Abstract
Background
The β-lactam antibiotics have traditionally been the main treatment of Enterobacteriaceae infections, nonetheless, the emergence of species producing β- Lactamases has rendered this class of antibiotics largely ineffective. There are no published data on etiology of urinary tract infections (UTI) and antimicrobial resistance profile of uropathogens among children in Qatar. The aim of this study is to determine the phenotypic and genotypic profiles of antimicrobial resistant Enterobacteriaceae among children with UTI in Qatar.
Methods
Bacteria were isolated from 727 urine positive cultures, collected from children with UTI between February and June 2017 at the Pediatric Emergency Center, Doha, Qatar. Isolated bacteria were tested for antibiotic susceptibility against sixteen clinically relevant antibiotics using phoenix and Double Disc Synergy Test (DDST) for confirmation of extended-spectrum beta-lactamase (ESBL) production. Existence of genes encoding ESBL production were identified using polymerase chain reaction (PCR). Statistical analysis was done using non-parametric Kappa statistics, Pearson chi-square test and Jacquard’s coefficient.
Results
201 (31.7%) of samples were confirmed as Extended Spectrum β -Lactamases (ESBL) Producing Enterobacteriaceae. The most dominant pathogen was E. coli 166 (83%) followed by K. pneumoniae 22 (11%). Resistance was mostly encoded by bla CTX-M (59%) genes, primarily bla CTX-MG1 (89.2%) followed by bla CTX-MG9 (7.7%). 37% of isolated bacteria were harboring multiple bla genes (2 genes or more). E. coli isolates were categorized into 11 clusters, while K. pneoumoniae were grouped into five clonal clusters according to the presence and absence of seven genes namely bla TEM, bla SHV, bla CTX-MG1, bla CTX-MG2, bla CTX-MG8 bla CTX-MG9, bla CTX-MG25.
Conclusions
Our data indicates an escalated problem of ESBL in pediatrics with UTI, which mandates implementation of regulatory programs to reduce the spread of ESBL producing Enterobacteriaceae in the community. The use of cephalosporins, aminoglycosides (gentamicin) and trimethoprim/sulfamethoxazole is compromised in Qatar among pediatric population with UTI, leaving carbapenems and amikacin as the therapeutic option for severe infections caused by ESBL producers.
Keywords: Urinary tract infection (UTI), Enterobacteriaceae, Antibiotic resistance, Children, ESBL, Qatar
Background
Enterobacteriaceae carrying extended-spectrum β-lactamases (ESBLs) is a global concern that demands global attention due to the limited available treatment options [1–4]. Over the past two decades, there has been an exponential increase in β-lactamase resistance worldwide accompanied with a significant escalation in the prevalence of ESBL-producing Enterobacteriaceae [5]. Unfortunately, ESBL-producing bacteria in children have come to the forefront of emerging antibiotic-resistant bacteria worldwide [6]. β-lactamases are divided into four functional groups: penicillinases, ESBLs, carbapenemases, and AmpC-type cephalosporinases [7]. Specifically, ESBLs is a group of plasmid-encoded enzymes that confer resistance to third generation cephalosporins [8–10]. ESBLs are divided into three groups according the encoding of TEM, SHV and CTX-M genes [11, 12]. CTX-M enzymes are the most common and are further classified into five major phylogenetic groups based on gene sequences namely, CTX-M -1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 [11]. ESBL-producing Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) are the predominant organisms in childhood infections, and they pose significant threat to human health [13]. These organisms are listed among the pathogens for which there are few potentially effective drugs [1]. About 57% of bloodstream infections are caused by ESBL-producing Enterobacteriaceae, which are more likely to result in death compared to the infections caused by a non ESBL-producing strains [14].
Urinary Tract Infections (UTIs) continue to be one of the most common cause of illness in young children worldwide [9]. It distresses the child, concern the parent, and may cause permanent renal sequelae. The β-lactam antibiotics have traditionally been the main antibiotics for treatment of infections caused by Enterobacteriaceae, but the emergence of ESBL has rendered this class of antibiotics largely ineffective. It is therefore important to run epidemiological studies to define the epidemiology and profiles of these resistant bacteria for its impact on the development and implementation of stewardship programs.
High prevalence of extended-spectrum-β-lactamase (ESBL) and carbapenemase producing gram negative bacteria (GNB) has been reported in the Arabian Peninsula [15]. Nonetheless, little is known about the prevalence and profile of these bacteria in pediatric population [16] in the region. Prevalence of ESBL in urine among pediatric population has been gradually increasing in Qatar (HMC annual antibiogram). For example, the percentage of E.coli ESBL producers have gradually increased from 18% in 2010 to 24% in 2014, and reached 31.7% in 2017 (January–June) as reported in this study.
This is the first study that describes at the molecular level the genotypic profile of ESBL producing bacteria among children with UTIs in Qatar. Our data indicates rapid increase in ESBL resistance among Enterobacteriaceae in pediatrics with UTI, which mandates rapid regulatory and monitoring reforms at the State level.
Methods
Clinical isolates and controls strains
Ethical approval for this study was obtained from the Medical Research Centre (MRC), Hamad Medical Corporation (HMC), Doha, Qatar, protocol no. 16434/16. A total of 727 urine Samples were collected between February and June of 2107 from children (0–15 years of age) hospitalized with lower UTI at the Pediatric Emergency Center-HMC. All urine analysis were performed on patients presented with symptoms, mainly fever and dysuria. Urinary catheter was applied for all patients less than or equal 2 years of age, cerebral palsy (CP) patients and patients under intermittent catheterization. Otherwise, urine was obtained from mid-stream catch. Samples that did not yield significant bacterial growth, those that had multiple organisms and samples with suspected contamination as per lab report, were excluded from the study, and no duplicate samples were collected. All of the reported cases had UTI as their primary diagnosis. For each patient, demographic data such as age, nationality, and gender were collected. Out of the above samples, 635 (87.3%) were positive for Enterobacteriaceae species which were then isolated using readymade Cystine Lactose Electrolyte-Deficient media (IMES, Doha, Qatar). Isolated bacteria were identified by MALDI-TOF (Bruker Daltonik GmbH, Leipzig, Germany) and initial antimicrobial susceptibility testing was performed by Phoenix using the NMIC/ID-5 panel (BD Biosciences, Heidelberg, Germany) according to the manufacturer’s recommendations. Both automated tests were performed at Hamad General Hospital Microbiology laboratory. All intermediate resistant isolates were considered as susceptible. Initial testing with Phoenix revealed 201 (31.7%) isolates as Extended Spectrum β -Lactamases (ESBL) producer Enterobacteriaceae. Susceptibility testing was done for 16 clinically relevant antibiotics. 110 of these samples were randomly selected for further genotypic analysis.
Standard strains, E. coli ATCC® 25,922 and E. coli ATCC® 35,218, were used as controls for antimicrobial drug susceptibility testing. E. coli NCTC® 13,461™, E. coli NCTC® 13,462™, E. coli NCTC ®13,463™, Enterobacter cloacae NCTC ®13,464™ and K. pneumonia NCTC® 13,465™, E. coli ATCC® 35,218™ and E. coli NCTC ®13,368™ were used as positive controls, for CTX-M G1, CTX-M G 2, CTX-M G 8, CTX-M G 9, CTX-M G25 25, bla TEM and bla SHV, polymerase chain reaction (PCR) assays, respectively.
ESBL phenotype confirmation
Isolates that were tested positive for ESBL by Phoenix were consequently confirmed by Double Disc Synergy Test (DDST) as previously described [17, 18]. Briefly, synergy was determined between a 20/10 μg disc of amoxicillin-clavulanate (BD- Sensi Disc™) and 30-μg disc of ceftazidime and ceftriaxone (BD- Sensi Disc™), placed onto Mueller–Hinton agar (Oxoid Ltd., Basingstoke, Hampshire, England) inoculated with a microbial suspension of 0.5 McFarland turbidity, at a distance of 15 mm apart from the edge of the amoxicillin-clavulanate disc. The cefoxitin (30 μg, BD- Sensi Disc™) disc was placed in any available space remaining on the plate. Extension of the edge of the exhibition zone by > 5 mm towards the disc of amoxicillin-clavulanate disc, together with susceptibility to cefoxitin was interpreted as positive for the ESBL production [18].
Molecular genotyping of ESBL genes
DNA was extracted from bacterial cultures using QIAamp® UCP pathogen mini Kit (Qiagen, Germany) following manufacturer’s instructions. Extracted DNA was then used to run PCR for seven genes and using previously published primers [19, 20] . The conditions used for blaTEM and blaSHV reactions were as follows: PCR mixture was made in volume of 20 μl containing 0.5 μM of each primer, 50 ng DNA, 1× master mix (Hot star Taq plus master mix (Qiagene, Germany)) and DPEC H2O up to 20 μl. The reaction was amplified in GeneAmp* PCR system 9700 thermocycler under the following conditions: 1. Initial denaturation at 96 °C for 5 min.; 2. 32 cycles consisting of denaturation at 96 °C for 30 s., annealing at 44 °C (bla TEM) and at 58 °C (bla SHV) for 45 s, and extension for 60 s. at 72 °C; and 3. A final extension cycles at 72 °C for 10 min. Multiplex PCR (MPCR) was performed in a final volume of 30 μl containing 0.23 μM of each primer (bla CTX-M-G(1,2,8,9 &25)), 50 ng DNA, 1× master mix (Hot star Taq plus master mix (Qiagene, Germany) [19]) and DPEC H2O up to 30 μl to screen for bla CTX-M- G (1,2,8,9 & 25) genes. Amplified products were subjected to electrophoresis in 1.2% agarose (Agarose- LE, Ambion®, USA), stained with ethidium bromide (Promega, Madison, USA) and visualized using Bio-Rad gel doc system (Bio - rad, Gel Doc tm XR System 170–8170, Canada).
Clustering of ESBL-positive isolates
An agglomerative hierarchical algorithm was used to derive a cluster analysis dendrogram to establish the relationship between individual E. coli (n = 95) and Klebsiella pneumoniae (n = 13) isolates based on the presence and absence of 7 genes (blaTEM, blaSHV, bla CTXM -G 1, bla CTXM -G2, bla CTXM -G8, bla CTXM -G9 & bla CTXM –G25), whichare reported in the literature to encode for ESBL. The scores ‘1’ and ‘0’ were given for the presence and absence of bands respectively [21–23]. The data obtained by scoring of the genetic profiles of different ESBL genes were subjected to cluster analysis, and hierarchical cluster dendrogram was created using Past software version 1.91 [24]. A similarity matrix values were used for cluster analysis.
Data analysis
Data were introduced into Microsoft Excel 2010 (Microsoft Corporation, New York, USA) to generate figures and run initial analysis and further statistical analysis were done using SPSS statistics 24 (Statistical Package for the Social Science; SPSS Inc., Chicago, IL, USA). Relation between gene type and resistance of each antibiotic was cross-tabulated using non-parametric Kappa statistics; on the other hand, relation between gene type, nationality and age grouping was calculated using Pearson chi-square test. Probability value (P value) less than 0.05 was considered statistically significant. Past software, version 1.91, was used to construct hierarchical clustering dendrogram and Jacquard’s coefficient was applied to generate the similarity values for generation of the cluster analysis [24].
Results
Demography of the study population
The demographic profile of the studied population is summarized in Table 1. Sixteen percent (n = 34) of samples were collected from males compared to 83.% (n = 167) from females (0–15 years of age), with Male to female ratio of approximately 1:5. ESBL producing Enterobacteriaceae were more prevalent among Qataris 46 (22.8%), Egyptians 37 (18.4%), Indians 27 (13.4%), and to lesser extent in Pakistani 21(10.4%). Most of the ESBL was detected among children between 0 and 5 years of age, n = 142 (70.6%).
Table 1.
Demographic profile of the study population (n = 201) with ESBL UTI in the State of Qatar
| Gender | Total number/percentage | Nationality | |
|---|---|---|---|
| Qatari | Non Qatari (na=24) | ||
| Male | 34 (16.9%) | 5 (2.5%) | 29 (14.42%) |
| Female | 167(83%) | 41 (20.4%) | 126 (62.7%) |
| Total no./percentage | 201 (100%) | 46 (22.9%) | 155 (77.11%) |
| Age group (years) | |||
| < 2 | 60 (29.9%) | 7 (36.8%) | 53 (26.4%) |
| 2–5 | 82 (40.8%) | 18 (9%) | 64 (31.8%) |
| 6–15 | 59 (29.4%) | 20 (10%) | 39 (19.4) |
aRepresent the number of nationalities tested
Etiology of ESBL-associated UTI infections
Out of 635 Enterobacteriaceae positive urine cultures, 201 (31.7%) were found to be ESBL producing bacteria. E. coli species was the most prominent with prevalence rate of 83% (n = 166), followed by Klebsiella pneumoniae 11% (n = 22) and the rest 6% included Citrobacter koseri, Enterobacter cloacae, Serratia marcescens, Citrobacter amalonaticus.
Phenotypic resistance profile of ESBL isolates
Antibiotics resistance profile of ESBL-producing pathogens is depicted in Fig. 1. All ESBL-producing isolates showed 100% resistance to ampicillin, and to all cephalosporins including cephalothin, cefazolin, ceftriaxone and cefepime. Low resistance was recorded to carbapenems. Resistance ranged between 2.5% to meropenem, ertapenem and 10% to imipenem. Among the β-lactam/β-lactamase inhibitor combinations, 9% were resistant to piperacillin/tazobactam, whereas 99% were resistant to amoxicillin/clavulanic acid. Regarding aminoglycosides, all the isolates were susceptible to amikacin, and 24.4% of which were resistant to gentamicin. The resistance prevalence to other classes of antibiotics namely, cefoxitin, nitrofurantoin, trimethoprim/sulfamethoxazole and ciprofloxacin was 19.4, 13, 59.7 and 36%, respectively.
Fig. 1.
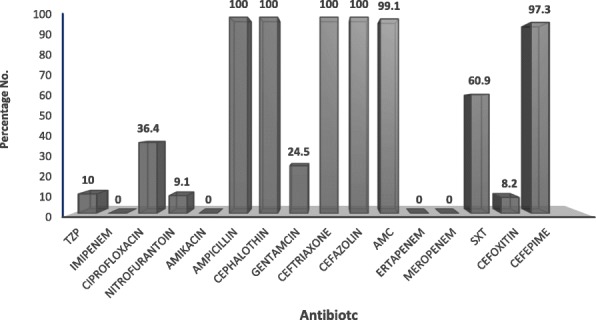
Antimicrobial resistance profile of 201 ESBL producing bacteria isolated from children (age 0 to 15 years) with UTI. Isolates were tested for antibiotics resistance against 16 clinically relevant antibiotics using phoenix NMIC/ID-5 panel (BD Biosciences, Heidelberg, Germany). The figure depicts the percentage of isolates with resistance to each of the antibiotics. TZP: piperacillin/tazobactam; SXT: trimethoprim/sulfamethoxazole; AMC: Amoxicillin/clavulanic acid
Molecular genotyping profile of ESBL isolates
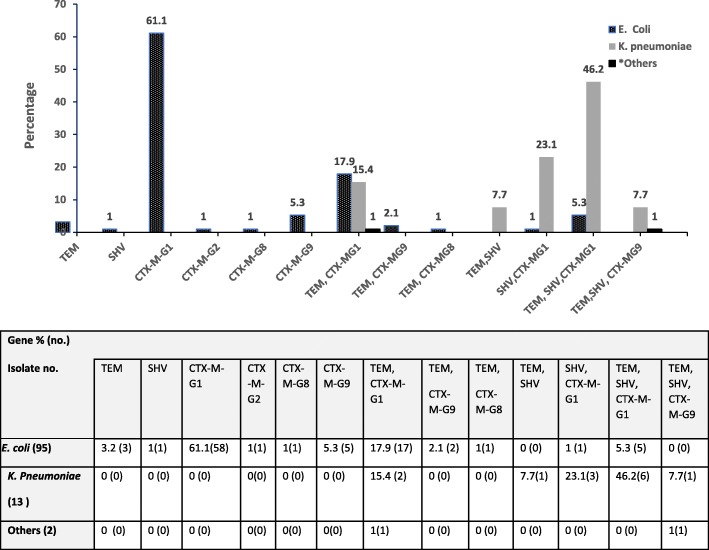
110 ESBL producing bacterial isolates representing 95 E. coli, 13 K. Pneumonia, one Citrobacter koseri and one Enterobacter cloacae were randomly selected and characterized with PCR for genes encoding resistance (Fig. 2). Of these, the highest resistance (n = 65, 59%) was encoded by bla CTX-M genes: bla CTX-MG1 (89.2%), bla CTX-MG9 (7.7%), and bla CTX-MG2 (0.9%) and bla CTX-MG8 (1.5%). bla TEM and bla SHV genes were detected in 2.7 and 0.9% of the isolates, respectively. 37.3% of bacteria harbored multiple bla genes (≥ two genes). Two bla genes were detected in 20 (18.2%) E. coli and 3 (3.2%) of K. pneoumoniae isolates. 77% of K. pneoumoniae and 6.3% of E. coli isolates were harboring three bla genes. While majority of the ESBL E. coli resistance was encoded by one gene, blaCTX-M-G1 58 (61.1%), resistance in k. pneoumoniae isolates was encoded by blaSHV, blaTEM and blaCTX-MG1 genes (46.2%) concurrently (Fig. 3). blaCTX-M-G25 was not detected in any isolate.
Fig. 2.
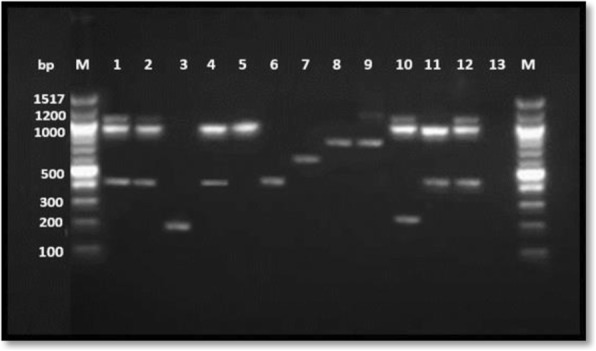
Detection of blaSHV, blaTEM and blaCTX-M-G (1,2,8,9, &25) antibiotic resistance genes in 110 ESBL Enterobacteriaceae pathogens isolated from children with UTIs. Representative samples are shown. Multiplex PCR was performed for detection of CTX-M groups while monoplex PCR was used for detection of TEM and SHV. The amplification products of each isolate were run on the same lane for detection of bla genes. Lane 1: blaTEM, blaSHV& blaCTX-MG1; Lane 2: blaTEM, blaSHV & bla CTX-M-G1: Lane 3: blaCTXM-G9; Lane 4: blaSHV & blaCTXM-G1; Lane 5: blaSHV; Lane6: blaCTX-MG1; Lane 7: blaCTXM-G2; Lane 8; blaCTXM-G8, Lane 9: blaTEM &blaCTXM-G8; Lane 10, bla TEM, blaSHV &blaCTXM-G8; Lane 11: blaSHV & blaCTXM-G1; Lane 12: blaNCTC 13,351 E. coli Positive control for blaTEM, NCTC 13368 K. pneumonia positive control for blaSHV, NCTC 13461 E. coli positive control for blaCTX-MG1: Lane 13: ATCC 25922 E. coli negative control
Fig. 3.
Distribution of bla genes among ESBL Enterobacteriaceae obtained from urine samples of children with lower urinary tract infection
Correlation between phenotypic and genotypic profiles
The antibiotic resistance outcomes (resistance or susceptible [i.e., binary]) were cross- tabulated with the six detected ESBL genes using kappa statistics (Table 2). A significant association (p < 0.05) were found between the presence of TEM, nitrofurantoin and trimethoprim/sulfamethoxazole; SHV and nitrofurantoin; CTXM-G2 and piperacillin/tazobactam. The presence of individual ESBL genes was not significantly different (P > 0.05, Pearson Chi- square test) by nationality or age.
Table 2.
Measure of agreement between resistance of antibiotics and presence of ESBL genes by cross tabulation Kappa statistics
| Antibiotic | Number of resistance / Kappa significant value | |||||
|---|---|---|---|---|---|---|
| TEM | SHV | CTXM-G1 | CTXM-G2 | CTXM-G8 | CTXM-G9 | |
| Piperacillin/Tazobactam | 11/0.186 | 11/0.253 | 11/0.5 | 11/0.003 | 11/0.634 | 11/0.328 |
| Ciprofloxacin | 40/0.06 | 40/0.654 | 40/0.401 | 40/0.2 | 40/0.9 | 40/0.9 |
| Nitrofurantoin | 12/0.021 | 12/0.000 | 12/0.825 | 12/0.7 | 12/0.074 | 12/0.3 |
| Gentamicin | 28/0.2 | 28/0.843 | 28/0.08 | 28/0.4 | 12/404 | 12/0.9 |
| Trimethoprim/Sulfamethoxazole | 67/0.02 | 67/0.727 | 67/0.2 | 67/0.08 | 67/0.75 | 67/0.396 |
| Cefoxitin | 11/0.180 | 11/0.538 | 11/0.7 | 11/0.057 | 11/0.057 | 11/0.8 |
Clustering and similarity of ESBL-positive isolates
Cluster analysis was used to study similarity among individual E.coli (n = 94) and K. pneumoniae (n = 13) isolates according to presence and absence of 7 genes (bla TEM, bla SHV, bla CTXM -G 1, bla CTXM- G2, bla CTXM G8, bla CTXM G9 & bla CTXM G25). The E. coli positive isolates were distributed into one of the three main branches A, B and C (Fig. 4a), then sub grouped into 11 clusters (A1, A2, A3, A4 A5, B1, B2, B3, B4, B5 and C1). Most of the E. coli isolates clustered in A2 (61.7%) which includes only CTXM-G1 enzyme.Around 20.2% clustered in A5 (which includes a combination of TEM and CTXM-G1 enzymes. On the other hand, K. pneoumoniae were distributed into one of the two main branches A and B (Fig. 4b), then sub grouped into five clusters (A1, A2, B1, B2 and B3), with the main cluster being B2 (46.2%) which represent combination of TEM, SHV and CTXM-G1 type enzymes.
Fig. 4.
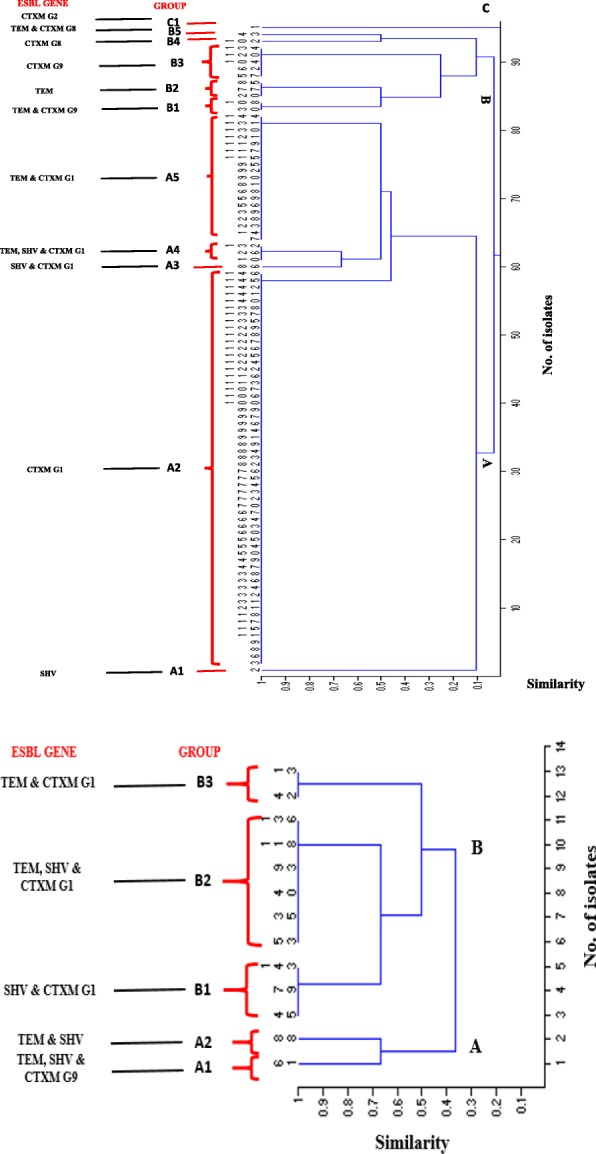
Agglomerative hierarchical algorithm illustrating the similarity of E. coli (a) and K. pneumoniae (b) based on the presence and absence of ESBL genes, using past software version, 1.91 and Jaccard’s coefficient. A similarity value = 0 indicates dissimilarity between organisms, while similarity value of 1 means the organisms are identical or very close to each other
Discussion
Out of a 635 Enterobacteriaceae isolated from urine samples obtained from a cohort of children with UTI, 201 (31.7%) were found to be ESBL. E. coli species was the most prevalent representing 83% (n = 166) of the isolates, followed by K. pneumoniae which represented 11% (n = 22). This is in agreement with other studies including a recent one from Sri Lanka [25] in which E. coli and Klebsiella species represented 86.8 and 13.1% of UTI infections among adult patients. Expectedly, ESBL was more predominant among females than males with a ratio of 4.9 to 1. It has been frequently reported thatUTIs occur far more frequently in girls than in boys during the first few months of life, presumably due to the shorter length of the female urethra [26, 27]. The most affected group of our study were the children ranging in age from 0 to 5 years (70.7%), wherecolonization with E. coli and enterococci is known to diminish after 5 years of age [26].
Molecular genotyping of ESBL-positive isolates showed that the highest resistance was due to the presence of bla CTX-M genes (59%), particularly bla CTX-MG1 (89.2%). bla TEM and bla SHV genes contributed to only 2.7 and 0.9% of ESBL resistance, which is consistent with previous reports from the region and around the globe [15, 28–31]. The high occurrence of bla CTX-MG1 gene among pediatric population suggests high dynamic transmission-ability of the plasmid-carried gene, that contributes to the dissemination of CTX-M enzymes to the community and could cause extra horizontal transmission in healthcare facilities [32]. In busy health care facilities, especially in developing countries, the rapid turnover of patients to accommodate multiple patients’ treatment could lead to breaching of infection control measures, such as compliance with hand hygiene, which could lead to this horizontal transmission of resistant bacteria from one patient to another. Unfortunately, we could not compare our findings to others from the region due to the scarcity of published data. However, it has been globally reported that the predominant genotype of ESBL-producing E. coli and K. pneumonia has changed from TEM and/or SHV to CTX-M-1 since 2006 [33, 34], which seems to be the trend in Qatar. Interestingly, our results indicated that three bla genes (blaSHV, blaTEM and blaCTX-MG1/ blaCTX-MG9) were concurrently detected in 77% of K. pneoumoniae isolates, whereas only 6.3% of the E. coli isolates had combinations of the three genes (Fig. 3). On the other hand, two out of seven bla genes were simultaneously detected in 18.2 and 3.2% of E. coli and K. pneumonia, respectively. These results indicate different genotypic profile of ESBL resistance between E. coli and K. pneumoniae in urine samples which might indicate a high transfer of genes among K. pneumoniae than E. coli. Concerning the cephalosporins, all ESBL-producing isolates showed 100% resistance to ampicillin, cephalothin, cefazolin, ceftriaxone and 97.3% resistance to cefepime. This finding coincided with a study carried in intensive care unit (ICU) among adult patients at HMC, Qatar, on respiratory tract and blood samples [35]. No resistance in ESBL-producing isolates were observed against amikacin, ertapenem, meropenem and imipenem. Among the β-lactam/β-lactamase inhibitor combinations, 10% were resistant to piperacillin/tazobactam, whereas 99.1% were resistant to amoxicillin/clavulanic acid. The clinical isolates presented 24.5% resistance to gentamicin compared with 100% susceptibility to amikacin among aminoglycosides. Similarly, these findings concur with the findings from the aforementioned study in ICU patients, [35] with an exception of piperacillin/tazobactam resistance which was reported at 22% in ICU adult patients compared to 10% in pediatrics with UTI. Resistance to piperacillin/tazobactam and ciprofloxacin were significantly lower in our study (10 and 36.4% respectively) compared with another one from Kingdom Saud Arabia (KSA) in a hospital at Dammam-Eastern province, where resistance reached63.2 and 84.6%, respectively [30]. Authors attributed high resistance to these antibiotics due to the misuse of ciprofloxacin and restriction of aminoglycosides among adult in Dammam hospital. Whereas, ciprofloxacin is restricted for use among adult patients at HMC and very rarely used in pediatric population (personal communication, 2017).
The resistance to oral antibiotics namely, nitrofurantoin, trimethoprim/sulfamethoxazole and ciprofloxacin was found to be 9.1, 60.9 and 36.4%, respectively. Obtained results were comparable to those observed in the Middle East region [27], which indicate rapid dissemination of multidrug resistance bacteria in the communities [15, 36]. Worldwide, previous studies have reported higher prevalence of ESBL resistance in Europe compared to United States (US), but lower than South America and Asia [37]. Our results indicate that the patterns of ESBL resistance in Qatar, at least in pediatric population, is more similar to Asia. It is worth noting that 80% of population in Qatar is expatriates; arriving mostly from countries in South East Asia. This could partially explain our findings. Cumulatively, our findings mandates the establishment of the antimicrobial stewardship program and formulation of guidelines for empirical use of prescription of antibiotics for UTI infections among pediatric population.
We found that the majority of ESBL (156, 77.6%) producers were multidrug resistant (resistant to 3 or more different classes of antibiotics), with the most common resistance pattern being to amoxicillin-clavulanate, ampicillin, ciprofloxacin, Sulfamethoxazole/trimethoprim in addition to cephalosporines. Plasmids encoding ESBL resistance often carry multiple genes conferring multiple resistance, thus simultaneous resistance to fluoroquinolones, aminoglycosides, tetracyclines, and trimethoprim sulfamethoxazole [12]. Our study revealed a significant association (p < 0.05) between the presence of TEM, nitrofurantoin and trimethoprim/sulfamethoxazole; SHV and nitrofurantoin; CTXM-G2 and piperacillin/tazobactam resistance. Thus, in our study, the use of extended-spectrum cephalosporins, ciprofloxacin, aminoglycosides (gentamicin) and trimethoprim/sulfamethoxazole is compromised, leaving carbapenems as the therapeutic option for severe infections caused by ESBL producers. In agreement with our findings, [38] found that plasmid-mediated quinolone resistance has been associated with bla CTX-M genes, where genes conferring resistance to aminoglycosides and tetracycline and other bla genes have been found on the same plasmids as the bla CTXM [39]. Isolates in this study were all from pediatric patients in the community who attended emergency setting This highlights the therapeutic challenges posed by ESBL producers, particularly in the UTI treatment of community-onset [40].
Using seven genes to run agglomerative hierarchical algorithm analysis [21–24] our study revealed eleven clonal clusters among the 94 tested E. coli isolates. Clonally related strains of cluster A2 were responsible for the predominant UTI in pediatrics population 67.1% (58/94), which produce only CTXM-G1 type enzyme, followed by A5 (20.2%), which produce a combination of TEM and CTXM-G1type enzymes. Five clonal clusters were detected among K. pneumoniae with the main cluster B2 producing concurrently a combination of three enzyme types SHV, TEM and CTXM-G1. Whole plasmid sequencing for representative isolates from each cluster can be very helpful to better understand the relatedness and typing differences between the clusters and elucidating further information on the mechanism of resistance in these isolates. Unfortunately, similar molecular analysis of ESBL bacteria are lacking in the surrounding country. This limit our understanding of the trends and distribution of the strains, noting that more than 80% of Qatari population are expats that arrive from MENA region and South East Asia.
Conclusions
Although ESBL-producing members of Enterobacteriaceae have been reported in all Arabian Gulf region, very limited data is available about the genetic makeup encoding for such resistance. This is the first study among pediatric population in the state of Qatar that demonstrate the correlation between genetic and phenotypic profile of ESBL producing Enterobacteriaceae. The study highlights an escalated problem of ESBL resistance in Enterobacteriaceae causing UTI in pediatric population in Qatar. Resistance was predominantly mediated by CTX-MG1-type enzymes. The use of cephalosporins, aminoglycosides (gentamicin) and trimethoprim/sulfamethoxazole is compromised in Qatar among pediatric population with UTI, leaving carbapenems and amikacin as the therapeutic option for severe infections caused by ESBL producers. The negative impact of extensive use of carpabenemes could lead to carbapenamase resistant Enterobacteriaceae. This mandates further epidemiological studies and implementation of regulatory programs to reduce spread of ESBL producing Enterobacteriaceae in the community. Further next generation sequencing (NGS) studies are necessary for a more comprehensive analysis of ESBL variants.
Acknowledgements
This work was funded by Qatar University Grant No. QUUG-BRC-2017-2 to Dr. Nahla Omer Ahmed Eltai.
Abbreviations
- CP
Cerebral palsy
- DDST
Double disc synergy test
- ESBL
Extended-spectrum beta-lactamase
- GCC
Gulf cooperation council
- GNB
Negative bacteria
- HMC
Hamad medical corporation
- MRC
Medical research centre
- NGS
Next generation sequencing
- UTI
Urinary tract infection
Authors’ contributions
NOE, HMY, AAA, KA, and AD designed the study and drafted the manuscript. NOE, EW, SH and AD performed the experimental work. NO and HMY analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethical approval for this study was obtained from the Medical Research Centre (MRC), Hamad Medical Corporation (HMC), Doha, Qatar, protocol no. 16434/16.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nahla O. Eltai, Email: nahla.eltai@qu.edu.qa
Asmaa A. Al Thani, Email: aaja@qu.edu.qa
Khalid Al-Ansari, Email: kalansari1@hamad.qa.
Anand S. Deshmukh, Email: adeshmukh@hamad.qa
Eman Wehedy, Email: eabdelsalam@hbku.edu.qa.
Sara H. Al-Hadidi, Email: saraalhadidi@yahoo.com
Hadi M. Yassine, Email: hyassine@qu.edu.qa
References
- 1.Talbot GH, Bradley J, Edwards Jr JE, Gilbert D, Scheld M, Bartlett JG: Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the Infectious Diseases Society of America. Clin Infect Dis 2006, 42(5):657–668. [DOI] [PubMed]
- 2.Goyal A, Prasad K, Prasad A, Gupta S, Ghoshal U, Ayyagari A. Extended spectrum beta-lactamases in Escherichia coli & Klebsiella pneumoniae & associated risk factors. 2009. [PubMed] [Google Scholar]
- 3.Sharma M, Pathak S, SrivaStava P. Prevalence and antibiogram of extended Spectrum β-lactamase (ESBL) producing gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J Clin Diagn Res. 2013;7(10):2173. doi: 10.7860/JCDR/2013/6460.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura T, Komatsu M, Yamasaki K, Fukuda S, Miyamoto Y, Higuchi T, Ono T, Nishio H, Sueyoshi N, Kida K. Epidemiology of Escherichia coli, Klebsiella species, and Proteus mirabilis strains producing extended-spectrum β-lactamases from clinical samples in the Kinki region of Japan. Am J Clin Pathol. 2012;137(4):620–626. doi: 10.1309/AJCP48PDVKWQOXEZ. [DOI] [PubMed] [Google Scholar]
- 5.Birgy A, Mariani-Kurkdjian P, Bidet P, Doit C, Genel N, Courroux C, Arlet G, Bingen E. Characterization of extended-spectrum-beta-lactamase-producing Escherichia coli strains involved in maternal-fetal colonization: prevalence of E. Coli ST131. J Clin Microbiol. 2013;51(6):1727–1732. doi: 10.1128/JCM.03255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukac PJ, Bonomo RA, Logan LK. Extended-Spectrum β-lactamase–producing Enterobacteriaceae in children: old foe, emerging threat. Clin Infect Dis: Offic Public Infect Dis Soc Am. 2015;60(9):1389–1397. doi: 10.1093/cid/civ020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol. 2010;13(5):558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitout JD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 10.Stedt J, Bonnedahl J, Hernandez J, Waldenström J, McMahon BJ, Tolf C, Olsen B, Drobni M. Carriage of CTX-M type extended spectrum β-lactamases (ESBLs) in gulls across Europe. Acta Vet Scand. 2015;57(1):74. doi: 10.1186/s13028-015-0166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahlaoui H, Khalifa A, Haj B, Moussa MB. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum β-lactamase (ESBL) Medecine et maladies infectieuses. 2014;44(9):400–404. doi: 10.1016/j.medmal.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Malloy AM, Campos JM. Extended-spectrum beta-lactamases: a brief clinical update. Pediatr Infect Dis J. 2011;30(12):1092–1093. doi: 10.1097/INF.0b013e31823c0e9d. [DOI] [PubMed] [Google Scholar]
- 13.Bakshi R, Walia G, Shikha J. Prevalence of extended spectrum β-lactamases in multidrug resistant strains of gram negative bacilli. J Acad Indus Res. 2013;1:558–560. [Google Scholar]
- 14.CDC . Centers for disease control and prevention USA. 2013. Antibiotic resistance threats in the United States, 2013. [Google Scholar]
- 15.Zowawi HM, Balkhy HH, Walsh TR, Paterson DL. β-Lactamase production in key gram-negative pathogen isolates from the Arabian peninsula. Clin Microbiol Rev. 2013;26(3):361–380. doi: 10.1128/CMR.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandramohan L, Revell PA. Prevalence and molecular characterization of extended-spectrum-β-lactamase-producing Enterobacteriaceae in a pediatric patient population. Antimicrob Agents Chemother. 2012;56(9):4765–4770. doi: 10.1128/AAC.00666-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eltai NO, Abdfarag EA, Al-Romaihi H, Wehedy E, Mahmoud MH, Alawad OK, Al-Hajri MM, Al Thani AA, Yassine HM. Antibiotic resistance profile of commensal Escherichia coli isolated from broiler chickens in Qatar. J Food Prot. 2017;81(2):302–307. doi: 10.4315/0362-028X.JFP-17-191. [DOI] [PubMed] [Google Scholar]
- 18.CLSI Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing. M100S. 2016;26:1–129. [Google Scholar]
- 19.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2005;57(1):154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 20.Bora A, Hazarika NK, Shukla SK, Prasad KN, Sarma JB, Ahmed G. Prevalence of blaTEM, blaSHV and blaCTX-M genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Northeast India. Indian J Pathol Microbiol. 2014;57(2):249. doi: 10.4103/0377-4929.134698. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed SF, Ali MMM, Mohamed ZK, Moussa TA, Klena JD. Fecal carriage of extended-spectrum β-lactamases and AmpC-producing Escherichia coli in a Libyan community. Ann Clin Microbiol Antimicrob. 2014;13(1):22. doi: 10.1186/1476-0711-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamatchi C, Magesh H, Sekhar U, Vaidyanathan R. Identification of clonal clusters of Klebsiella pneumoniae isolates from Chennai by extended spectrum beta lactamase genotyping and antibiotic resistance phenotyping analysis. Am J Infect Dis. 2009;5(2):74–82. [Google Scholar]
- 23.Chapman TA, Wu X-Y, Barchia I, Bettelheim KA, Driesen S, Trott D, Wilson M, Chin JJ-C. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl Environ Microbiol. 2006;72(7):4782–4795. doi: 10.1128/AEM.02885-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer Ø, Harper D, Ryan P. Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electron. 2001;4:9.
- 25.Fernando M, Luke W, Miththinda J, Wickramasinghe R, Sebastiampillai B, Gunathilake M, Silva F, Premaratna R. Extended spectrum beta lactamase producing organisms causing urinary tract infections in Sri Lanka and their antibiotic susceptibility pattern–a hospital based cross sectional study. BMC Infect Dis. 2017;17(1):138. doi: 10.1186/s12879-017-2250-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellerstein S. Urinary tract infections in children: why they occur and how to prevent them. Am Fam Physician. 1998;57(10):2440–2446. [PubMed] [Google Scholar]
- 27.Zeyaullah M, Kaul V. Prevalence of urinary tract infection and antibiotic resistance pattern in Saudi Arabia population. Global J Biol, Agricult Health Sci. 2015;4(1):206–214. [Google Scholar]
- 28.Zhao W-H, Hu Z-Q. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in gram-negative bacteria. Crit Rev Microbiol. 2013;39(1):79–101. doi: 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potz NA, Hope R, Warner M, Johnson AP, Livermore DM. Prevalence and mechanisms of cephalosporin resistance in Enterobacteriaceae in London and south-East England. J Antimicrob Chemother. 2006;58(2):320–326. doi: 10.1093/jac/dkl217. [DOI] [PubMed] [Google Scholar]
- 30.Hassan H, Abdalhamid B. Molecular characterization of extended-spectrum beta-lactamase producing Enterobacteriaceae in a Saudi Arabian tertiary hospital. J Infect Dev Countries. 2014;8(03):282–288. doi: 10.3855/jidc.3809. [DOI] [PubMed] [Google Scholar]
- 31.Bindayna K, Khanfar HS, Senok AC, Botta GA. Predominance of CTX-M genotype among extended spectrum beta lactamase isolates in a tertiary hospital in Saudi Arabia. Saudi Med J. 2010;31(8):859–863. [PubMed] [Google Scholar]
- 32.Mendonça N, Leitão J, Manageiro V, Ferreira E, Caniça M. Portugal ARSPi: spread of extended-spectrum β-lactamase CTX-M-producing Escherichia coli clinical isolates in community and nosocomial environments in Portugal. Antimicrob Agents Chemother. 2007;51(6):1946–1955. doi: 10.1128/AAC.01412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantón R, Coque TM. The CTX-M β-lactamase pandemic. Curr Opin Microbiol. 2006;9(5):466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59(2):165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed AMS, Sultan AA, Deshmukh A, Acharya A, Elmi AA, Bansal D, Ibrahim E, Hamid JM, Ahmed MAS, Bilal NE. Antimicrobial susceptibility and molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae from intensive care units at Hamad Medical Corporation, Qatar. Antimicrob Resist Infect Control. 2016;5(1):4. doi: 10.1186/s13756-016-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balkhy HH, Assiri AM, Al Mousa H, Al-Abri SS, Al-Katheeri H, Alansari H, Abdulrazzaq NM, Aidara-Kane A, Pittet D, Erlacher-Vindel E. The strategic plan for combating antimicrobial resistance in gulf cooperation council states. J Infect Public Health. 2016;9(4):375–385. doi: 10.1016/j.jiph.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Giamarellou H. Multidrug resistance in gram-negative bacteria that produce extended-spectrum β-lactamases (ESBLs) Clin Microbiol Infect. 2005;11(s4):1–16. doi: 10.1111/j.1469-0691.2005.01160.x. [DOI] [PubMed] [Google Scholar]
- 38.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006;50(4):1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother. 2004;48(10):3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heffernan H, Dyet K, Woodhouse R, Williamson D. Antimicrobial susceptibility and molecular epidemiology of extended-spectrum β-lactamase producing Enterobacteriaceae in New Zealand, 2013. 2014. [Google Scholar]