Abstract
The synthesis of a fully degradable, bio-based, sustained release, pro-antimicrobial polymer network comprised of degradable acetals (PANDA) is reported. The active antimicrobial agent – p-anisaldehyde (pA) (an extract from star anise) – was converted into a UV curable acetal containing pro-antimicrobial monomer and subsequently photopolymerized into a homogenous thiol-ene network. Under neutral to acidic conditions (pH < 8), the PANDAs undergo surface erosion and exhibit sustained release of pA over 38 days. The release of pA from PANDAs was shown to be effective against both bacterial and fungal pathogens. From a combination of confocal microscopy and transmission electron microscopy, we observed that the released pA disrupts the cell membrane. Additionally, we demonstrated that PANDAs have minimal cytotoxicity towards both epithelial cells and macrophages. Although a model platform, these results point to promising pathways for the design of fully degradable sustained-release antimicrobial systems with potential applications in agriculture, pharmaceuticals, cosmetics, household/personal care, and food industries.
Keywords: pro-antimicrobial, acetal, thiol-ene, degradable, sustained release
Graphical Abstract
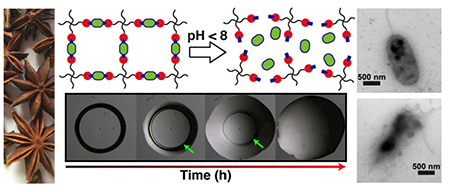
1. Introduction
With the increasing prevalence of antimicrobial resistance (AMR), opportunistic pathogens pose a significant global crisis. Compounding the issue, over 23.5 million immunocompromised patients (i.e. patients with rheumatoid arthritis, HIV, organ transplants, etc.) are prescribed immunosuppressants, making them more prone to both fungal and bacterial infections [1]. For instance, the fungus Histoplasma capsulatum aggressively infects immunocompromised patients with over 500,000 cases annually in the United States, 300,000 of which are HIV-related infections resulting in 10,000 deaths per year [2]. Additionally, exposure to other common pathogens such as Pseudomonas aeruginosa, Escherichia coli, Salmonella Typhi, and Staphylococcus aureus can escalate into life threating infections resulting in sepsis and death [3, 4]. To combat AMR, new antimicrobial strategies with low propensity to trigger resistance have been developed including quorum sensing inhibitors [5], synergistic pairing to induce sensitivity [6, 7], antibiotic adjuvants [8], antibiotic cycling [9], and multimodal antibiotics [10, 11]. Of the aforementioned methods, essential oils (EOs) – a unique class of naturally occurring, plant-derived extracts – hold promise in eradicating resistant microbial populations via the multimodal approach. EOs exhibit broad spectrum antimicrobial activity by interfering with membrane integrity, ATP synthesis, protein stability, and quorum sensing pathways [12, 13]. Despite positive results, the use of EOs is still restricted due to their limited water solubility, high volatility, and chemical instability [14]. Many approaches have been reported to sequester EO derivatives within polymeric materials; however, these strategies often utilize slow or non-degradable linkages which slowly release or permanently entrap EOs.
Poly(actives), or polymeric pro-drugs that rely on degradable linkages for the release of therapeutic agents, can improve chemical stability, enable tunable release profiles, and reduce volatility [15]. Multiple linear poly(actives) have been reported including poly(esters) [16, 17], poly(anhydride-esters) [18–25], poly(anhydride-amides) [26], poly(carbonate-amides) [27], poly(carbonate esters) [28], poly(ketals) [29], and poly(oxalate-acetals) [30]. While linear poly(actives) successfully entrap drugs and allow for tunable release profiles, crosslinked poly(actives) represent a relatively unexplored class of degradable drug-releasing materials where opportunities for greater control over degradation profiles, thermomechanical properties, and sample geometry (particles, thin films, or coatings) are available. Recently, crosslinked poly(actives) including poly(esters) [31–33], poly(β-amino esters) [34], and poly(acetals) [35] have utilized degradation rates, crosslink density, and hydropathy as mechanisms to control the release of actives. Of the degradable linkages available, both acetals and ketals have gained recent attention because of their charge-neutral and potentially nontoxic byproducts (alcohols and ketones/aldehydes) upon cleavage [36].
We have recently demonstrated the sequestration of p-chlorobenzaldehyde within a proantimicrobial network via degradable acetals (PANDA) [35]. While p-chlorobenzaldehyde is a relatively potent antimicrobial drug, we wanted to shift focus away from oil derived compounds and towards bio-based aldehydes found in nature to offer a fully degradable antimicrobial system based on essential oils. In this work, we turned towards p-anisaldehyde (pA), an extract from star anise (Pimpinella anisum) seeds, to fabricate a new class of antimicrobial PANDAs. We demonstrate that incorporation and subsequent release of pA from a PANDA exhibits potent antimicrobial activity against a variety of clinically relevant pathogens with minimal cytotoxicity.
2. Materials and methods
2.1. Materials
The chemicals p-anisaldehyde (pA), trimethylol propane diallyl ether, 2-hydroxy-2-methylpropiophenone (Darocur 1173), trimethylsilyl trifluoromethanesulfonate (TMSOTf), dichloromethane (DCM), allyloxytrimethylsilane, pyridine, sodium bicarbonate, diethyl ether, magnesium sulfate (MgSO4), dextrose, 2,3,5,6-tetrachloronitrobenzene, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), hexane, 0.5 M Tris-HCl in H2O, acetonitrile-d3, Aqua Dead Cell Stain™, LIVE/DEAD BacLight Bacterial Viability Kit staining kit, BacLight RedoxSensor CTC Vitality Kit and ethyl acetate were acquired from Thermo Fisher Scientific. Pentaerythritol tetra(3-mercaptopropionate) (PETMP) was provided by Bruno Bock. Difco Agar, yeast extract, Bacto Tryptone, Mueller Hinton broth and agar, were from Becton, Dickinson and Company. Ham’s F-12 and trypsin-EDTA were obtained from GIBCO. Hemin chloride (type II), protoporphyrin IX, histidine (free base), proline, 37% formaldehyde in H2O, dimethyl sulfoxide (DMSO), fluconazole, fetal bovine serum (FBS), Dulbecco’s Modified Eagle’s Medium (DMEM) and N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) was purchased from Sigma Aldrich. All the materials were obtained at the highest purity available and used without further purification unless otherwise specified.
2.2. Characterization
A Bruker Ascend 600 MHz (TopSpin 3.5) spectrometer was used to record 1H/13C NMR spectra with either chloroform-d or acetonitrile-d3. High resolution mass spectroscopy (HRMS) was performed with positive electrospray ionization on a Bruker 12 Tesla APEX-Qe FTICR-MS with an Apollo II ion source. Dynamic mechanical analysis (DMA) was performed using a TA Instruments Q800 dynamic mechanical analyzer in tension mode equipped with a gas cooling accessory. Samples were clamped, evaluated at a strain of 1 %, and heated from −80 °C to 80 °C at a ramp rate of 3 °C min−1. Kinetic data was obtained using real-time FTIR (RT-FTIR) spectroscopy by determining the conversions of the thiol and ene functional groups. The RT-FTIR studies were conducted using a Nicolet 8700 FTIR spectrometer with a KBr beam splitter and a MCT/A detector with a 320–500 nm filtered ultraviolet light source. Each sample was exposed to a UV light with an intensity of 400 mW cm−2. Series scans were recorded, where spectra were taken approximately 2 scan s−1 with a resolution of 4 cm−1. Thiol conversion was monitored via integration of the SH peak between 2500–2620 cm−1 while the conversion of the alkene was monitored between 3050–3125 cm−1. Optical density (OD) and fluorescence readings were performed in a BioTek Synergy 2 programmable microplate reader (BioTek Instruments).
2.3. Synthesis of p-anisaldehyde diallylacetal (pAA)
The synthesis of pAA was done using a slightly modified procedure from Noyori and coworkers [37]. To a flame dried 250 mL round bottom equipped with a stirbar, TMSOTf (200 μL, 1.1 mmol) was added along with 30 mL dry DCM under nitrogen at −84 °C. While maintaining the same temperature, a mixture of allyloxytrimethylsilane (37 mL, 208 mmol) and pA (12 g, 88 mmol) in 25 mL of dry DCM was added dropwise into the round bottom. The reaction turned yellow and stirred for 3 h. After, the reaction was warmed to −30 °C and stirred for 1 h (reaction turned red). The deep red mixture was then quenched with the addition of pyridine (15 mL, 74.5 mmol) and poured into 100 mL of saturated sodium bicarbonate prior to extraction with (3 × 100 mL) of diethyl ether. The organic layer was dried over anhydrous MgSO4, then the excess solvent was removed under reduced pressure. The yellow oil was then purified via column chromatography using a mixture of hexane: ethyl acetate (9:1) and yielded a clear oil (13.6 g, 65.9 % yield). 1H NMR (CDCCl3) δ 7.45 (dd), 6.93 (dd), 5.97 (m), 5.62 (s), 5.35 (dd), 5.17 (dd), 4.07 (d), 3.82 (s). 13C NMR (CDCCl3) δ 159.66, 134.16, 130.68, 127.97, 116.67, 113.52, 100.29, 66.00, and 55.22. HRMS (ESI+) m/z calculated for C14H18O3 [M+Na]+ 257.114816; Found 257.114817.
2.4. General preparation of PANDA disks
The PANDA disks were prepared by adding PETMP and pAA at 1:1 ratio of SH:alkene, along with 4 wt.% of Darocur 1173. The mixtures were well mixed and 25 μL of the monomer formulations were aliquoted onto a glass slide and covered with another glass slide spaced with Teflon spacers (0.76 ± 0.02 mm in thickness). The samples were then cured using an Omnicure S1000–1B with a 100W mercury lamp λmax = 365 nm, 320–500 nm filter) at an intensity of 400 mW cm−2 for 40 seconds. Control disks were prepared following the same procedure by using PETMP and trimethylol propane diallyl ether at 1:1 ratio of SH to alkene with addition of 4 wt.% of Darocur 1173.
2.5. Degradation of PANDAs
The hydrolysis of PANDA disks was analyzed via 1H NMR under a pH 7.4 Tris-HCl buffered water:acetonitrile-d3 solution. First 2,3,5,6-tetrachloronitrobenzene (internal standard) was weighed into a vial along with 500 μL of acetonitrile-d3. Then 125 μL of 100 mM Tris-HCl in deionized water was added and mixed. A 5 mm3 disk and 500 μL of the acetonitrile:buffer solution was added to an NMR tube and sealed. Three replicates were prepared for each data point (with mean and standard deviation reported) and NMR spectra were taken over 38 days. To calculate the concentration released of different sized PANDA disks (5–100 mm3), the grams of pA released (at 24 and 48 h from a 5 mm3 disk) were multiplied by a factor to equal the desired PANDA disk size (e.g. ×10 for a 50 mm3 disk) and divided by the total volume the disk was placed into. Since the thickness of the PANDA disks does not change, this assumption is valid, however the degradation profile in the presence of microbes may not be the same as pH 7.4 PBS, yet remains our best estimate.
2.6. Evaluation of antibacterial activity of PANDAs
2.6.1. Zone of inhibition (ZOI) assay
The antimicrobial activity of PANDA disks was tested against several species of bacteria via the zone of inhibition method. The indicator microorganisms included Escherichia coli ATCC 43895 (serotype O157:H7), Staphylococcus aureus RN6390, Salmonella enterica serovar Typhi ATCC 6539 (S. Typhi), and Pseudomonas aeruginosa PAO1. The testing was done on Mueller Hinton II agar (MHA) plates that have been overlaid with soft agar seeded with individual bacterial strains. The soft agar contained (per liter): 10 g of Bacto Tryptone, 6 g of Difco agar, and 8 g of sodium chloride. To create an overlay, the indicator organisms were grown overnight at 37 °C in Mueller Hinton II broth (MHB). The overnight cultures were diluted 1:5 with fresh MHB, and mixed with molten soft agar to achieve a ~108 CFU mL−1 density. From this mixture, 4 mL aliquots were overlaid onto MHA base plates and allowed to completely solidify. After solidification of the soft agar, 50 mm3 disks were overlaid on the plates and incubated at 37 °C. The zones of inhibition (ZOI) were measured after 24 h and reported as the distance from the edge of the disk to the edge of the zone. Three replicates were carried out for each disk/bacterial strain. with the mean and standard deviation reported (experiment was repeated twice).
2.6.2. Minimum inhibitory assay
Minimum size of PANDA disks required to inhibit bacterial growth were determined using a modified broth macrodilution method. Briefly, overnight bacterial cultures in MHB were adjusted to ~105 CFU mL−1. PANDA disks of different sizes (5–100 mm3) were added to 4 mL of bacterial solutions. The tubes were then incubated at 37 °C and shaken at 200 rpm for 24 h. Bacteria suspended in MHB served as a positive control, while MHB without bacterial inoculum served as a negative control. The inoculated tubes were assessed by measuring optical density at 600 nm after 24 h where OD < 0.05 considered negative for bacterial growth. Three replicates were carried out for each disk size and bacterial strain. with the mean and standard deviation reported (experiment repeated twice).
2.6.3. Kill kinetics via track dilution assays.
To compare the rate of bacterial killing by PANDA disks, the test organisms were exposed to 100 mm3 PANDA disk and the number of viable bacteria was determined by a modified kinetic track dilution method [38]. Individual disks were submerged in 4 mL aliquots of bacterial cultures adjusted to ~105 CFU mL−1 as described above and incubated at 37 °C with shaking (200 rpm). Bacterial populations were determined immediately after the addition of disks (0 h), and at 4, 8, 16, 24, 30, and 48 h of exposure. For the track-dilution technique, Falcon 15-mm square plates with 13-mm grids (Becton Dickinson Labware) were used. At each time point, six 20 μL aliquots of the bacterial suspensions exposed to PANDAs were transferred into 96-well microplates prefilled with 200 μL of MHB and serially diluted 1 to 10 to reach a final dilution of 10−8. From inoculated samples, 10 μL were spotted on the agar surface along one side of the square plate using a multichannel micropipette. The plate was tipped onto its side (at a 45°−90° angle), to allow inoculum to migrate in parallel tracks across the agar and to dry for 1 min. The plate was then inverted and incubated for 24 h at 37 °C prior to colony counting. Bacteria suspended in 4 mL MHB served as a positive control, while MHB without bacterial inoculum served as a negative control. Three replicates were carried out for each time point and bacterial strain with the mean and standard deviation reported (experiment repeated twice).
2.6.4. Determination of respiratory activity
A BacLight RedoxSensor CTC Vitality Kit was used to evaluate respiratory activity of bacteria (adjusted to ~105 CFU mL−1) exposed to either PANDA disks or control disks (100 mm3) after a 24 h incubation at 37°C at 200 rpm. According to the manufacturer’s protocol, 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) (15 mg) was dissolved in 1 mL of sterile deionized water. Next, 180 μL of each sample was mixed with 20 μL of the CTC solution in a Corning™ 96-well solid black microplate and incubated at 37 °C for 30 min prior to reading with a BioTek reader equipped with a monochromator at 450 nm excitation and 630 nm emission.
2.6.5. Assessment of cell membrane integrity
Bacterial cell membrane integrity was evaluated using a LIVE/DEAD BacLight Bacterial Viability Kit. A 4 mL of ~105 CFU mL−1 P. aeruginosa was prepared and exposed to a 100 mm3 PANDA disk, prior to incubation at 37 °C with constant shaking (200 rpm). From each inoculated sample, 100 μL aliquots of the bacteria from the PANDA treatment were transferred into a Corning™ 96-well solid black microplate, and mixed with 100 μL of the LIVE/ DEAD BacLight staining reagent. The samples were taken at 0 and 30 h and incubated in the dark for 15 min. The fluorescence was measured using a 485/20 nm excitation filter (for both SYTO9 and propidium iodide) and two separate emission filters: 528/20 nm (SYTO9 emission wavelength) and a 620/40 nm (PI emission wavelength). For confocal imaging, 5 μL of the stained samples were cast onto separate microscope slides. Each sample was assayed in triplicate, and the experiment was repeated twice. From the inoculated sample, transmission electron micrographs were taken with a Zeiss 900 electron microscope operating at 50 kV and outfitted with a Model 785 Erlangshen ES1000 WCCD camera (Gatan). Samples were applied to 200 mesh copper grids (3.05 mm, 200 lines per inch square mesh, Electron Microscopy Sciences) coated with Formvar (5% polyvinyl formal resin).
2.7. Determination of antifungal activity
2.7.1. Zone of inhibition assay
The wild-type Histoplasma capsulatum G217B (ATCC 26032) strain was used in this study. Liquid H. capsulatum yeast cultures were grown to mid-log (OD600 ~1.4-1.9) in Histoplasma macrophage medium (HMM) [39] and enumerated using a hemocytometer (Incyto C-Chip). Once quantified, 1.0 × 107 cells were spread in triplicate onto pre-warmed solid HMM plates and allowed to dry for 1 h before PANDA disks (50 mm3) were overlaid in the center of each plate. After 4 days of incubation at 37 °C in 5% CO2 / 95% room air, images were captured using a DinoLite microscope camera and zones of inhibition were measured using the DinoLite software. For a negative control, a control disk was added, whereas, the positive control contained 4 mg fluconazole. Three biological replicates were used to calculate mean and standard deviation (experiment repeated twice).
2.7.2. Determination of minimum inhibitory concentration (MIC)
Four-day-old H. capsulatum yeast cultures were diluted 1:5 and grown overnight at 37 °C in 5% CO2 / 95% room air with shaking (200 rpm). Cells were then diluted with pre-aerated, pre-warmed HMM to a final OD600 of 0.1. Various sizes of PANDA disks (0–45 mm3) were added to 50 mL aliquots of the diluted yeasts as described above. The cultures were then incubated for a total of 7 days. Every 24 h, 1 mL aliquots of each culture were transferred to disposable plastic cuvettes and OD600 was measured using a BioTek reader. After 7 days, a growth curve of OD600 vs. time was generated to determine MICs. The experiment was repeated twice.
2.7.3. Fungal viability
For LIVE/DEAD staining of H. capsulatum yeast-phase cells, 1 mL aliquots of a mid-log phase culture of H. capsulatum challenged with either 45 mm3 control or PANDA disks was centrifuged at 1000 × g for 5 min, washed once with PBS, and re-suspended in PBS. Next, cells were enumerated via hemocytometry and diluted to 1 × 106 cells per mL. At this point Aqua Dead Cell Stain™ was added to the cell suspension in a final dilution of 1:1000 and incubated for 30 min at room temperature in the dark. Once labelled, the cells were washed with PBS and fixed with 37% formaldehyde for 15 min at room temperature. Flow cytometry was performed on a BD Accuri C6 Flow Cytometer using an excitation wavelength of 405 nm with emission capture at ~525 nm. A gate was then applied and viability assessed by analyzing the shift in fluorescent intensity. To qualitatively assess fungal viability, the same labelled cells were analyzed on a Zeiss LSM 510 Meta confocal microscope at 40x magnification using both differential interference contrast and 405 nm laser excitation.
2.8. Direct contact mammalian cell viability
To assess the effects of the PANDAs directly on host cells, a direct contact assay based on ISO 10993–5 with MTT was used to quantify viability of RAW 264.7 murine macrophages and VERO epithelial cells (isolated from kidney) after the addition of either control (1.5 mm3) or PANDA disks (0.5, 1.0, and 1.5 mm3) [40]. Cells with no additional supplementations were used as a control. Six replicates were measured to calculate percent survival and standard deviation. The MTT assay was carried out per the manufacturer’s protocol (Thermo Fisher) with modifications to adapt the assay from the standard 96-well plate to a 24-well cell culture plate. Both RAW 264.7 and VERO cell lines were maintained in 20 mL DMEM supplemented with 10% FBS, 50 μg mL−1 ampicillin, 4.5 mg mL−1 glucose, 584 μg mL−1 L-glutamine, and 100 μg mL−1 streptomycin in T-150 culture flasks (Sigma). Cells were incubated at 37 °C in 5% CO2. Briefly, cells were grown to confluence and detached from the culture flask with trypsin-EDTA and centrifuged briefly at 200 × g for 5 min. They were then re-suspended in 1 mL DMEM and enumerated using a hemocytometer. The cells were diluted to 5.0 × 104 cells mL−1 and 1 mL aliquots were added to each well of a 24-well cell culture plate (Sigma). Once the culture reached subconfluency (~80% confluency) in the 24-well plate, 1.5 mm3 disks were overlayed on the cells and allowed to incubate for 24 h at 37 °C in 5% CO2. After the incubation, all media were removed from the wells and replaced with 500 μL of Dulbecco’s phosphate-buffered saline (DPBS), pH 7.5. 50 μL of 12 mM MTT was then added to each well and incubated at 37 °C for 2 h. After labelling the cultures with MTT, all but 250 μL of DPBS/MTT mixture was removed from the wells. To solubilize the MTT dye, 500 μL of DMSO was added to each well and mixed thoroughly before incubating at 37°C for 10 min. Lastly, each 24-well plate was mixed and absorbance was read at 540 nm. These experiments were repeated twice.
2.9. Statistical Analysis
All measurements were made in triplicate unless otherwise stated and each experiment was performed on at least two separate occasions; the results are expressed as the average of the parallel assays with error bars representing a standard deviation from the mean.
3. Results and Discussion
3.1. Monomer synthesis and PANDA fabrication/properties
As shown in Scheme 1A, the acyclic diallyl p-anisaldehyde acetal (pAA) was synthesized in modest yields (66% after column) through Noyori’s acetal forming procedure by the reaction of allyloxytrimethylsilane with pA in the presence of TMSOTf (1H NMR, Fig. 1A) [37]. To assess the pro-antimicrobial nature of pAA, 10 μL of pure pAA and pA were placed on plates overlayed with S. Typhi (Scheme 1A). After 24 h of incubation, pAA showed no apparent zone of inhibition indicating no antimicrobial activity. In contrast, pA showed a 1 cm zone of inhibition indicating that pAA functions as a pro-antimicrobial compound only exhibiting antimicrobial activity upon hydrolysis back to pA.
Scheme 1.
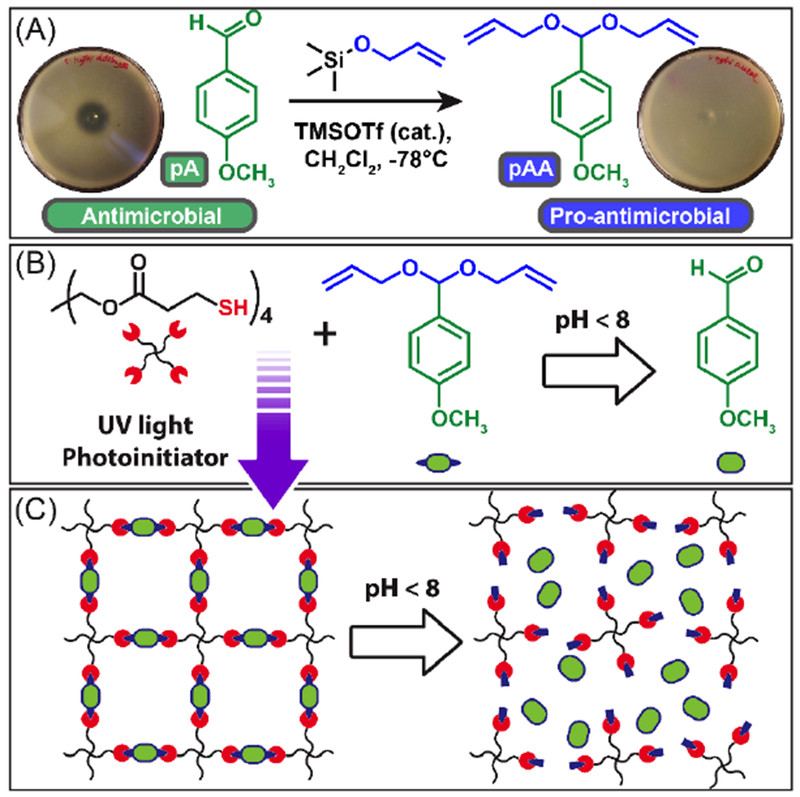
Overview of monomer synthesis and PANDA fabrication. (A) Synthesis of pro-antimicrobial pAA from antimicrobial pA. Antimicrobial properties of pAA/pA are indicated by a zone of inhibition assay against S. Typhi. (B) Monomers used to synthesize the PANDAs via thiol-ene photopolymerization and acid mediated transformation of pAA to pA. (C) Schematic depiction of PANDA degradation mechanism
Figure 1.
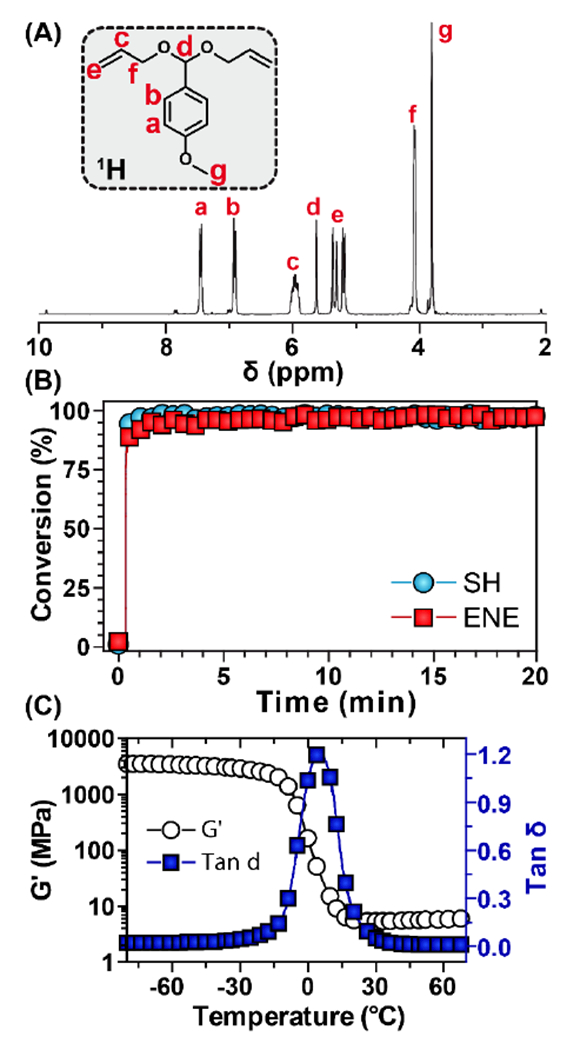
Monomer structure, cure kinetics, and final network properties. (A) 1H NMR spectra of pAA. (B) Real-time FT-IR of SH and ene conversion during photopolymerization. (C) Dynamic mechanical analysis of the resulting PANDA.
To fabricate PANDAs, pentaerythritol tetrakis(3-mercaptopropionate) (PETMP), photoinitiator and pAA were copolymerized with 1:1 alkene/thiol mole stoichiometry via radically mediated step-growth thiol-ene photopolymerization (Scheme 1B). The fabricated PANDAs are comprised of 47 wt.% pAA which corresponds to 27 wt.% pA loading. The polymerization kinetics are rapid and exhibit nearly quantitative monomer conversion (>95%, Fig. 1B) for both thiol and alkene functional groups within seconds under a medium pressure UV light (400 mW cm−2). The step-growth polymerization ensures that each crosslink junction contains a hydrolytically cleavable acetal linkage for the release of pA from the network (Scheme 1C). Additionally, the resulting PANDAs possess a narrow tan δ (indicative of a homogenous network), a glass transition temperature of −0.5 °C (Fig. 1C), and are optically transparent – typical of thiol-ene materials [41].
3.2. PANDA release kinetics
The release kinetics of pA from the PANDAs were monitored via 1H NMR spectroscopy over 38 days at pH 7.4 by submerging disks in buffered solutions in acetonitrile-d3 within sealed NMR tubes. An internal standard, 2,3,5,6-tetrachloronitrobenzene (δ = 7.75 ppm), was added to the buffered solution and integrated relative to the benzylic aldehyde proton (δ = 9.85 ppm) to determine the concentration of pA over time (Fig. 2A). Figure 2B shows the release profile of pA from the integrated 1H NMR results. At physiological pH, no burst release was observed and 90% pA was released after 38 days. The release profile was then fit to the cylindrical Hopfenberg model [42] described as:
where, Mt is the concentration of released pA at time t, M∞ is the theoretical maximum of pA released, k0 is the erosion rate constant, C0 is the initial concentration of drug in the matrix, and a is the initial radius of the cylinder. From the Hopfenberg model, the half-life was determined to be 14.9 days and the fit was in good agreement with the release data as indicated by an r2 value of 0.989. To verify the surface erosion of the PANDAs, control (PETMP and trimethylolpropane diallyl ether) and PANDA disks were fabricated, immediately submerged in a 1N aqueous HCl solution, and imaged over time. Within 1 h, significant surface erosion was visually observed as noted by the degradation front highlighted by the green arrows in Figure 2C. Due to the low solubility of the degradation products (e.g. p-anisaldehyde and tetrafunctional alcohol) in water, the solid disk gradually transitions to an oil-like residue within 3 h. A plot of the remaining disk area at time t (At) relative to the initial area (A∞) was plotted overtime and then fit by the Hopfenberg model, where Mt/M∞ were replaced by At/A∞. In comparison to the degradation at pH 7.4, the degradation at pH 0.1 was 380× faster (half-life = 0.04 days), indicating that PANDA degradation is acid sensitive. While small molecule model studies show that acetal degradation is generally 10 times faster with each unit of pH decrease [43], Liu et al. reported that crosslinked acetal network degradation follows small molecule degradation kinetics qualitatively [36]. Moreover, Garripelli and coworkers showed that acetal-based material degradation does not follow first order kinetics relative to the hydronium ion concentration [44]. The differences between network and small molecule degradation are likely attributed to differences in accessibility of the acid to each acetal as well as the hydropathy of the crosslinked network.
Figure 2.
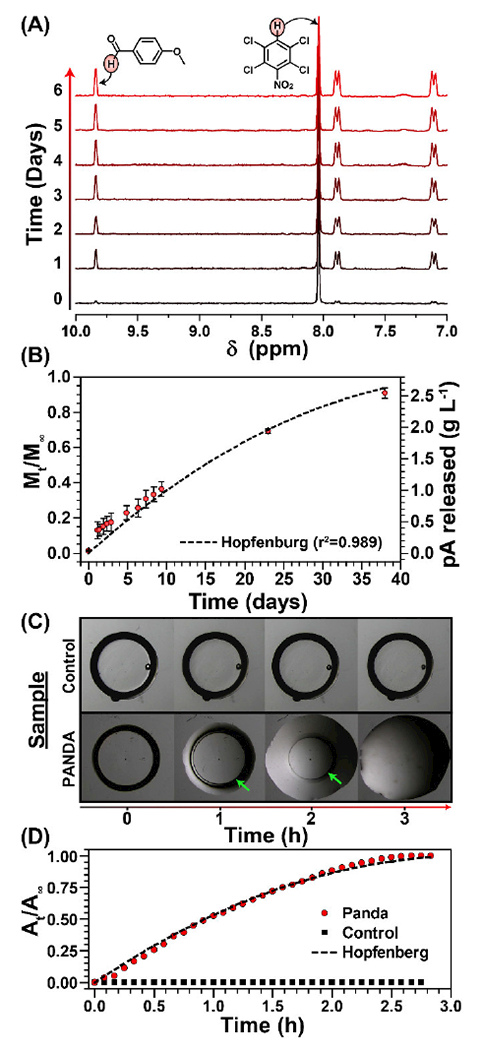
Degradation of PANDAs. (A) 1H NMR degradation kinetics of PANDA at pH 7.4. (B) Cumulative release of pA from PANDA subjected to pH 7.4. (C) Kinetic optical microscopy images of a PANDA disk submerged in 1 N HCl. Green arrows indicate remaining PANDA disk. (D) Calculated cumulative area from 1N HCl degradation kinetics determined by imageJ analysis.
3.3. PANDA antibacterial activity
3.3.1. Bacterial inhibitory assays
The use of pA was motivated by its potent activity exhibited in screening experiments against pathogenic bacteria. The antimicrobial activity of 50 mm3 PANDAs and control disks was initially evaluated via a ZOI assay with clinically isolated strains: S. aureus RN6390 and P. aeruginosa PAO1, and foodborne pathogens: E. coli ATCC 43895 (serotype O157:H7) and S. Typhi ATCC 6539 (Fig. 3A). In all cases, the control disks showed no inherent antimicrobial activity towards any bacteria, whereas zones of inhibition (>1 cm) were present for the PANDA disks. The ZOI assay showed that the order of antimicrobial inhibition was S. Typhi > S. aureus > E. coli O157:H7 > P. aeruginosa. To determine the PANDA disk size required to inhibit bacterial growth, a minimum inhibitory disk size assay was performed with exponentially grown bacteria (105 CFU mL−1 in 4 mL media) challenged with PANDA disks (0-100 mm3). As shown in Figure 3b, an increase in disk size correlates with an increase in bacterial inhibition with a 100 mm3 PANDA disk [910 μg mL−1 released after 24 h (determined from Fig. 2B)] completely inhibiting the growth of all bacteria. The trend in bacterial susceptibility from the ZOI assay was also observed in the minimum inhibition assay with E. coli O157:H7 and P. aeruginosa requiring the largest PANDA disk (100 mm3), while S. Typhi and S. aureus were inhibited by a 50 mm3 PANDA disk [455 μg mL−1 released after 24 h (determined from Fig. 2B). It should be noted that the specified MIC values are estimated using concentrations determined at 24 h from the previously described NMR release profiles. While the release profiles were not conducted in MHB growth media due to challenges quantifying pA concentrations under such conditions, these MICs are in good agreement with values reported in literature for pA involving a range of bacteria [45].
Figure 3.
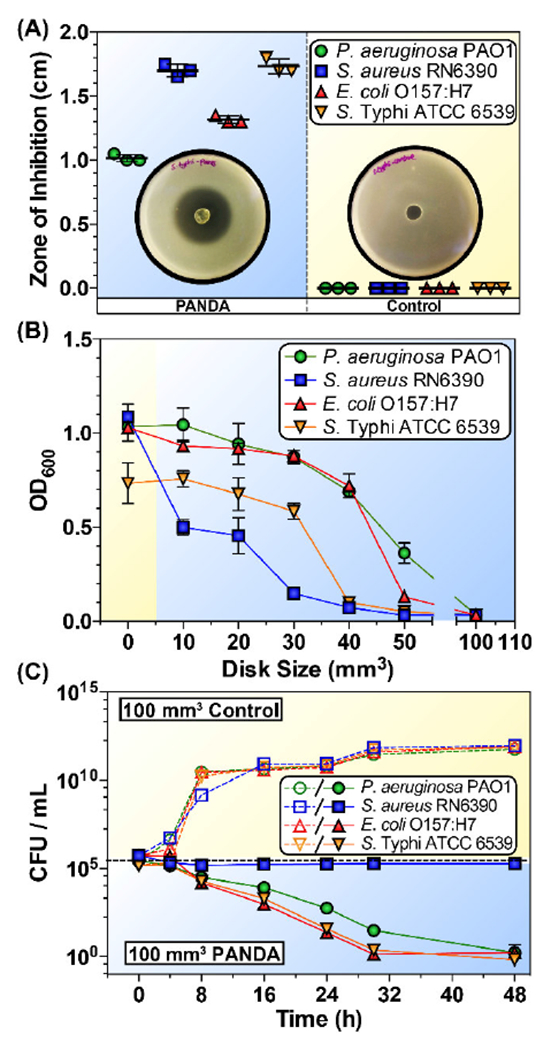
Antibacterial activity of PANDAs. (A) ZOI of PANDA and control disks. Plotted data points represent individual measurements. (B) Minimum inhibition assay of different sized PANDA disks. (C) Time-dependent killing of pathogens by 100 mm3 PANDA disks. Data are representative of 2 independent experiments ± s.d.
3.3.2. Determination of bacteriostatic/bactericidal activity of PANDA disks
To examine the bactericidal activity of 100 mm3 PANDA disks, the number of viable bacteria was quantified via a kinetic track dilution assay at 0, 4, 8, 16, 24, 30, and 48 h. The PANDA disk (1085 μg mL−1 pA released within 48 h) exhibited bactericidal activity with a >4 log reduction in bacteria count (>99.99% elimination) against P. aeruginosa, E. coli, and S. Typhi. However, the concentration of gram-positive S. aureus remained constant at 105 CFU mL−1, indicating a bacteriostatic effect. Our results are in agreement with previous studies which have shown that the bactericidal/bacteriostatic activity is dependent on both EO concentration and bacterial species [46, 47].
3.3.3. Evaluation of bacterial respiratory activity and membrane integrity
To investigate the antibacterial efficacy of PANDA disks (100 mm3), a series of physiological assays along with micrographs were performed. The fluorogenic probe, 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) was used to measure bacterial respiratory activity. Bacteria capable of respiration reduce CTC into CTC-formazan (CTF), an insoluble, fluorescent product that can be quantified via fluorescence readings. As shown in Figure 4A, in comparison to the control disks, bacteria treated with PANDAs showed a 90–95% loss of fluorescence indicating a significant decrease in respiratory activity. Interestingly, with S. aureus, PANDAs had a bacteriostatic effect and the population remained unchanged despite the significant decrease in metabolic activity. The different response of bacteria to pA is not unprecedented as some antibiotics can act as both bactericidal or bacteriostatic agents [48]. Lobritz et al. [48] attributed the differences between the bactericidal or bacteriostatic modes of actions to the interference with cellular respiration. Bactericidal antibiotics cause an acceleration in respiration and basal metabolism that ultimately lead to cell death. In contrast, bacteriostatic antibiotics suppress cellular respiration and translation thus interfering with killing. It is plausible that the altered responses of S. aureus and the tested gram-negative species to pA are caused by the different effect of pA on metabolism in these microorganisms. We also investigated the effect of PANDA disks (100 mm3) on membrane integrity of P. aeruginosa stained with SYTO9 and propidium iodide at 0 h and 30 h by confocal imaging. Green-fluorescing SYTO9 can penetrate intact and damaged bacterial membranes, whereas red propidium iodide enters only cells with damaged cytoplasmic membranes and displaces SYTO9, leading to red fluorescence when both dyes are present [49]. Prior to PANDA exposure (0 h), a majority of cells fluoresce green indicating intact membranes while after 30 h, a large number of cells fluoresce red denoting damage of the cytoplasmic membranes (Fig. 4B & 4C). To compliment the membrane integrity assay, transmission electron microscopy (TEM) was also used to image P. aeruginosa after 0 and 30 h of incubation with a PANDA disk. At 0 h, an intact membrane is clearly observed (Fig. 4D), however after 30 h, TEM shows the complete destruction of the cell membrane with crumpled cell envelopes (Fig. 4E). These results show that PANDAs lead to a loss of respiratory activity and damage of cell membrane integrity, indicating that the pA released from PANDAs, can disrupt the membrane and damage other metabolic functions as previously reported in literature [50, 51]. Additionally, other labs have begun to elucidate the specific antimicrobial mechanism of pA with evidence demonstrating up/down regulation of genes critical for cellular survival [52], and rapid shiff-base formation with amines potentially present in peptides, proteins, and DNA/RNA [53].
Figure 4.
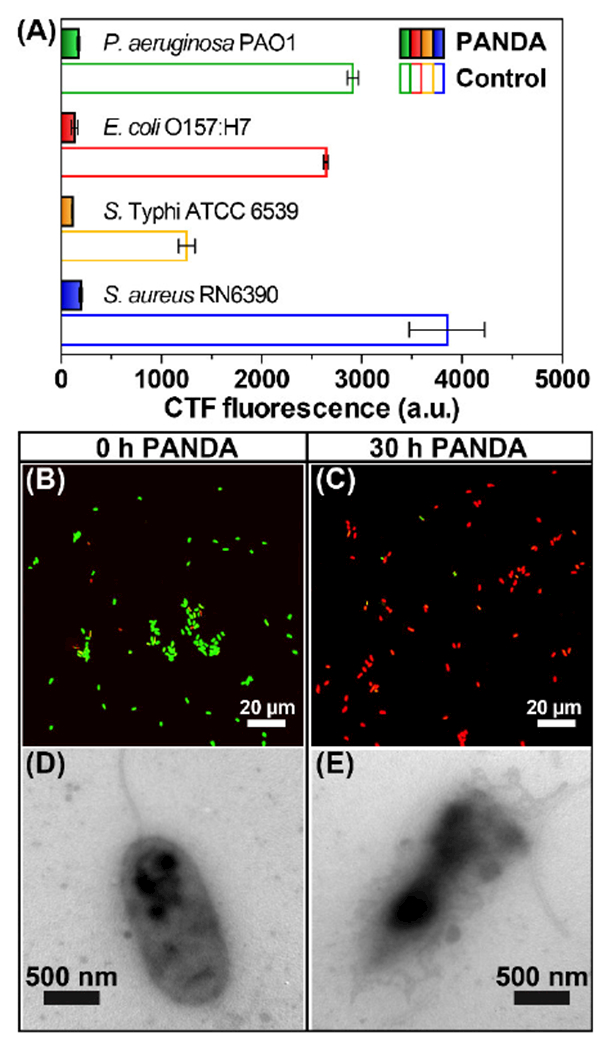
PANDA antibacterial mode of action. (A) CTC fluorescence assay indicating bacterial respiratory activity. Experiment done in triplicate ± s.d. (B) Confocal microscopy image of P. aeruginosa incubated with 100 mm3 PANDA for 0 h and 30 h (C). (D) TEM of P. aeruginosa challenged by 100 mm3 PANDA for 0 h and 30 h (E).
3.4. Antifungal activity of PANDAs.
3.4.1. Fungal inhibitory assays
To further probe the broad spectrum antimicrobial activity of PANDAs, we performed a series of assays against the pathogenic fungus H. capsulatum. The invasive nature of H. capsulatum coupled with its resistance to most azole-based antifungal treatments makes the highly pathogenic H. capsulatum strain G217B, a model fungal system to assess PANDA activity [54]. As a preliminary experiment, a ZOI assay was performed with a 50 mm3 PANDA and control disk on yeast-phase H. capsulatum. Complete inhibition of growth was observed with the PANDA treatment, whereas no measureable antifungal activity was detected for the control disk (Fig. 5A). To determine the minimum PANDA disk size required to inhibit H. capsulatum, PANDAs of various disk sizes (0–45 mm3) were added to freshly passed cells, incubated, and every 24 h, optical density measurements were recorded to assess the growth kinetics. As the size of the PANDA disk increased, a reduction in H. capsulatum growth rate was observed, with 45 mm3 disks (32.8 μg mL−1 released in 24 h) completely inhibiting growth. In contrast, the control disk, as expected, displayed a normal growth pattern with initial log phase (1–3 days) followed by lag phase (3–7 days), which is consistent with healthy H. capsulatum growth. Interestingly, the MIC for the fungus (32.8 μg mL−1) was significantly less than the MIC for the bacteria (910 μg mL−1) indicating that the fungus is more sensitive to the PANDA disks than the pathogenic bacteria tested.
Figure 5.
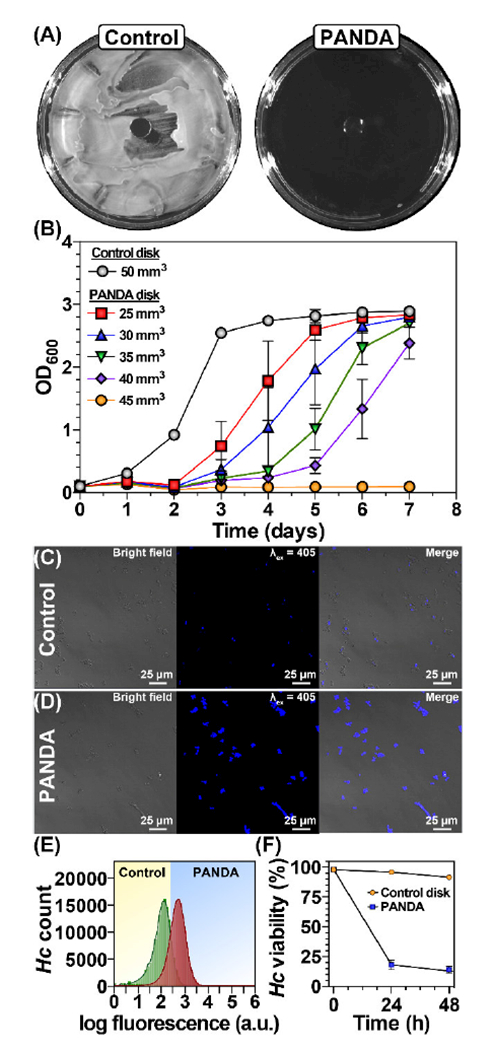
Antifungal activity of PANDAs. (A) ZOI of PANDA and control disks. (B) Minimum inhibition assay of different sized PANDA disks. Confocal microscopy images of H. capsulatum challenged with control (C) and PANDA disks (D) after 48 h. (E) Flow cytometry data of control and PANDA disks after 48 h incubation F) Time-dependent killing of pathogens by PANDA. Data are representative of 3 independent experiments ± s.d.
3.4.2. Fungicidal activity of PANDAs
To examine the fungicidal activity of the PANDA disks, we utilized an Aqua live/dead cell stain and flow cytometry. In brief, an exponential-growing culture of H. capsulatum was split in half where one half of the culture received a control disk and the other half received a 50 mm3 PANDA disk. At 24 h and 48 h after PANDA addition, aliquots were taken and subjected to staining, fixation (37% formaldehyde), confocal microscopy, and flow cytometry to assess viability. Once complete gating was performed on the sample data, the viability was determined as a percent of the survived cells. The Aqua fluorescent dye reacts with amines yielding fluorescence, but is unable to penetrate living cells leading to weakly fluorescent cells. However, compromised cellular membranes allow the dye to permeate the cell, reacting with both cell-wall bound amines and intracellular amines, leading to intense fluorescence. As shown in Figure 5C, at 48 h post addition the control disk cells showed very little, if any fluorescence, indicating that the cell walls are functional and intact. In contrast, treatment with the PANDA disks resulted in intense fluorescence from all cells, indicating membrane destabilization and cell death (Fig. 5D). The difference in fluorescence intensity is sufficient to quantify living from non-living cells using flow cytometry. As shown in Figure 5F, roughly 25% of the H. capsulatum population were viable within 24 h for the PANDA treatment, whereas virtually no change in viability was observed for the control disk. After 48 h only 15% of PANDA treated H. capsulatum cells remained viable whereas there was again no significant change in viability in the control disk. We can conclude from this assay that the release of pA from PANDAs results in potent membrane disruption and that the PANDA disk is not only bactericidal but also fungicidal.
3.5. Cytotoxicity analysis
Successful antimicrobial agents should possess both potency against pathogens as well as low cytotoxicity to host cells. Clinically speaking, pathogens like H. capsulatum are difficult to treat because they sequester themselves inside alveolar macrophages, one of the most valuable immune cell needed to eradicate them. To evaluate PANDA cytotoxicity, two cell lines, including RAW 264.7 murine macrophages and VERO epithelial cells (isolated from kidney), were analyzed by directly exposing cells to multiple sizes of PANDA disks [0.5 mm3 (18.2 μg mL−1), 1.0 mm3 (36.4 μg mL−1), and 1.5 mm3 (54.6 μg mL−1)], 1.5 mm3 control disks, and a native control (blank). Cell viabilities were determined using a MTT assay over a 24 h period. MTT is a colorimetric dye that assesses cellular metabolic activity. In brief, live viable cells can reduce MTT to an insoluble formazan by their native oxidase system, which is accompanied by a change in color to purple that can be observed via spectrophotometry. As shown in Figure 6A, both cell lines showed no significant decrease in viability (90–100 % viable compared to the native control) after exposure to PANDA disks of various sizes (0.5–1.5 mm3), indicating minimal cytotoxicity towards host cells. We also qualitatively investigated cell morphologies to ensure no changes were observed after PANDA treatments. After 24 h, the cells treated with the PANDA disk exhibited indistinguishable morphologies and confluency compared to the control disk and native control. Macrophage cells displayed a dominant spherical morphology with minimal pseudopodia (Figure 6B) whereas epithelial cells displayed a spindle like morphology of closely aggregated cells (Figure 6C), both without any signs of cytoplasmic vacuolation or granularity around the nucleus. Of significance, the low cytotoxicity of PANDAs against macrophages at a concentration that is shown to eradicate H. capsulatum holds great promise for clinical applications moving forward.
Figure 6.
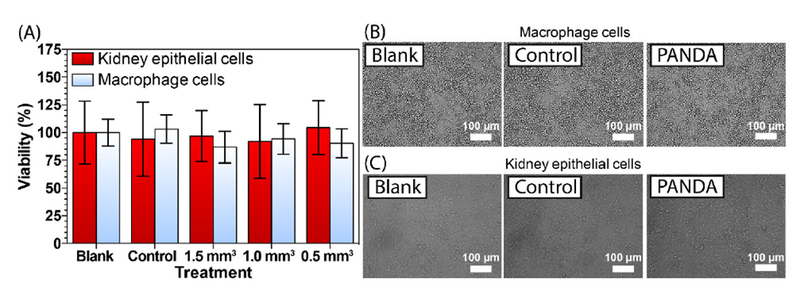
Cytocompatability of PANDAs. (A) MTT assay of PANDAs incubated with VERO kidney epithelial and RAW 264.7 macrophage cells. (B) Bright-field images of macrophage and epithelial cells (C), incubated with either nothing (blank), a 1.5 mm3 control disk, or a 1.5 mm3 PANDA disk.
4. Conclusion
In summary, we have demonstrated a potent bio-based PANDA that has the versatility to treat both prokaryotic and eukaryotic pathogens through the sustained release of pA over time. The sustained release of pA allows many generations of the microbes to be exposed, thus successfully clearing even slow growing pathogens such as H. capsulatum. Both inhibitory and biocidal activity was measured against bacteria and fungi and found that the fungi were significantly more susceptible to the PANDA treatment. Through a combination of confocal microscopy and transmission electron microscopy, we showed PANDAs primary mode of action is via membrane disruption. Additionally, we demonstrated that PANDAs have minimal cytotoxicity towards both epithelial and macrophages which are both relevant in immunosuppressed patient response. The PANDA network design coupled with the potency of pA has the potential to be a streamline target to add to the arsenal of defenses against a broad spectrum of pathogens and bares potential for future in vivo experiments.
Statement of significance.
With the increasing number of patients prescribed immunosuppressants coupled with the rise in antibiotic resistance – life-threatening microbial infections are a looming global threat. With limited success within the antibiotic pipeline, nature-based essential oils (EOs) are being investigated for their multimodal effectiveness against microbes. Despite the promising potential of EOs, difficulties in their encapsulation, limited water solubility, and high volatility limit their use. Various studies have shown that covalent attachment of these EO derivatives to polymers can mitigate these limitations. The current study presents the synthesis of a fully-degradable, sustained release, cytocompatible, pro-antimicrobial acetal network derived from p-anisaldehyde. This polymer network design provides a pathway toward application-specific EO releasing materials with quantitative encapsulation efficiencies, sustained release, and broad-spectrum antimicrobial activity.
Acknowledgements
The authors acknowledge the National Science Foundation (CHE-1710589 and OIA-1430364) for partial support of this research. DVA and DNA acknowledges traineeship support from the NSF NRT program “Interface” (DGE-1449999) through the University of Southern Mississippi. Confocal microscopy was supported by MS INBRE funded by NCRR (5P20RR-016476–11) and NIGMS/NIH (8 P20 GM103476–11). D.V.M. acknowledges startup funds from the University of Southern Mississippi. The authors thank Bruno Bock for the kind donation of PETMP used in this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].White TC, Marr KA, Bowden RA, Clinical, Cellular, and Molecular Factors That Contribute to Antifungal Drug Resistance, Clinical Microbiology Reviews 11(2) (1998) 382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Armstrong-James D, Meintjes G, Brown GD, A neglected epidemic: fungal infections in HIV/AIDS, Trends in Microbiology 22(3) (2014) 120–127. [DOI] [PubMed] [Google Scholar]
- [3].Ribeiro M, Monteiro FJ, Ferraz MP, Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions, Biomatter 2(4) (2012) 176–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cianflone NFC, Salmonellosis and the GI Tract: More than Just Peanut Butter, Current gastroenterology reports 10(4) (2008) 424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho M-C, Almo SC, Schramm VL, Transition state analogs of 5[prime]-methylthioadenosine nucleosidase disrupt quorum sensing, Nat Chem Biol 5(4) (2009) 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stone LK, Baym M, Lieberman TD, Chait R, Clardy J, Kishony R, Compounds that select against the tetracycline-resistance efflux pump, Nat Chem Biol 12(11) (2016) 902–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Baym M, Stone LK, Kishony R, Multidrug evolutionary strategies to reverse antibiotic resistance, Science 351(6268) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wright GD, Antibiotic Adjuvants: Rescuing Antibiotics from Resistance, Trends in Microbiology 24(11) (2016) 862–871. [DOI] [PubMed] [Google Scholar]
- [9].Yoshida M, Reyes SG, Tsuda S, Horinouchi T, Furusawa C, Cronin L, Time-programmable drug dosing allows the manipulation, suppression and reversal of antibiotic drug resistance in vitro, Nature Communications 8 (2017) 15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yarlagadda V, Samaddar S, Paramanandham K, Shome BR, Haidar J, Membrane Disruption and Enhanced Inhibition of Cell-Wall Biosynthesis: A Synergistic Approach to Tackle Vancomycin-Resistant Bacteria, Angewandte Chemie International Edition 54(46) (2015) 13644–13649. [DOI] [PubMed] [Google Scholar]
- [11].Yarlagadda V, Sarkar P, Samaddar S, Haldar J, A Vancomycin Derivative with a Pyrophosphate-Binding Group: A Strategy to Combat Vancomycin-Resistant Bacteria, Angewandte Chemie 128(27) (2016) 7967–7971. [DOI] [PubMed] [Google Scholar]
- [12].Nazzaro F, Fratianni F, De Martino L, Coppola R, De Feo V, Effect of essential oils on pathogenic bacteria, Pharmaceuticals 6(12) (2013) 1451–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yap PSX, Yiap BC, Ping HC, Lim SHE, Essential Oils A New Horizon in Combating Bacterial Antibiotic Resistance, The Open Microbiology Journal 8 (2014) 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Turek C, Stintzing FC, Stability of Essential Oils: A Review, Comprehensive Reviews in Food Science and Food Safety 12(1) (2013) 40–53. [Google Scholar]
- [15].Stebbins ND, Faig JJ, Yu W, Guliyev R, Uhrich KE, PolyActives: Controlled and Sustained Bioactive Release Via Hydrolytic Degradation, Biomaterials science 3(8) (2015) 1171–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wattamwar PP, Mo Y, Wan R, Palli R, Zhang Q, Dziubla TD, Antioxidant activity of degradable polymer poly (trolox ester) to suppress oxidative stress injury in the cells, Advanced Functional Materials 20(1) (2010) 147–154. [Google Scholar]
- [17].Rosario-Meléndez R, Yu W, Uhrich KE, Biodegradable Polyesters Containing Ibuprofen and Naproxen As Pendant Groups, Biomacromolecules 14(10) (2013) 3542–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ouimet MA, Griffin J, Carbone-Howell AL, Wu W-H, Stebbins ND, Di R, Uhrich KE, Biodegradable ferulic acid-containing poly (anhydride-ester): degradation products with controlled release and sustained antioxidant activity, Biomacromolecules 14(3) (2013) 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Carbone-Howell AL, Stebbins ND, Uhrich KE, Poly (anhydride-esters) comprised exclusively of naturally occurring antimicrobials and EDTA: Antioxidant and antibacterial activities, Biomacromolecules 15(5) (2014) 1889–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rosenberg L, Carbone A, Römling U, Uhrich K, Chikindas M, Salicylic acid-based poly (anhydride esters) for control of biofilm formation in Salmonella enterica serovar Typhimurium, Letters in applied microbiology 46(5) (2008) 593–599. [DOI] [PubMed] [Google Scholar]
- [21].Ouimet MA, Stebbins ND, Uhrich KE, Biodegradable Coumaric Acid-Based Poly (anhydride-ester) Synthesis and Subsequent Controlled Release, Macromolecular rapid communications 34(15) (2013) 1231–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Johnson ML, Uhrich KE, Concurrent release of admixed antimicrobials and salicylic acid from salicylate-based poly (anhydride-esters), Journal of Biomedical Materials Research Part A 91(3) (2009) 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Carbone-Howell A, Ouimet M, Uhrich K, Biodegradable, Bioactive-Based Poly (anhydride-esters) for Personal Care and Cosmetic Applications, Polymers for Personal Care and Cosmetics, ACS Publications 2013, pp. 145–155. [Google Scholar]
- [24].Prudencio A, Faig JJ, Song M, Uhrich KE, Phenolic Acid-based Poly (anhydride-esters) as Antioxidant Biomaterials, Macromolecular bioscience 16(2) (2016) 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stebbins ND, Yu W, Uhrich KE, Linear, Mannitol-Based Poly (anhydride-esters) with High Ibuprofen Loading and Anti-Inflammatory Activity, Biomacromolecules 16(11) (2015) 3632–3639. [DOI] [PubMed] [Google Scholar]
- [26].Prudencio A, Stebbins ND, Johnson M, Song M, Langowski BA, Uhrich KE, Polymeric prodrugs of ampicillin as antibacterial coatings, Journal of Bioactive and Compatible Polymers 29(3) (2014) 208–220. [Google Scholar]
- [27].Noel A, Borguet YP, Raymond JE, Wooley KL, Poly(carbonate-amide)s Derived from Bio-Based Resources: Poly(ferulic acid-co-tyrosine), Macromolecules 47(9) (2014) 2974–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Faig JJ, Moretti A, Joseph LB, zhang Y, Nova MJ, Smith K, Uhrich KE, Biodegradable Kojic Acid-Based Polymers: Controlled Delivery of Bioactives for Melanogenesis Inhibition, Biomacromolecules 18(2) (2017) 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Guo S, Nakagawa Y, Barhoumi A, Wang W, Zhan C, Tong R, Santamaria C, Kohane DS, Extended Release of Native Drug Conjugated in Polyketal Microparticles, Journal of the American Chemical Society 138(19) (2016) 6127–6130. [DOI] [PubMed] [Google Scholar]
- [30].Kwon J, Kim J, Park S, Khang G, Kang PM, Lee D, Inflammation-Responsive Antioxidant Nanoparticles Based on a Polymeric Prodrug of Vanillin, Biomacromolecules 14(5) (2013) 1618–1626. [DOI] [PubMed] [Google Scholar]
- [31].Chandorkar Y, Bhagat RK, Madras G, Basu B, Cross-Linked, Biodegradable, Cytocompatible Salicylic Acid Based Polyesters for Localized, Sustained Delivery of Salicylic Acid: An In Vitro Study, Biomacromolecules 15(3) (2014) 863–875. [DOI] [PubMed] [Google Scholar]
- [32].van Lith R, Gregory EK, Yang J, Kibbe MR, Ameer GA, Engineering biodegradable polyester elastomers with antioxidant properties to attenuate oxidative stress in tissues, Biomaterials 35(28) (2014) 8113–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dasgupta Q, Madras G, Chatterjee K, Controlled release kinetics of p-aminosalicylic acid from biodegradable crosslinked polyesters for enhanced anti-mycobacterial activity, Acta Biomaterialia 30 (2016) 168–176. [DOI] [PubMed] [Google Scholar]
- [34].Wattamwar PP, Biswal D, Cochran DB, Lyvers AC, Eitel RE, Anderson KW, Hilt JZ, Dziubla TD, Synthesis and characterization of poly(antioxidant β-amino esters) for controlled release of polyphenolic antioxidants, Acta Biomaterialia 8(7) (2012) 2529–2537. [DOI] [PubMed] [Google Scholar]
- [35].Amato DN, Amato DV, Mavrodi OV, Martin WB, Swilley SN, Parsons KH, Mavrodi DV, Patton DL, Pro-Antimicrobial Networks via Degradable Acetals (PANDAs) Using Thiol-Ene Photopolymerization, ACS Macro Letters 6(2) (2017) 171–175. [DOI] [PubMed] [Google Scholar]
- [36].Liu B, Thayumanavan S, Substituent Effects on the pH Sensitivity of Acetals and Ketals and Their Correlation with Encapsulation Stability in Polymeric Nanogels, Journal of the American Chemical Society 139(6) (2017) 2306–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tsunoda T, Suzuki M, Noyori R, A facile procedure for acetalization under aprotic conditions, Tetrahedron letters 21(14) (1980) 1357–1358. [Google Scholar]
- [38].Jett BD, Hatter KL, Huycke MM, Gilmore MS, Simplified agar plate method for quantifying viable bacteria, Biotechniques 23(4) (1997) 648–650. [DOI] [PubMed] [Google Scholar]
- [39].Worsham P, Goldman W, Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores, Journal of Medical and Veterinary Mycology 26(3) (1988) 137–143. [PubMed] [Google Scholar]
- [40].I. ISO, 10993–5: 2009 Biological Evaluation of Medical Devices—Part 5: Tests for in Vitro Cytotoxicity, International Organization for Standardization, Geneva: (2009). [Google Scholar]
- [41].Hoyle CE, Bowman CN, Thiol—Ene Click Chemistry, Angew. Chem., Int. Ed. 49(9) (2010) 1540–1573. [DOI] [PubMed] [Google Scholar]
- [42].Hopfenberg HB, Controlled Release from Erodible Slabs, Cylinders, and Spheres, Controlled Release Polymeric Formulations, AMERICAN CHEMICAL SOCIETY; 1976, pp. 26–32.. [Google Scholar]
- [43].Fife TH, Jao LK, Substituent Effects in Acetal Hydrolysis, The Journal of Organic Chemistry 30(5) (1965) 1492–1495. [Google Scholar]
- [44].Garripelli VK, Kim JK, Namgung R, Kim WJ, Repka MA, Jo S, A novel thermosensitive polymer with pH-dependent degradation for drug delivery, Acta Biomaterialia 6(2) (2010) 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Si W, Gong J, Tsao R, Zhou T, Yu H, Poppe C, Johnson R, Du Z, Antimicrobial activity of essential oils and structurally related synthetic food additives towards selected pathogenic and beneficial gut bacteria, Journal of Applied Microbiology 100(2) (2006) 296–305. [DOI] [PubMed] [Google Scholar]
- [46].Mayaud L, Carricajo A, Zhiri A, Aubert G, Comparison of bacteriostatic and bactericidal activity of 13 essential oils against strains with varying sensitivity to antibiotics, Letters in Applied Microbiology 47(3) (2008) 167–173. [DOI] [PubMed] [Google Scholar]
- [47].Smith P, Stewart, Fyfe, Antimicrobial properties of plant essential oils and essences against five important food-borne pathogens, Letters in Applied Microbiology 26(2) (1998) 118–122. [DOI] [PubMed] [Google Scholar]
- [48].Lobritz MA, Belenky P, Porter CBM, Gutierrez A, Yang JH, Schwarz EG, Dwyer DJ, Khalil AS, Collins JJ, Antibiotic efficacy is linked to bacterial cellular respiration, Proceedings of the National Academy of Sciences 112(27) (2015) 8173–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Berney M, Hammes F, Bosshard F, Weilenmann H-U, Egli T, Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry, Applied and environmental microbiology 73(10) (2007) 3283–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chen X, Zhang X, Meng R, Zhao Z, Liu Z, Zhao X, Shi C, Guo N, Efficacy of a combination of nisin and p-Anisaldehyde against Listeria monocytogenes, Food Control 66 (2016) 100–106. [Google Scholar]
- [51].Shi C, Zhao X, Meng R, Liu Z, Zhang G, Guo N, Synergistic antimicrobial effects of nisin and p-Anisaldehyde on Staphylococcus aureus in pasteurized milk, LWT - Food Science and Technology 84(Supplement C) (2017) 222–230. [Google Scholar]
- [52].Yu L, Guo N, Yang Y, Wu X, Meng R, Fan J, Ge F, Wang X, Liu J, Deng X, Microarray analysis of p-anisaldehyde-induced transcriptome of Saccharomyces cerevisiae, Journal of industrial microbiology & biotechnology 37(3) (2010) 313–322. [DOI] [PubMed] [Google Scholar]
- [53].Natsch A, Gfeller H, Haupt T, Brunner G, Chemical Reactivity and Skin Sensitization Potential for Benzaldehydes: Can Schiff Base Formation Explain Everything?, Chemical Research in Toxicology 25(10) (2012) 2203–2215. [DOI] [PubMed] [Google Scholar]
- [54].Kauffman CA, Histoplasmosis: a Clinical and Laboratory Update, Clinical Microbiology Reviews 20(1) (2007) 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]


