellular dNTP pool.
Development
Since ARD1 was first identified to have important roles in yeast growth and sporulation, many studies on ARD1 focused on its functions related to cell growth or cell cycle. However, ARD1 is expressed in many types of cells; thus, ARD1 was believed to have additional biological functions in addition to the regulation of cell proliferation. Yoon et al. investigated the role of ARD1 during the developmental stage and found a novel function of ARD1 in bone development65. During osteogenesis, ARD1 expression was induced by Runt-related transcription factor 2 (Runx2), which led to osteoblast differentiation by the transactivation of osteoblast marker genes and negative control of Runx2 activity by ARD1. ARD1 acetylates Runx2 at the K225 residue, and this acetylation interferes with the binding of Runx2 to CBFβ, inhibiting the transcriptional activity of Runx2. ARD1 transgenic neonatal mice showed delayed bone development relative to their littermates, whereas many bones of ARD1 knockout mice were denser and more extended. The osteoblast differentiation of primary osteoblasts stimulated by BMP-2 was also negatively regulated by ARD1 in vitro. Moreover, the depletion of ARD1 augmented bone regeneration in a critical-size calvarial defect rat model, suggesting a new strategy for facilitating fracture healing by modulating ARD1 activity. In addition to bone development, ARD1 knockout mice displayed severe developmental defects, including partial embryonic lethality, growth retardation, brain disorders, and maternal effect lethality, although the relation between the KAT activity of ARD1 and these defects is unclear66. In humans, a Ser37Pro mutation in ARD1, which results in an ARD1 that lacks NAT activity, is known to cause genetic diseases, but the effect of this mutation on the KAT activity of ARD1 is also unknown67. The roles of ARD1 in development and genetic disease are addressed at some length in other articles written by Oh’s group and Lyon’s group in this issue.
Cell motility
On the basis of previous studies that reported that ARD1 is closely related to the cell cycle, Vo et al. traced the cellular location of ARD1 during the cell cycle and observed that ARD1 is colocalized with Aurora kinase A (AuA) in the centrosome during cell division68. AuA is a mitotic serine/threonine kinase that functions in centrosome maturation and separation and spindle assembly during mitosis69. Vo et al. showed that ARD1 directly acetylates the K75 and K125 residues of AuA and that this event is essential for the kinase activity of AuA68. Double mutations at K75R/K125R decreased the kinase activity of AuA and caused severe defects in cell migration. Since AuA is known to be overexpressed in many cancers and is correlated with metastasis and poor prognosis, Vo et al. suggested that ARD1-mediated AuA probably contributes to cancer development. In contrast to this suggestion, Shin et al. proposed that ARD1 downregulates tumor cell migration and invasion by myosin light chain kinase (MLCK) acetylation. MLCK is a calcium/calmodulin-dependent serine/threonine kinase that phosphorylates MLC and is implicated in a variety of cellular functions, including cell contraction and migration70. Shin et al. suggested that ARD1 acetylates the K608 residue of MLCK in vitro and in vivo, reducing the kinase activity of MLCK. Even though the effect of ARD1 on cell motility is not clear, since AuA and MLCK are expressed in diverse types of human cells, such as endothelial cells, neuronal cells, and smooth muscle cells, it is possible that ARD1 regulates diverse biological events related to cell motility, such as angiogenesis, vascular permeability, neurite extension, transmitter release on synapse formation, and muscle contraction.
Regulation of the KAT activity of ARD1
Many kinases and acetyltransferases catalyze themselves to stimulate enzymatic activities in a process called autophosphorylation or autoacetylation71,72. Seo et al. found that ARD1 acetylates itself at the K136 residue, and this modification is essential for the functional activation of ARD1 as a KAT58. The K136R mutation, which inhibits the autoacetylation of ARD1, abolished the KAT activity of ARD1. A number of studies have linked ARD1 to cancers, and human ARD1 has been increasingly considered to be a therapeutic target for cancer treatment47. Notably, the inhibition of human ARD1 autoacetylation by the K136R mutation blocked cancer growth in vitro and in vivo by the downregulation of cyclin D1, suggesting modulation of the autoacetylation of ARD1 for cancer therapy58. Autoacetylation is a key switch for regulating the KAT activity of ARD1; however, the question still remains whether the autoacetylation of K136 also affects the NAT activity of ARD1. If this modification regulates only the KAT activity and not the NAT activity of ARD1, comparison studies of the cellular functions of K136R mutant ARD1 and the dominant negative mutant (DN), which lacks both NAT and KAT activities, could offer clues on how the KAT and NAT functions of ARD1 are differentially regulated in cells.
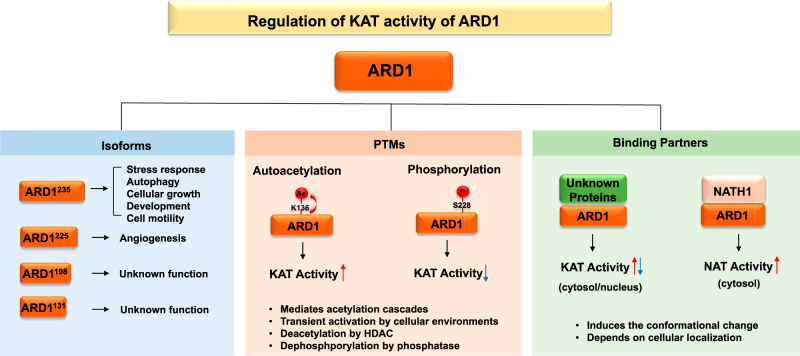
The intriguing part is that autoacetylation is conserved among diverse ARD1 variants derived from alternative RNA splicing73; however, it regulates differentially the roles of ARD1 variants in tumorigenesis. The characteristics and biological function of ARD1 variants are reviewed in other articles in this issue written by Chung’s group. Seo et al. found that among several ARD1 variants, human ARD1235, mouse ARD1235, and mouse ARD1225 recombinants undergo similar autoacetylation with the target site conserved at K136 in vitro74. While autoacetylation of human ARD1235 and mouse ARD1235 contributed to cancer growth in normoxic conditions, mouse ARD1225 autoacetylation had no effect on cellular growth. Instead, autoacetylation of mouse ARD1225 prevented tumor angiogenesis by the degradation of hypoxia-inducible factor-1α (HIF-1α) protein under hypoxic conditions. Autoacetylation stimulated the catalytic activity of mouse ARD1225 to acetylate the K532 residue of HIF-1α, leading to the proteasomal degradation of HIF-1α. The autoacetylation of ARD1 variants selectively regulates cancer growth and angiogenesis in an isoform-specific manner (Fig. 2).
Fig. 2. Regulation of the KAT activity of ARD1.
The KAT activity of ARD1 is regulated by PTMs. While autoacetylation at the K136 residue stimulates the KAT activity of ARD1, phosphorylation at the S228 residue inhibits the KAT activity of ARD1. Autoacetylation is conserved among ARD1 isoforms, including ARD1235 and ARD1225; however, autoacetylation regulates differential cellular functions depending on the isoform. While the autoacetylation of ARD1235 enhances cellular growth, the autoacetylation of ARD1225 inhibits angiogenesis. The KAT activity of ARD1 might also be regulated by unknown binding proteins
Whereas autoacetylation stimulates the KAT activity of ARD1, phosphorylation inhibits the KAT activity of ARD1. Qian et al. found that mTOR directly phosphorylates the S228 residue of ARD1 in vitro, and this phosphorylation inhibits the KAT activity of ARD1, which is especially required for PGK1 acetylation54. Under glutamine deprivation or hypoxic conditions, mTOR-induced ARD1 phosphorylation was downregulated, and ARD1 was activated and consequently acetylated PGK1, leading to autophagy promotion. Therefore, the KAT activity of ARD1 may be balanced by various post-translational modifications, including acetylation and phosphorylation, which are stimulated by certain cellular stimuli (Fig. 2). Further studies are needed to elucidate how these modifications coordinate with each other to regulate the KAT activity of ARD1.
Regarding the KAT activity of ARD1, we should not overlook the fact that the KAT activity of ARD1 requires delicate handling; it seems to be influenced by various experimental conditions. Indeed, previous reports presented conflicting data about the KAT activity of ARD1. Murray-Rust et al. observed that recombinant ARD1 underwent self-mediated N-terminal acetylation in vitro, suggesting that ARD1 has NAT activity rather than KAT activity75. Magin et al. also argued for the KAT activity of ARD1 because they observed no difference in the lysine acetylation of substrate proteins, such as MSRA, MLCK, and RUNX2, with or without recombinant ARD1 in vitro, suggesting that the substrates may be acetylated chemically76. However, Sanchez-Puig et al. characterized the fibrillar conformation of recombinant ARD177. Size exclusion chromatography and electron micrograph revealed the presence of soluble aggregates of recombinant ARD1. Consistent with this finding, Seo et al. found that the KAT activity of recombinant ARD1 easily disappeared and that only freshly prepared recombinant ARD1 showed KAT activity, suggesting that recombinant ARD1 formed aggregates over time58. Therefore, the labile catalytic activity of recombinant ARD1 could be one explanation for the conflicting results on the KAT activity of recombinant ARD1. Since autoacetylation is essential step to stimulate the KAT activity of ARD1, the presence of KAT activity could be determined by the autoacetylation status of recombinant ARD1.
In cells, the basal level of KAT activity of ARD1 might be not sufficient for detection because the KAT activity of ARD1 was upregulated briefly by a specific cellular stimulus. Qian et al. found that the KAT activity of ARD1 required to acetylate the lysine residue of PGK1 was usually inhibited under normal conditions and that this activity was stimulated only after glutamine deprivation or hypoxia stimulation54. Similarly, Seo et al. showed that the autoacetylation of ARD1 rapidly increased with hydrogen peroxide or 1-methyl-4-phenylpyridinium (MPP+) treatment and that it disappeared soon after49. Therefore, the KAT activity of ARD1 seems to be transiently upregulated through autoacetylation in response to certain cellular stimuli. Furthermore, the autoacetylation of ARD1 stimulated Hsp70 acetylation, which was followed by the HDAC4-mediated deacetylation of Hsp7049. These results propose the existence of an “acetylation cascade” in which proteins are sequentially activated by acetylation to amplify the original signal and cross-talk with different signals, similar to the phosphorylation cascade (Fig. 2). Like the phosphorylation and dephosphorylation induced by kinases and phosphatases, acetylation cascades could be regulated by reversible acetylation and deacetylation reactions mediated by KATs and HDACs, respectively. As most PTMs are transient, ARD1-mediated-protein acetylation is probably maintained briefly. Therefore, to detect the lysine acetylation of ARD1 substrate protein or discover the acetylation cascades regulated by ARD1 in cells, the specific physiological conditions that transiently stimulate the KAT activity or autoacetylation of ARD1 should be determined first. The labile and transient characteristics of the KAT activity of ARD1 in vitro and in vivo should be treated with caution when investigating the biological functions of ARD1 as a KAT.
As the NAT function of ARD1 is regulated by binding with NATH, the KAT functions of ARD1 could be regulated by binding with diverse proteins (Fig. 2). Unlike its NAT activity, the KAT activity of ARD1 does not seem to need NATH binding since recombinant ARD1 acetylates the internal lysine residues of its substrate proteins in vitro without NATH. In addition, NATH protein is located in only the cytosol, whereas ARD1 is localized in both the nucleus and cytosol, suggesting independent roles of ARD1 that do not involve NATH. Beyond NATH, little is known about the binding proteins of ARD1 so far; thus, the identification of unknown binding partners of ARD1 that regulate KAT activity might be challenging. For example, proteome analysis under various cellular conditions that stimulate the KAT activity of ARD1 could be helpful for identifying critical molecules for the KAT functions of ARD1.
ARD1 and human diseases
The expression of ARD1 is universal across cells. Furthermore, ARD1 displays distinct activity dependent on its sole activation or its activation as a subunit of NatA or its subcellular localization. Numerous substrates of ARD1 have been identified. Hence, ARD1 is a critical player in pathological progression.
Cancer
Many studies have demonstrated the association between the overexpression of ARD1 and cancer progression and poor prognosis. A previous study on ARD1 expression in 19 common types of cancers in human tissues revealed that ARD1 is highly expressed in cancer tissues: adenocarcinoma, squamous cancer and especially breast cancer, bladder cancer, stomach carcinoma, cervical carcinoma, hypothyroid carcinoma, lung cancer, and intestinal cancer. Furthermore, there is a significant difference in ARD1 expression between cancer tissues and adjacent tissues in the same tissue section78. ARD1 has also been found to be overexpressed in colorectal cancer79, hepatocellular carcinoma80, lung cancer81, and prostate cancer59. In a previous study, targeting of ARD1 by microRNA repressed the tumorigenesis of colon cancer in vitro, suggesting that ARD1 acts as an oncogene in colon cancer82. The KAT activity of ARD1 is probably the mechanism underlying the contribution of ARD1 to cancer progression. Indeed, ARD1 has been reported to promote cell proliferation by regulating cyclin D1. The first study that reported the link between ARD1 and cyclin D1 used lung cancer cells. This study showed that ARD1 acetylated β-catenin and activated the binding of β-catenin/TCF4 to the cyclin D1 promoter. The depletion of ARD1 by siRNA in H1299 and A549 cells caused the transcriptional repression of cyclin D1, leading to the arrest of G1 and inhibition of cell proliferation57. ARD1 also exhibits autoacetylation at the K136 residue, resulting in the activation of AP-1, which is a transcriptional regulator of cyclin D1 during tumorigenesis, thereby increasing cyclin D1 expression. Consequently, A549 cell proliferation is promoted and tumorigenesis is enhanced in a xenograft model with H460 lung cancer cells58. Similarly, the overexpression of ARD1 in lung cancer cells showed the positive regulation of ARD1 in DNMT1 enzymatic activity, mediating E-cadherin silencing and thus inducing cell proliferation and xenograft tumor formation81. The androgen receptor (AR) has been reported to be a substrate of ARD1. AR acetylation by ARD1 generates AR-mediated gene transcription involved in tumor formation, promoting prostate tumorigenesis59. In addition, a previous study showed that the acetylation of SAMHD1 by ARD1 intensifies the dNTPase activity of SAMHD1, which promotes cell cycle progression in cancer cells and increases the expression level of cyclin D1 and cyclin B1 and promotes cell proliferation61. In addition, ARD1 also participates in governing cell cycle progression by regulating AuA acetylation. AuA, which overrides mitosis in cancer cells, is acetylated by ARD1; its activity is enhanced, and the G2/M transition is accelerated, contributing to cell proliferation. Moreover, the acetylation of AuA stimulates cell mobility, which is a crucial trait of metastasis in cancer development, by activating the p38/AKT pathway68.
ARD1 also contributes to tumorigenesis by inhibiting cell death. Silencing of ARD1 causes the cell to become susceptible to daunorubicin, triggering apoptosis83. Park et al. found that the interaction of ARD1 and RIP1 via the acetyltransferase domain is involved in doxorubicin-induced NF-κB activation, preventing cell death84. In addition, Xu et al. elucidated that the depletion of ARD1 sensitizes cancer cells to stress-inducing agents, including oxidative stress-inducer H2O2, mitotic stress-inducer Taxol, and DNA-damage inducer 5-fluorouracil, because of the requirement of ARD1 to interact with RelA/p56 and activate MCL1 transcription for the prevention of apoptosis85. Another function of ARD1 is protecting cells from death via the acetylation of Hsp70. The acetylation of Hsp70 by ARD1 stimulates the formation of Hsp70/Apaf-1 and Hsp70/AIF complexes, consequently preventing caspase-dependent and caspase-independent apoptosis, respectively. ARD1-mediated Hsp70 acetylation also attenuates autophagic cell death, which is linked to programmed cell death86. However, autophagy plays binary roles in cancer. On one hand, autophagy functions as a pro-death mechanism under stress conditions, as it can trigger apoptosis via several proteins with dual roles in autophagy and apoptosis, such as Beclin1, Bcl-2, Atg5, and p5387,88. On the other hand, autophagy can enable tumor cell survival under metabolic stress88,89. Recently, ARD1 was found to regulate PGK1-phosphorylated Beclin1 by acetylating PGK1, which in turn stimulates autophagy activation under glutamine deprivation and promotes brain tumorigenesis54. ARD1 has been demonstrated as a tumor suppressor in breast cancer, and the occurrence of ARD1 is correlated with a better clinical outcome90,91; this finding is possibly associated with the NAT activity of ARD1 rather than its KAT activity90. Moreover, ARD1 is required for caspase activation under stress conditions92. Thus, ARD1 acts as an oncogene or tumor suppressor because of its localization, stress conditions, and tissue specificity.
Neuronal diseases
Many studies on the functions of ARD1 in neuronal regulation have been performed. The overexpression of mutant ARD1 and silencing of ARD1 limit the dendritic development of cerebellar Purkinje cells, which means that ARD1 is crucial for neuronal dendritic development93. Amyloid β-protein (Aβ) is a product of amyloid precursor protein (APP) proteolysis, and the accumulation of Aβ in the brain is a well-known cause of Alzheimer’s disease (AD)94. ARD1 in a complex with NAT1 has been investigated to suppress APP endocytosis correlated with the inhibition of Aβ secretion, suggesting the protective role of ARD1 in AD95. However, the mechanism underlying Aβ suppression is still unclear. Further studies are required to clarify how the ARD1/NAT1 complex suppresses Aβ production. Huntington’s disease (HD) is another progressive neurodegenerative disorder. A hallmark of HD is the aggregation of huntingtin (HTT) and formation of cytoplasmic and nuclear inclusions in the brain96. Arnesen et al. proposed that ARD1 in a complex with NatA interacts with HTT interacting protein (HYPK), which displays chaperon-like activity at the ribosome, and prevents HTT aggregation. Knockdown of either HYPK or ARD1 increases the aggregation of HTT97. Thus, ARD1 as a subunit of NatA could possibly contribute to preventing the progression of HD, although the underlying mechanism has not yet been verified.
Genetic diseases
The link between genetic diseases and protein acetylation was unknown until Rope et al. showed the association of an X-linked disorder with NAT in 2011. Lethality in male infants with severe global developmental delays, craniofacial anomalies, hypotonia, and cardiac arrhythmia is a distinct feature of this X-linked syndrome, which is termed Ogden syndrome. The cause could be the mutation c.109 T > C in ARD1, accounting for p.Ser37Pro, which results in a reduction of ARD1/NatA enzymatic activity67. This assumption was confirmed using a Saccharomyces cerevisiae model20. Later, it was revealed that the ARD1 S37P mutant shortens helix α2 and changes ARD1 flexibility, negatively affecting NatA complex activity and thereby impairing the acetylation of ARD1/NatA substrates, including THOC798. Recently, Ogden syndrome was found in a female patient99.
In addition, Lenz microphthalmia syndrome, characterized by microphthalmia or anophthalmia, developmental delay, intellectual disability, skeletal abnormalities, and malformations of the teeth, fingers, and toes, is attributed to ARD1 mutation-induced retinoic acid pathway dysregulation. The splice donor mutation c.471 + 2 T > A generates ARD1 aggregation in the cytoplasm and the downregulation of retinol uptake100.
ARD1 also plays a role in different developmental delay phenotypes because of its mutated location. The mutations at c.319 G > T (p.Val107Phe) and c.346 C > T (p.Arg116Trp) in ARD1 cause severe non-syndromic developmental delay101. The missense variant c.128 A > C (p.Tyr43Ser) of ARD1 at the X chromosome induces developmental delay with long QT in patients102. A clinical study has shown that X-link recessive mutations p.Arg83Cys and p.Phe128Leu of ARD1 are correlated with ARD1 acetyltransferase activity103. It has been noted that a reduction in NAT activity in ARD1 mutants seems to produce more symptoms during developmental delay101.
Conclusion
In the past 50 years, our understanding of protein acetylation has increased greatly since the study by Phillips. It is now widely known that protein acetylation plays a pivotal role in regulating gene transcription by modulating histone acetylation and physiological and metabolic processes by mediating the acetylation of either chromatin or non-chromatin proteins in various cell compartments from the nucleus to the membrane. ARD1, as a member of the KAT proteins, displays its vital roles in cellular regulation via its enzymatic activity towards its substrates. The precise subcellular localization of ARD1 is also a considerable issue. ARD1 and activation protein acetylation are administered by multiple delicate modifications. Overexpression, dysregulation or depletion of ARD1 functionally impacts cellular homeostasis, which is the cause of oncogenesis and neurodegeneration. Further studies are required to understand by which catalytic mechanism ARD1 acetylates its partners and how the conformation of ARD1 changes when it is regulated by other modifications, such as autoacetylation and phosphorylation, since the structure of human ARD1 has not yet been validated. However, the crystal structures of its homologs in lower evolutional species, such as Thermoplasma volcanium104, Schizosaccharomyces pombe105, Sulfolobus solfataricus106, and Chaetomium thermophilum107, have been investigated. The implication of ARD1 in tumorigenesis has been progressively studied, showing that ARD1 is a promising target for anti-tumor therapeutic approaches. The development of an ARD1 inhibitor is still required. Achievements in generating chemicals that target ARD1 would provide information on ARD1 and ARD1-related disease treatment.
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1A6A1A03011325) and was also supported by Global Core Research Center (GCRC) Program and National Research Foundation of Korea (NRF) grants funded by the Korean Government (MSIP) (2011-0030001 and 2018R1A2B6001590).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims inpublished maps and institutional affiliations.
References
- 1.PHILLIPS D. The presence of acetyl groups in histones. Biochem. J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proc. . Natl Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gershey EL, Vidali G, Allfrey VG. Chemical studies of histone acetylation: the occurrence of ε-N-acetyllysine in the f2a1 histone. J. Biol. Chem. 1968;243:5018–5022. [PubMed] [Google Scholar]
- 4.Gallwitz D. Acetylation of histones by a kinase from rat liver nuclei. Biochem. Biophys. Res. Commun. 1968;32:117–121. doi: 10.1016/0006-291x(68)90355-0. [DOI] [PubMed] [Google Scholar]
- 5.Gallwitz D, Sures I. Histone acetylation. Purification and properties of three histone-specific acetyltransferases from rat thymus nuclei. Biochim. Biophys. Acta. 1972;263:315–328. doi: 10.1016/0005-2795(72)90085-2. [DOI] [PubMed] [Google Scholar]
- 6.Kelner DN, McCarty KS. Porcine liver nuclear histone acetyltransferase. Partial purification and basic properties. J. Biol. Chem. 1984;259:3413–3419. [PubMed] [Google Scholar]
- 7.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone h4 acetyltransferase. J. Biol. Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 8.Sterner R, Vidali G, Heinrikson RL, Allfrey VG. Postsynthetic modification of high mobility group proteins. Evidence that high mobility group proteins are acetylated. J. Biol. Chem. 1978;253:7601–7604. [PubMed] [Google Scholar]
- 9.L’Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 10.Imhof A, et al. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 11.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 12.Mullen JR, et al. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong JW, et al. Regulation and destabilization of HIF-1α by ARD1-mediated acetylation. Cell. 2002;111:709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 14.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. . Natl Acad. Sci. USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetze S, et al. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 2009;7:e1000236. doi: 10.1371/journal.pbio.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helsens K, et al. Bioinformatics analysis of a Saccharomyces cerevisiae N-terminal proteome provides evidence of alternative translation initiation and post-translational N-terminal acetylation. J. Proteome Res. 2011;10:3578–3589. doi: 10.1021/pr2002325. [DOI] [PubMed] [Google Scholar]
- 17.Van Damme P, et al. Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of n(α)-acetyltransferases and point to hNaa10p as the post-translational actin N(α)-acetyltransferase. Mol. Cell. Proteom. 2011;10:M110.004580. doi: 10.1074/mcp.M110.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim. Biophys. Acta. 2016;1864:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15:536. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 21.Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other post-translational modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownell JE, et al. Tetrahymena histone acetyltransferase a: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 24.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo MH, et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 26.Sakaguchi K, et al. DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sapountzi V, Côté J. MYST-family histone acetyltransferases: beyond chromatin. Cell. Mol. Life Sci. 2011;68:1147–1156. doi: 10.1007/s00018-010-0599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015;115:2419–2452. doi: 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Y, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat. Struct. Mol. Biol. 2008;15:738. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Ruijter AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10:32. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakrabarti A, et al. HDAC8: a multifaceted target for therapeutic interventions. Trends Pharmacol. Sci. 2015;36:481–492. doi: 10.1016/j.tips.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 34.Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]
- 35.Ran J, Yang Y, Li D, Liu M, Zhou J. Deacetylation of α-tubulin and cortactin is required for HDAC6 to trigger ciliary disassembly. Sci. Rep. 2015;5:12917. doi: 10.1038/srep12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, et al. HDAC10 promotes lung cancer proliferation via AKT phosphorylation. Oncotarget. 2016;7:59388–59401. doi: 10.18632/oncotarget.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hai Y, Shinsky SA, Porter NJ, Christianson DW. Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nat. Commun. 2017;8:15368. doi: 10.1038/ncomms15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Peng L, Seto E. Histone deacetylase 10 regulates the cell cycle G(2)/M phase transition via a novel let-7–HMGA2–cyclin A2 pathway. Mol. Cell Biol. 2015;35:3547–3565. doi: 10.1128/MCB.00400-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanginlar C, Logie C. HDAC11 is a regulator of diverse immune functions. Biochim. Biophys. Acta. 2018;1861:54–59. doi: 10.1016/j.bbagrm.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Deubzer HE, et al. HDAC11 is a novel drug target in carcinomas. Int. J. Cancer. 2013;132:2200–2208. doi: 10.1002/ijc.27876. [DOI] [PubMed] [Google Scholar]
- 41.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiorino E, et al. The sirtuin class of histone deacetylases: regulation and roles in lipid metabolism. IUBMB Life. 2014;66:89–99. doi: 10.1002/iub.1246. [DOI] [PubMed] [Google Scholar]
- 43.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD + -dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 44.Vaquero A, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Gene Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorfel MJ, Lyon GJ. The biological functions of Naa10 - From amino-terminal acetylation to human disease. Gene. 2015;567:103–131. doi: 10.1016/j.gene.2015.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalvik TV, Arnesen T. Protein N-terminal acetyltransferases in cancer. Oncogene. 2013;32:269–276. doi: 10.1038/onc.2012.82. [DOI] [PubMed] [Google Scholar]
- 48.Hohfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2:885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seo JH, et al. ARD1-mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nat. Commun. 2016;7:12882. doi: 10.1038/ncomms12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin SH, et al. Arrest defective 1 regulates the oxidative stress response in human cells and mice by acetylating methionine sulfoxide reductase A. Cell Death Dis. 2014;5:e1490. doi: 10.1038/cddis.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moskovitz J. Roles of methionine suldfoxide reductases in antioxidant defense, protein regulation and survival. Curr. Pharm. Des. 2005;11:1451–1457. doi: 10.2174/1381612053507846. [DOI] [PubMed] [Google Scholar]
- 53.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 54.Qian X, et al. Phosphoglycerate kinase 1 phosphorylates beclin1 to induce autophagy. Mol. Cell. 2017;65:917–931. doi: 10.1016/j.molcel.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qian X, Li X, Lu Z. Protein kinase activity of the glycolytic enzyme PGK1 regulates autophagy to promote tumorigenesis. Autophagy. 2017;13:1246–1247. doi: 10.1080/15548627.2017.1313945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trend Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim JH, Park JW, Chun YS. Human arrest defective 1 acetylates and activates β-catenin, promoting lung cancer cell proliferation. Cancer Res. 2006;66:10677–10682. doi: 10.1158/0008-5472.CAN-06-3171. [DOI] [PubMed] [Google Scholar]
- 58.Seo JH, et al. Arrest defective 1 autoacetylation is a critical step in its ability to stimulate cancer cell proliferation. Cancer Res. 2010;70:4422–4432. doi: 10.1158/0008-5472.CAN-09-3258. [DOI] [PubMed] [Google Scholar]
- 59.Wang Z, et al. Inactivation of androgen-induced regulator ARD1 inhibits androgen receptor acetylation and prostate tumorigenesis. Proc. . Natl Acad. Sci. USA. 2012;109:3053–3058. doi: 10.1073/pnas.1113356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DePaolo JS, et al. Acetylation of androgen receptor by ARD1 promotes dissociation from HSP90 complex and prostate tumorigenesis. Oncotarget. 2016;7:71417–71428. doi: 10.18632/oncotarget.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee EJ, et al. SAMHD1 acetylation enhances its deoxynucleotide triphosphohydrolase activity and promotes cancer cell proliferation. Oncotarget. 2017;8:68517–68529. doi: 10.18632/oncotarget.19704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 63.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoon H, et al. NAA10 controls osteoblast differentiation and bone formation as a feedback regulator of Runx2. Nat. Commun. 2014;5:5176. doi: 10.1038/ncomms6176. [DOI] [PubMed] [Google Scholar]
- 66.Lee CC, et al. The Role of N-alpha-acetyltransferase 10 protein in DNA methylation and genomic imprinting. Mol. Cell. 2017;68:89–103 e7. doi: 10.1016/j.molcel.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rope AF, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vo TTL, et al. ARD1-mediated aurora kinase A acetylation promotes cell proliferation and migration. Oncotarget. 2017;8:57216–57230. doi: 10.18632/oncotarget.19332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 70.Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005;96:379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beenstock J, Mooshayef N, Engelberg D. How do protein kinases take a selfie (autophosphorylate)? Trend Biochem. Sci. 2016;41:938–953. doi: 10.1016/j.tibs.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 72.McCullough CE, Marmorstein R. Molecular basis for histone acetyltransferase regulation by binding partners, associated domains, and autoacetylation. ACS Chem. Biol. 2016;11:632–642. doi: 10.1021/acschembio.5b00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim SH, et al. Characterization of ARD1 variants in mammalian cells. Biochem. Biophys. Res. Commun. 2006;340:422–427. doi: 10.1016/j.bbrc.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 74.Seo JH, et al. Autoacetylation regulates differentially the roles of ARD1 variants in tumorigenesis. Int. J. Oncol. 2015;46:99–106. doi: 10.3892/ijo.2014.2708. [DOI] [PubMed] [Google Scholar]
- 75.Murray-Rust TA, Oldham NJ, Hewitson KS, Schofield CJ. Purified recombinant hARD1 does not catalyse acetylation of Lys532 of HIF-1alpha fragments in vitro. FEBS Lett. 2006;580:1911–1918. doi: 10.1016/j.febslet.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 76.Magin RS, March ZM, Marmorstein R. The N-terminal acetyltransferase Naa10/ARD1 does not acetylate lysine residues. J. Biol. Chem. 2016;291:5270–5277. doi: 10.1074/jbc.M115.709428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanchez-Puig N, Fersht AR. Characterization of the native and fibrillar conformation of the human N-alpha-acetyltransferase ARD1. Protein Sci. 2006;15:1968–1976. doi: 10.1110/ps.062264006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu M, et al. Immunohistochemical analysis of human arrest-defective-1 expressed in cancers in vivo. Oncol. Rep. 2009;21:909–915. doi: 10.3892/or_00000303. [DOI] [PubMed] [Google Scholar]
- 79.Jiang B, et al. Peptide mimic isolated by autoantibody reveals human arrest defective 1 overexpression is associated with poor prognosis for colon cancer patients. Am. J. Pathol. 2010;177:1095–1103. doi: 10.2353/ajpath.2010.091178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shim JH, et al. Clinical Implications of Arrest-Defective Protein 1 Expression in Hepatocellular Carcinoma: A Novel Predictor of Microvascular Invasion. Dig. Dis. 2012;30:603–608. doi: 10.1159/000343090. [DOI] [PubMed] [Google Scholar]
- 81.Lee CF, et al. hNaa10p contributes to tumorigenesis by facilitating DNMT1-mediated tumor suppressor gene silencing. J. Clin. Invest. 2010;120:2920–2930. doi: 10.1172/JCI42275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang H, et al. microRNA-342-5p and miR-608 inhibit colon cancer tumorigenesis by targeting NAA10. Oncotarget. 2016;7:2709–2720. doi: 10.18632/oncotarget.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arnesen T, et al. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-α-acetyltransferase complex. Oncogene. 2006;25:4350. doi: 10.1038/sj.onc.1209469. [DOI] [PubMed] [Google Scholar]
- 84.Park J, Kanayama A, Yamamoto K, Miyamoto Y. ARD1 binding to RIP1 mediates doxorubicin-induced NF-κB activation. Biochem. Biophys. Res. Commun. 2012;422:291–297. doi: 10.1016/j.bbrc.2012.04.150. [DOI] [PubMed] [Google Scholar]
- 85.Xu H, et al. N -α-Acetyltransferase 10 protein inhibits apoptosis through RelA/p65-regulated MCL1 expression. Carcinogenesis. 2012;33:1193–1202. doi: 10.1093/carcin/bgs144. [DOI] [PubMed] [Google Scholar]
- 86.Park YH, Seo JH, Park JH, Lee HS, Kim KW. Hsp70 acetylation prevents caspase-dependent/independent apoptosis and autophagic cell death in cancer cells. Int. J. Oncol. 2017;51:573–578. doi: 10.3892/ijo.2017.4039. [DOI] [PubMed] [Google Scholar]
- 87.Gump JM, Thorburn A. Autophagy and apoptosis- what’s the connection? Trend Cell Biol. 2011;21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choi KS. Autophagy and cancer. Exp. Mol. Med. 2012;44:109. doi: 10.3858/emm.2012.44.2.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuo HP, et al. ARD1 stabilization of TSC2 suppresses tumorigenesis through the mTOR signaling pathway. Sci. Signal. 2010;3:ra9–ra9. doi: 10.1126/scisignal.2000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zeng Y, et al. Inhibition of STAT5a by Naa10p contributes to decreased breast cancer metastasis. Carcinogenesis. 2014;35:2244–2253. doi: 10.1093/carcin/bgu132. [DOI] [PubMed] [Google Scholar]
- 92.Yi CH, et al. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J. Cell Biol. 2007;179:619–626. doi: 10.1083/jcb.200708090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohkawa N, et al. N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes Cell. 2008;13:1171–1183. doi: 10.1111/j.1365-2443.2008.01235.x. [DOI] [PubMed] [Google Scholar]
- 94.O’Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Asaumi M, et al. Interaction of N-terminal acetyltransferase with the cytoplasmic domain of β-amyloid precursor protein and its effect on Aβ secretion. J. Biochem. 2005;137:147–155. doi: 10.1093/jb/mvi014. [DOI] [PubMed] [Google Scholar]
- 96.Labbadia J, Morimoto RI. Huntington’s disease: underlying molecular mechanisms and emerging concepts. Trend Biochem. Sci. 2013;38:378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arnesen T, et al. The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of huntingtin aggregation. Mol. Cell. Biol. 2010;30:1898–1909. doi: 10.1128/MCB.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Myklebust LM, et al. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Hum. Mol. Genet. 2015;24:1956–1976. doi: 10.1093/hmg/ddu611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sidhu M, Brady L, Tarnopolsky M, Ronen GM. Clinical manifestations associated with the N-terminal-acetyltransferase NAA10 gene mutation in a girl: Ogden syndrome. Pediatr. Neurol. 2017;76:82–85. doi: 10.1016/j.pediatrneurol.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 100.Esmailpour T, et al. A splice donor mutation in NAA10 results in the dysregulation of the retinoic acid signaling pathway and causes Lenz microphthalmia syndrome. J. Med. Genet. 2014;51:185–196. doi: 10.1136/jmedgenet-2013-101660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Popp B, et al. De novo missense mutations in the NAA10 gene cause severe non-syndromic developmental delay in males and females. Eur. J. Hum. Genet. 2015;23:602–609. doi: 10.1038/ejhg.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Casey JP, et al. NAA10 mutation causing a novel intellectual disability syndrome with Long QT due to N-terminal acetyltransferase impairment. Sci. Rep. 2015;5:16022. doi: 10.1038/srep16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saunier C, et al. Expanding the phenotype associated with NAA10‐related N‐terminal acetylation deficiency. Hum. Mutat. 2016;37:755–764. doi: 10.1002/humu.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ma C, et al. Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A. Biochim. Biophys. Acta. 2014;1844:1790–1797. doi: 10.1016/j.bbapap.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 105.Liszczak G, et al. Molecular basis for amino-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 2013;20:1098–1105. doi: 10.1038/nsmb.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chang YY, Hsu CH. Multiple conformations of the loop region confers heat-resistance on SsArd1, a thermophilic NatA. ChemBioChem. 2016;17:214–217. doi: 10.1002/cbic.201500568. [DOI] [PubMed] [Google Scholar]
- 107.Weyer FA, et al. Structural basis of HypK regulating N-terminal acetylation by the NatA complex. Nat. Commun. 2017;8:15726. doi: 10.1038/ncomms15726. [DOI] [PMC free article] [PubMed] [Google Scholar]