Abstract
Resolving the temporal dynamics of cell signaling pathways is essential for regulating numerous downstream functions, from gene expression to cellular responses. Mapping these signaling pathways requires the exposure of cells to time-varying chemical signals; these are difficult to generate and control over a wide temporal range. Herein, we present an acoustofluidic chemical signal generator based on sharp-edge-based micromixing strategy. The device, simply by modulating the driving signals of acoustic transducer including the ON/OFF switching frequency, actuation time and duty cycle, is capable of generating both single-pulse and periodic chemical signals that are temporally controllable in terms of stimulation period, stimulation duration and duty cycle. We also demonstrate the device’s applicability and versatility for cell signaling studies by probing the calcium (Ca2+) release dynamics for three different types of cells stimulated by ionomycin signals of different shapes. Upon a short, single-pulse of ionomycin stimulation (~100 ms) generated by our device, we discover that cells tend to dynamically adjust the intracellular level of Ca2+ through constantly releasing and accepting Ca2+, respectively, to the cytoplasm and from the extracellular environment. With advantages such as simple fabrication and operation, compact device design, and reliability and versatility, our device will enable decoding of the temporal characteristics of signaling dynamics for various physiological processes.
Graphic Content Entry
A sharp-edge-based acoustofluidic device capable of generating temporally controllable chemical signals is presented to enable cell-signaling studies.

Introduction
Cell signaling pathways act as signal processors that dynamically convert input signals – chemical cues from surrounding environments – to appropriate output responses.1–3 Signaling dynamics are linked to specific activations of downstream signaling molecules that trigger cellular responses.4–6 Time-varying chemical signals have been found to significantly influence on whole cell level and are physiologically relevant;7–11 such signals can be used to resolve temporal dynamics of signaling pathways and therefore, probe cellular responses. Delivering time-varying chemical signals to which cells respond can help researchers determine the dynamic properties of specific signaling pathways, and also help to elucidate specific downstream transcriptional responses.
To probe a signaling pathway’s response to different chemical cues, biologists conventionally switch between media and stimulants with pipettes, which is a time-consuming procedure and demands trained personnel. As a result, the temporal resolution of chemical cues provided by the conventional setup is very limited. Microfluidics, due to its capability of handling tiny sample volumes, simplicity in setup, and amenability to automation, has recently emerged as a promising tool to generate time-varying chemical cues for studying signaling dynamics.12–15 Various microfluidic chemical signal generators have been developed based on different mechanisms, including interface shifting of laminar flow,16–20 alternating flow pumping,21–25 pneumatic-valve control,26–32 flow photolysis,33–35 diffusion through microgrooves36 and micro/nanoporous membranes,37,38 and microflow injection.39 Of these developments, interface shifting and alternating pumping are the most common methods; however, they have limited temporal resolution, require external components such as solenoid or pneumatic valves, and necessitate precise control over inlet and outlet pressure drops. To improve temporal resolution, oscillating microbubble-based mixing40,41 was recently proposed to generate temporally-controllable chemical waveforms, based on the rapid mixing of buffer and stimulant. While this approach does provide an improvement in temporal resolution, it is unstable in terms of the size changes of the bubbles over time and inherent size-dependent operation.41–46 This instability makes the microbubbles-based approach less attractive to be implemented for resolving signaling dynamics where long-term cell-culture/monitoring are necessary; therefore, the applicability and versatility is limited. Though focused-travelling surface acoustic waves could partially mix two solutions to rapidly generate concentration gradients, their ability to completely mix two solutions and obtain a uniform concentration profile across the channel’s width – a requirement that ensures that all the cells in the channel can experience the same chemical signals – has yet to be proven.
Here, we present an acoustofluidics (i.e., the fusion of acoustics and microfluidics)48–57 chemical signal generator based on the rapid mixing of buffer and stimulant which is enabled by acoustic streaming. This acoustofluidic chemical signal generator is built upon a sharp-edge-based58–66 micromixer, which allows rapid, homogeneous mixing of fluids with a mixing time less than 100 ms. With this mixing strategy, we can generate chemical signals that are temporally controllable over stimulation period, duration, and duty cycle. We also demonstrate the applicability and versatility of our device by monitoring the intracellular calcium (Ca2+) release dynamics of cells in response to the ionomycin stimulus generated from our device. The implementation of an active mixing strategy to synthesize time-varying chemical signals and to temporally adjust microchemical environments for cells presents a technical innovation. We remove the need for time-consuming processes and trained-personnel that are required in conventional setups,5,6 and remove the need for sophisticated pneumatic-valve control and precise pressure-drop control employed in other existing microfluidic cell-stimulation devices.24,25,28 Owing to its short mixing time, on-demand control, simplicity, and stability, our sharp-edge-based acoustofluidic chemical signal generator is an easy-to-use and efficient tool for cell signaling studies where determining temporal characteristics of signaling pathways is critical to unveil downstream responses in gene expression and regulation.
Device design and concept
Fig. 1A schematically shows the design and working concept of our acoustofluidic chemical signal generator. The device consists of a single-layer polydimethylsiloxane (PDMS) microfluidic channel constructed with two pairs of sharp-edge structures on the sidewalls and an acoustic transducer. Each of the sharp-edge structures is 300 μm apart from the adjacent structure and identical in dimensions: 600 μm in length with a tip angle of 15°. Phosphate buffered saline (PBS; 1X-PBS, Life Technology, USA) and PBS containing fluorescein isothiocyanate-dextran (FITC-Dextran, 1 mg/mL, MW = 10 kDa, Sigma-Aldrich, USA), which are modelled as the medium and stimulant, respectively, are simultaneously infused into the channel through two separate inlets. The fluorescence intensity shown in Fig. 1 is normalized to that of 100 % FITC-Dextran. Two regions of interest (ROI), ROI 1 and ROI 2, are set up and positioned 1 mm after the last sharp-edge structure, such that we can avoid both the flow fluctuation when switching the transducer ON and OFF, and also the shear stress developed by the acoustic streaming.
Figure 1.
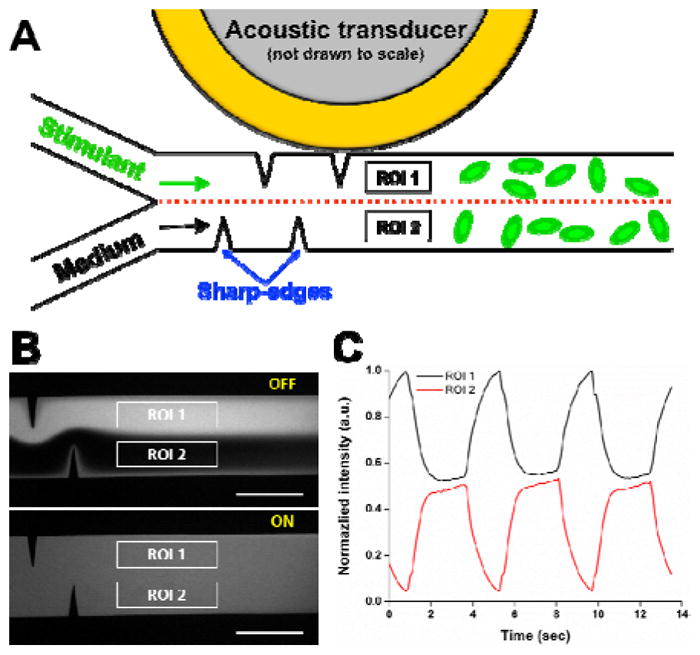
(A) Schematic showing the top view of the sharp-edge-based acoustofluidic chemical signal generator. Two regions of interest (ROI), ROI 1 and ROI 2, are set up and positioned about 1 mm after the last sharp-edge structure. (B) Fluorescent images showing unmixed, laminar flow of PBS and FITC-dextran (Top) when the transducer is switched OFF, and completely mixed fluid (Bottom) when the transducer is switched ON. Scale bar: 500 μm. (C) Periodic fluorescence intensity profiles generated in the region of interest by periodically switching the transducer ON and OFF at a switching frequency of 0.25 Hz.
Our sharp-edge-based acoustofluidic chemical signal generator is operated by controlling an acoustic transducer. While the transducer is switched OFF, PBS and FITC-dextran form a side-by-side laminar flow (Fig. 1B, top), leading to two flat fluorescence intensity curves of different intensities, 1 for ROI 1 and 0 for ROI 2 (not shown here). Upon switching the transducer ON, the sharp-edge structures acoustically oscillate, perturbing the surrounding flow and therefore, generating acoustic streaming around the tip of each sharp-edge structure. The generated acoustic streaming then greatly enhances the mass transport of the two solutions to rapidly and efficiently mix the two solutions (Fig. 1B, bottom). This complete mixing yields a flat fluorescence intensity curve at 0.5 for both ROI 1 and ROI 2. Because our mixing strategy is of active, on-demand mixing, if we further switch the transducer ON and OFF periodically at a given switching frequency (fswitching = 0.25 Hz for Fig. 1C), we can generate a periodic fluorescence intensity profile in both ROI 1 and ROI 2. In other words, by alternating the mixing ON and OFF, we can produce chemical signals that are temporally controllable in terms of stimulation period, stimulation duration, and duty cycle. It is important to note that because our sharp-edge-based acoustofluidic micromixing can completely mix fluids in less than 100 ms, in this work the stimulation duration is defined and referred to as the “activation time” of the transducer.
Materials and methods
Device fabrication
Our device was of a single-layer configuration and was simply prepared using standard photolithography, deep ion etching (DRIE), and PDMS replica molding. First, a single-side polished silicon wafer was patterned by standard photolithography and then anisotropically etched to have a channel height of 100 μm by DRIE, forming a silicon master mold. The surface of the silicon master mold was coated with silane vapor using 1H,1H,2H,2H-perfluorooctyl-trichlorosilane (Sigma-Aldrich, MO, USA) for 1 hour. After silane coating, a 10:1 mixture of PDMS-Sylgard 184 Silicone Elastomer Base and Sylgard 184 Curing Agent (Dow Corning, MI, USA) was prepared and placed under vacuum to remove air bubbles, then was poured onto the silicon master mold and baked at 65 °C for 1 hour to form PDMS channels. Upon completely curing, the PDMS channel was punched with two inlets and one outlet. Its surface and a glass slide (Cat. no. 48404-454, VWR) were treated with oxygen plasma (BD-10AS, Electro-Technic Products, IL, USA) for 60 seconds and 10 seconds, respectively. Following plasma treatment, they were bonded together and then baked at 65 °C for 24 hours. The last step was to bond the acoustic transducer (Model no. 273-073, RadioShack, TX, USA) beside the PDMS channel onto the glass slide using epoxy (84101, Permatex, CT, USA).
Experimental setup and operation
The device was mounted on the stage of an inverted microscope (TE2000-U, Nikon, Japan) and all the experiments were conducted on the microscope. Additionally, the microscope was equipped with a cell incubation system (Chamlide TC, Live Cell Instrument, South Korea), which was employed for on-chip cell culture in a humidified environment at 37 °C with a CO2 level of 5%. Each solution was injected with separate 1 mL syringes (BD Bioscience, NJ, USA), and the injection was administered by an automated syringe pump (neMESYS, Germany).
The control of the acoustic transducer, including ON/OFF switching, driving frequency and voltage, and actuation time and frequency, was realized with a function generator (AFG3011C, Tektronix, OR, USA) and an amplifier (25A250A, Amplifier Research, PA, USA). Since our acoustofluidic chemical signal generator works by actuating an acoustic transducer, we experimentally determine a proper driving frequency for the transducer by sweeping frequencies with a 100 Hz increment from 1 kHz to 50 kHz. The chosen driving frequency was 4.5 kHz, at which the sharp-edge structures generated the strongest acoustic streaming effect to rapidly and completely mix two fluids. In addition, the driving voltage of the transducer was set to 20 VPP, at which the generated acoustic streaming was strong enough to mix the two fluids at a total flow rate of 4 μL min−1 in the channel. As a result, the driving frequency of 4.5 kHz and the driving voltage of 20 VPP were used in all the experiments.
Cell preparation
Human dermal microvascular endothelial cells (HMVEC-d, ATCC, VA, USA) were the main cell type to be stimulated in our device. To validate the ability of our devices to stimulate different cell types, we also examined HeLa (ATCC, VA, USA), a human cell line derived from cervical cancer, and U-251, a human glioblastoma cell line. HMVEC-d, HeLa, and U-251 cells were grown, respectively, in EndoGRO-LS complete media (Millipore, MA, USA), DMEM/F12 (Gibco, Life Technologies, MA, USA) and MEM (Gibco, Life Technologies, MA, USA) at 37 °C and a CO2 level of 5 % in a CO2 incubator (Nu-4750, NuAire, MN, USA). Once the cells were grown to 80–90% confluency, they were harvested with trypsin-EDTA-0.05% (Gibco, Life Technologies, NY, USA) and re-suspended in fresh culture media at desired cell concentrations, followed by on-chip cell culture for experimentation.
On-chip cell culture, staining and stimulation
To ensure our device could generate chemical signals that could be encoded by cells, we performed cell stimulation experiments where HMVEC-d, HeLa, and U-251 cells were stimulated by ionomycin stimulation delivered by our device. These three types of cells were separately cultured, stained, and stimulated on-chip inside the channel of our device in their respective experiments. The channel’s surface was first treated with fibronectin-coating (100 μg/mL; Cat. No. 356008, BD Biosciences, NJ, USA) for 1 hour. Afterwards, the channels were rinsed with PBS several times to remove residual, un-bonded fibronectin. Following the fibronectin coating, suspended HMVEC-d, HeLa, and U-251 cells (1 × 106 cells mL−1) were injected into the channel through the outlet, ensuring that the cells could evenly disperse across the channel. The cells were then allowed for 60 minutes to spread and adhere to the bottom of the channel. To visualize and quantitatively characterize the calcium release behavior of the cells, Fluo-4 AM dye (Cat. no. F14201, ThermoFisher Scientific, MA USA; here called “Fluo-4 AM”), a cell-permeant fluorescent calcium indicator, was used to measure intracellular calcium concentration. The cells were loaded with Fluo-4 AM dye by injecting Hank’s Balanced Salt Solution (HBSS; Cat. no. 14025092, ThermoFisher Scientific, MA, USA) containing 1 μM Fluo-4 AM dye into the channel through the outlet at a flow rate ratio of 5 μL/min for 60 mins, followed by washing with a pure culture medium containing anion transport inhibitor for 30 mins. The cellular fluorescence expressed by each cell was characterized over the cytoplasm area, which covers at least 70% of the entire cell area (Fig. S1).
After the staining was completed, the stimulant, which was a pure culture medium containing 2.0 μM ionomycin (Cat. No. 9995, Cell Signaling Technology, MA, USA), was injected into the channel through the ionomycin inlet (the top inlet in Fig. 1A) at a flow rate of 2 μL min−1, while the pure medium was injected through the medium inlet also at a flow rate of 2 μL min−1 (the bottom inlet in Fig. 1A). Upon complete mixing, the entire channel was filled with a uniform mixture of the culture medium containing 1.0 μM ionomycin; cells seeded in ROI 1 were subject to ionomycin stimulation with concentrations between 1.0 and 2.0 μM, while those seeded in ROI 2 were subject to ionomycin stimulation with concentrations between 0 and 1.0 μM.
Image/video acquisition and data analysis
All the experimental images and videos were acquired, processed, and analyzed using Nikon NIS-Elements Advanced Research (AR) software and ImageJ (NIH, MD, USA). We started video recording 5 seconds before the chemical stimulation was generated to record the entire cell stimulation process and cell responses.
Data was presented as group means ± standard deviation (SD), and was analyzed by the one-way ANOVA test using OriginPro 8.0 (OriginLab Corporation, MA, USA). Tukey’s Multiple Comparison test was employed to compare differences among treatment groups, where a p-value of less than 0.05 was considered statistically significant.
Results and discussion
Generation of chemical signals
We first validated the generation of chemical signals with various shapes; to do so, PBS and FITC-dextran solutions were injected into the channel at the same flow rate (QPBS = 2 μl min−1 & QFITC = 2 μl min−1). The flow rates, unless otherwise specified, were constant throughout the experiments. To quantitatively characterize the chemical signals that can be generated by our device, videos were taken at the location accommodating ROI 1 and ROI 2. Afterwards, the fluorescence intensity over time at each region was normalized to that of the completely mixed solution (i.e., 50% FITC-dextran).
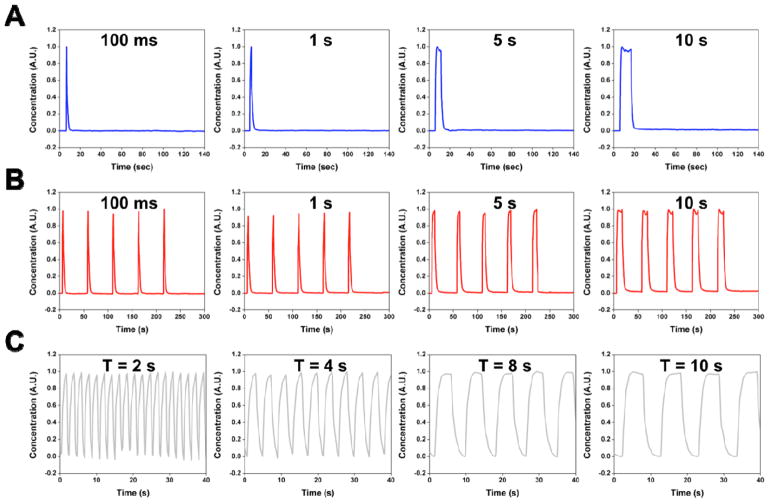
Fig. 2 shows the chemical signals of various shapes obtained at ROI 2. Note that herein only chemical signals generated at ROI 2 were characterized, because the chemical signals at ROI 1 were merely complementary shapes with normalized intensities ranging from 1 to 2. By simply activating the transducer for given amount of time (e.g., 100 ms, 1, 5 and 10 sec), we could generate single-pulse chemical signals of different durations, as shown in Fig. 2A. When repeatedly activating the transducer, similarly, we could obtain periodic chemical signals of different durations (Fig. 2B), where the chemical signals had an identical period (50 sec) but different duty cycles (e.g., 0.2, 2, 10 and 20%). Other than duty cycle, periodic chemical signals with the same duty cycle (50%) but different periods (e.g., 2, 4, 8, and 10 sec) were also produced by switching the transducer ON and OFF at different switching frequencies (fswitching), as shown in Fig. 2C. The results demonstrate that by using our acoustofluidic-based chemical signal generator, we can generate chemical signals that are temporally controllable.
Figure 2.
Examples of chemical signals that can be generated from our acoustofluidic chemical signal generator. Chemical signals with a wide temporal range can be generated solely by modulating the driving signals of the transducer. (A) Single-pulse signals with stimulation durations of 100 ms, and 1, 5 and 10 sec. (B) Periodic chemical signals with a constant period of 50 sec and various durations of 100 ms, and 1, 5 and 10 sec. (C) Periodic chemical signals with a fixed duty cycle of 50 % and varying periods of 2, 4, 8 and 10 sec.
The chemical signals generated by our acoustofluidic devices can also be spatially controllable – their concentrations can be varied relative to the position in the channel – by either sweeping the driving frequency of the transducer or adjusting its driving voltage (Fig. S2). The generation of spatially controllable chemical signals can be very useful. For example, within a single experiment, one could expose the same cells to stimulants of different concentrations without manually changing the stimulant concentration. Simply speaking, by modulating the driving signals of the transducer, one may generate chemical signals with both spatially and temporally-controllable characteristics. In addition to modulating the driving signals of the transducer, rearranging the sharp-edge structures may lead to different mixing schemes, which in turn, would allow for the generation of spatiotemporally controllable chemical signals. For example, by rearranging the sharp-edge structures similar to what was reported previously (Fig. S2),60 our device can, along with the signal modulation of the transducer, generate such chemical signals. Additionally, it is worth mentioning that the stimulation duration may be further reduced by raising the flow rate. When further shortening the stimulation duration by increasing the flow rate, however, one would need to take into account the high flow shear stress induced by the increased flow rate, which may initiate undesired cellular responses.
Calcium (Ca2+) responses to single-pulse ionomycin stimulation
To demonstrate the utility of our device in stimulating cells for cell-signaling related studies, we investigated how HMVEC-d, HeLa, and U-251 cells respond to the time-varying, single-pulse ionomycin stimulation produced by our device, in terms of calcium release dynamics. Ionomycin, a calcium ionophore, is able to enter cells through plasma membranes to initiate Ca2+ depletion from cells’ internal calcium stores. The oscillation dynamics of Ca2+, as a universal signaling messenger in both excitable and non-excitable cells, have contributed to regulating many cellular processes from cell adhesion and motility to cell immune response and gene expression.68–72 As such, with an experimental tool capable of modulating calcium oscillation dynamics, one can regulate cellular processes.
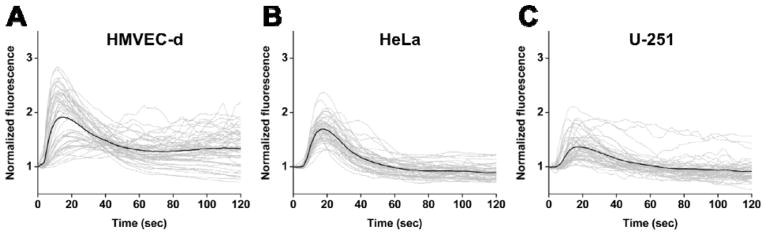
Fig. 3A, B and C show, respectively, the calcium responses of different cells when single-pulse ionomycin stimulus of 1, 5 and 10 sec (as depicted in Fig. 2A) were applied. Each panel in Fig. 3 shows the individual (gray) and average (black) calcium responses of 50 representative cells from at least three independent experiments. Regardless of cell type, the cells started to release Ca2+ from internal stores within just a few seconds of ionomycin stimulation, then constantly released Ca2+ and eventually reached a peak Ca2+ concentration. The dynamic process showing the change in Ca2+ fluorescence intensity over time for HMVEC-d cells can be found in Video S1. This result demonstrates that the cells faithfully responded to the single-pulse ionomycin stimulation generated using our device, as exhibited by single Ca2+ transients with differences in peak Ca2+ concentrations. These differences were in large part due to the inherent heterogeneity of cells, which could be further elucidated in Fig. S3. Averaging the responses, we observed that the cell populations also had single Ca2+ transients. As a comparison, the calcium release behavior of cells before the culture medium containing ionomycin was infused into the channel were also characterized; apparently, no calcium transient was observed in the absence of ionomycin (Fig. S3).
Figure 3.
Plots showing the individual (gray curves) and average (black curve) calcium response of 50 representative cells when (A) HMVEC-d cells, (B) HeLa cells and (C) U-251 cells were challenged by single-pulse ionomycin stimulation with stimulation duration of 1, 5 and 10 sec. All the cells began to release Ca2+ from internal stores within just a few seconds of being exposed to ionomycin stimulus, constantly released calcium and eventually reached a peak calcium concentration, regardless of cell types. The prolonged stimulation seemed to have significant influence on the recovery time of cells from after reaching the peak intensity to the time point when recovering to calcium baseline level. Each plot is the representative results from at least three independent experiment where more than 200 cells were examined.
It is also important to point out that the degradation in fluorescence intensity over time due to photobleaching might be possible (Fig. S4). A previous study also showed that photobleaching of Fluo-4 AM fluorescence was observed over a given period of time in other cell types.73 Nevertheless, photobleaching can be considered negligible in this work, owing to the short exposure time and the intensity of excitation light used in this work. To further verify if photobleaching significantly affected the fluorescence intensity response, we took the fluorescence curve shown in Fig. S4A (HMVEC-d) as a baseline/background curve and subtracted it from the fluorescence intensity curves under single-pulse stimulation (Fig. 3A). The subtracted curves (Fig. S5) still show similar trends of Ca2+ response to those shown in Fig. 3A, indicating that photobleaching has a negligible influence on the Ca2+ response curves.
To further quantitatively characterize the difference in calcium response when stimulating the cells with different stimulation durations, we statistically analyzed the fold increase in peak Ca2+ fluorescence intensity (Fig. 4). The representative population of HMVEC-d cells, when exposed to 1-sec single-pulse ionomycin stimulant, exhibited an average peak fluorescence intensity of 1.78 ± 0.26. As the stimulation duration was prolonged to 5 sec and 10 sec, the HMVEC-d cells showed, respectively, average peak fluorescence intensities of 2.48 ± 0.40 and 2.73 ± 0.24. The peak fluorescence intensities of the HMVEC-d cells differed among stimulation durations, which suggests that the cells responded differently to the stimuli in terms of peak calcium concentration.
Figure 4.
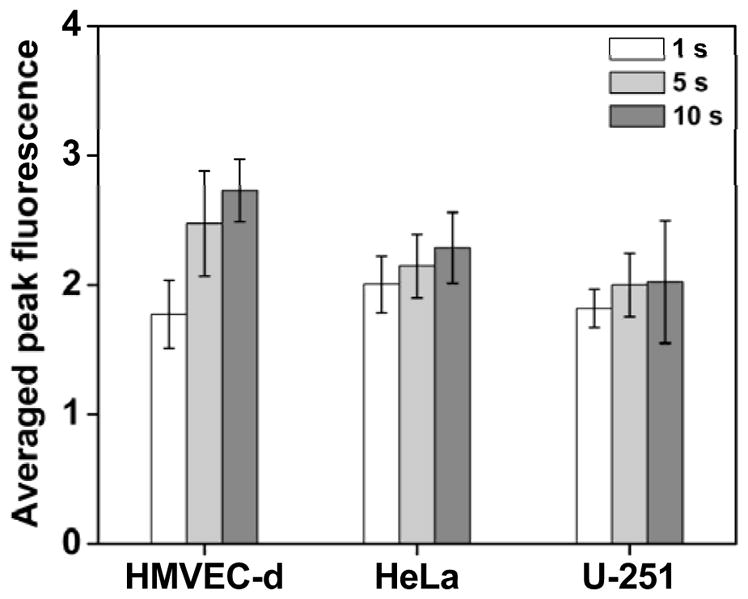
Statistical analysis of the peak fluorescence intensity exhibited by different cells under single-pulse ionomycin stimulation of 1 sec, 5 sec and 10 sec. As the stimulation duration was increased, the peak fluorescence intensity of calcium rose accordingly. This result suggests that under prolonged ionomycin stimulation, the cells released more calcium ions from the internal stores to the cytoplasm. Data is represented as the average of at least n = 3 independent experiments with standard deviation (*p < 0.05, **p < 0.01).
HeLa cells, similarly, released more calcium when the stimulation duration was extended. Peak fluorescence intensities of 2.01 ± 0.22, 2.15 ± 0.25 and 2.29 ± 0.28 were attained under the single-pulse stimulations of, respectively, 1, 5, and 10 sec, which were all statistically different (Fig. 4). Although the U-251 cells depleted more Ca2+ from their internal stores as the stimulation duration was increased, the peak calcium concentration barely changed between the treatments of 5 and 10 sec. For all of the experiments with U-251 cells exposed to the prolonged ionomycin stimulation, at least one-third of the U-251 cells changed their morphologies, from a “spread-out” shape to a round shape. The morphologically changed cells then detached from the surface of the channel. The U-251 cells’ altered morphology and detachment in response to the prolonged ionomycin stimulation are the behavior of early apoptosis; this observation is consistent with a previous report where ionomycin markedly induced U-251 cell apoptosis.74 As such, the phenomenon of unchanged peak calcium concentration may be attributed to the early apoptosis of U-251 cells in response to the prolonged ionomycin stimulation, which further influences the Ca2+ release. Taken as a whole, our statistical analysis indicates that when triggered by the prolonged ionomycin stimulation, the cells depleted more Ca2+ from their internal stores, regardless of cell type.
Aside from the peak Ca2+ concentration, we also considered the dependence of the recovery time on the stimulation duration, which was defined as the time that the cells recover to the baseline Ca2+ levels after the peak intensity was reached. The Ca2+ fluorescence intensity of HMVEC-d cells, for example, reached a peak intensity after 10 – 20 sec (the rising time) under 1-sec single-pulse stimulation, and recovered to the calcium baseline level about 50 – 70 sec after reaching the peak. As the stimulation duration was prolonged, the rising time was hardly changed, whereas the recovery time was extended by about 20 sec (70 – 90 sec) and 40 sec (90 – 100 sec) under the stimulation durations of 5 sec and 10 sec, respectively. We observed similar trends from the calcium responses of HeLa and U-251 cells; when we increased the stimulation duration, the rising time changed insignificantly, while the recovery time extended noticeably. These results suggest that prolonged ionomycin stimulation may cause a prolonged recovery time, which is in agreement with previous stuies.20
Collectively, the results presented in this section provide evidence that the cells, regardless of cell type, experienced changes in their surrounding microenvironment, and differently, yet faithfully responded to different chemical cues provided by our device. The results also prove our device capable of generating chemical signals that cells can receive and in turn, initiate appropriate cellular responses.
Ca2+ responses to 100 ms, single-pulse ionomycin stimulation
After monitoring the calcium response of cells under normal, single-pulse ionomycin stimulation (> 1 sec), we examined cells that were exposed to rapid, single-pulse ionomycin stimulation (i.e., 100-ms stimulation), to see if their responses would differ greatly from when exposed to normal, single-pulse stimulation. Fig. 5 shows the Ca2+ response of three different types of cells under rapid, single-pulse ionomycin stimulation. Again, each panel contains individual (gray) and average (black) calcium responses of 50 representative cells from at least three independent experiments, in which more than 200 cells were assayed. Upon the 100-ms stimulation, all three different types of cells exhibited the first components of rapidly-rising calcium transients with peak intensities that were lower than those under normal, single-pulse stimulation. It is surprising that after the first calcium transient occurred, the cells exhibited slowly-rising components as well as rapidly-rising components. The dynamic process exhibiting multiple calcium transients for HMVEC-d cells can be found in Video S2. The slowly-rising components may be attributed to that extracellular Ca2+ enter the cells through cell membranes. According to a previous study,75 the calcium released from internal stores was the principal cause of the rapidly-rising component, whereas the entry of calcium into cells from surrounding microenvironments predominately contributed to the slowly-rising component. Simply speaking, the cells may constantly release and accept Ca2+, respectively, to cytoplasm and from surrounding environments, so that they can maintain a given internal Ca2+ concentration through this process. In other words, unlike when cells were exposed to normal, single-pulse stimulation, they could potentially undergo a “self-equilibrium” process in response to rapid stimulation. In the future, we can use our acoustofluidic devices to shed light on how this phenomenon is initiated. For example, by chemically blocking calcium ion channels that control calcium transport from the extracellular environment, our devices will be able to accurately analyze these cellular responses. These results suggest that under rapid stimulation, the Ca2+ response of cells, besides the rising time and recovery time, may be significantly different. In addition, these results also demonstrate that our acoustofluidic device could produce rapid stimulation signals that could trigger uncommon cellular responses, allowing for studies where cell signaling dynamics under rapid stimulation is yet to be explored.
Figure 5.
Plots showing the individual (gray curves) and average (black curve) calcium response of 50 representative cells when (A) HMVEC-d, (B)HeLa, and (C) U-251 cells were subject to rapid, single-pulse ionomycin stimulation of duration 100 ms. All three different types of cells, challenged by the 100-ms ionomycin stimulation, first exhibited rapidly-rising calcium transients with peak intensities that were lower than those under normal, single-pulse stimulation. After the first calcium transient was probed, the cells were also found to feature slowly-rising components as well as rapidly-rising components. These results suggest that the cells seemed to undergo a process where they, to maintain a certain calcium concentration, constantly released calcium from internal stores and received calcium from surrounding environments. Each plot is a representative result from at least three independent experiments where more than 200 cells in total were examined.
Ca2+ responses to periodic ionomycin stimulation
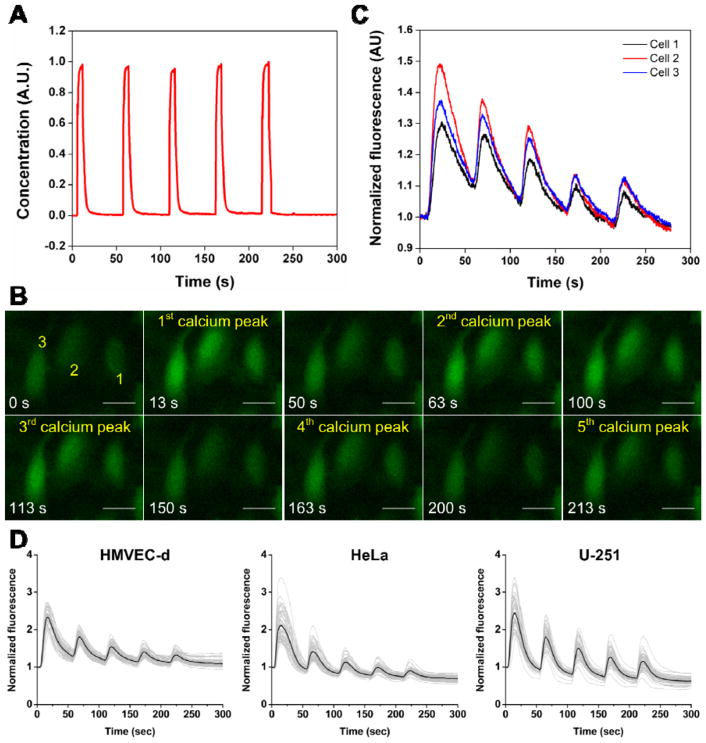
As a demonstration, we also monitored the Ca2+ responses of different types of cells under periodic ionomycin stimulation. The periodic stimulation was composed of 5 single-pulse stimulations, in which the period and stimulation duration for each single-pulse stimulation were 50 sec and 5 sec, respectively (Fig. 6A). To present the dynamic process of calcium response under the periodic stimulation, we first provided the response curves and fluorescent images of three representative HMVEC-d cells. These cells rapidly and consistently released Ca2+ after each single-pulse of the periodic stimulation, responding faithfully to the periodic stimulation (Fig. 6B). Upon each single-pulse stimulation, the fluorescence intensity of the cells reached a maximum in about 10 – 15 sec and then declined gradually towards the baseline level (Fig. 6C). Note that though the cells responded predictably to each single-pulse stimulation, their peak fluorescence intensity decreased with the number of single-pulse stimulations (Fig. 6C; Video S3). This observation is evidence that the periodic ionomycin stimulation would gradually deplete the internal calcium store, which agrees with previous conclusions that ionomycin enters cells through plasma membranes and facilitates the depletion of Ca2+ from internal calcium stores.20,76,77 Additionally, as a comparison, the calcium release behavior of cells over the similar period of time before the introduction of the culture medium containing ionomycin into the channel were also characterized (Fig. S6A). In the absence of ionomycin, the cells exhibited no calcium release behavior over a period of time of 300 sec.
Figure 6.
(A) Profile of the periodic stimulation used in stimulating cells with periodic ionomycin stimulation, which has a fixed period of 50 sec and a stimulation duration of 5 sec. (B) Fluorescent images showing the change in fluorescence intensity of three representative HMVEC-d cells over the entire stimulation process. (C) Plot showing the calcium response curves corresponding to the three representative cells in (B). (D) Plots showing the individual (gray curves) and average (black curve) calcium response of 50 representative cells when HMVEC-d, HeLa and U-251 cells were stimulated by the periodic ionomycin stimulation plotted in (A). Each plot in (D) shows the representative results from at least three independent experiments where more than 200 cells in total were examined. The cells responded predictably to the periodic stimulation; however, the peak fluorescence intensity was reduced with the number of single-pulse stimulations. This phenomenon is a result of that the periodic ionomycin stimulation gradually depleted the internal calcium store. Scale bar: 25 μm
Fig. 6D shows the Ca2+ responses of three different types of cells under the periodic ionomycin stimulation, in which individual (gray) and average (black) calcium response curves of 50 representative cells from at least three independent experiments are provided (more than 200 cells were assayed). Despite the heterogeneity of the cells, they exhibited five fluorescence peaks under the periodic stimulation, regardless of cell type. The heterogeneity between cells only resulted in a difference in the amount of Ca2+ (which corresponded to the magnitude of fluorescence intensity) that each cell released after each single-pulse stimulation. By averaging the response curves, we confirmed that the depletion of internal calcium store was occurring under the periodic stimulation. These results also validate that our device could generate periodic chemical signals to which cells faithfully respond.
We also tested if the degradation due to photobleaching significantly influenced the fluorescence intensity response over this period of time (300 sec) and verified whether or not photobleaching leads to the decrease of fluorescence intensity with the number of single-pulse stimulations. The degraded fluorescence intensity curve of HMVEC-d cells (Fig. S6A) was taken as a baseline/background curve and subtracted from the fluorescence intensity curve under 5-sec periodic stimulation (Fig. 6D: HMVEC-d). The subtracted curve still displays a similar trend of Ca2+ response to that shown in Fig. 6D (HMVEC-d), demonstrating that photobleaching has an insignificant influence on the Ca2+ response curve. In other words, the decrease of the peak fluorescence intensity with an increasing number of single-pulse stimulation does indeed result from the gradual depletion of Ca2+ from the internal calcium stores of cells. As we mentioned above, previous studies have also confirmed the gradual depletion of Ca2+ under periodic ionomycin stimulation, and they concluded that cells take less time to release Ca2+ from their internal calcium stores to the cytoplasm than to recover Ca2+ from the cytoplasm back to the internal calcium stores.20,76,77
Other than being challenged by periodic stimulation with normal, single-pulse stimulation profiles, the cells were also assayed by periodic stimulation with rapid, single-pulse stimulation profiles (Fig. 2B; 100 ms). The cells, when exposed to a periodic stimulation with rapid, single-pulse stimulation profiles, constantly released and took in Ca2+ from the internal calcium store and surrounding environment, respectively, thereby displaying more than one calcium transients between each calcium peak (Fig. S7; Video S4). This result is in accordance with that obtained under a rapid, single-pulse ionomycin stimulation and once again, suggests that when the cells are subject to rapid chemical stimulation, they may respond differently.
Conclusions
In summary, we have developed an acoustofluidic chemical signal generator based on sharp-edge-based, active, on-demand mixing strategy. Our acoustofluidic chemical signal generator can synthesize temporally-controllable, single and periodic chemical signals with varying durations, periods, and duty cycles, through modulating the driving signals of the transducer. The applicability and versatility of our acoustofluidic device have been verified by cell stimulation experiments using three different types of cells, where the cells could respond faithfully to the chemical signals generated by our acoustofluidic device. And interestingly, cells responded to the short, single-pulse ionomycin stimulation (~100 ms) in a different manner from when exposed to the long, single-pulse stimulation (> 1 sec). Because all the cells in each experiment were simultaneously subject to the same chemical signal, our device allows one to investigate cell-to-cell variations in a cell population due to inherent heterogeneity. In addition, concentration-varying chemical signals with our device may be produced by, for example, changing the flow rate ratio of buffer and stimulant and reconfiguring the arrangement of sharp-edge structures. With these characteristics along with features including ease of device preparation and operation, reliability, and versatility, our acoustofluidic device is a promising tool to shed lights on cells’ signaling dynamics in physiological processes such as immune response and transcription-factor-dependent gene expression.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health (R01 GM112048-01A1 and R33EB019785-01), National Science Foundation (CBET-1438126 and IIP-1534645), and the Penn State Center for Nanoscale Science (MRSEC) under grant DMR-1420620. Components of this work were conducted at the Penn State node of the NSF-funded National Nanotechnology Infrastructure Network.
Footnotes
Electronic Supplementary Information (ESI) available: Video1-Acoustic streaming patterns generated in our device. Video2-Generation of chemical signal at sharp-edge region. Video3-Generation of chemical signal at downstream regions. Video4-Generation of different types of chemical signal. Video5-spatiotemporally controlled generation of chemical signals. Video6-The movement of HMVEC-d cells in response to a VEGF signal. See DOI: 10.1039/x0xx00000x
References
- 1.Kholodenko BN. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kholodenko BN, Hancock JF, Kolch W. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behar M, Hoffmann A. Curr Opin Genet Dev. 2010;20:684–693. doi: 10.1016/j.gde.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann A. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 5.Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, Cox CD, Simpson ML, Weinberger LS. Proc Natl Acad Sci. 2012;109:17454–17459. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locke JCW, Young JW, Fontes M, Jiménez MJH, Elowitz MB. Science. 2011;334:366–369. doi: 10.1126/science.1208144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashall L, Horton Ca, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, Kell DB, Rand Da, See V, White MRH. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett MR, Pang WL, Ostroff NA, Baumgartner BL, Nayak S, Tsimring LS, Hasty J. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. Science. 2008;319:482–484. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson DE, et al. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 11.Hersen P, McClean MN, Mahadevan L, Ramanathan S. Proc Natl Acad Sci. 2008;105:7165–7170. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irimia D. Annu Rev Biomed Eng. 2010;12:259–284. doi: 10.1146/annurev-bioeng-070909-105241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JY, Takayama S, Lee SH. Integr Biol. 2010;2:229. doi: 10.1039/c000442a. [DOI] [PubMed] [Google Scholar]
- 14.Jovic A, Howell B, Takayama S. Microfluid Nanofluidics. 2009;6:717–729. [Google Scholar]
- 15.Bennett MR, Hasty J. Nat Rev Genet. 2009;10:628–638. doi: 10.1038/nrg2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Meiners JC. Appl Phys Lett. 2004;84:2193. [Google Scholar]
- 17.Morel M, Galas JC, Dahan M, Studer V. Lab Chip. 2012;12:1340–6. doi: 10.1039/c2lc20994b. [DOI] [PubMed] [Google Scholar]
- 18.King KR, Wang S, Jayaraman A, Yarmush ML, Toner M. Lab Chip. 2008;8:107–16. doi: 10.1039/b716962k. [DOI] [PubMed] [Google Scholar]
- 19.Kuczenski B, LeDuc PR, Messner WC. Lab Chip. 2007;7:647. doi: 10.1039/b617065j. [DOI] [PubMed] [Google Scholar]
- 20.Kuczenski B, Ruder WC, Messner WC, LeDuc PR. PLoS One. 2009;4:e4848. doi: 10.1371/journal.pone.0004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jovic A, Wade SM, Miyawaki A, Neubig RR, Linderman JJ, Takayama S. Mol Biosyst. 2011;7:2238–2244. doi: 10.1039/c1mb05031a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J, Wang J, Chen P, Feng X, Du W, Liu BF. Anal Bioanal Chem. 2011;400:2973–2981. doi: 10.1007/s00216-011-4987-2. [DOI] [PubMed] [Google Scholar]
- 23.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niman CS, Beech JP, Tegenfeldt JO, Curmi PMG, Woolfson DN, Forde NR, Linke H. Lab Chip. 2013;13:2389. doi: 10.1039/c3lc50194a. [DOI] [PubMed] [Google Scholar]
- 25.Kawada J, Kimura H, Akutsu H, Sakai Y, Fujii T. Lab Chip. 2012;12:4508. doi: 10.1039/c2lc40268h. [DOI] [PubMed] [Google Scholar]
- 26.Jovic A, Howell B, Cote M, Wade SM, Mehta K, Miyawaki A, Neubig RR, Linderman JJ, Takayama S. PLoS Comput Biol. 2010;6:e1001040. doi: 10.1371/journal.pcbi.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie DC, Easley CJ, Seker E, Karlinsey JM, Utz M, Begley MR, Landers JP. Nat Phys. 2009;5:231–235. [Google Scholar]
- 28.Taylor RJ, Falconnet D, Niemistö A, Ramsey SA, Prinz S, Shmulevich I, Galitski T, Hansen CL. Proc Natl Acad Sci U S A. 2009;106:3758–63. doi: 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azizi F, Mastrangelo CH. Lab Chip. 2008;8:907. doi: 10.1039/b716634f. [DOI] [PubMed] [Google Scholar]
- 30.Irimia D, Liu SY, Tharp WG, Samadani A, Toner M, Poznansky MC. Lab Chip. 2006;6:191–198. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chingozha L, Zhan M, Zhu C, Lu H. Anal Chem. 2014;86:10138–10147. doi: 10.1021/ac5019843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu HW, Lin CC, Hwang SM, Chang YJ, Lee GB. Microfluid Nanofluidics. 2011;11:545–556. [Google Scholar]
- 33.Westendorf C, Negrete J, Bae AJ, Sandmann R, Bodenschatz E, Beta C. Proc Natl Acad Sci U S A. 2013;110:3853–8. doi: 10.1073/pnas.1216629110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beta C, Wyatt D, Rappel WJ, Bodenschatz E. Anal Chem. 2007;79:3940–3944. doi: 10.1021/ac070033y. [DOI] [PubMed] [Google Scholar]
- 35.Bae AJ, Beta C, Bodenschatz E. Lab Chip. 2009;9:3059. doi: 10.1039/b905521e. [DOI] [PubMed] [Google Scholar]
- 36.Cohen MS, Orth CB, Kim HJ, Jeon NL, Jaffrey SR. Proc Natl Acad Sci. 2011;108:11246–11251. doi: 10.1073/pnas.1012401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandersarl JJ, Xu AM, Melosh Na. Lab Chip. 2011;11:3057–3063. doi: 10.1039/c1lc20311h. [DOI] [PubMed] [Google Scholar]
- 38.Scott A, Weir K, Easton C, Huynh W, Moody WJ, Folch A. Lab Chip. 2013;13:527–535. doi: 10.1039/c2lc40826k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabounchi P, Ionescu-Zanetti C, Chen R, Karandikar M, Seo J, Lee LP. Appl Phys Lett. 2006;88:1–4. [Google Scholar]
- 40.Ahmed D, Muddana HS, Lu M, French JB, Ozcelik A, Fang Y, Butler PJ, Benkovic SJ, Manz A, Huang TJ. Anal Chem. 2014;86:11803–11810. doi: 10.1021/ac5033676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tovar AR, Lee AP. Lab Chip. 2009;9:41–3. doi: 10.1039/b812435c. [DOI] [PubMed] [Google Scholar]
- 42.Huang P-H, Ian Lapsley M, Ahmed D, Chen Y, Wang L, Huang TJ. Appl Phys Lett. 2012;101:141101. doi: 10.1063/1.4742864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang PH, Chen HM, Huang TC, Yang SY, Sun YT. J Vac Sci Technol B Microelectron Nanom Struct. 2013;31:31602. [Google Scholar]
- 44.Ahmed D, Mao X, Juluri BK, Huang TJ. Microfluid Nanofluidics. 2009;7:727–731. [Google Scholar]
- 45.Hashmi A, Yu G, Reilly-Collette M, Heiman G, Xu J. Lab Chip. 2012;12:4216–27. doi: 10.1039/c2lc40424a. [DOI] [PubMed] [Google Scholar]
- 46.Xie Y, Nama N, Li P, Mao Z, Huang PH, Zhao C, Costanzo F, Huang TJ. Small. 2016;12:902–910. doi: 10.1002/smll.201502220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Destgeer G, Im S, Hang Ha B, Ho Jung J, Ahmad Ansari M, Jin Sung H. Appl Phys Lett. 2014;104:23056. [Google Scholar]
- 48.Bruus H, Dual J, Hawkes J, Hill M, Laurell T, Nilsson J, Radel S, Sadhal S, Wiklund M. Lab Chip. 2011;11:3579–3580. doi: 10.1039/c1lc90058g. [DOI] [PubMed] [Google Scholar]
- 49.Li S, Ren L, Huang PH, Yao X, Cuento RA, McCoy JP, Cameron CE, Levine SJ, Huang TJ. Anal Chem. 2016;88:5655–5661. doi: 10.1021/acs.analchem.5b03383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lata JP, Guo F, Guo J, Huang PH, Yang J, Huang TJ. Adv Mater. 2016;28:8632–8638. doi: 10.1002/adma.201602947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding X, Li P, Lin SCS, Stratton ZS, Nama N, Guo F, Slotcavage D, Mao X, Shi J, Costanzo F, Huang TJ. Lab Chip. 2013;13:3626–3649. doi: 10.1039/c3lc50361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeo LY, Friend JR. Annu Rev Fluid Mech. 2014;46:379–406. [Google Scholar]
- 53.Ahmed D, Ozcelik A, Bojanala N, Nama N, Upadhyay A, Chen Y, Hanna-Rose W, Huang TJ. Nat Commun. 2016;7:1–11. doi: 10.1038/ncomms11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmed D, Chan CY, Lin SCS, Muddana HS, Nama N, Benkovic SJ, Huang TJ. Lab Chip. 2013;13:328–331. doi: 10.1039/c2lc40923b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, Li H, Li P, Quinn D, Dao M, Suresh S, Sadovsky Y, Huang TJ. Proc Natl Acad Sci. 2017;114:10584–10589. doi: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y, Nawaz AA, Zhao Y, Huang PH, McCoy JP, Levine SJ, Wang L, Huang TJ. Lab Chip. 2014;14:916–923. doi: 10.1039/c3lc51139a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Ding X, Steven Lin S-C, Yang S, Huang P-H, Nama N, Zhao Y, Nawaz AA, Guo F, Wang W, Gu Y, Mallouk TE, Huang TJ. ACS Nano. 2013;7:3306–14. doi: 10.1021/nn4000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang PH, Xie Y, Ahmed D, Rufo J, Nama N, Chen Y, Chan CY, Huang TJ. Lab Chip. 2013;13:3847–3852. doi: 10.1039/c3lc50568e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nama N, Huang PH, Huang TJ, Costanzo F. Lab Chip. 2014;14:2824–2836. doi: 10.1039/c4lc00191e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang PH, Chan CY, Li P, Nama N, Xie Y, Wei CH, Chen Y, Ahmed D, Huang TJ. Lab Chip. 2015;15:4166–4176. doi: 10.1039/c5lc00868a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang PH, Ren L, Nama N, Li S, Li P, Yao X, Cuento RA, Wei CH, Chen Y, Xie Y, Nawaz AA, Alevy YG, Holtzman MJ, McCoy JP, Levine SJ, Huang TJ. Lab Chip. 2015;15:3125–3131. doi: 10.1039/c5lc00539f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nama N, Huang PH, Huang TJ, Costanzo F. Biomicrofluidics. 2016;10:24124. doi: 10.1063/1.4946875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozcelik A, Nama N, Huang PH, Kaynak M, McReynolds MR, Hanna-Rose W, Huang TJ. Small. 2016;12:5230. doi: 10.1002/smll.201601760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bachman H, Huang PH, Zhao S, Yang S, Zhang P, Fu H, Huang TJ. Lab Chip. 2018;18:433–441. doi: 10.1039/c7lc01222e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang PH, Nama N, Mao Z, Li P, Rufo J, Chen Y, Xie Y, Wei CH, Wang L, Huang TJ. Lab Chip. 2014;14:4319–4323. doi: 10.1039/c4lc00806e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leibacher I, Hahn P, Dual J. Microfluid Nanofluidics. 2015;19:923–933. [Google Scholar]
- 67.Kawada J, Kimura H, Akutsu H, Sakai Y, Fujii T. Lab Chip. 2012;12:4508. doi: 10.1039/c2lc40268h. [DOI] [PubMed] [Google Scholar]
- 68.Dolmetsch RE, Xu K, Lewis RS. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 69.Dolmetsch RE, Lewis RS, Goodnowt CC, Healyt JI. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 70.Arrol HP, Church LD, Bacon PA, Young SP. Clin Exp Immunol. 2008;153:86–95. doi: 10.1111/j.1365-2249.2008.03677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu B, Chen W, Evavold BD, Zhu C. Cell. 2014;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Faley S, Seale K, Hughey J, Schaffer DK, VanCompernolle S, McKinney B, Baudenbacher F, Unutmaz D, Wikswo JP. Lab Chip. 2008;8:1700. doi: 10.1039/b719799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lopez-Ayon GM, Oliver DJ, Grutter PH, Komarova SV. Microsc Microanal. 2012;18:808–815. doi: 10.1017/S1431927612000402. [DOI] [PubMed] [Google Scholar]
- 74.Han S, Tie X, Meng L, Wang Y, Wu A. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0076717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshida S, Plant S. J Physiol. 1992;458:307–318. doi: 10.1113/jphysiol.1992.sp019419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morgan AJ, Jacob R. Biochem J. 1994;300:665–672. doi: 10.1042/bj3000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lau BW, Colella M, Ruder WC, Ranieri M, Curci S, Hofer AM. Gastroenterology. 2005;128:695–707. doi: 10.1053/j.gastro.2004.12.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.