Abstract
Sleep and wake quality, quantity and architecture become modified with aging. Sleep and wake quality decline coinciding with increased fragmentation of both states across aging. We have previously shown that this age-related decline in sleep-wake quality is associated with increased endoplasmic reticular (ER) stress and decreased expression of the major ER chaperone BiP (Binding immunoglobulin Protein). BiP, also known as glucose-regulated protein 78 (GRP78), plays a key role in controlling the cellular response to ER stress, acting as a regulator of a protein homeostatic signaling pathway known as the unfolded protein response (UPR). Induction of BiP during cellular stress is part of an adaptive pro-survival mechanism. Here, using mice heterozygous for BiP, we investigated the effect of reduced BiP expression on sleep-wake behavior across aging; complete knockdown of BiP is embryonic lethal. We report that BiP heterozygosity accentuates the aging sleep-wake phenotype. Sleep and wake fragmentation was more pronounced in the BiP heterozygotes across the three ages examined. In mice lacking one functional copy of BiP, we observed an age-related significant reduction in wake bout duration and increase in wake bout numbers during the active period, as well as an increase in NREM and REM bout numbers accompanied by reduced bout durations of both NREM and REM during the sleep period. In addition, we observed increased ER stress in orexin neurons as well as occurrence of aggregates immunopositive for orexin at the terminals and projections of orexin neurons in the middle aged BiP heterozygotes. Taken together, our data indicates that a reduction in the molecular chaperone BiP impacts sleep architecture across aging and that orexin processing is likely to be affected.
Keywords: Aging, sleep, unfolded protein response, chaperone, BiP
Graphical abstract
1. INTRODUCTION
Binding immunoglobulin Protein (BiP), also known as glucose regulated protein 78 (GRP78) and Hspa5, is a molecular chaperone and a member of the heat shock 70 family of proteins. BiP is an ATPase and the major chaperone resident in the endoplasmic reticulum (ER). Until very recently, BiP has been considered exclusively an ER luminal protein, where it plays a major role in protein homeostasis (Luo and Lee, 2013). BiP is a key regulator of the protein quality control and signaling pathway known as the unfolded protein response (UPR) [for reviews, see (Naidoo, 2009; Ron and Walter, 2007; Schroder and Kaufman, 2005)]. Emerging evidence indicates that BiP can also be detected in other cellular locations, including the cell surface, cytosol, mitochondria and the nucleus, and assumes novel functions that control signaling, proliferation, invasion, apoptosis, inflammation and immunity (Luo and Lee, 2013; Ni et al., 2011). We have found that brain BiP levels decline in both mice and Drosophila during aging (Brown et al., 2014; Naidoo et al., 2008). Similar declines with age have been described in rat hippocampus and peripheral tissues (Hussain and Ramaiah, 2007; Paz Gavilan et al., 2006). Our studies have shown that reduction in BiP expression is accompanied by increased endoplasmic reticular stress and deterioration in the efficacy and efficiency of the UPR (Brown et al., 2014; Naidoo et al., 2008). At the same time, while BiP levels decline with age, the levels of CHOP, a marker of the maladaptive apoptotic response to ER stress, increase (Naidoo et al., 2008; Naidoo et al., 2011). In younger animals, CHOP is increased when there is sustained ER stress (Brown and Naidoo, 2012; Oyadomari and Mori, 2004; Ron and Walter, 2007; Rozpedek et al., 2016). We have also established that BiP plays a role in sleep homeostasis. Sleep loss leads to upregulation of BiP transcript levels in several species, including Drosophila, mice, rats and sparrows (Cirelli et al., 2004; Jones et al., 2008; Mackiewicz et al., 2007; Naidoo et al., 2007). Overexpression and expression of dominant negative BiP both alter recovery sleep following sleep deprivation in Drosophila (Naidoo et al., 2007). We have also established that supplementing chaperone levels with a chemical chaperone consolidates sleep in aged flies (Brown et al., 2014). Flies, like mice, display fragmented sleep with aging (Brown et al., 2014; Koh et al., 2006). Thus, reduction in BiP levels with age might contribute to the changes in sleep and wake behavior with age.
Given that BiP levels decline with age and that BiP expression is correlated with sleep-wake behavior, we wanted to determine the effect of reduced BiP on sleep-wake behavior across the lifespan. We hypothesized that the transgenic mouse line expressing reduced BiP (BiP heterozygote) would display an accentuated effect of age on sleep and wake behavior compared to wildtype mice. To test this hypothesis, we examined the effect of reduced BiP expression on sleep-wake behavior in a BiP heterozygous mouse model at three different ages: 2–3, 12 and 18 months. BiP heterozygous mice were chosen, as complete knockout of BiP is embryonic lethal (Luo et al., 2006). Analyses concentrated on the known effects of age on sleep and wake behavior, i.e., fragmentation of state and changes in theta power in the electroencephalogram (EEG) (Hasan et al., 2012). To examine potential mechanisms, we compared other features between wildtype and BiP heterozygous mice in the intermediate age group (12 months). We report that the BiP heterozygosity accentuates the age related decline in these sleep-wake phenotypes. In addition, we find that BiP heterozygous mice at 12 months of age display more ER stress, as assessed by increased CHOP expression in orexin neurons using immunohistochemistry, and accumulate protein aggregates immunopositive for orexin at the terminals and projections of orexin neurons, which may impact the function of these neurons. This is likely to contribute to the effects of reduced levels of BiP on sleep and wake behavior.
2. MATERIALS AND METHODS
2.1 Mice
The BiP heterozygous (HET) and wildtype (WT) littermate mice were bred from a pair of heterozygous mice generously provided to us by Dr. Amy Lee (Luo et al., 2006). The heterozygous mice had been back crossed into the C57 BL/6 background (Luo et al, 2006). Mice were genotyped using the REDExtract-N-Amp Tissue PCR kit (Sigma) and nested primers as described in Luo et al (2006). Primers used were PF2: 5′- GTT GAT ATT GGA GGT GGG CAA ACC AAG -3′; PF3: 5′- GAT TTG AAC TCA GGA CCT TCG GAA GAG CAG -3′; and PTR: 5′- TTG TTA GGG GTC GTT CAC CTA GA -3′
Mice were housed under conditions of 23.5±1°C and 35–45% humidity in a 12:12 light-dark cycle with lights on at 7am. Food and water were provided ad libitum. The methods and study protocols were approved in full by the Institutional Animal Care and Use Committee of the University of Pennsylvania and conformed to the revised NIH Office of Laboratory Animal Welfare Policy. Sleep-wake behavior was assessed by EEG/EMG (electromyogram) in BiP heterozygous and wildtype littermate mice at 3 ages: 2–3 months (n=7 WT, 8 HET), 12 months (n=13 WT, 9 HET) and 18 months (n= 8 WT, 6 HET), as described below. Mice studied in this and other protocols were obtained from the same litter.
2.2 Surgical Procedures
Mice were implanted with EEG/EMG electrodes under deep anesthesia [i.p. injection of Ketamine (100 mg/kg)/Xylazine (2.5mg/kg)/Acepromazine (2.5mg/kg)]. For EEG recordings, the skull was exposed and three burr holes were drilled in the following locations: (1) right frontal cortex (1.7 mm lateral to midline and 1.5 mm anterior to Bregma), (2) right parietal cortex (1.7 mm lateral to midline and 1 mm anterior to lambda); and (3) a reference electrode over the cerebellum (1 mm posterior lambda on the midline). Three silver wire electrodes were made by holding one end of each wire over an open flame to form a silver ball. The ball end of the wire was placed inside each of the three holes and spot glued in place with dental acrylic. A pair of Teflon coated stainless steel wires with exposed ends were sutured onto the dorsal surface of the nuchal muscles immediately posterior to the skull. All electrodes were connected to a 6-pin plastic connector/pedestal (Plastics One, Inc, VA) and then bonded to the skull with dental acrylic. After surgery mice were housed in individual cages for a 7–10 day recovery period, before study.
2.3 Sleep/Wake Recording
The mice were connected to the recording cable and commutator system (Plastics One Inc., Roanoke, VA) and habituated to the recording environment for 3–4 days before signal collection. EEG and EMG signals were amplified using a Neurodata amplifier system (Models M15, Astro-Med. Inc., West Warwick, RI). Signals were amplified (2000×) and conditioned using the following settings for EEG signals: low cut off frequency (−6dB), 0.3 Hz and high cut-off frequency (−6dB), 30 Hz; for EMG signals: low cut-off frequency (−6dB), 10 Hz and the high cut-off frequency (−6dB), 100 Hz. Signals were digitized at 100 Hz. All data were acquired and analyzed using Gamma software (Astro–Med) and converted to European Data Format (EDF) for manual scoring and analysis in the Somnologica science software (Medcare).
2.4 EEG Data Analysis
Wake, NREM and REM sleep were manually scored in consecutive 4-second epochs using standard criteria for rodent sleep (Hasan et al., 2012). All 4-second epochs containing artifact were identified and excluded from EEG spectral analysis. Spectral power changes in the EEG were determined in 0.25 Hz bins using Discrete-Fourier Transform (DFT), with a hamming window function, for the three behavioral states. EEG spectra were expressed individually as a percentage of the average power of frequencies between 0.25 and 25 Hz. Sleep and wakefulness spectra were assessed from the 12-hour baseline light or dark period, respectively.
The baseline spectral changes were determined as described by Hasan et al (Hasan et al., 2012). Briefly, EEG spectra were expressed individually as a percentage of the average power of frequencies between 0.25 and 25 Hz. Sleep and wake spectra were assessed from the 12-hour baseline lights on or lights off period, respectively. Theta-peak frequency in REM sleep and waking EEG were determined by the frequency at which EEG power density in these states in the theta range (6–10Hz) was maximal.
2.5 Immunohistochemistry
We used immunohistochemical approaches to examine the orexin neurons in the 12 month old BiP wildtype and heterozygous mice to probe the mechanism underlying the wake response: neurostereology was used to determine orexin neuron numbers (West and Gundersen, 1990); immunofluorescence to assess the c-fos response to prolonged waking, as described previously (Naidoo et al., 2011; Panossian et al., 2011); and Nickel-DAB staining to determine CHOP induction in orexin neurons and orexin aggregation at the terminals.
2.5.1 Neurostereology
Briefly, coronal sections (60 μM) were obtained. All consecutive sections encompassing the lateral hypothalamus (Bregma −0.82 to −2.30 mm) were collected. Every other section was placed in 12 well plates for free floating immunohistochemistry staining. The sections were incubated in 3%H2O2 solution for 30 minutes followed by 120 minutes incubation in blocking solution (4% donkey serum, 1% BSA, 0.4% Triton-100 in 1X PBS). The sections were then incubated for 48 hours at 4°C with the primary antibody for Orexin-A (1:5000) (SC-8070, Santa Cruz Biotechnology). After washing, the sections were incubated with biotinylated secondary antibody (1:1000, Vector Laboratories) for 90 minutes and then incubated in avidin-biotin substrate (ABC kit, Vector Laboratories) for 60 minutes. The signals were visualized with DAB. Next, the sections were counterstained with a modified Giemsa stain (Sigma) for detection of all neurons and their nuclei within the counting frame (see Figure S1). The orexin positive neurons with clearly visible stained nuclei were counted by neurostereology. Briefly, using a Nikon Eclipse 600 microscope and a Stereo Investigator workstation, version 10.3 (MicroBrightField), an optical fractionator approach (West and Gundersen, 1990) was used to estimate the total number of orexinergic neurons per mouse. The sampling was optimized to count at least 300 cells per animal with error coefficients less than 0.07. Each counting frame for optical fractionator (50×50μm) was used and grid space used was 100×100μm allowing 2 μm guard zones on either side. For orexin neuron counts, all Giemsa labeled somata >15 μm diameter with a nucleus that came into focus within the counting frame (probe) were counted using a 100 × oil objective. Section thickness was measured in all counting sites.
2.5.2 Immunoflorescence and c-Fos staining
Immunofluorescence was used to characterize the waking c-fos response in orexinergic neurons in BiP wildtype and heterozygous 12 month old mice as previously described (Naidoo et al., 2011).
To measure the c-fos response to wakefulness, mice were randomized to either undisturbed sleep to act as controls or to enforced wakefulness for 3 hours by gentle handling across the same time span (10am-1pm; lights on) as previously described (Naidoo et al., 2011). For the enforced wakefulness group, wake was electrographically confirmed for the entire 3-hour period. For the undisturbed /sleep control only mice that had experienced at least 75% sleep during the lights on period - were selected by online monitoring. At the end of the 3 hour conditions (1pm) all mice (enforced wakefulness and controls) were deeply anesthetized and perfused; brains were post-fixed, cryopreserved and sectioned using previously published techniques with 40 μm sections (Zhu et al., 2007). Orexinergic neurons in the lateral hypothalamus were labeled with monoclonal mouse anti-Orexin-A antibody (Orex-A; MAB763, R&D Systems, Minneapolis, MN). Neurons were double-labeled with the above neuronal markers for orexin-A and c-fos protein (Ab-5, EMD Calbiochem). Orex-A was labeled with Alexa fluor 488 (green), and the c-fos secondary antibody was tagged with Alexa fluor 594 (red) (Molecular Probes, Carlsbad, CA). All immunopositive orexin-A positive neurons with visible nuclei in 6 rostral-caudal sections across the nucleus/mouse were analyzed. Cells were scored as c-fos positive if c-fos labeling in the nucleus was more intense than background (Image J, NIH). Three scorers, blinded to condition, counted numbers of orexin neurons and c-fos labeling, and scores were averaged (90–98% Agreement between scorers). For each mouse >150 neurons/region were examined.
2.6 Determination of CHOP in orexin neurons and orexin aggregates at terminals
Orexin terminals, distal from the orexin neuronal cell bodies, in the medial septal region of the basal forebrain were immunostained in 40 μm sections using the orexin antibody and orexin immunopositive foci (aggregates) were quantified using Image-Pro Plus 6.1.
2.6.1 Immunostaining
Goat polyclonal orexin-A antibody was used to identify orexin-A like fibers and terminals in the basal forebrain. 1/6th basal forebrain sections from each animal were used for free-floating immunohistochemistry staining. The sections were incubated overnight at room temperature with goat antibody against orexin-A (1:5000, orexin-A (C-19), SC-8070, Santa Cruz). The next day, the sections were washed with PBS 5minX6. The sections were then incubated with biotinylated anti-goat IgG at 1:1000 for 90min (Vector Laboratories) followed by incubation in the ABC complex at 1:1000 for 60min (Vector Laboratories). The color reaction was carried out by incubating the sections in 0.175M sodium acetate with 0.02% DAB for 10 to 15 minutes. The sections were then dehydrated and cover-slipped with Permount.
For CHOP staining, free-floating sections of lateral hypothalamus were incubated overnight at room temperature with CHOP antibody (Santa Cruz; 1:200) as described previously (Naidoo et al, 2011). All orexin immunopositive neurons in 6–8 rostral-caudal sections across the nucleus/mouse were analyzed. For each mouse >220 neurons/region were examined. Cells were scored as CHOP positive if CHOP labeling in the cell and nucleus was more intense than background (Image J, NIH). Three scorers, blinded to condition, counted numbers of orexin neurons and CHOP labeling, and scores were averaged (90–93% Agreement between scorers).
2.6.2 Quantification of Aggregates
Aggregate object areas (identified by orexin immunopositivity) were determined as follows; two images of Medial Septal Nucleus area (MS) of each sample around Bregma 0.62mm were taken with Leica microscope (Leica DM 5500B). The images were quantified using Image-Pro Plus 6.1 analysis software using the online protocol provided by the software program, (https://www.youtube.com/watch?v=pSC12G7vWs0) and https://www.youtube.com/watch?v=EkU10F_t2tE). Briefly, the image was opened and the green channel was chosen for measuring the objects. Prior to determining object areas, background flattening was applied Objects were binned into area sizes for classification of the mean areas. Once binned, the program determined the number or percentages of objects in each bin. In our study, object areas fell into 2 categories with greater than 75% of the object areas in the 41–46 pixel range and the remaining corresponding to greater than 143 pixels. Thus, small objects were defined as an area of 41–46 pixels, while larger objects (aggregates) corresponded to an area over 143 pixels.
2.7 Tissue preparation and Western Blots analyses
2.7.1 Tissue preparation
Frozen brain tissue (basal forebrain and cortex) was sonicated on ice, in a lysis buffer (20 mM Tris-HCl pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 10% glycerol) in the presence of protease (1mM PMSF, 2 μg/ml pepstatin and 4 μg/ml aprotinin) and phosphatase inhibitors (1mM orthovanadate). The lysate was centrifuged at 13,000 rpm for 10 minutes and the supernatant was collected. Protein was determined by the Pierce micro-BCA assay.
2.7.2 Western Blots
Homogenized lysates (20μg protein) from individual mice were run under reducing conditions on SDS-PAGE gels (Bio-Rad, 10% Tris-HCl) according to Laemmli (Laemmli, 1970), and then transferred to nitrocellulose membranes (Bio-Rad). Following transfer onto nitrocellulose, blots were incubated with primary antibody. Primary antibodies and dilutions used were: BiP/anti-KDEL (Enzo) at 1:1000; Orexin A (C-19; Santa Cruz Biotechnology) at 1:500; CHOP (Santa Cruz Biotechnology) at 1:200 and B-Actin at 1:1000 (C-4; Santa Cruz Biotechnology). After incubation with IR conjugated secondary secondary antibody, protein bands were detected and quantified by infra-red imaging on an Odyssey (LiCor). For detection of orexin aggregates samples were also run under non-reducing conditions.
2.8 Statistical Analyses
Continuous measures are summarized using means and appropriate measures of variability (including standard deviations, standard errors and/or confidence intervals, as indicated). Categorical measures are summarized using frequencies and percentages. Analyses were performed using SAS Version 9.4 (SAS Institute, Cary, NC), and Stata/SE Version 14.1 (StataCorp LP, College Station, TX).
2.8.1 Sleep-wake
Sleep and wake characteristics were summarized and assessed over the entire 24-hour period in each mouse, and separately during lights on (7AM-7PM) and lights off (7PM-7AM), using similar methods. Wake-related characteristics were the primary outcomes of interest during the active lights off period and sleep-related traits of primary interest during lights on.
Two complementary methods were used to summarize sleep/wake characteristics. First, we used summary statistics to calculate the mean bout duration, total number of bouts and total minutes for wake, NREM, and REM sleep. Second, we utilized the more sophisticated spike and slab methodology (McShane et al., 2010), which uses a mixture model to more accurately account for the non-normal distribution of short and long sleep/wake bout lengths. Briefly, the duration of unique sleep/wake states are modeled accounting for the prior state, with short bouts (defined as ≤40 seconds) modeled by a spike and long bouts (>40 seconds) by a slab component of the model. This model results in 12 summary statistics: 10 numbers representing the proportion of short bouts of 1, 2, 3,… 10 epochs in duration (the spike) and the α and β parameters of the gamma distribution representing the slab. From these 12 parameters, for each mouse three key measures of interest are generated for each sleep/wake transition state: 1) the number of bouts for a given sleep/wake state combination; 2) the size of the spike (e.g., the total proportion of short bouts); and 3) the average size of the slab (e.g., the average duration of long bouts). For the purposes of this manuscript, sleep was defined as Wake to NREM bouts and wake was assessed as NREM to Wake and REM to Wake bouts, separately. These approaches provide comprehensive and accurate assessment of sleep/wake characteristics (McShane et al., 2010; Naidoo et al., 2012; Wimmer et al., 2013). Changes in the duration of long bouts of sleep and wake with age have been previously described (Hasan et al., 2012; Naidoo et al., 2008; Naidoo et al., 2011; Wimmer et al., 2013). In addition to bout characteristics, we examined changes in peak theta frequency based on previous studies of the effects of age in mice (Hasan et al., 2012).
The primary goal of these analyses was to assess the effect of BiP genotype on age-related associations in sleep/wake characteristics, as well as other related outcome measures. To perform this assessment in a comprehensive manner, we first tested whether associations between age-groups and sleep/wake characteristics differed by BiP genotype, or vice versa, by evaluating the significance of an interaction between age group (2–3, 12 or 18 months) and BiP genotype (wildtype vs. heterozygous) in the context of a linear regression model including both main effects and the product term. As interaction tests are typically underpowered, regardless of the significance of this term we subsequently performed specific between group comparisons within strata defined by BiP genotype and age. Specifically, we first assessed differences among age groups and for all outcomes within BiP wildtype and heterozygous animals, separately. Analyses were performed using an analysis of variance (ANOVA) testing the global null hypothesis of no differences among age groups. If a difference was observed (p≤0.05) in this overall comparison, we conducted pairwise comparisons to examine which ages may be driving the overall associations, including calculating the pairwise mean difference, 95% confidence interval (CI) and p-value. Similarly, within each age group separately, we evaluated whether outcome measures differed between BiP wildtype and heterozygous animals using two-tailed T-tests. Together, this set of analyses provides a comprehensive evaluation of the BiP genotype and age-group relationships to sleep/wake characteristics.
2.8.2 Cell count number, c-Fos response, CHOP induction and Orexin aggregate analyses
We also examined differences in cell counts, orexin aggregates, CHOP induction and c-Fos response to sleep deprivation between 12 month-old wildtype and BiP heterozygous mice to explore possible mechanisms of effect of reduction in BiP. Pairwise comparisons between groups (e.g., wildtype vs. BiP heterozygous or undisturbed vs. sleep deprivation) were performed using two-tailed T-tests. Differences in the c-Fos response to sleep deprivation were assessed by examining the interaction between BiP genotype (wildtype vs. heterozygous) and group (undisturbed vs. sleep deprivation). A significant interaction suggests that the difference between the undisturbed and sleep deprivation groups is different within BiP heterozygous and wildtype mice.
3. RESULTS
Targeted disruption of one copy of the BiP gene has been shown to reduce BiP mRNA and protein by 50% in mouse embryonic fibroblasts (Luo et al, 2006). We have also observed a similar reduction in BiP expression in young adult cortex (Figure 1A). Further, we have observed no discernable differences in size (data not shown) and no differences in body weight between the heterozygous mice and their littermates at each of the ages studied (Figure 1B). We did find that BiP protein expression levels decreased with age in both genotypes (Figure 1C), but, as expected, were lower in the 2–3 and 12 month old heterozygous mice when compared to the wildtype littermates (n=10–12/genotype/age; Figure 1D). There was no difference in BiP expression between the genotypes at 18 months.
Figure 1.
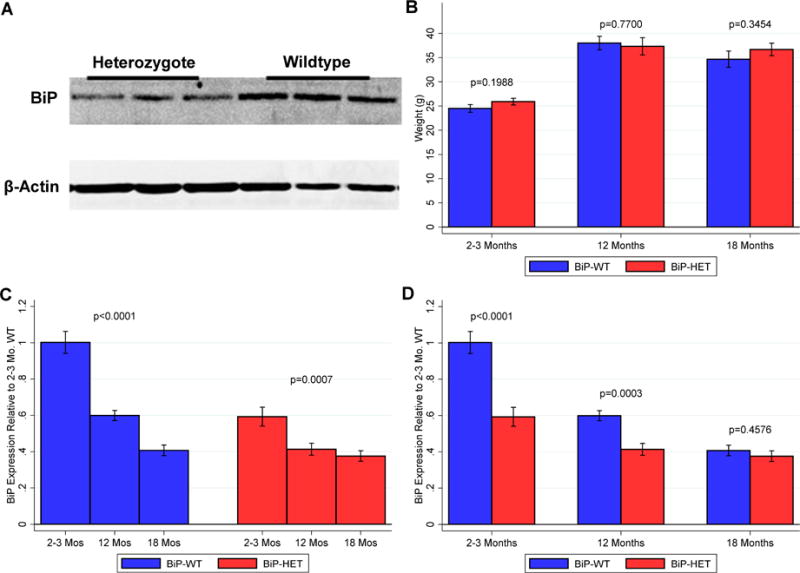
A. Representative western blot showing BiP protein expression in young (2–4mo old) wildtype (n=3) and heterozygous (n=3) mice. Each lane has 20μg of protein loaded. β-actin shown as loading control. B. Histogram showing weights of WT and Het mice at all ages (n = 12–15 /genotype/age) C. BiP expression is decreased across the 3 ages in WT and Het mice (n = 11–13/genotype/age). D. Comparison of BiP expression in WT and Het mice at 2–3months,12 months and 18 months.
We summarize below the wake and sleep characteristics during lights off and lights on; results across 24 hours are presented in the online supplement. We then present data examining theta peak frequency across both BiP genotypes and age groups. Finally, among 12 month-old animals, we evaluate the effect of BiP heterozygosity on characteristics of orexin cells, including number, neuronal function and varicosities, to examine potential mechanisms of effect.
3.1 BiP heterozygosity may accentuate age-related sleep/wake characteristics, particularly for REM sleep-related traits
3.1.1 Wake Bout Summary Measures
To determine whether there was statistical evidence for differences in age-related associations between BiP genotypes, or, whether associations with BiP heterozygosity differ by age group, we first tested for an interaction between age group and genotype for each outcome. We observed no significant interactions between BiP genotype and age group for number of wake bouts, average wake bout duration, or total wake during lights on, lights off or 24 hours (see Table S1). Despite this, as specified in the Methods, we next examined differences in age groups by BiP genotype, as well as associations with BiP heterozygosity within each age group, to better understand potential differences.
When examining wake characteristics during the active lights off period, we observed trending, but non-significant, age-related associations in BiP wildtype mice, compared to statistically significant age effects among BiP heterozygotes (see Table 1 and Figure 2). Specifically, in the lights off period there were borderline non-significant differences in wake bout duration (p=0.0797) and number (p=0.0680) among the wildtype age-groups, with evidence for a shorter bout duration and more bouts, on average, in 18 month-old compared to 2–3 month-old mice. Conversely, among BiP heterozygotes, statistically significant age-related associations were seen for average wake bout duration (p=0.0092) and number of wake bouts (p=0.0020) during lights off (see Table 1 and Figure 2). Large differences were observed between 18-month and 2–3 month mice, including a 5.1 (1.9, 8.3) minute shorter average wake bout duration (p=0.0034) and 102 (51, 153) more wake bouts in the older mice (p=0.0005); 18-month mice also had 54 (4, 104) more wake bouts than 12 month-old mice (p=0.0366), but no significant difference in bout duration (p=0.2575). In contrast to the BiP wildtype mice, we also observed increased fragmentation in 12 month-old vs. 2–3 month-old heterozygous mice, including a 3.3 (0.5, 6.2) minute shorter average wake bout duration (p=0.0248) and 48 (2, 94) more wake bouts (p=0.0426). Thus, both the 12 and 18 month-old BiP heterozygous mice show increased sleep fragmentation when compared to the 2–3 month-old heterozygous mice, as compared to only a trending difference between 2–3 and 18 month-old wildtype mice, and no difference between 2–3 and 12 month-old BiP wildtype mice. This result suggests that, despite the lack of statistically significant interactions, the age-related effect on sleep fragmentation may be more accentuated within BiP heterozygotes. Similar results were found in the lights on period, where mice are less active (see Table 1 and Figure 3), as well as when pooled across 24 hours (see Table S2 in the online supplement).
Table 1.
Sleep and wake characteristics among age groups within BiP genotypes during lights off and lights on
| Characteristic | BiP Wildtype | BiP Heterozygous | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | ANOVA p† | Mean ± SD | ANOVA p† | |||||
| 2–3 Months (N=7) |
12 Months (N=13) |
18 Months (N=8) |
2–3 Months (N=8) |
12 Months (N=9) |
18 Months (N=6) |
|||
| Lights Off | ||||||||
| Wake | ||||||||
| Bout Duration‡ | 5.62 ± 2.14 | 3.88 ± 2.47 | 3.08 ± 1.3 | 0.0797 | 7.84 ± 4.36 | 4.50 ± 1.75 | 2.77 ± 0.65 | 0.0092 |
| Bout Number | 94.6 ± 37.2 | 138.9 ± 60.6 | 169.1 ± 70.4 | 0.0680 | 77.8 ± 26.9 | 125.8 ± 60.7 | 179.7 ± 37.7 | 0.0020 |
| Total Minutes | 469.9 ± 50.6 | 426.2 ± 79.7 | 456.0 ± 74.6 | 0.4018 | 517.3 ± 61.9 | 482.6 ± 74.1 | 477.8 ± 33.4 | 0.4117 |
| NREM | ||||||||
| Bout Duration‡ | 2.35 ± 0.69 | 2.12 ± 0.79 | 1.55 ± 0.67 | 0.1040 | 2.41 ± 0.53 | 1.93 ± 0.80 | 1.15 ± 0.26 | 0.0039 |
| Bout Number | 103.0 ± 36.1 | 142.2 ± 62.8 | 173.0 ± 72.2 | 0.1025 | 78.1 ± 26.7 | 129.6 ± 67.1 | 180.8 ± 37.0 | 0.0034 |
| Total Minutes | 225.5 ± 50.5 | 267.4 ± 78.9 | 233.5 ± 67.9 | 0.3710 | 181.7 ± 58.7 | 222.7 ± 67.5 | 201.9 ± 35.8 | 0.3636 |
| REM | ||||||||
| Bout Duration‡ | 1.08 ± 0.34 | 1.08 ± 0.19 | 0.91 ± 0.27 | 0.2974 | 1.11 ± 0.231 | 0.74 ± 0.36 | 0.99 ± 0.320 | 0.0417 |
| Bout Number | 26.9 ± 16.7 | 24.5 ± 7.1 | 32.5 ± 15.4 | 0.3804 | 19.5 ± 8.7 | 17.9 ± 11.5 | 42.0 ± 11.9 | 0.0007 |
| Total Minutes | 24.5 ± 7.0 | 26.3 ± 8.5 | 30.6 ± 13.8 | 0.4824 | 21.0 ± 9.7 | 14.7 ± 11.0 | 40.4 ± 7.9 | 0.0003 |
| Lights On | ||||||||
| Wake | ||||||||
| Bout Duration‡ | 1.63 ± 0.58 | 1.58 ± 0.56 | 1.16 ± 0.41 | 0.1574 | 1.97 ± 0.65 | 1.57 ± 0.46 | 1.14 ± 0.16 | 0.0180 |
| Bout Number | 163.6 ± 65.1 | 178.9 ± 63.0 | 288.8 ± 173.4 | 0.0497 | 144.1 ± 41.3 | 185.9 ± 67.2 | 285.3 ± 38.6 | 0.0003 |
| Total Minutes | 239.8 ± 45.7 | 256.7 ± 51.7 | 281.4 ± 54.1 | 0.2955 | 269.3 ± 61.9 | 271.3 ± 63.9 | 320.1 ± 14.3 | 0.1887 |
| NREM | ||||||||
| Bout Duration‡ | 2.51 ± 0.9 | 2.32 ± 0.87 | 1.54 ± 0.71 | 0.0672 | 2.76 ± 0.76 | 2.25 ± 0.77 | 1.10 ± 0.17 | 0.0006 |
| Bout Number | 182.9 ± 56.5 | 193.5 ± 60.0 | 302.9 ± 189.8 | 0.0720 | 151.6 ± 41.6 | 200.2 ± 80.2 | 294.3 ± 39.8 | 0.0010 |
| Total Minutes | 417.1 ± 48.2 | 409.9 ± 55.7 | 376.4 ± 71.6 | 0.3497 | 394.0 ± 56.3 | 410.8 ± 65.8 | 318.7 ± 22.6 | 0.0125 |
| REM | ||||||||
| Bout Duration‡ | 1.12 ± 0.32 | 1.03 ± 0.31 | 0.89 ± 0.33 | 0.3954 | 1.20 ± 0.17 | 0.85 ± 0.39 | 0.86 ± 0.18 | 0.0349 |
| Bout Number | 62.9 ± 31.3 | 56.2 ± 18.7 | 72.6 ± 33.0 | 0.4041 | 48.4 ± 18.1 | 43.1 ± 22.8 | 97.5 ± 24.1 | 0.0003 |
| Total Minutes | 63.2 ± 15.9 | 53.5 ± 11.4 | 62.2 ± 27.5 | 0.4278 | 56.7 ± 18.3 | 37.8 ± 20.2 | 81.2 ± 12.5 | 0.0007 |
p-value testing the global null hypothesis of no differences in sleep and wake characteriscs across age groups;
Average bout duration in minutes; Nominally significant ANOVA p-values (p<0.05) shown in bold.
Figure 2.
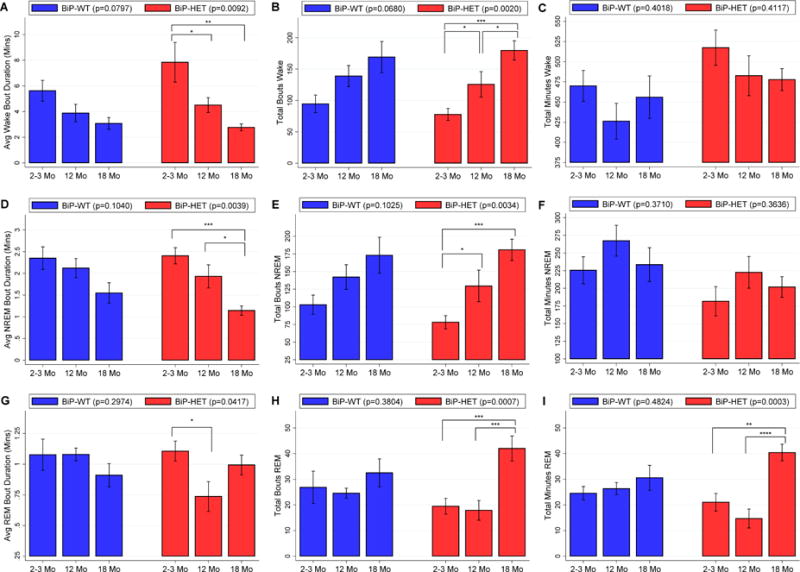
Average ± SEM of A) wake bout duration in minutes, B) number of wake bouts, C) total minutes of wake, D) NREM bout duration in minutes, E) number of NREM bouts, F) total minutes of NREM, G) REM bout duration in minutes, H) number of REM bouts and I) total minutes of REM during the lights off active period in BiP wild-type and BiP HET mice at 3 ages. Statistically significant differences are indicated by * (p<0.05), ** (p<0.01) and *** (p<0.001).
Figure 3.
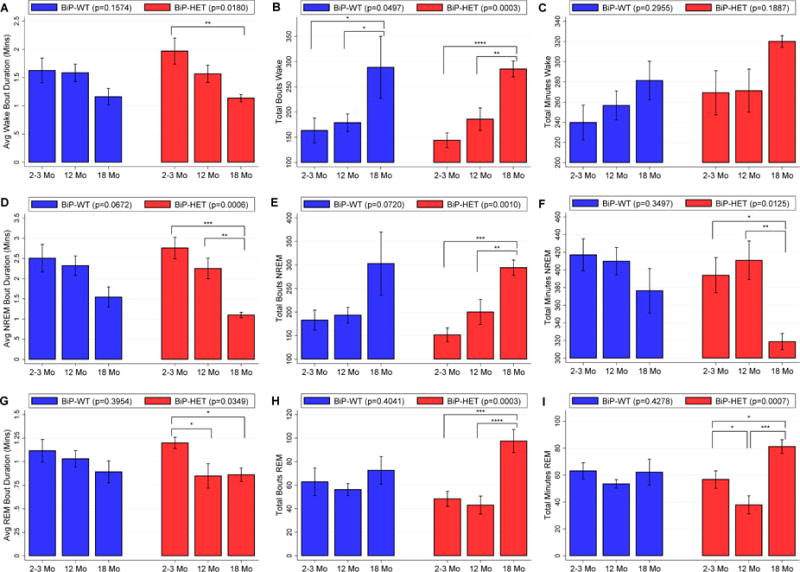
Average ± SEM of A) wake bout duration in minutes, B) number of wake bouts, C) total minutes of wake, D) NREM bout duration in minutes, E) number of NREM bouts, F) total minutes of NREM, G) REM bout duration in minutes, H) number of REM bouts and I) total minutes of REM during the lights on period in BiP wild-type and BiP HET mice at 3 ages. Statistically significant differences are indicated by * (p<0.05), ** (p<0.01) and *** (p<0.001).
As a complementary analysis, we examined the differences in wake characteristics between BiP wildtype and heterozygous mice within each of our three age groups during the active lights off period (see Table 2). Despite the evidence for accentuated age-related associations within the BiP heterozygous mice, we observed no significant genotype differences in wake characteristics within age groups. Similarly, no differences in wake characteristics were observed during lights on (Table 2) or when examining the entire 24-hour period (Table S3).
Table 2.
Sleep and wake characteristics between BiP genotypes within age groups during lights off and lights on
| Characteristic | 2–3 Months | 12 Months | 18 Months | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BiP-WT (N=7) |
BiP-HET (N=8) |
p† | BiP-WT (N=13) |
BiP-HET (N=9) |
p† | BiP-WT (N=8) |
BiP-HET (N=6) |
p† | |
| Lights Off | |||||||||
| Wake | |||||||||
| Bout Duration‡ | 5.62 ± 2.14 | 7.84 ± 4.36 | 0.2451 | 3.88 ± 2.47 | 4.50 ± 1.75 | 0.5235 | 3.08 ± 1.3 | 2.77 ± 0.65 | 0.6053 |
| Bout Number | 94.6 ± 37.2 | 77.8 ± 26.9 | 0.3297 | 138.9 ± 60.6 | 125.8 ± 60.7 | 0.6246 | 169.1 ± 70.4 | 179.7 ± 37.7 | 0.7467 |
| Total Minutes | 469.9 ± 50.6 | 517.3 ± 61.9 | 0.1322 | 426.2 ± 79.7 | 482.6 ± 74.1 | 0.1089 | 456.0 ± 74.6 | 477.8 ± 33.4 | 0.5201 |
| NREM | |||||||||
| Bout Duration‡ | 2.35 ± 0.69 | 2.41 ± 0.53 | 0.8673 | 2.12 ± 0.79 | 1.93 ± 0.80 | 0.5887 | 1.55 ± 0.67 | 1.15 ± 0.26 | 0.1883 |
| Bout Number | 103.0 ± 36.1 | 78.1 ± 26.7 | 0.1497 | 142.2 ± 62.8 | 129.6 ± 67.1 | 0.6574 | 173.0 ± 72.2 | 180.8 ± 37.0 | 0.8134 |
| Total Minutes | 225.5 ± 50.5 | 181.7 ± 58.7 | 0.1477 | 267.4 ± 78.9 | 222.7 ± 67.5 | 0.1815 | 233.5 ± 67.9 | 201.9 ± 35.8 | 0.3228 |
| REM | |||||||||
| Bout Duration‡ | 1.08 ± 0.34 | 1.11 ± 0.23 | 0.8463 | 1.08 ± 0.19 | 0.74 ± 0.36 | 0.0248 | 0.91 ± 0.27 | 0.99 ± 0.320 | 0.5334 |
| Bout Number | 26.9 ± 16.7 | 19.5 ± 8.7 | 0.2946 | 24.5 ± 7.1 | 17.9 ± 11.5 | 0.1081 | 32.5 ± 15.4 | 42.0 ± 11.9 | 0.2349 |
| Total Minutes | 24.5 ± 7.0 | 21.0 ± 9.7 | 0.4444 | 26.3 ± 8.5 | 14.7 ± 11.0 | 0.0112 | 30.6 ± 13.8 | 40.4 ± 7.9 | 0.1470 |
| Lights On | |||||||||
| Wake | |||||||||
| Bout Duration‡ | 1.63 ± 0.58 | 1.97 ± 0.65 | 0.3043 | 1.58 ± 0.56 | 1.57 ± 0.46 | 0.9374 | 1.16 ± 0.41 | 1.14 ± 0.16 | 0.8933 |
| Bout Number | 163.6 ± 65.1 | 144.1 ± 41.3 | 0.4958 | 178.9 ± 63.0 | 185.9 ± 67.2 | 0.8044 | 288.8 ± 173.4 | 285.3 ± 38.6 | 0.9583 |
| Total Minutes | 239.8 ± 45.7 | 269.3 ± 61.9 | 0.3189 | 256.7 ± 51.7 | 271.3 ± 63.9 | 0.5584 | 281.4 ± 54.1 | 320.1 ± 14.3 | 0.0878 |
| NREM | |||||||||
| Bout Duration‡ | 2.51 ± 0.9 | 2.76 ± 0.76 | 0.5647 | 2.32 ± 0.87 | 2.25 ± 0.77 | 0.8544 | 1.54 ± 0.71 | 1.10 ± 0.17 | 0.1264 |
| Bout Number | 182.9 ± 56.5 | 151.6 ± 41.6 | 0.2402 | 193.5 ± 60.0 | 200.2 ± 80.2 | 0.8230 | 302.9 ± 189.8 | 294.3 ± 39.8 | 0.9046 |
| Total Minutes | 417.1 ± 48.2 | 394.0 ± 56.3 | 0.4126 | 409.9 ± 55.7 | 410.8 ± 65.8 | 0.9709 | 376.4 ± 71.6 | 318.7 ± 22.6 | 0.0614 |
| REM | |||||||||
| Bout Duration‡ | 1.12 ± 0.32 | 1.20 ± 0.17 | 0.5292 | 1.03 ± 0.31 | 0.85 ± 0.39 | 0.2406 | 0.89 ± 0.33 | 0.86 ± 0.18 | 0.8400 |
| Bout Number | 62.9 ± 31.3 | 48.4 ± 18.1 | 0.2842 | 56.2 ± 18.7 | 43.1 ± 22.8 | 0.1548 | 72.6 ± 33.0 | 97.5 ± 24.1 | 0.1460 |
| Total Minutes | 63.2 ± 15.9 | 56.7 ± 18.3 | 0.4852 | 53.5 ± 11.4 | 37.8 ± 20.2 | 0.0309 | 62.2 ± 27.5 | 81.2 ± 12.5 | 0.1432 |
p-value from T-test comparing sleep and wake characteristics between wildtype and heterozygous animals within each age group;
Average bout duration in minutes; nominally significant genotype differences (p<0.05) shown in bold.
3.1.2 BiP heterozygosity is associated with reduced REM
As with wake characteristics, we examined associations between BiP genotype and age groups for NREM and REM sleep during lights on and lights off. Taken together, results indicate more robust associations with age for NREM among BiP heterozygous mice when compared to wildtype animals. Moreover, supported by significant interaction effects for REM characteristics, there were strong differences in age-related effects on REM associated with BiP heterozygosity, compared to no age effects among the wildtype mice. We also observed significant differences in specific REM characteristics between BiP wildtype and heterozygous 12 month-old mice.
We observed significant interactions between BiP genotype and age group (see Table S1) for REM characteristics, but not for NREM sleep. During the less active lights on period, there was evidence for interactive effects on REM bout duration (p=0.0419) and total minutes (p=0.0123), as well as a suggestive interaction for the number of REM bouts (p=0.0997). Similarly, during lights off, there was a significant interaction for total REM sleep (p=0.0275) and a suggestive interaction for number of REM bouts (p=0.0559), although no evidence for an interaction for REM bout duration (p=0.4290). Supporting these results, when examining data pooled across 24 hours, we found significant BiP genotype by age group interactions for both REM bout number (p=0.0461) and total REM minutes (p=0.0124). Altogether, these results indicate that BiP heterozygosity results in stronger differences in age-related associations for changes in REM characteristics. Relatedly, there are also greater differences in REM characteristics between BiP genotypes in particular age groups.
To better understand these associations, we next examined differences in NREM and REM characteristics across age groups within BiP wildtype and heterozygous mice, separately (see Table 1). Results described below are focused on the less active lights on period, however, generally similar associations were observed during lights off (see Table 1) and when pooling across 24 hours (see Table S2). Among BiP wildtype animals, borderline non-significant associations with age were seen for NREM sleep bout duration (p=0.0672) and number (p=0.0720), but there were no differences total NREM (p=0.3497) or associations with REM characteristics. When examining these NREM characteristics between age groups, 18 month-old wildtype mice tended to show more fragmentation compared to both 2–3 month and 12 month-old mice.
In contrast to these borderline age-related effects in BiP wildtype animals, significant differences among age groups were observed for all summary measure of NREM (all p≤0.0125) and REM (all p≤0.035) sleep among BiP heterozygous mice (see Table 1); this observed difference in age-related effects in REM characteristics between BiP wildtypes and heterozygotes was suggested by the significant interactions seen for these traits. For NREM characteristics, 18 month-old BiP heterozygotes showed significant differences compared to 12 and 2–3 month-old mice for all summary measures. In particular, 18 month-old mice had 1.7 (0.9, 2.4) minute shorter NREM bouts (p=0.0002), 143 (75, 210) more bouts (p=0.0003), and 75 (14, 137) minutes less total NREM (p=0.0187) compared to the youngest animals. When compared to 12 month-old heterozygotes, the 18 month-old mice exhibited 1.2 (0.4, 1.9) minute shorter bouts (p=0.0037), 94 (28, 160) more NREM bouts (p=0.0073) and 92 (32, 152) fewer minutes of NREM (p=0.0044). When examining between age group differences for REM characteristics, which were not seen in BiP wildtypes, 2–3 month-old animals had longer average bout duration than both the 12 month-old (0.35 [0.07, 0.63] minutes; p=0.0180) and 18 month-old (0.34 [0.02, 0.65] minutes; p=0.0365), as well as more total REM than the middle-aged mice (19 [1, 37] minutes; p=0.0419). On the other hand, 18 month-old mice had a higher number of REM bouts than both the 2–3 month-old (49 [25, 73] bouts; p=0.0004) and 12 month-old (54 [31, 78] bouts; p=0.0001) mice, as well as 25 (4, 45) minutes more REM than 2–3 month-old mice (p=0.0197) and 43 (24, 63) minutes more REM than 12 month-old heterozygotes (p=0.0002).
Finally, we again examined NREM and REM differences associated with BiP heterozygosity within each age group (see Table 2 and Table S3). No genotype differences were observed for NREM characteristics. However, when examining REM characteristics, we found statistically significant differences among middle aged animals. Specifically, 12 month-old BiP heterozygous mice had less total REM than wildtypes during lights on (p=0.0309), as well as less total REM (p=0.0112) and shorter REM bout durations (p=0.0248) during lights off (see Table 2). When examining results pooled across 24 hours (see Table S3), BiP heterozygosity was associated with significantly less REM sleep (52.5 vs. 79.8 minutes, p=0.0121), as well as non-significant trends towards shorter bout durations (0.82 vs. 1.04 minutes, p=0.1120) and fewer REM bouts (61 vs. 81, p=0.0961). Thus, there is evidence that BiP heterozygosity affects REM sleep, particularly in middle-aged animals.
3.1.3 Spike and Slab Characteristics
In addition to the standard descriptive summaries of Wake, NREM and REM described above, we performed a spike and slab analyses (McShane et al., 2010) to derive bout characteristics relative to specific state transitions of interest. This addresses the non-normal distribution of bout durations and allows one to more accurately examine characteristics of short and long bouts (McShane et al., 2010). The spike and slab results generally support the effects seen in summary measures, but estimates account for the expected non-normal distribution of bout durations.
When examining interactions between BiP genotype and age groups for measures derived using the spike and slab methodology, we observed no significant interactions for the total number, proportion short bouts, or average duration of long bouts for wake to NREM or NREM to wake transitions (see Table S1). For REM to wake transitions, there was a suggestive interaction between age group and BiP genotype for the total number of bouts during lights off (p=0.0730) and a significant interaction for total number of REM to wake bouts during lights on (p=0.0187); this interaction was statistically significant when examining the number of REM to wake bouts over all 24 hours (p=0.0226). No interactions were observed for the proportion of short bouts or average long bout duration in either time-period. Thus, there is some evidence for differential age effects within BiP genotypes or vice versa for REM to wake-related transitions, but not with those associated with NREM.
Following our comprehensive analysis strategy, we next examined the age-related differences within BiP wildtype and heterozygous mice, separately (see Table 3). During the active lights off period in BiP wildtype mice, we observed a significant difference in the average durations of long REM to wake bouts (p=0.0360), but no differences in other characteristics associated with transitions from NREM or REM to wake. When comparing the duration of long REM to wake bouts between age groups in wildtypes, 2–3 month animals sustained longer bouts, including a borderline 6.4 (95% CI: 0.0, 12.9) minute longer duration than 12 month-old BiP wildtypes (p=0.0513) and a 9.3 (2.2, 16.5) minutes longer bout duration than 18 month-old mice (p=0.0122). When examining wake to NREM transition bout characteristics during lights off, there was a borderline, non-significant difference in the number of bouts across age groups (p=0.0694) and a significant difference in average long bout duration (p=0.0448); older animals had evidence for more fragmentation, including 75 (11, 139) more wake to NREM bouts (p=0.0234) and a 1.1 (0.2, 1.9) minute shorter long bout duration (p=0.0158) in 18 month-old compared to 2 month-old wildtype mice. Similar results were seen for wake to NREM bouts in the less active lights on period, including significant age related effects for number of bouts (p=0.0364) and average long bout duration (p=0.0342). The 18 month-old wildtype mice showed similarly increased sleep fragmentation compared to both the young and middle aged wildtype mice, with 125 (12, 239) more bouts (p=0.0320) and 1.2 (0.1, 2.2) minute shorter long bouts (p=0.0346) than the 2–3 month-old mice and 121 (23, 220) more bouts (p=0.0179) and 1.2 (0.2, 2.1) minute shorter bouts (p=0.0154) compared to 12 month-old wildtypes. There were no differences in NREM to wake or REM to wake characteristics among BiP wildtype mice during lights on. Differences in the characteristics of wake to NREM bouts, but not NREM to wake or REM to wake bouts, remained significant when examining data across 24 hours (see Table S4).
Table 3.
Spike and Slab characteristics among age groups within BiP genotypes during lights off and lights on
| Characteristic | BiP-Wildtype | BiP-Heterozygous | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | ANOVA p† | Mean ± SD | ANOVA p† | |||||
| 2–3 Months (N=7) |
12 Months (N=13) |
18 Months (N=8) |
2–3 Months (N=8) |
12 Months (N=9) |
18 Months (N=6) |
|||
| Lights Off | ||||||||
| Wake to NREM | ||||||||
| Number of bouts | 93.9 ±37.2 | 141.5 ± 62.5 | 168.9 ± 70.6 | 0.0694 | 77.1 ± 27.0 | 117.6 ± 44.1 | 178.7 ± 37.7 | 0.0003 |
| Proportion short‡ | 0.111 ± 0.029 | 0.106 ± 0.055 | 0.141 ± 0.086 | 0.4358 | 0.123 ± 0.044 | 0.115 ± 0.055 | 0.203 ± 0.101 | 0.0484 |
| Average duration§ | 2.76 ± 0.89 | 2.38 ± 0.78 | 1.71 ± 0.69 | 0.0448 | 2.70 ± 0.56 | 2.17 ± 0.72 | 1.32 ± 0.26 | 0.0010 |
| NREM to Wake | ||||||||
| Number of bouts | 75.9 ± 39.2 | 120.5 ± 62.0 | 140.0 ± 78.4 | 0.1501 | 58.4 ± 22.8 | 102.1 ± 44.3 | 139.0 ± 37.2 | 0.0020 |
| Proportion short‡ | 0.607 ± 0.188 | 0.618 ± 0.156 | 0.626 ± 0.203 | 0.9799 | 0.664 ± 0.109 | 0.568 ± 0.148 | 0.702 ± 0.085 | 0.1093 |
| Average duration§ | 15.05 ± 15.19 | 7.31 ± 4.21 | 9.67 ± 7.35 | 0.1977 | 21.32 ± 12.68 | 11.82 ± 6.30 | 8.49 ± 5.10 | 0.0312 |
| REM to Wake | ||||||||
| Number of bouts | 18.3 ± 6.3 | 21.0 ± 8.3 | 28.9 ± 14.0 | 0.1063 | 19.0 ± 9.0 | 15.4 ± 11.6 | 40.0 ± 11.3 | 0.0008 |
| Proportion short‡ | 0.396 ± 0.154 | 0.475 ± 0.222 | 0.470 ± 0.204 | 0.6833 | 0.426 ± 0.169 | 0.535 ± 0.321 | 0.399 ± 0.176 | 0.5104 |
| Average duration§ | 15.57 ± 9.60 | 9.16 ± 5.833 | 6.23 ± 4.70 | 0.0360 | 18.70 ± 11.19 | 6.21 ± 6.05 | 6.65 ± 3.11 | 0.0067 |
| Lights On | ||||||||
| Wake to NREM | ||||||||
| Number of bouts | 163.0 ± 65.4 | 166.9 ± 63.3 | 288.4 ± 173.5 | 0.0364 | 143.5 ± 41.1 | 169.0 ± 23.95 | 284.7 ± 38.7 | <0.0001 |
| Proportion short‡ | 0.075 ± 0.026 | 0.088 ± 0.048 | 0.132 ± 0.103 | 0.2047 | 0.071 ± 0.027 | 0.088 ± 0.026 | 0.122 ± 0.056 | 0.0475 |
| Average duration§ | 2.84 ± 1.01 | 2.85 ± 1.15 | 1.68 ± 0.68 | 0.0342 | 3.01 ± 0.89 | 2.56 ± 0.78 | 1.20 ± 0.15 | 0.0006 |
| NREM to Wake | ||||||||
| Number of bouts | 119.4 ± 61.5 | 128.3 ± 57.6 | 229.6 ± 189.5 | 0.1027 | 103.0 ± 41.2 | 139.3 ± 28.6 | 196.7 ± 33.2 | 0.0003 |
| Proportion short‡ | 0.651 ± 0.125 | 0.677 ± 0.170 | 0.708 ± 0.163 | 0.7835 | 0.672 ± 0.109 | 0.662 ± 0.162 | 0.737 ± 0.100 | 0.5298 |
| Average duration§ | 4.77 ± 3.01 | 5.15 ± 4.00 | 4.30 ± 2.50 | 0.8553 | 5.75 ± 3.72 | 4.96 ± 2.69 | 3.16 ± 1.28 | 0.2588 |
| REM to Wake | ||||||||
| Number of bouts | 43.3 ± 12.3 | 38.4 ± 16.6 | 58.4 ± 32.8 | 0.1376 | 40.1 ± 11.0 | 29.6 ± 15.9 | 87.8 ± 21.8 | <0.0001 |
| Proportion short‡ | 0.651 ± 0.183 | 0.663 ± 0.179 | 0.669 ± 0.213 | 0.9834 | 0.576 ± 0.143 | 0.574 ± 0.271 | 0.564 ± 0.128 | 0.9934 |
| Average duration§ | 4.41 ± 2.76 | 7.69 ± 6.40 | 3.54 ± 2.56 | 0.1370 | 4.73 ± 3.83 | 4.99 ± 3.19 | 2.97 ± 1.02 | 0.4390 |
p-value testing the global null hypothesis of no differences in spike and slab characteristics across age groups;
Proportion of bouts ≤40 seconds;
Average long bout duration in minutes; nominally significant ANOVA p-values (p<0.05) shown in bold.
In contrast to these marginal effects in the BiP wildtype animals, statistically significant age-related differences were seen for a number of spike and slab derived traits among the BiP heterozygous animals (see Table 3). During lights off, there were age-related differences in the number and average long bout durations for both NREM to wake and REM to wake transitions, with older animals generally showing more bouts and shorter durations. Specifically, for NREM to wake transitions, the 2 month-old BiP heterozygotes exhibited 44 (7, 80) fewer bouts (p=0.0220) and 9.5 (0.5, 18.5) minute longer long bouts (p=0.0395) than 12 month-old mice and 81 (40, 121) fewer bouts (p=0.0005) and 12.8 (2.8, 22.8) minute longer long bouts (p=0.0144) than the 18 month-old heterozygotes. When examining wake to NREM bouts during lights off, there were significant age-group differences for number of bouts (p=0.0003), proportion short bouts (p=0.0484) and the average long bout duration (p=0.0010). As with characteristics of wake transitions, the oldest animals showed increased fragmentation, compared to similar values in the 2–3 month and 12 month-old mice. In particular, the 18 month-old BiP heterozygotes exhibited 102 (60, 144) more bouts (p<0.0001), an 8% (1%, 16%) higher proportion of short bouts (p=0.0387), and 1.4 (0.7, 2.0) minute shorter long bouts (p=0.0002) than 2–3 month-old mice, as well as 61 (20, 102) more bouts (p=0.0055), a 9% (1%, 16%) higher proportion of short bouts (p=0.0216), and 0.9 (0.2, 1.5) minute shorter long bouts (p=0.0111) than the 12 month-old heterozygotes. As shown in Table 3, age-related associations with spike and slab characteristics were generally similar during lights on, with the exception of no significant differences in the average long bout durations of NREM to wake and REM to wake bouts. There were again significant differences in all 3 wake to NREM transition characteristics, with 18 month-old animals exhibiting 141 (102, 180) more bouts (p<0.0001), a 5% (1%, 9%) higher proportion of short bouts (p=0.0154) and 1.8 (1.0, 2.6) minute shorter long bout duration (p=0.0002) than 2–3 month-old heterozygotes. Similarly, 18 month-old mice had 116 (78, 154) more wake to NREM bouts (p<0.0001) and 1.4 (0.6, 2.2) minute shorter long bout duration (p=0.0020) than 12 month-old BiP heterozygotes. For the number of NREM to wake bouts, 18 month-old animals had 94 (55, 133) more bouts than 2–3 month-old mice (p<0.0001) and 57 (19, 95) more bouts than 12 month-old heterozygotes (p=0.0051). There was also evidence for 36 (1, 71) fewer NREM to wake bouts in 2–3 month versus 12 month-old animals (p=0.0431). Relatedly, 18 month-old heterozygotes had significantly more REM to wake bouts than both 2–3 month-old mice (48 [29, 66]; p<0.0001) and 12 month-old mice (58 [40, 76]; p<0.0001). Results were similar when examining the entire 24-hour period (see Table S4).
Finally, comparisons of spike and slab characteristics between BiP genotypes within age groups during lights on and lights off are shown in Table 4 (see Table S5 for results over 24 hours). Despite the strong differences in observed between age groups associated with BiP heterozygosity, and significant interactions for certain REM to wake transition traits, we observed no statistically significant differences between the BiP genotypes within any age groups.
Table 4.
Spike and slab characteristics between BiP genotypes within age groups during lights off and lights on
| Characteristic | 2–3 Months | 12 Months | 18 Months | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BiP-WT (N=7) |
BiP-HET (N=8) |
p† | BiP-WT (N=13) |
BiP-HET (N=9) |
p† | BiP-WT (N=8) |
BiP-HET (N=6) |
p† | |
| Lights Off | |||||||||
| Wake to NREM | |||||||||
| Number of bouts | 93.9 ±37.2 | 77.1 ± 27.0 | 0.3323 | 141.5 ± 62.5 | 117.6 ± 44.1 | 0.3339 | 168.9 ± 70.6 | 178.7 ± 37.7 | 0.7645 |
| Proportion short‡ | 0.111 ± 0.029 | 0.123 ± 0.044 | 0.5493 | 0.106 ± 0.055 | 0.115 ± 0.055 | 0.7092 | 0.141 ± 0.086 | 0.203 ± 0.101 | 0.2394 |
| Average duration§ | 2.76 ± 0.89 | 2.70 ± 0.56 | 0.8785 | 2.38 ± 0.78 | 2.17 ± 0.72 | 0.5293 | 1.71 ± 0.69 | 1.32 ± 0.26 | 0.1723 |
| NREM to Wake | |||||||||
| Number of bouts | 75.9 ± 39.2 | 58.4 ± 22.8 | 0.3027 | 120.5 ± 62.0 | 102.1 ± 44.3 | 0.4538 | 140.0 ± 78.4 | 139.0 ± 37.2 | 0.9776 |
| Proportion short‡ | 0.607 ± 0.188 | 0.664 ± 0.109 | 0.4816 | 0.618 ± 0.156 | 0.568 ± 0.148 | 0.4652 | 0.626 ± 0.203 | 0.702 ± 0.085 | 0.4081 |
| Average duration§ | 15.05 ± 15.19 | 21.32 ± 12.68 | 0.3992 | 7.31 ± 4.21 | 11.82 ± 6.30 | 0.0568 | 9.67 ± 7.35 | 8.49 ± 5.10 | 0.7419 |
| REM to Wake | |||||||||
| Number of bouts | 18.3 ± 6.3 | 19.0 ± 9.0 | 0.8628 | 21.0 ± 8.3 | 15.4 ± 11.6 | 0.2027 | 28.9 ± 14.0 | 40.0 ± 11.3 | 0.1379 |
| Proportion short‡ | 0.396 ± 0.154 | 0.426 ± 0.169 | 0.7261 | 0.475 ± 0.222 | 0.535 ± 0.321 | 0.6087 | 0.470 ± 0.204 | 0.399 ± 0.176 | 0.5080 |
| Average duration§ | 15.57 ± 9.60 | 18.70 ± 11.19 | 0.5744 | 9.16 ± 5.833 | 6.21 ± 6.05 | 0.2641 | 6.23 ± 4.70 | 6.65 ± 3.11 | 0.8539 |
| Lights On | |||||||||
| Wake to NREM | |||||||||
| Number of bouts | 163.0 ± 65.4 | 143.5 ± 41.1 | 0.4954 | 166.9 ± 63.3 | 169.0 ± 23.95 | 0.9155 | 288.4 ± 173.5 | 284.7 ± 38.7 | 0.9548 |
| Proportion short‡ | 0.075 ± 0.026 | 0.071 ± 0.027 | 0.7451 | 0.088 ± 0.048 | 0.088 ± 0.026 | 0.9984 | 0.132 ± 0.103 | 0.122 ± 0.056 | 0.8361 |
| Average duration§ | 2.84 ± 1.01 | 3.01 ± 0.89 | 0.7340 | 2.85 ± 1.15 | 2.56 ± 0.78 | 0.5130 | 1.68 ± 0.68 | 1.20 ± 0.15 | 0.0913 |
| NREM to Wake | |||||||||
| Number of bouts | 119.4 ± 61.5 | 103.0 ± 41.2 | 0.5489 | 128.3 ± 57.6 | 139.3 ± 28.6 | 0.6032 | 229.6 ± 189.5 | 196.7 ± 33.2 | 0.6433 |
| Proportion short‡ | 0.651 ± 0.125 | 0.672 ± 0.109 | 0.7397 | 0.677 ± 0.170 | 0.662 ± 0.162 | 0.8403 | 0.708 ± 0.163 | 0.737 ± 0.100 | 0.7088 |
| Average duration§ | 4.77 ± 3.01 | 5.75 ± 3.72 | 0.5854 | 5.15 ± 4.00 | 4.96 ± 2.69 | 0.8986 | 4.30 ± 2.50 | 3.16 ± 1.28 | 0.3322 |
| REM to Wake | |||||||||
| Number of bouts | 43.3 ± 12.3 | 40.1 ± 11.0 | 0.6075 | 38.4 ± 16.6 | 29.6 ± 15.9 | 0.2261 | 58.4 ± 32.8 | 87.8 ± 21.8 | 0.0819 |
| Proportion short‡ | 0.651 ± 0.183 | 0.576 ± 0.143 | 0.3853 | 0.663 ± 0.179 | 0.574 ± 0.271 | 0.3583 | 0.669 ± 0.213 | 0.564 ± 0.128 | 0.3055 |
| Average duration§ | 4.41 ± 2.76 | 4.73 ± 3.83 | 0.8577 | 7.69 ± 6.40 | 4.99 ± 3.19 | 0.2581 | 3.54 ± 2.56 | 2.97 ± 1.02 | 0.6190 |
p-value from T-test comparing sleep and wake characteristics between wildtype and heterozygous animals within each age group;
Proportion of bouts ≤40 seconds;
Average long bout duration in minutes
3.2 Theta Peak Frequency differences with age are more pronounced in the BiP Heterozygotes
We used fast Fourier transform (FFT) of EEG recordings to examine the EEG spectral profiles during each behavioral state for the three age groups. We examined theta peak frequency by recording the frequency at which absolute power was highest in the theta range (6–10 Hz) during REM and wake. While we did not observe significant interactions between BiP genotype and age group for peak frequency during either wake (p=0.8225) or REM (p=0.2184; see Table S1), there was evidence of stronger age-related associations among BiP heterozygous animals (see Table 5), as well as a genotype difference in REM peak frequency for the youngest animals (see Table 6).
Table 5.
Theta peak frequencies among and between age groups within BiP genotypes
| Characteristic | BiP Wildtype | BiP Heterozygous | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | ANOVA p† | Mean ± SD | ANOVA p† | |||||
| 2–3 Months (N=7) |
12 Months (N=13) |
18 Months (N=8) |
2–3 Months (N=8) |
12 Months (N=9) |
18 Months (N=6) |
|||
| Wake Peak Hz | 7.39 ± 0.56 | 6.50 ± 0.76 | 6.71 ± 0.23 | 0.0166 | 7.31 ± 0.77 | 6.28 ± 0.44 | 6.75 ± 0.42 | 0.0051 |
| REM Peak Hz | 6.82 ± 0.12 | 6.85 ± 0.47 | 6.46 ± 0.47 | 0.1408 | 6.50 ± 0.23 | 6.97 ± 0.48 | 6.29 ± 0.25 | 0.0036 |
p-value testing the global null hypothesis of no differences in theta peak frequencies across age groups; Nominally significant ANOVA p-values (p<0.05) shown in bold.
Table 6.
Theta peak frequencies between BiP genotypes within age groups
| Characteristic | 2–3 Months | 12 Months | 18 Months | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BiP-WT (N=7) |
BiP-HET (N=8) |
p† | BiP-WT (N=13) |
BiP-HET (N=9) |
p† | BiP-WT (N=7) |
BiP-HET (N=6) |
p† | |
| Wake Peak Hz | 7.39 ± 0.56 | 7.31 ± 0.77 | 0.8221 | 6.50 ± 0.76 | 6.28 ± 0.44 | 0.4392 | 6.71 ± 0.23 | 6.75 ± 0.42 | 0.8481 |
| REM Peak Hz | 6.82 ± 0.12 | 6.50 ± 0.23 | 0.0059 | 6.85 ± 0.47 | 6.97 ± 0.48 | 0.5467 | 6.46 ± 0.47 | 6.29 ± 0.25 | 0.4340 |
p-value from T-test comparing values between wild-type and heterozygous mice within each age group; nominally significant genotype differences (p<0.05) shown in bold.
Specifically, when comparing values across age groups among BiP wildtype animals (Table 5), we observed a significant difference between age groups for the theta peak frequency during wake (p=0.0166). Younger mice experienced their peak theta at higher frequency (7.39 Hz) than both 12 month (6.50 Hz; p=0.0048) and 18 month (6.71 Hz; p=0.0495) old mice. There were no differences in theta peak frequency during REM across age groups in wildtype animals. In contrast, we found significant age-related differences in Theta peak frequency during both wake (p=0.0051) and REM (p=0.0036) among BiP heterozygotes, suggesting stronger age differences in these animals. In particular, during wake, we found that younger animals had significantly higher peak frequency (7.31 Hz) compared to 12 month animals (6.28 Hz; p=0.0013) and a non-significantly higher average peak frequency than 18 month animals (6.75 Hz; p=0.0832). While 12 month animals had the lowest peak frequency during wake, these animals showed the highest peak frequency during REM (6.97 Hz) when compared to either the 2–3 month (6.50 Hz; p=0.0121) or 18 month (6.29 Hz; p=0.0015) old mice.
This difference in the age-related associations for peak theta during REM may be driven by a lower peak frequency among younger BiP heterozygous animals when compared to the wildtype mice of the same age (see Table 6). Specifically, 2–3 month BiP heterozygous mice had a peak frequency of REM of 6.50 Hz, compared to a higher frequency of 6.82 Hz in REM for wildtype mice (p=0.0059). Peak frequencies during REM were similar between the two genotype groups within both the 12 month-old and 18 month-old animals and there were no genotype differences in peak frequency during wake within age groups.
3.3 Effect of reduced levels of BiP on orexin cell number
We wanted to determine whether the increased fragmentation of behavioral state in BiP heterozygous mice, described above, was due to changes in orexin neuron number or function. Orexinergic signaling is known to be required for state stability (Chemelli et al., 1999; Sawai et al., 2010). Therefore, using a neurostereological approach we compared orexin neuron numbers between 12 month aged BiP wildtype (n=7) and BiP heterozygous (n=6) mice. Neurostereology counting indicated no significant difference (p=0.6406) in the orexin neuron numbers in the BiP heterozygous mice (3101 ± 289 orexin positive neurons) when compared to that in wildtype mice (3236 ± 631 neurons).
3.4 Functional assessment of orexin neurons in BiP heterozygous mice
Given that neurostereology indicated that orexin neuron counts were not changed in the BiP heterozygous mice, we next assessed the functionality of these neurons in these mice. We used c-fos staining as a proxy for orexin neuron function. C-fos is an immediate early gene product that is expressed when neurons are activated (Hoffman et al., 1993). We examined undisturbed and sleep deprived mice in both the BiP wildtype and heterozygous genotypes at 12 months of age, resulting in 4 groups of mice studied (n=6 per group; see also Figure 4). We observed a significant increase in c-fos immunoreactivity in the orexin neurons with sleep deprivation in both the wildtype (p=0.0006) and heterozygous mice (p<0.0001) when compared to their undisturbed controls. However, there was no evidence of a difference in this c-Fos immunoreactivity increase with sleep deprivation between the two BiP genotypes (p=0.9749), as also evidenced by no differences between BiP wildtype and heterozygous animals in either the undisturbed (p=0.6125) or sleep deprived (p=0.5517) conditions.
Figure 4.
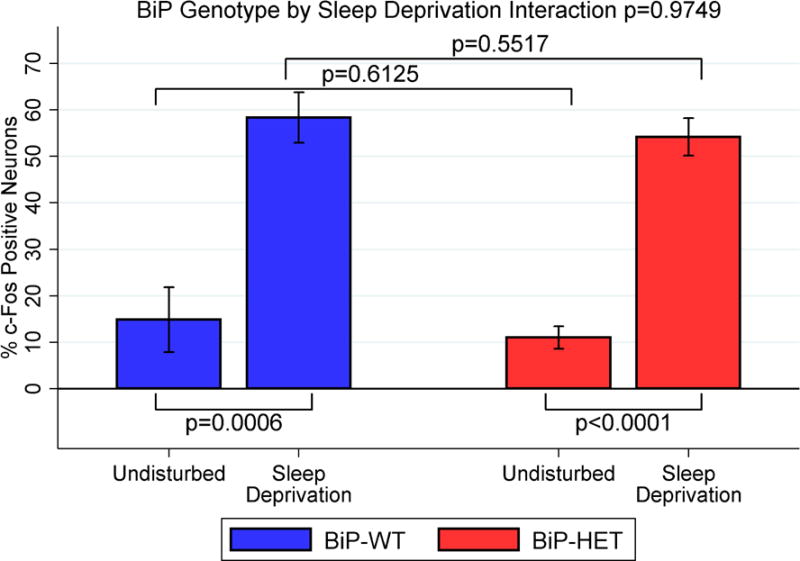
Percent c-fos positive orexin neurons in undisturbed and sleep deprived BiP WT and HET mice. Mean ± SE shown.
3.5 BiP heterozygous mice display orexin aggregates at terminals
During the course of the immunohistochemical studies, we observed that the terminals of the orexin neurons in the 12 month old BiP heterozygous mice were enlarged with protein deposits that stained positive for orexin. Therefore, we systematically compared the terminals of orexin neurons in the median septum of basal forebrain (Khakpai et al., 2013) (Ballinger et al., 2016) in wildtype and BiP heterozygous mice at 12 months of age. This region was chosen as it is most distal from the orexin cell bodies and as such is expected to experience more metabolic stress. We used pixel density to define these enlarged areas. We determined the number of terminals displaying varicosities within this region and also determined the size of the varicosity in both wildtype and heterozygous strains (n=12 per group). We found that the varicosities or aggregates segregated into two populations, those that had an area of approximately 41–46 pixels and those that had areas greater than 143 pixels. We therefore defined a small varicosity as having an area between 41–46 pixels while a large varicosity had an area greater than 143 pixels. We found that the BiP heterozygous mice had significantly more terminals displaying both large (489 ± 50 vs. 430 ± 46; p=0.0060) and small (1578 ± 247 vs. 1396 ± 168; p=0.0462) varicosities and that the immunopositive staining of these areas was also greater in the heterozygous mice compared to the wildtype mice (Figure 5; Figure 6). We further quantified these orexin aggregates in the basal forebrain using western analyses and found that the BiP heterozygous mice had more high molecular weight species with immune-reactivity to the orexin-A antibody than in the wildtype mice (Figure 7), supporting the presence of orexin aggregates. We also detected orexin-A-immunoreactive bands with molecular weights greater than those of mature orexin-A peptide (3.5 kDa) or its precursor prepro-orexin (13.5 kDa) in BiP heterozygotes.
Figure 5.
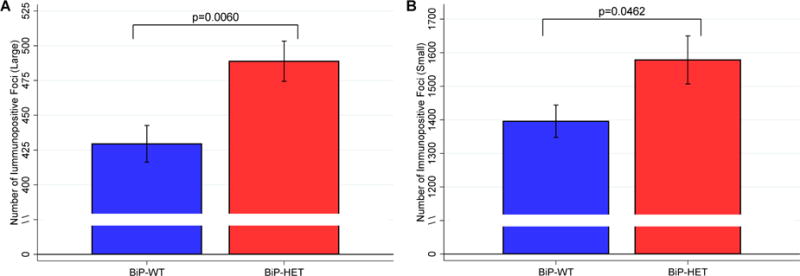
Number of large(A) and small (B) orexin positive varicosities in the BiP WT and HET mice. Mean ± SE shown.
Figure 6.
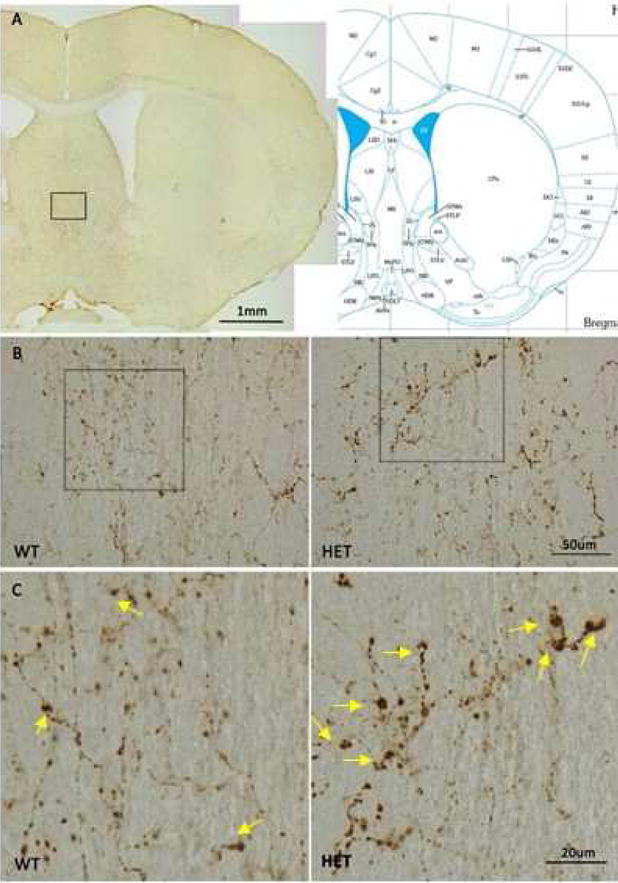
A) Diagram showing medial septal brain region in study. B. Orexin positive aggregates at terminals in wild-type and heterozygous mice. Lower magnification (20×) shown in B, higher magnification (60×) shown in C. BiP heterozygous mice display many more enlarged terminals staining positive for orexin compared to wild-type mice. Arrows in the bottom panels) indicate the large varicosities.
Figure 7.
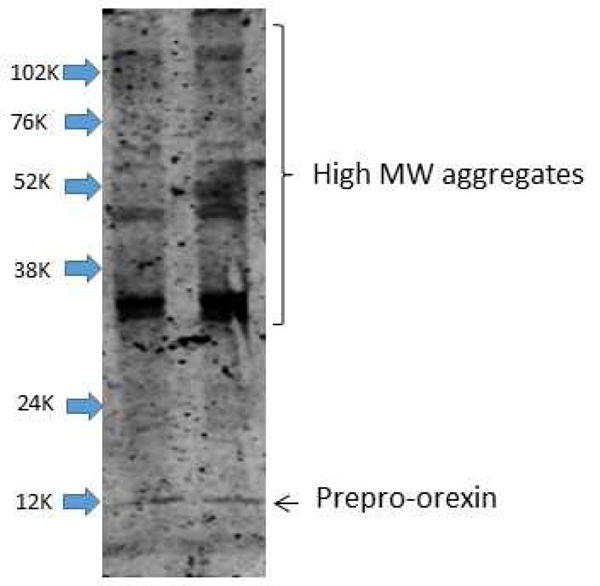
Representative western blot of samples run under non-reducing conditions showing higher molecular weight orexin-A immuno-reactive bands in basal forebrain homogenates of 12 month old heterozygous and wild-type mice.
3.6 BiP heterozygous mice have more ER stress in orexin neurons and in the basal forebrain
Having observed the increased varicosities in the axonal projections of the orexin neurons, we then surveyed the orexin neurons for ER stress by staining for CHOP, a marker of the maladaptive ER stress response (Oyadomari and Mori, 2004; Ron and Walter, 2007). We also determined CHOP expression in the basal forebrain by westerns. The heterozygous mice at 12 months of age had significantly more CHOP expressed in the basal forebrain (Figure 8A; p=0.018) as well as orexin neurons staining positive for CHOP than wildtype mice, 58% vs 47% (Figure 8B; p=0.050). Further, the heterozygous mice displayed more nuclear CHOP than the wildtype mice (28% vs 19%; p=0.016). CHOP in the nucleus indicates increased cellular stress and transcriptional activation (Marciniak et al., 2004; Voccoli et al., 2007).
Figure 8.
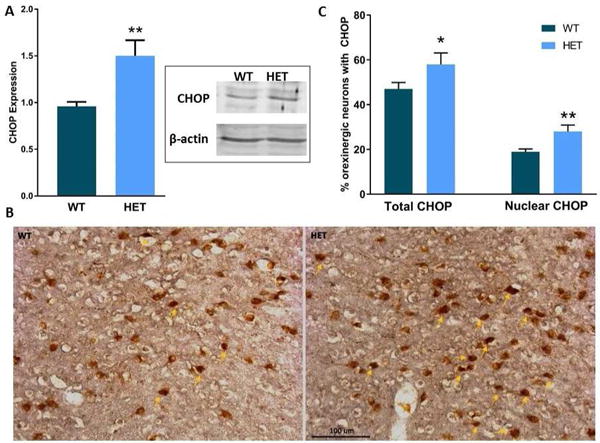
A) Histogram of CHOP expression in basal forebrain of WT and Het mice (n=6/genotype) as determined by western analyses. Representative western blot of CHOP in WT and Het mice shown in inset. Each lane has 15 μg of protein loaded. β-actin shown as loading control. B) Representative images of CHOP, (GADD153), a pro-apoptotic protein in orexinergic neurons in WT and Het mice. Arrows delineate nuclei with CHOP translocation. light-brown DAB indicates orexin immunopositivity and dark brown Nickel-DAB shows CHOP staining.
C) Histogram of percentage orexin neurons with CHOP immunostaining and nuclear CHOP. Mean ± SE shown. (n=3/genotype; 8 sections per mouse). Statistically significant differences are indicated by * (p≤0.05), ** (p≤0.01)
4. DISCUSSION
In this study, we compared sleep-wake behavior in mice heterozygous for the molecular chaperone BiP and their wildtype littermates at 3 different ages: 2–3, 12 and 18 months. We found that BiP heterozygosity was associated with reduced REM. Both REM bout duration and total REM minutes were reduced in mice with one functional copy of BiP. While there were no significant interactions between genotype and age observed for wake or NREM, we did find significant differences across age-groups within genotype. Sleep and wake fragmentation across aging was more pronounced in the BiP heterozygous mice. The most robust changes were observed in the number and duration of wake bouts during the active period; mice lacking one functional copy of BiP had many more bouts of shorter duration across aging. Similarly, there were significantly more NREM bouts of shorter duration in the heterozygous mice during the sleep period across aging. Normal aging has been associated with increased sleep fragmentation and impaired ability to sustain wakefulness in humans (Bliwise, 1993; Dijk et al., 1989; Dijk et al., 2000; Ehlers and Kupfer, 1989), rodents (Colas et al., 2005; Hasan et al., 2012; Naidoo et al., 2008; Naidoo et al., 2011; Welsh et al., 1986; Wimmer et al., 2013) and Drosophila (Brown et al., 2014; Bushey et al., 2010; Koh et al., 2006; Metaxakis et al., 2014). Changes in sleep architecture are well documented in aged animals. In rats, hippocampal theta activity during wakefulness is indicative of arousal, exploratory behavior, and spatial navigation (Wyble et al., 2004) and we found that aging slowed theta peak frequency in the wake EEG spectra in both the wildtype and heterozygous mice. This is consistent with previously published data (Hasan et al., 2012; Wimmer et al., 2013) that showed a slowing of wake theta peak frequency with age. While age-related differences in theta peak frequency were significant only during wake for BiP wildtype mice, the heterozygous mice displayed significant changes in peak theta frequency during both wake and REM. Hasan et al (Hasan et al., 2012) have reported only an effect of age on theta peak frequency during wake for B6 mice, which is the background strain of the mice used in this study, while Wimmer et al (Wimmer et al., 2013) have reported an effect of age on both wake and REM theta peak frequency. Why reduced levels of BiP would specifically alter theta during REM sleep is not clear, however, it is possible that some of the differences may be a consequence of developmental changes that result from reduced BiP.
We and others have previously shown that BiP levels decrease with age (Brown et al., 2014; Naidoo et al., 2008; Paz Gavilan et al., 2006). Similarly, in this study we find that BiP protein expression is decreased with age in both the wildtype and heterozygous mice. Even though the heterozygous mice express 50% lower BiP levels than the wildtype mice at a young age (2–3 months) they display a further reduction in BiP at 12 months. At 18 months of age, BiP levels have been reduced to a level that is similar between genotypes. Age-related loss of BiP is expected to result in significant impairment of cellular protein homeostasis, which is mediated by the UPR (Brown and Naidoo, 2012). Aged flies also display reduced BiP expression and both sleep and wake fragmentation with aging. Our studies in Drosophila have indicated that restoring chaperone function ameliorated changes in sleep and wake with age (Brown et al., 2014). Specifically, those studies indicated that supplementing chaperones consolidated sleep and wake in older flies, suggesting that maintaining protein homeostasis promotes improved sleep quality. Interestingly, we also showed in that study that disrupting protein homeostasis by inducing protein misfolding was sufficient to fragment sleep in young Drosophila (Brown et al., 2014).
Given our observation of enhanced wake fragmentation across aging in the mice with reduced BiP expression, we ascertained whether there were any observable effects of age on orexin neurons in these mice and their littermate controls. Loss of orexin neurons is associated with changes in wake state stability (Chemelli et al., 1999; Sawai et al., 2010). We first counted the number of orexin neuron numbers in the 12 month old heterozygous mice, but did not find significant differences when compared with wildtype mice of the same age. Previous rodent studies have indicated that the number of orexin neurons and their projection densities decrease with age (Kessler et al., 2011; Sawai et al., 2010). In the macaque monkey, however, orexinergic neuronal counts across the entire lateral hypothalamus reveal no differences with age, although orexinergic axons to the locus coeruleus are diminished with age (Downs et al., 2007). Our study on orexin numbers was carried out on middle-aged 12 month old mice, unlike the earlier studies which showed a decline in number in aged rats 24 months or older. Whether or not this effect of age on number of orexin neurons is species dependent is not known. Another important measure aside from neuron number is neuronal functionality. This could be altered with age. We had previously shown that aged (24 month) mice do not increase c-fos immunoreactivity when subjected to 3 hours of enforced wakefulness (Naidoo et al., 2011). Thus, we examined c-fos induction as a proxy for neuronal functional activity of orexin neurons in 12 month old mice, as this was an age at which there were still differences in BiP expression levels between the genotypes. Here again, while we noted marginal changes in c-fos induction with heterozygosity across aging, there were no significant differences between the wildtype and heterozygous mice in the percentage of c-fos immunoreactive orexin neurons during waking.
We did, however, find that the orexin neurons in the BiP het mice displayed increased CHOP immunoreactivity, suggestive of heightened ER stress, and that projections from these neurons to the basal forebrain displayed varicosities staining positive for orexin. Increased CHOP expression, particularly within the nucleus, is a sign of prolonged cellular stress (Szegezdi et al., 2006; Voccoli et al., 2007). Varicosities similar to those observed in this study have been reported at the terminals of orexin fibers in the LC of aged cat brains (Zhang et al., 2002) and it is thought that they represent degeneration of orexin positive fibers in the aged cats. Our data also indicate that orexin is likely misfolded and aggregated in BiP heterozygous mice as a result of improper processing because of the appearance of orexin immunoreactive higher molecular weight species. BiP, in addition to playing a role in folding as a chaperone, also plays a key role in escorting proteins to the proteasome for degradation (Gething, 1999; Hegde et al., 2006; Jin et al., 2000). The increase in orexin positive higher molecular weight species suggests that protein degradation may also being hampered in the BiP heterozygous mice.
Age-related alterations in the orexin system in rats and other species have been previously reported and include decreased orexin innervations of target regions (Downs et al., 2007; Zhang et al., 2002), decreased expression of the peptide or its receptors (Porkka-Heiskanen et al., 2004; Terao et al., 2002), diminished behavioral responses to centrally-administered orexin (Kotz et al., 2005; Takano et al., 2004) and altered patterns of orexin release across the diurnal cycle (Desarnaud et al., 2004). Whether or not declines in the activity of chaperones such as BiP would impact these outcomes is not known and requires future studies.
This study has a number of strengths and limitations worth mentioning. A strength of this study is that it includes three age-groups not typically studied – 2–3 months, 12 months, and 18 months – adding important information to the literature on age-related sleep and wake characteristics. Moreover, this is the first to implicate a molecular chaperone in age-related changes of sleep and wake behavior. Finally, this study includes a number of important mechanistic phenotypes, including BiP expression levels and identification of protein aggregation or altered protein homeostasis in a wake active neuronal population (orexin) that may impact upon the stability of wake behavior. While we found consistent evidence of stronger age-related associations on sleep and wake within BiP heterozygous mice, the lack of significance of interactions terms and limited differences between BiP genotypes within age groups mean these results should be replicated in larger studies. Similarly, the relatively small number of animals means we are underpowered for smaller effect sizes. Finally, analyses on possible mechanisms of effect were performed within 12 month old mice only, as this was an aged group where BiP expression still differed; these assessments should be performed in future studies in other age groups.
To summarize, we have found that loss of one functional copy of BiP, which resulted in reduced BiP expression at 2–3 and 12 months of age, was associated with reduced REM and altered theta during REM and wake. Further, we found that the expected age-related sleep and wake fragmentation was more pronounced in the BiP heterozygous mice as compared to wildtype animals, suggesting reduction in BiP accentuates age-related effects. When examining possible mechanisms, we found that there was considerably more ER stress in orexin neurons, as well as greater orexin aggregates at terminals of neurons in 12 month old BiP heterozygotes when compared to wildtype mice of the same age. Whether or not these contributed directly to the sleep and wake phenotype remains to be established, and future studies should be performed to validate these effects of reduction in BiP on age-related sleep and wake characteristics.
Supplementary Material
Highlights.
Sleep and wake fragmentation is more pronounced in aging mice with reduced BiP
The ability to maintain long wake bouts is lost
Mice with one functional copy of BiP display more ER stress
There is aggregation of orexin in mice with reduced BiP expression
Acknowledgments
This project was supported by funding from NIA P01 AG17628. The authors would like to thank Dr. Kristan Singletary for the preliminary studies on the BiP heterozygous mice and Dr. Lin Zhang and May Chan for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors state that there are no actual or potential conflicts of interest.
References
- Ballinger EC, Ananth M, Talmage DA, Role LW. Basal Forebrain Cholinergic Circuits and Signaling in Cognition and Cognitive Decline. Neuron. 2016;91:1199–1218. doi: 10.1016/j.neuron.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliwise DL. Sleep in normal aging and dementia. Sleep. 1993;16:40–81. doi: 10.1093/sleep/16.1.40. [DOI] [PubMed] [Google Scholar]
- Brown MK, Chan MT, Zimmerman JE, Pack AI, Jackson NE, Naidoo N. Aging induced endoplasmic reticulum stress alters sleep and sleep homeostasis. Neurobiology of Aging. 2014;35:1431–1441. doi: 10.1016/j.neurobiolaging.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MK, Naidoo N. The endoplasmic reticulum stress response in aging and age-related diseases. Front Physiol. 2012;3:263. doi: 10.3389/fphys.2012.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Hughes KA, Tononi G, Cirelli C. Sleep, aging, and lifespan in Drosophila. BMC Neurosci. 2010;11:56. doi: 10.1186/1471-2202-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999;98:437–451. doi: 10.1016/s0092-8674(00)81973-x. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Colas D, Cespuglio R, Sarda N. Sleep wake profile and EEG spectral power in young or old senescence accelerated mice. Neurobiol Aging. 2005;26:265–273. doi: 10.1016/j.neurobiolaging.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Desarnaud F, Murillo-Rodriguez E, Lin L, Xu M, Gerashchenko D, Shiromani SN, Nishino S, Mignot E, Shiromani PJ. The diurnal rhythm of hypocretin in young and old F344 rats. Sleep. 2004;27:851–856. doi: 10.1093/sleep/27.5.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk DJ, Beersma DG, van den Hoofdakker RH. All night spectral analysis of EEG sleep in young adult and middle-aged male subjects. Neurobiol Aging. 1989;10:677–682. doi: 10.1016/0197-4580(89)90004-3. [DOI] [PubMed] [Google Scholar]
- Dijk DJ, Duffy JF, Czeisler CA. Contribution of circadian physiology and sleep homeostasis to age-related changes in human sleep. Chronobiol Int. 2000;17:285–311. doi: 10.1081/cbi-100101049. [DOI] [PubMed] [Google Scholar]
- Downs JL, Dunn MR, Borok E, Shanabrough M, Horvath TL, Kohama SG, Urbanski HF. Orexin neuronal changes in the locus coeruleus of the aging rhesus macaque. Neurobiol Aging. 2007;28:1286–1295. doi: 10.1016/j.neurobiolaging.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Kupfer DJ. Effects of age on delta and REM sleep parameters. Electroencephalogr Clin Neurophysiol. 1989;72:118–125. doi: 10.1016/0013-4694(89)90172-7. [DOI] [PubMed] [Google Scholar]
- Gething MJ. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- Hasan S, Dauvilliers Y, Mongrain V, Franken P, Tafti M. Age-related changes in sleep in inbred mice are genotype dependent. Neurobiol Aging. 2012;33:195 e113–126. doi: 10.1016/j.neurobiolaging.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Hegde NR, Chevalier MS, Wisner TW, Denton MC, Shire K, Frappier L, Johnson DC. The role of BiP in endoplasmic reticulum-associated degradation of major histocompatibility complex class I heavy chain induced by cytomegalovirus proteins. J Biol Chem. 2006;281:20910–20919. doi: 10.1074/jbc.M602989200. [DOI] [PubMed] [Google Scholar]
- Hoffman GE, Smith MS, Verbalis JG. c-Fos and related immediate early gene products as markers of activity in neuroendocrine systems. Front Neuroendocrinol. 1993;14:173–213. doi: 10.1006/frne.1993.1006. [DOI] [PubMed] [Google Scholar]
- Hussain SG, Ramaiah KV. Reduced eIF2alpha phosphorylation and increased proapoptotic proteins in aging. Biochem Biophys Res Commun. 2007;355:365–370. doi: 10.1016/j.bbrc.2007.01.156. [DOI] [PubMed] [Google Scholar]
- Jin T, Gu Y, Zanusso G, Sy M, Kumar A, Cohen M, Gambetti P, Singh N. The chaperone protein BiP binds to a mutant prion protein and mediates its degradation by the proteasome. J Biol Chem. 2000;275:38699–38704. doi: 10.1074/jbc.M005543200. [DOI] [PubMed] [Google Scholar]
- Jones S, Pfister-Genskow M, Benca RM, Cirelli C. Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J Neurochem. 2008;105:46–62. doi: 10.1111/j.1471-4159.2007.05089.x. [DOI] [PubMed] [Google Scholar]
- Kessler BA, Stanley EM, Frederick-Duus D, Fadel J. Age-related loss of orexin/hypocretin neurons. Neuroscience. 2011;178:82–88. doi: 10.1016/j.neuroscience.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakpai F, Nasehi M, Haeri-Rohani A, Eidi A, Zarrindast MR. Septohippocampo-septal loop and memory formation. Basic and clinical neuroscience. 2013;4:5–23. [PMC free article] [PubMed] [Google Scholar]
- Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci U S A. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotz CM, Mullett MA, Wang C. Diminished feeding responsiveness to orexin A (hypocretin 1) in aged rats is accompanied by decreased neuronal activation. American journal of physiology. Regulatory, integrative and comparative physiology. 2005;289:R359–R366. doi: 10.1152/ajpregu.00717.2004. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, Baldwin DA, Jensen ST, Churchill GA, Pack AI. Macromolecule biosynthesis: a key function of sleep. Physiol Genomics. 2007;31:441–457. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane BB, Galante RJ, Jensen ST, Naidoo N, Pack AI, Wyner A. Characterization of the bout durations of sleep and wakefulness. J Neurosci Methods. 2010;193:321–333. doi: 10.1016/j.jneumeth.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Tain LS, Gronke S, Hendrich O, Hinze Y, Birras U, Partridge L. Lowered insulin signalling ameliorates age-related sleep fragmentation in Drosophila. PLoS Biol. 2014;12:e1001824. doi: 10.1371/journal.pbio.1001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N. Cellular stress/the unfolded protein response: relevance to sleep and sleep disorders. Sleep Med Rev. 2009;13:195–204. doi: 10.1016/j.smrv.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Casiano V, Cater J, Zimmerman J, Pack AI. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep. 2007;30:557–565. doi: 10.1093/sleep/30.5.557. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Galante RJ, McShane B, Hu JH, Zimmerman J, Maislin G, Cater J, Wyner A, Worley P, et al. Role of Homer proteins in the maintenance of sleep-wake states. PLoS ONE. 2012;7:e35174. doi: 10.1371/journal.pone.0035174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Master M, Zhu Y, Pack AI. Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J Neuroscience. 2008;28:6539–6548. doi: 10.1523/JNEUROSCI.5685-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Zhu J, Zhu Y, Fenik P, Lian J, Galante R, Veasey S. Endoplasmic reticulum stress in wake-active neurons progresses with aging. Aging Cell. 2011;10:640–649. doi: 10.1111/j.1474-9726.2011.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochemical Journal. 2011;434:181–188. doi: 10.1042/BJ20101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Panossian L, Fenik P, Zhu Y, Zhan G, McBurney MW, Veasey S. SIRT1 regulation of wakefulness and senescence-like phenotype in wake neurons. Journal of Neuroscience. 2011;31:4025–4036. doi: 10.1523/JNEUROSCI.5166-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz Gavilan M, Vela J, Castano A, Ramos B, del Rio JC, Vitorica J, Ruano D. Cellular environment facilitates protein accumulation in aged rat hippocampus. Neurobiol Aging. 2006;27:973–982. doi: 10.1016/j.neurobiolaging.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Porkka-Heiskanen T, Kalinchuk A, Alanko L, Huhtaniemi I, Stenberg D. Orexin A and B levels in the hypothalamus of female rats: the effects of the estrous cycle and age. Eur J Endocrinol. 2004;150:737–742. doi: 10.1530/eje.0.1500737. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The Role of the PERK/eIF2alpha/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr Mol Med. 2016;16:533–544. doi: 10.2174/1566524016666160523143937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai N, Ueta Y, Nakazato M, Ozawa H. Developmental and aging change of orexin-A and -B immunoreactive neurons in the male rat hypothalamus. Neuroscience Letters. 2010;468:51–55. doi: 10.1016/j.neulet.2009.10.061. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annual Reviews in Biochemistry. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Reports. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano S, Kanai S, Hosoya H, Ohta M, Uematsu H, Miyasaka K. Orexin-A does not stimulate food intake in old rats. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1182–1187. doi: 10.1152/ajpgi.00218.2004. [DOI] [PubMed] [Google Scholar]
- Terao A, Apte-Deshpande A, Morairty S, Freund YR, Kilduff TS. Age-related decline in hypocretin (orexin) receptor 2 messenger RNA levels in the mouse brain. Neuroscience Letters. 2002;332:190–194. doi: 10.1016/s0304-3940(02)00953-9. [DOI] [PubMed] [Google Scholar]
- Voccoli V, Mazzoni F, Garcia-Gil M, Colombaioni L. Serum-withdrawal-dependent apoptosis of hippocampal neuroblasts involves Ca++ release by endoplasmic reticulum and caspase-12 activation. Brain Research. 2007;1147:1–11. doi: 10.1016/j.brainres.2007.01.145. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Richardson GS, Dement WC. Effect of age on the circadian pattern of sleep and wakefulness in the mouse. J Gerontol. 1986;41:579–586. doi: 10.1093/geronj/41.5.579. [DOI] [PubMed] [Google Scholar]
- West MJ, Gundersen HJ. Unbiased stereological estimation of the number of neurons in the human hippocampus. J Comp Neurol. 1990;296:1–22. doi: 10.1002/cne.902960102. [DOI] [PubMed] [Google Scholar]
- Wimmer ME, Rising J, Galante RJ, Wyner A, Pack AI, Abel T. Aging in mice reduces the ability to sustain sleep/wake states. PLoS One. 2013;8:e81880. doi: 10.1371/journal.pone.0081880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyble BP, Hyman JM, Rossi CA, Hasselmo ME. Analysis of theta power in hippocampal EEG during bar pressing and running behavior in rats during distinct behavioral contexts. Hippocampus. 2004;14:662–674. doi: 10.1002/hipo.20012. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Sampogna S, Morales FR, Chase MH. Age-related changes in hypocretin (orexin) immunoreactivity in the cat brainstem. Brain Research. 2002;930:206–211. doi: 10.1016/s0006-8993(02)02240-0. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Fenik P, Zhan G, Mazza E, Kelz M, Aston-Jones G, Veasey SC. Selective loss of catecholaminergic wake active neurons in a murine sleep apnea model. Journal of Neuroscience. 2007;27:10060–10071. doi: 10.1523/JNEUROSCI.0857-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


