Abstract
DA closure is crucial for the transition from fetal to neonatal life. This closure is supported by changes to the DA’s signaling and structural properties that distinguish it from neighboring vessels. Examining transcriptional differences between these vessels is key to identifying genes or pathways responsible for DA closure. Several microarray studies have explored the DA transcriptome in animal models but varied experimental designs have led to conflicting results. Thorough transcriptomic analysis of the human DA has yet to be performed. A clear picture of the DA transcriptome is key to guiding future research endeavors, both to allow more targeted treatments in the clinical setting, and to understand the basic biology of DA function. In this review, we use a cross-species cross-platform analysis to consider all available published rodent microarray data and novel human RNAseq data in order to provide high priority candidate genes for consideration in future DA studies.
Keywords: ductus arteriosus, PDA, microarray, RNA-seq, transcriptional profiling, aorta
Introduction
The ductus arteriosus (DA) is an essential vascular shunt connecting the pulmonary artery and aorta, allowing oxygenated blood from the placenta to bypass the developing lungs in utero. After birth, DA closure is required for a proper transition to neonatal life. Often, the postnatal DA fails to close, resulting in persistent patency of the ductus arteriosus (PDA). PDA accounts for nearly 10% of congenital heart defects1,2, including more than 30% of preterm infants with a birth weight of <1500g3,4.
Effective DA closure is dependent on a combination of signaling and structural changes which support constriction and eventual remodeling of the vessel5–7. Despite the proximity and common neural crest lineage of their smooth muscle cells8, the ascending aorta (Ao) doesn’t undergo these changes, suggesting transcriptional differences between these vessels may define the DA’s function. Numerous attempts to understand these differences at the transcriptome level have provided insight, but varied experimental design and statistical analyses have created contradictions and ambiguity in the literature. Further, the transcriptome of the human DA has not been explored with the advanced genomic techniques now available, such as RNA-seq. A clear picture of the DA’s transcriptional profile is key to guiding future research endeavors, both to allow more targeted treatments in the clinical setting, and to understand the basic biology underlying DA function.
The goals of this study were to: 1) define differentially expressed genes (DEGs) in DA versus Ao samples that were commonly identified in previously published microarray datasets using rodent models, 2) identify human DA-enriched transcripts using RNA-seq analysis, and 3) explore transcriptional commonalities between the rodent and human DA. Although cross-species and cross-platform comparisons are fraught with limitations, identification of robust markers of DA identity or novel DA-enriched pathways promises to provide unique insights into DA development and function.
Methods
Microarray Meta-Analysis
Microarray data were obtained from NCBI’s Gene Expression Omnibus (GEO) database (Table 1). All available studies were considered, but only investigations that included a DA/Ao comparison were selected for analysis. For the array data from Bokenkamp et al.9, values for E21 laser micro-dissected endothelium and smooth muscle cells were pooled. For the array data from Hseih et al.10, only the F344 control samples were considered. CEL files of selected data sets were evaluated in Partek Genomics Suite version 7.17.1222 (Partek Inc.). All data were normalized using the Robust Multi-Array (RMA) method. One-way ANOVA was used to analyze contrasts of interest, namely vessel type, to generate lists of DEGs between DA and Ao. Permissive DEG lists (fold change ≥1.2) were then separated by increased or decreased DA/Ao expression to generate UP and DOWN lists respectively. A naïve vote counting strategy was used to evaluate consistency between studies. Shared genes between UP lists or DOWN lists from each study were determined using Partek Genomics Suite.
Table 1.
Summary of Included and Excluded Studies using Microarray to Compare DA and Aorta
| Study (ref) | Species | Strain | Description | GSE# | Platform | Probe# | Analysis | Samples | Age | Genes UP |
Genes DOWN |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Jin 201113 | Rattus norwegicus | Wistar | Term and preterm DA | 3422 | Affy Rat U34 A Array | 8799 | ANOVA | 2 DA vs 2 Ao | E21 | 328 | 366 |
| Bokenkamp 20149 | Rattus norwegicus | Wistar | Isolated SMC vs endothelium | 51248 | Affy Rat 230 2.0 Array | 12088 | ANOVA | 6 DA vs 6 Ao | E21 | 2453 | 2347 |
| Hseih 201410 | Rattus norwegicus | F344 | PDA in Brown-Norway rat | 40534 | Affy Rat 1.0 ST Array | 29215 | ANOVA | 2 DA vs 2 Ao | E21 | 2027 | 2336 |
| Shelton 201414 | Mus musculus | CD1 WT | Term DA expression | 51664 | Affy Mouse 430 2.0 Array | 45101 | ANOVA | 4 DA vs 4 Ao | E19 | 1532 | 1958 |
| Costa 200642 | Rattus norwegicus | Long-Evans | Oxygen's effect on preterm DA | 3290 | Affy Rat U34 A,B,C Array | 26379 | Excluded (preterm only) | – | – | – | – |
| Yokoyama 200743 | Rattus norwegicus | Wistar | Vitamin A and DA maturation | 3420 | Affy Rat U34 A Array | 8799 | Excluded (Ao data unavailable) | – | – | – | – |
| Gruzdev 201226 | Mus musculus | 129S6 | DA from EP4 null mice | NA | Illumina Mouse Ref8 v1.1 Array | 24613 | Excluded (Ao data unavailable) | – | – | – | – |
| Liu 201344 | Rattus norwegicus | Wistar | Term DA endothelium | 40500 | Affy Rat 1.0 ST Array | 29215 | Excluded (endo only) | – | – | – | – |
| Goyal 201612 | Ovis aries | – | Term and preterm DA | 87840 | Agilent 019921 Sheep Array | 15068 | Excluded (incomplete annotation) | – | – | – | – |
Abbreviations: Affy – Affymetrix, ANOVA – analysis of variance, Ao – aorta, DA – ductus arteriosus, E21 – embryonic day 21, NA – not applicable, PDA – persistent patency of the ductus arteriosus, SMC – smooth muscle cell
RNA-seq Analysis
Previable (21 - 21 5/7 weeks gestation) tissue samples for human RNA-seq analysis were obtained as previously described11. Tissues were homogenized in Trizol with the IKA T10 basic Ultra-Turrax. Total RNA was isolated using the RNeasy Mini kit (Qiagen). RNA quality was assessed using the Agilent 2100 Bioanalyzer. RNA samples with RIN score ≥6.5 were considered for further study. 1 microgram of total RNA was used for library construction using the ScriptSeq Complete Gold Kit (Illumina). The libraries, four biological replicates per vessel type, were sequenced on an Illumina HiSeq 2000 by 100bp pair-end sequencing. RNA-seq data was uploaded to Partek Flow (Partek Inc.). Trimming of raw reads (both ends) was based on a minimum read length of 25 and discarded bases after 85. Trimmed reads were aligned to Human Genome Version 38 (hg38) using STAR – 2.5.3a and quantified to Refseq Transcripts 83 using Partek’s E/M method. Aligned counts were FPKM normalized with an offset of 1. Gene specific analysis (GSA) was used to detect DA vs. Ao DEGs. DEGs with an FDR of ≤0.1 and a fold change ≥2 were considered significant. Volcano plot and dendogram heat map figures were generated using Partek Flow.
Comparison of Microarray and RNA-seq DEGs
Rodent gene symbols were converted to human orthologues using biological Database network (bioDBnet) to identify genes common between microarray and RNA-seq DEG lists. Lists were manually aligned to determine genes differentially expressed in both microarray and RNA-seq analyses. Microarray and RNA-seq gene lists were submitted independently for functional annotation using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). Functional terms from Gene Ontology (GO) Biological Process (BP), GO Cellular Component (CC), GO Molecular Function (MF), Kyoto Encyclopedia of Genes and Genomes (KEGG), and UniProt (UP) Keywords databases were evaluated for similarities. Lists of functional terms and keywords from each database were then manually aligned. In order to create comparison diagrams, significant terms (p≤0.05) were chosen based on count (defined as number of genes identified in each term), and ordered by –Log(p value).
Results
Comparison of Published Rodent Microarrays
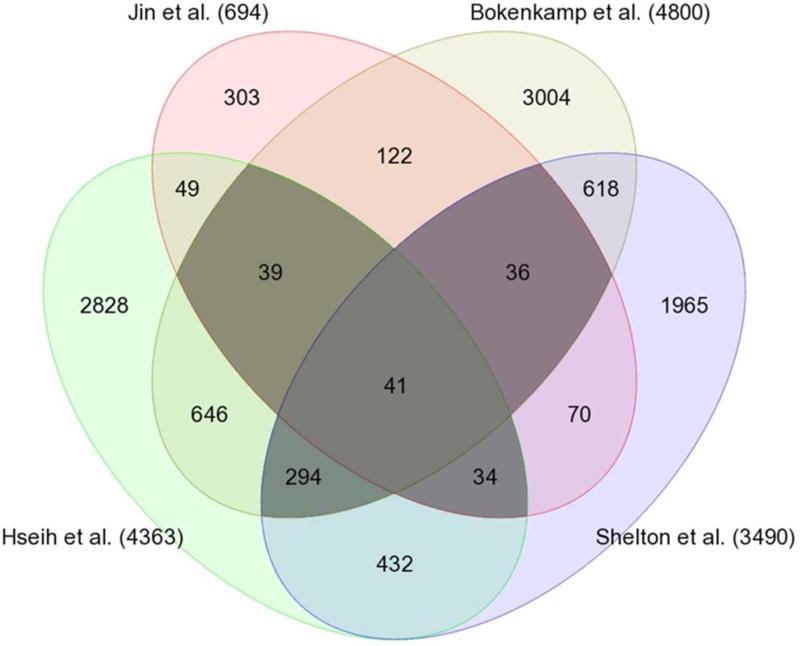
Our microarray meta-analysis focused on studies containing DA to Ao comparisons in term animals, since there were too few preterm studies for comparison. Comparison of DA to Ao expression allowed DA-specific genes to be distinguished from temporally-regulated genes that are important for generalized vessel development. Differential expression was recalculated from raw data using a uniform statistical approach. Array data from three mammalian species (rat, mouse, sheep) were available, but differences in experimental design and genome annotation limited the use of data from sheep12. Four rodent studies met pre-specified criteria (vessel type, gestational stage) and were included for analysis9,10,13,14 (Table 1). There were 444 genes identified as differentially expressed in at least three of four arrays. Of these, 87 genes were consistently increased in DA versus Ao (DA enriched), while 189 genes were consistently decreased in DA versus Ao (Ao enriched) (Figure 1). Complete gene lists are provided for both DA enriched (Table S1) and Ao enriched (Table S2) gene sets. Interestingly, many of the genes that were common to at least 3 studies, such as Abcc911,15–17, Cacna1c18, Edn111,13,19, Pde4b12,20, Ptger411,21–26, and Tfap2b11,27–33, have previously been identified as significant for DA function (Table S1, blue typeface). A more thorough listing of previously identified genes significant for DA function can be found in Lewis et al.34. Identification of these established genes suggests that this approach was sufficient to detect genes and pathways relevant for DA function.
Figure 1. Venn diagram of compared rodent microarray studies.
Darkly shaded areas represent 444 DEGs (p-value ≤ 0.05, fold change ≥ 1.2) identified by at least 3 of 4 studies. Of these, 168 genes had conflicting direction of expression and were excluded from further analysis. Of the remaining 276 genes, 87 were consistently increased, while 189 were consistently decreased in DA versus aorta (genes are listed in Tables S1 and S2, respectively).
Human RNA-seq Analysis
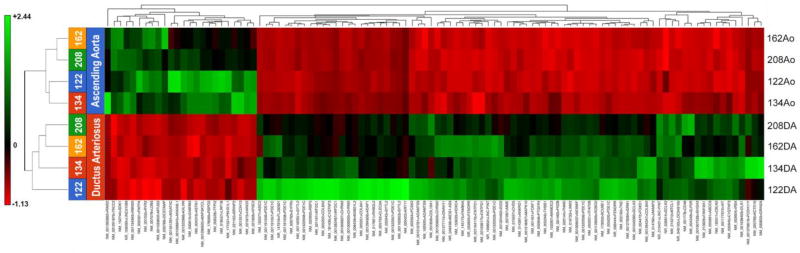
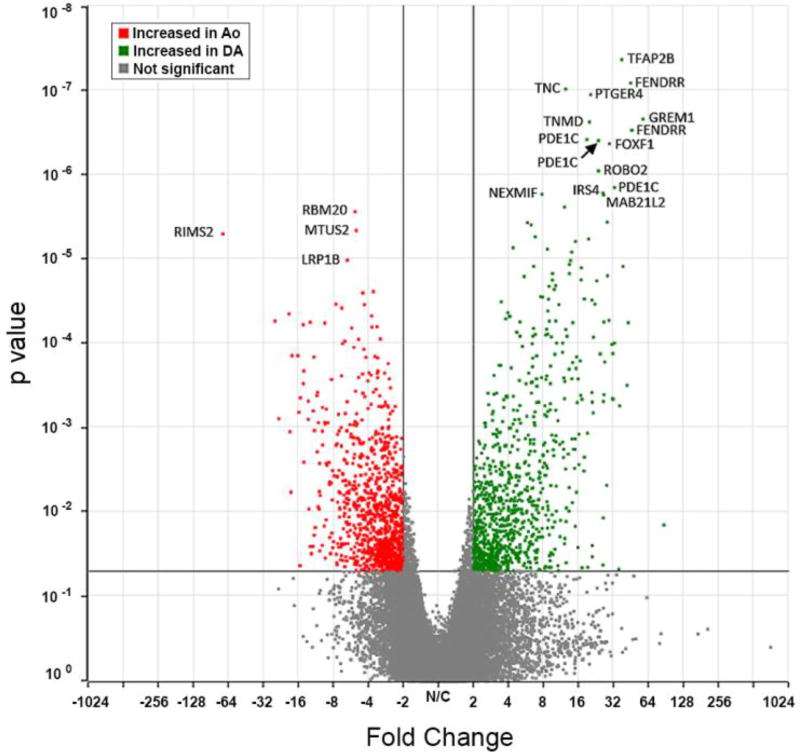
cDNA libraries were prepared for paired DA and Ao samples from 4 subjects. Libraries were sequenced to an average of 60 million genomic reads with an average genomic coverage rate of 67.7% (Table 2). Reads were aligned to human genome version 38 and quantified against Refseq Transcripts 83. GSA was then used to detect DEGs from counts normalized to Fragments Per Kilobase of transcript per Million (FPKM)-mapped reads. Hierarchical clustering analysis resulted in a heat map of RNA-seq samples (Figure 2). This heat map demonstrates that vessel identity was the primary determinant of clustered expression patterns. Interestingly, 77% of probes selected for hierarchical clustering were DA enriched, compared to 23% Ao enriched, suggesting the DA’s phenotype is driven by expression of DA-specific genes, as opposed to suppression of Ao-specific genes. Overall, 2082 genes showed differential expression between DA and Ao with a p-value of 0.05 and fold change of ≥2 (Figure 3). 186 of these genes met a permissive FDR criteria (Benjamini-Hochberg) of 0.10 or less, with 118 showing increased expression in the DA compared to Ao and 68 showing decreased expression in DA compared to Ao. Complete gene lists from this analysis are provided for both DA enriched (Table S4) and Ao enriched (Table S5) transcripts. The 20 most highly expressed (by FPKM) DEGs in the human DA were also identified (Table S6).
Table 2.
| Sample | Vessel | Genomic Reads |
Genomic Alignments |
Unique Alignments |
Non-unique Alignments |
Avg. Genomic Coverage |
Transcript Reads |
|---|---|---|---|---|---|---|---|
| 122 | Ao | 30428844 | 63437690 (87.7%) | 24463103 (80.4%) | 2215202 (7.3%) | 38.6% | 26761371 |
| 122 | DA | 60595040 | 97693434 (46.7%) | 23044387 (38.0%) | 5276853 (8.7%) | 73.9% | 28415074 |
| 134 | Ao | 52700686 | 121687422 (92.9%) | 43540599 (82.6%) | 5419786 (10.3%) | 85.8% | 49240999 |
| 134 | DA | 48552058 | 110777274 (91.1%) | 40162140 (82.7%) | 4064536 (8.4%) | 75.6% | 44456169 |
| 162 | Ao | 75583943 | 151451022 (85.6%) | 60853248 (80.5%) | 3811368 (5.0%) | 72.3% | 65157040 |
| 162 | DA | 61495042 | 124335202 (90.9%) | 53260107 (86.6%) | 2661866 (4.3%) | 54.4% | 56289111 |
| 208 | Ao | 79155461 | 160662212 (91.7%) | 69096983 (87.3%) | 3482646 (4.4%) | 65.2% | 73200814 |
| 208 | DA | 82789808 | 169365096 (90.9%) | 71228323 (86.0%) | 4003913 (4.8%) | 75.9% | 75899608 |
|
| |||||||
| Mean | 60375726 | 124051203 (87.2%) | 48909215 (80.6%) | 3764205 (6.6%) | 67.7% | 52427523 | |
Genome reads aligned to hg38
Transcriptome reads aligned to RefSeq Transcripts 83
Figure 2. Dendogram of Human RNA-seq samples.
Heat map analysis of RNA-seq data showing separation of samples by vessel identity. Gene identities are specified in Tables S4, S5.
Figure 3. Volcano plot of RNA-seq differentially expressed genes.
With p-value 0.05 and fold change >2, 2082 genes showed differential expression in the human DA compared to aorta: 1027 up-regulated, and 1055 down-regulated. Of these, 186 genes had an FDR (Benjamini-Hochberg) of 0.10 or less: 118 up-regulated, and 68 down-regulated (genes are listed in Tables S4 and S5, respectively).
Intersection of Human and Rodent DEGs
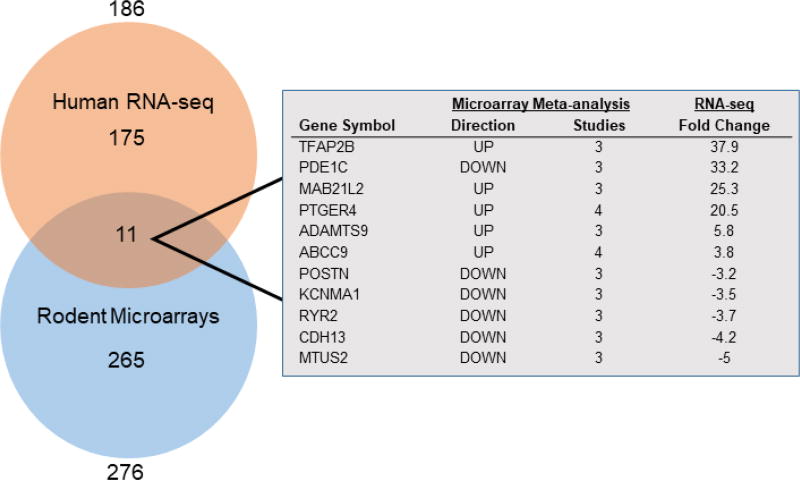
To compare the findings of the microarray and RNA-seq studies, rodent gene symbols were converted to human orthologues (bioDBnet). Of the 276 DEGs from the rodent microarray analysis and the 186 DEGs from the human RNA-seq, 11 genes were common to both studies (Figure 4). Several of these genes, including ABCC911,15–17, PDE1C11,20,35, PTGER411,21–26, and TFAP2B11,27–33 have previously been described as significant for DA identity. TFAP2B had a notably high fold change (37.9) similar to findings from individual microarray studies (Table S1)
Figure 4. Venn diagram of DA vs. Ao genes common between Microarray and RNA-seq analyses.
11 genes were identified as differentially expressed between DA and aorta in both the human RNA-seq and the rodent microarray comparison. 186 genes from RNA-seq are listed in Tables S4, S5; 276 genes from microarray comparisons are listed in Tables S1, S2.
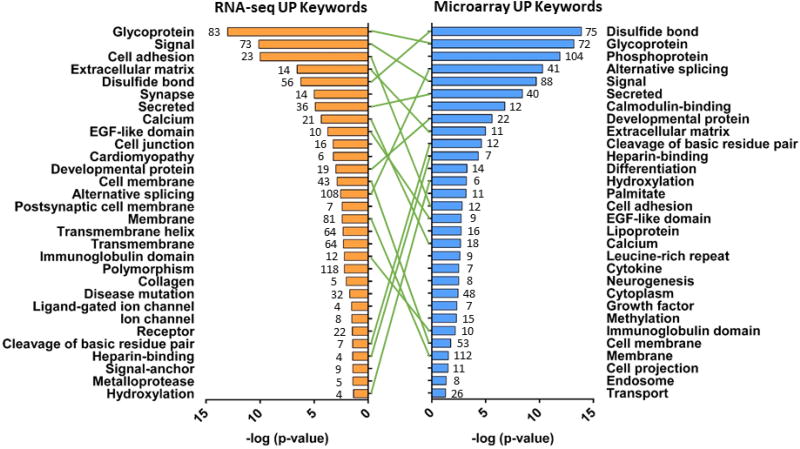
Genes differentially expressed between DA and Ao were categorized by functional annotation (DAVID). Overlap between rodent microarray and human RNA-seq was found for GO Biological Process (48.4%), GO Cellular Component (63.2%), GO Molecular Function (47.4%), KEGG (55.6%), and UniProt (UP) Keywords (59.5%) (Table S7). Of the top 30 UP Keyword terms from rodent microarray and human RNA-seq, 16 were found in common (Figure 5). Terms such as ‘Calcium’, ‘Cell adhesion’, ‘collagen’, ‘extracellular matrix’, and ‘metalloprotease’ align with pathways known to be important for DA constriction and remodeling5,30,36–41.
Figure 5. ‘Tornadogram’ showing top 30 UniProt (UP) Keywords common between Microarray and RNA-seq analyses.
Genes differentially expressed in DA vs. Ao were categorized by UP Keywords (DAVID), plotted by p-value, and compared across platforms. Number of genes represented in each category shown at the end of bars.
Discussion
There are currently nine published microarray studies of the DA9,10,12–14,26,42–44, each performed to answer a specific question, and consequently, each with a different experimental design. In order to extract the most meaningful information from these studies, criteria were selected that would include as many studies as possible while only considering comparable data sets. We focused on studies that included DA to Ao comparisons as they distinguish between DA-specific genes and developmental genes also expressed in neighboring vasculature, while also controlling for biases inherent to each study. In addition, studies that analyzed the term-gestation time point were chosen because this developmental time point was the most represented among available microarray data sets. Although individual array studies have identified differences between term and preterm DAs9,12,13, examination of the available data suggested that differences in study design and consideration of disparate preterm time points would make data alignment unreliable and preclude inter-study comparisons. There is one published microarray study of human DA samples, though it was not considered because it was conducted on abnormal tissue (patients requiring DA stents) and contained no Ao tissue for comparison45. Overall, 11 genes were differentially expressed in both the preterm human RNA-seq as well as the term rodent microarrays, suggesting that a small subset of genes may define DA identity over a broad developmental time span and between species. This number would be expected to increase in an analysis comparing differential time points in the same species or comparable time points across different species.
Among the DEGs identified as common between rodent microarray data sets and human RNA-seq analysis were several genes which have been previously identified as important for the DA. Two of these, PTGER4, which encodes the prostanoid receptor EP4, and TFAP2B, which encodes transcription factor AP-2β, have key roles in DA biology. EP4 is a G-protein coupled receptor and primary regulator of DA patency. EP4 is the predominant prostanoid receptor in the mammalian DA46–54. During late gestation, circulating prostaglandin E2 (PGE2) stimulates EP4 to maintain DA patency48–50. Catabolism of circulating prostaglandins by 15-hydroxy-prostaglandin dehydrogenase in the newly inflated lungs helps facilitate neonatal DA closure55,56. Interestingly, EP4 knockout mice die shortly after birth with a PDA21–25. Similarly, mice lacking both of the cyclooxygenase enzymes required for PGE2 production, also die with PDA57,58. Normally the removal of a dilatory stimulus would be expected to result in constriction, but these animals paradoxically present with a widely patent DA. Taken together, these findings suggest that prostaglandins, acting via the EP4 receptor pathway, may play a role in developmentally programming the DA in addition to their acute vasodilatory function21,26,57,59,60. The prevalence of EP4 among microarray studies and its high fold change in our human RNA-seq data support its role as an important regulator of DA development and function.
TFAP2B is thought to regulate proliferation and differentiation during development. Mutations in TFAP2B result in Char syndrome, a neural crest disorder typically associated with developmental abnormalities of the hands and face, as well as PDA27,29,32. Single nucleotide variants in TFAP2B have also been linked to non-syndromic PDA31,33. TFAP2B was highly enriched in DA vs. Ao expression in both human RNA-seq (38-fold increase) and three rodent microarrays (33-fold increase in Shelton et al.14). Tfap2b was not included in the probe set used by Jin et al., and thus was found significant in all possible sources. Interestingly, the array from Bokenkamp et al.9 found Tfap2b to be similarly enriched in both DA SMC and DA endothelium, despite only DA SMCs originating from the neural crest61. These findings are consistent with previous speculation that Tfap2b may act as a critical regulator of DA gene expression. Ivey et al. showed that Tfap2b expression was specific to DA SMCs and that knockout mice lacked proper SMC differentiation62. They also found that Tfap2b expression was required for the sequential expression of hypoxia inducible factor 2α (Hif2a) and endothelin-1 (Et-1). These data suggested the hypothesis that Tfap2b is a transcriptional regulator which interacts with Hif2a and Et-1 during development to drive differentiation of DA SMCs. In order to determine the true role of Tfap2b in DA development, transcriptional comparisons must be made between the DA of Tfap2b null and wild-type mice.
Of the eleven DEGs common to the human RNA-seq and rodent microarrays, one gene, phosphodiesterase 1C, (PDE1C) showed discordant results for DA vs. Ao expression between species. Pde1c had consistently decreased DA/Ao expression in three rodent microarray studies (−1.3, −2.4, and −2.9 fold change), but increased DA/Ao expression in human RNA-seq data (33.2 fold change). PDE1C catalyzes hydrolysis of the second messengers, cAMP and cGMP. Given the importance of both EP4 and nitric oxide signaling via cAMP and cGMP, respectively, in regulating DA tone60,63–65, PDE1C’s involvement in the DA is logical. PDE1C activity has been shown to affect DA tone66–68 and is thought to do this by attenuating the dilatory effects of EP4 stimulation and downstream cAMP production in the DA20. PDE1C is also associated with pathological vascular remodeling, driving proliferation and migration in vascular smooth muscle cells, processes relevant for DA functionality69,70. Due to this study’s limitations, it is unclear whether this discrepancy arises from gestational differences, species differences, or some unknown experimental factor.
Similarly, some genes shown to be relevant for DA function in prior studies were not identified in our combined approach. For example, KCNMA1, which encodes a subunit of the BKCA potassium channel, was identified in the microarray study by Shelton et al.14 as enriched in the DA (fold change 2.1). Those results were confirmed by RT-qPCR, in situ hybridization and functional analysis14. Despite these convincing results, KCNMA1 was enriched in the Ao by other microarrays and our human RNA-seq data. Considering Shelton et al. is the only study using mice, this may represent a species-specific difference. These findings highlight the importance of multi-model comparisons in DA research.
There are several limitations in our present analysis. Previous DA microarray studies have raised concerns about the efficacy and appropriateness of directly comparing different microarray data sets9,12,71,72. This is not field-specific, and there are well-known limitations to this type of comparison73–75. This is especially true for embryonic and pregnancy-associated tissues which have high internal variability even within tissue type76,77. The primary concern is a bias towards type 2 errors, or under-detection of truly differentially expressed and biologically relevant genes.
Direct gene comparison between species also suffers from differences in naming conventions and conservation of specific orthologues. To address this, we focused on functional annotation of our gene lists, to emphasize pathways that were in agreement rather than individual gene identity. This strategy has been shown to be more consistent across studies and yield more reproducible, biologically informative results75,78. In doing so, several aspects of vessel physiology which may be altered between the DA and neighboring vessels were identified. Genes associated with alterations in extracellular matrix (ECM) organization between DA and Ao were identified by GO biological process, GO cellular component, GO molecular function, and UP keywords in both rodent microarrays and human RNA-seq. Alterations in ECM composition play a critical role in remodeling of the DA in both late gestation and neonatal life5,30,36,39,40. Interestingly, several mouse knockout models of ECM-related genes have PDA79,80. There was limited direct alignment of ECM-associated genes between the microarray and RNA-seq studies. Only periostin (POSTN), an integrin binding protein which supports cell adhesion and migration, was identified in both rodent microarrays and human RNA-seq. However, both rodent arrays and human RNA-seq studies identified several collagen family members (Col11a1, Col3a1, Col5a2, Col6a3) (COL8A1, COL8A2, COL9A1, COL19A1) and ADAMTS family genes (Adamts1, Adamts9, Adamtsl2, Adam33, Adamtsl5) (ADAMTS9, ADAMT22, ADMATS8). These data suggest that though there are species differences in orthologues, similar pathways are important for both the rodent and human DA.
This study was also hindered by the limited number of preterm datasets. Because the human RNA-seq and rodent microarray data were obtained at different gestational time points, it is difficult to draw clear, unbiased comparisons. Ambiguity still exists concerning the correlation of critical events in DA development between rodents and humans. Thus, at this point, more information on developmental milestones is needed to identify parallel features of DA regulation.
Finally, microarray studies were performed on different platforms, resulting in different sets of transcripts that could possibly be detected for each array. Comparisons between these platforms were then limited to the sets of genes that are represented on all array platforms. This is a minor shortcoming, but is further compounded when comparing microarray results to RNA-seq, which is not restricted by predetermined probe sets as a sequencing-based approach. Thus, our list of genes common to both rodent microarrays and human RNA-seq data sets is capped by the set of genes which is represented on the most limited array platform, predisposing this analysis to under-report significant genes from the RNA-seq which may have been omitted from specific microarray platforms.
In conclusion, comparison of microarray studies from animal models with new information from human RNA-seq data generated high priority candidate genes to consider in future DA studies. Identification of DA-specific or highly enriched genes in the DA is a requisite step in finding new DA-selective drugs or providing a molecular address for homing of therapeutic agents to the target tissue. Although cross-platform and cross-species transcriptomic comparisons are fraught with limitations, they offer a unique opportunity to visualize pathways of interest to better understand DA development and function.
Supplementary Material
Acknowledgments
Supported by NIH HL128386 (J.R.), NHLBI HL109199 (R.I.C.), NHLBI HL46691 (R.I.C.), and NIH 5T32HD007502-20 (M.T.Y.). Special thanks to Xiaowen Wang (Partek Inc.) for contributions to microarray and RNA-seq analyses.
Abbreviations
- Ao
Aorta
- DEG
Differentially Expressed Gene
- bioDBnet
biological Database network
- BP
Biological Process
- CC
Cellular Component
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- ECM
extracellular matrix
- FDR
False Discovery Rate
- FPKM
Fragments Per Kilobase of transcript per Million mapped reads
- GO
Gene Ontology
- GEO
Gene Expression Omnibus
- hg38
human genome version 38
- RMA
Robust Multi-Array
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MF
Molecular Function
- UP
UniProt
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lloyd TR, Beekman RH., 3rd Clinically silent patent ductus arteriosus. Am Heart J. 1994;127:1664–1665. doi: 10.1016/0002-8703(94)90415-4. [DOI] [PubMed] [Google Scholar]
- 2.Hammerman C. Patent ductus arteriosus. Clinical relevance of prostaglandins and prostaglandin inhibitors in PDA pathophysiology and treatment. Clin Perinatol. 1995;22:457–479. [PubMed] [Google Scholar]
- 3.Reller MD, Rice MJ, McDonald RW. Review of studies evaluating ductal patency in the premature infant. J Pediatr. 1993;122:S59–62. doi: 10.1016/s0022-3476(09)90044-0. [DOI] [PubMed] [Google Scholar]
- 4.Yu VY. Patent ductus arteriosus in the preterm infant. Early Hum Dev. 1993;35:1–14. doi: 10.1016/0378-3782(93)90133-f. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama U, Minamisawa S, Ishikawa Y. Regulation of vascular tone and remodeling of the ductus arteriosus. J Smooth Muscle Res. 2010;46:77–87. doi: 10.1540/jsmr.46.77. [DOI] [PubMed] [Google Scholar]
- 6.Bokenkamp R, DeRuiter MC, van Munsteren C, Gittenberger-de Groot AC. Insights into the pathogenesis and genetic background of patency of the ductus arteriosus. Neonatology. 2010;98:6–17. doi: 10.1159/000262481. [DOI] [PubMed] [Google Scholar]
- 7.Stoller JZ, Demauro SB, Dagle JM, Reese J. Current Perspectives on Pathobiology of the Ductus Arteriosus. J Clin Exp Cardiolog. 2012;8 doi: 10.4172/2155-9880.S8-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaltzgraff ER, Shelton EL, Galindo CL, Nelms BL, Hooper CW, Poole SD, Labosky PA, Bader DM, Reese J. Embryonic domains of the aorta derived from diverse origins exhibit distinct properties that converge into a common phenotype in the adult. J Mol Cell Cardiol. 2014;69:88–96. doi: 10.1016/j.yjmcc.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bokenkamp R, van Brempt R, van Munsteren JC, van den Wijngaert I, de Hoogt R, Finos L, Goeman J, Groot AC, Poelmann RE, Blom NA, DeRuiter MC. Dlx1 and Rgs5 in the ductus arteriosus: vessel-specific genes identified by transcriptional profiling of laser-capture microdissected endothelial and smooth muscle cells. PLoS One. 2014;9:e86892. doi: 10.1371/journal.pone.0086892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh YT, Liu NM, Ohmori E, Yokota T, Kajimura I, Akaike T, Ohshima T, Goda N, Minamisawa S. Transcription profiles of the ductus arteriosus in Brown-Norway rats with irregular elastic fiber formation. Circ J. 2014;78:1224–1233. doi: 10.1253/circj.cj-13-1029. [DOI] [PubMed] [Google Scholar]
- 11.Waleh N, Barrette AM, Dagle JM, Momany A, Jin C, Hills NK, Shelton EL, Reese J, Clyman RI. Effects of Advancing Gestation and Non-Caucasian Race on Ductus Arteriosus Gene Expression. J Pediatr. 2015;167:1033–1041. e1032. doi: 10.1016/j.jpeds.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal R, Goyal D, Longo LD, Clyman RI. Microarray gene expression analysis in ovine ductus arteriosus during fetal development and birth transition. Pediatr Res. 2016;80:610–618. doi: 10.1038/pr.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin MH, Yokoyama U, Sato Y, Shioda A, Jiao Q, Ishikawa Y, Minamisawa S. DNA microarray profiling identified a new role of growth hormone in vascular remodeling of rat ductus arteriosus. J Physiol Sci. 2011;61:167–179. doi: 10.1007/s12576-011-0133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shelton EL, Ector G, Galindo CL, Hooper CW, Brown N, Wilkerson I, Pfaltzgraff ER, Paria BC, Cotton RB, Stoller JZ, Reese J. Transcriptional profiling reveals ductus arteriosus-specific genes that regulate vascular tone. Physiol Genomics. 2014;46:457–466. doi: 10.1152/physiolgenomics.00171.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grange DK, Nichols CG, Singh GK. Cantu Syndrome and Related Disorders. University of Washington; 2014. [PubMed] [Google Scholar]
- 16.Grange DK, Lorch SM, Cole PL, Singh GK. Cantu syndrome in a woman and her two daughters: Further confirmation of autosomal dominant inheritance and review of the cardiac manifestations. Am J Med Genet A. 2006;140:1673–1680. doi: 10.1002/ajmg.a.31348. [DOI] [PubMed] [Google Scholar]
- 17.Harakalova M, van Harssel JJ, Terhal PA, van Lieshout S, Duran K, Renkens I, Amor DJ, Wilson LC, Kirk EP, Turner CL, Shears D, Garcia-Minaur S, Lees MM, Ross A, Venselaar H, Vriend G, Takanari H, Rook MB, van der Heyden MA, Asselbergs FW, Breur HM, Swinkels ME, Scurr IJ, Smithson SF, Knoers NV, van der Smagt JJ, Nijman IJ, Kloosterman WP, van Haelst MM, van Haaften G, Cuppen E. Dominant missense mutations in ABCC9 cause Cantu syndrome. Nat Genet. 2012;44:793–796. doi: 10.1038/ng.2324. [DOI] [PubMed] [Google Scholar]
- 18.Boczek NJ, Miller EM, Ye D, Nesterenko VV, Tester DJ, Antzelevitch C, Czosek RJ, Ackerman MJ, Ware SM. Novel Timothy syndrome mutation leading to increase in CACNA1C window current. Heart Rhythm. 2015;12:211–219. doi: 10.1016/j.hrthm.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith GC. The pharmacology of the ductus arteriosus. Pharmacol Rev. 1998;50:35–58. [PubMed] [Google Scholar]
- 20.Liu H, Manganiello V, Waleh N, Clyman RI. Expression, activity, and function of phosphodiesterases in the mature and immature ductus arteriosus. Pediatr Res. 2008;64:477–481. doi: 10.1203/PDR.0b013e3181827c2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen M, Camenisch T, Snouwaert JN, Hicks E, Coffman TM, Anderson PA, Malouf NN, Koller BH. The prostaglandin receptor EP4 triggers remodelling of the cardiovascular system at birth. Nature. 1997;390:78–81. doi: 10.1038/36342. [DOI] [PubMed] [Google Scholar]
- 22.Segi E, Sugimoto Y, Yamasaki A, Aze Y, Oida H, Nishimura T, Murata T, Matsuoka T, Ushikubi F, Hirose M, Tanaka T, Yoshida N, Narumiya S, Ichikawa A. Patent ductus arteriosus and neonatal death in prostaglandin receptor EP4-deficient mice. Biochem Biophys Res Commun. 1998;246:7–12. doi: 10.1006/bbrc.1998.8461. [DOI] [PubMed] [Google Scholar]
- 23.Ushikubi F, Sugimoto Y, Ichikawa A, Narumiya S. Roles of prostanoids revealed from studies using mice lacking specific prostanoid receptors. Jpn J Pharmacol. 2000;83:279–285. doi: 10.1254/jjp.83.279. [DOI] [PubMed] [Google Scholar]
- 24.Fujino T, Yuhki K, Yamada T, Hara A, Takahata O, Okada Y, Xiao CY, Ma H, Karibe H, Iwashima Y, Fukuzawa J, Hasebe N, Kikuchi K, Narumiya S, Ushikubi F. Effects of the prostanoids on the proliferation or hypertrophy of cultured murine aortic smooth muscle cells. Br J Pharmacol. 2002;136:530–539. doi: 10.1038/sj.bjp.0704749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider A, Guan Y, Zhang Y, Magnuson MA, Pettepher C, Loftin CD, Langenbach R, Breyer RM, Breyer MD. Generation of a conditional allele of the mouse prostaglandin EP4 receptor. Genesis. 2004;40:7–14. doi: 10.1002/gene.20048. [DOI] [PubMed] [Google Scholar]
- 26.Gruzdev A, Nguyen M, Kovarova M, Koller BH. PGE2 through the EP4 receptor controls smooth muscle gene expression patterns in the ductus arteriosus critical for remodeling at birth. Prostaglandins Other Lipid Mediat. 2012;97:109–119. doi: 10.1016/j.prostaglandins.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satoda M, Zhao F, Diaz GA, Burn J, Goodship J, Davidson HR, Pierpont ME, Gelb BD. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat Genet. 2000;25:42–46. doi: 10.1038/75578. [DOI] [PubMed] [Google Scholar]
- 28.Zhao F, Weismann CG, Satoda M, Pierpont ME, Sweeney E, Thompson EM, Gelb BD. Novel TFAP2B mutations that cause Char syndrome provide a genotype-phenotype correlation. Am J Hum Genet. 2001;69:695–703. doi: 10.1086/323410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mani A, Radhakrishnan J, Farhi A, Carew KS, Warnes CA, Nelson-Williams C, Day RW, Pober B, State MW, Lifton RP. Syndromic patent ductus arteriosus: evidence for haploinsufficient TFAP2B mutations and identification of a linked sleep disorder. Proc Natl Acad Sci U S A. 2005;102:2975–2979. doi: 10.1073/pnas.0409852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider DJ, Moore JW. Patent ductus arteriosus. Circulation. 2006;114:1873–1882. doi: 10.1161/CIRCULATIONAHA.105.592063. [DOI] [PubMed] [Google Scholar]
- 31.Khetyar M, Syrris P, Tinworth L, Abushaban L, Carter N. Novel TFAP2B mutation in nonsyndromic patent ductus arteriosus. Genet Test. 2008;12:457–459. doi: 10.1089/gte.2008.0015. [DOI] [PubMed] [Google Scholar]
- 32.Zhao F, Bosserhoff AK, Buettner R, Moser M. A heart-hand syndrome gene: Tfap2b plays a critical role in the development and remodeling of mouse ductus arteriosus and limb patterning. PLoS One. 2011;6:e22908. doi: 10.1371/journal.pone.0022908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen YW, Zhao W, Zhang ZF, Fu Q, Shen J, Zhang Z, Ji W, Wang J, Li F. Familial nonsyndromic patent ductus arteriosus caused by mutations in TFAP2B. Pediatr Cardiol. 2011;32:958–965. doi: 10.1007/s00246-011-0024-7. [DOI] [PubMed] [Google Scholar]
- 34.Lewis TR, Shelton EL, Van Driest SL, Kannankeril PJ, Reese J. Genetics of the patent ductus arteriosus (PDA) and pharmacogenetics of PDA treatment. Semin Fetal Neonatal Med. 2018 doi: 10.1016/j.siny.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rybalkin SD, Rybalkina I, Beavo JA, Bornfeldt KE. Cyclic nucleotide phosphodiesterase 1C promotes human arterial smooth muscle cell proliferation. Circ Res. 2002;90:151–157. doi: 10.1161/hh0202.104108. [DOI] [PubMed] [Google Scholar]
- 36.Clyman RI. Mechanisms regulating the ductus arteriosus. Biol Neonate. 2006;89:330–335. doi: 10.1159/000092870. [DOI] [PubMed] [Google Scholar]
- 37.Yokoyama U, Minamisawa S, Quan H, Ghatak S, Akaike T, Segi-Nishida E, Iwasaki S, Iwamoto M, Misra S, Tamura K, Hori H, Yokota S, Toole BP, Sugimoto Y, Ishikawa Y. Chronic activation of the prostaglandin receptor EP4 promotes hyaluronan-mediated neointimal formation in the ductus arteriosus. J Clin Invest. 2006;116:3026–3034. doi: 10.1172/JCI28639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokoyama U, Minamisawa S, Shioda A, Ishiwata R, Jin MH, Masuda M, Asou T, Sugimoto Y, Aoki H, Nakamura T, Ishikawa Y. Prostaglandin E2 inhibits elastogenesis in the ductus arteriosus via EP4 signaling. Circulation. 2014;129:487–496. doi: 10.1161/CIRCULATIONAHA.113.004726. [DOI] [PubMed] [Google Scholar]
- 39.de Reeder EG, van Munsteren CJ, Poelmann RE, Patterson DF, Gittenberger-de Groot AC. Changes in distribution of elastin and elastin receptor during intimal cushion formation in the ductus arteriosus. Anat Embryol (Berl) 1990;182:473–480. doi: 10.1007/BF00178912. [DOI] [PubMed] [Google Scholar]
- 40.de Reeder EG, Poelmann RE, van Munsteren JC, Patterson DF, Gittenberger-de Groot AC. Ultrastructural and immunohistochemical changes of the extracellular matrix during intimal cushion formation in the ductus arteriosus of the dog. Atherosclerosis. 1989;79:29–40. doi: 10.1016/0021-9150(89)90030-0. [DOI] [PubMed] [Google Scholar]
- 41.Slomp J, van Munsteren JC, Poelmann RE, de Reeder EG, Bogers AJ, Gittenberger-de Groot AC. Formation of intimal cushions in the ductus arteriosus as a model for vascular intimal thickening. An immunohistochemical study of changes in extracellular matrix components. Atherosclerosis. 1992;93:25–39. doi: 10.1016/0021-9150(92)90197-o. [DOI] [PubMed] [Google Scholar]
- 42.Costa M, Barogi S, Socci ND, Angeloni D, Maffei M, Baragatti B, Chiellini C, Grasso E, Coceani F. Gene expression in ductus arteriosus and aorta: comparison of birth and oxygen effects. Physiol Genomics. 2006;25:250–262. doi: 10.1152/physiolgenomics.00231.2005. [DOI] [PubMed] [Google Scholar]
- 43.Yokoyama U, Sato Y, Akaike T, Ishida S, Sawada J, Nagao T, Quan H, Jin M, Iwamoto M, Yokota S, Ishikawa Y, Minamisawa S. Maternal vitamin A alters gene profiles and structural maturation of the rat ductus arteriosus. Physiol Genomics. 2007;31:139–157. doi: 10.1152/physiolgenomics.00007.2006. [DOI] [PubMed] [Google Scholar]
- 44.Liu NM, Yokota T, Maekawa S, Lu P, Zheng YW, Taniguchi H, Yokoyama U, Kato T, Minamisawa S. Transcription profiles of endothelial cells in the rat ductus arteriosus during a perinatal period. PLoS One. 2013;8:e73685. doi: 10.1371/journal.pone.0073685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller PP, Drynda A, Goltz D, Hoehn R, Hauser H, Peuster M. Common signatures for gene expression in postnatal patients with patent arterial ducts and stented arteries. Cardiol Young. 2009;19:352–359. doi: 10.1017/S1047951109004260. [DOI] [PubMed] [Google Scholar]
- 46.Leonhardt A, Glaser A, Wegmann M, Schranz D, Seyberth H, Nusing R. Expression of prostanoid receptors in human ductus arteriosus. Br J Pharmacol. 2003;138:655–659. doi: 10.1038/sj.bjp.0705092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rheinlaender C, Weber SC, Sarioglu N, Strauss E, Obladen M, Koehne P. Changing expression of cyclooxygenases and prostaglandin receptor EP4 during development of the human ductus arteriosus. Pediatr Res. 2006;60:270–275. doi: 10.1203/01.pdr.0000233066.28496.7c. [DOI] [PubMed] [Google Scholar]
- 48.Momma K, Toyoshima K, Takeuchi D, Imamura S, Nakanishi T. In vivo reopening of the neonatal ductus arteriosus by a prostanoid EP4-receptor agonist in the rat. Prostaglandins Other Lipid Mediat. 2005;78:117–128. doi: 10.1016/j.prostaglandins.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 49.Momma K, Toyoshima K, Takeuchi D, Imamura S, Nakanishi T. In vivo constriction of the fetal and neonatal ductus arteriosus by a prostanoid EP4-receptor antagonist in rats. Pediatr Res. 2005;58:971–975. doi: 10.1203/01.pdr.0000182182.49476.24. [DOI] [PubMed] [Google Scholar]
- 50.Kajino H, Taniguchi T, Fujieda K, Ushikubi F, Muramatsu I. An EP4 receptor agonist prevents indomethacin-induced closure of rat ductus arteriosus in vivo. Pediatr Res. 2004;56:586–590. doi: 10.1203/01.PDR.0000139409.25014.35. [DOI] [PubMed] [Google Scholar]
- 51.Fan FL, Zhu S, Chen LH, Zou YL, Fan LH, Kang JH, Ma AQ, Guan YF. Role of prostaglandin E and its receptors in the process of ductus arteriosus maturation and functional closure in the rabbit. Clin Exp Pharmacol Physiol. 2010;37:574–580. doi: 10.1111/j.1440-1681.2010.05354.x. [DOI] [PubMed] [Google Scholar]
- 52.Bouayad A, Kajino H, Waleh N, Fouron JC, Andelfinger G, Varma DR, Skoll A, Vazquez A, Gobeil F, Jr, Clyman RI, Chemtob S. Characterization of PGE2 receptors in fetal and newborn lamb ductus arteriosus. Am J Physiol Heart Circ Physiol. 2001;280:H2342–2349. doi: 10.1152/ajpheart.2001.280.5.H2342. [DOI] [PubMed] [Google Scholar]
- 53.Bouayad A, Bernier SG, Asselin P, Hardy P, Bhattacharya M, Quiniou C, Fouron JC, Guerguerian AM, Varma DR, Clyman RI, Chemtob S. Characterization of PGE2 receptors in fetal and newborn ductus arteriosus in the pig. Semin Perinatol. 2001;25:70–75. doi: 10.1053/sper.2001.23186. [DOI] [PubMed] [Google Scholar]
- 54.Bhattacharya M, Asselin P, Hardy P, Guerguerian AM, Shichi H, Hou X, Varma DR, Bouayad A, Fouron JC, Clyman RI, Chemtob S. Developmental changes in prostaglandin E(2) receptor subtypes in porcine ductus arteriosus. Possible contribution in altered responsiveness to prostaglandin E(2) Circulation. 1999;100:1751–1756. doi: 10.1161/01.cir.100.16.1751. [DOI] [PubMed] [Google Scholar]
- 55.de Reeder EG, Gittenberger-de Groot AC, van Munsteren JC, Poelmann RE, Patterson DF, Keirse MJ. Distribution of prostacyclin synthase, 6-keto-prostaglandin F1 alpha, and 15-hydroxy-prostaglandin dehydrogenase in the normal and persistent ductus arteriosus of the dog. Am J Pathol. 1989;135:881–887. [PMC free article] [PubMed] [Google Scholar]
- 56.Coggins KG, Latour A, Nguyen MS, Audoly L, Coffman TM, Koller BH. Metabolism of PGE2 by prostaglandin dehydrogenase is essential for remodeling the ductus arteriosus. Nat Med. 2002;8:91–92. doi: 10.1038/nm0202. [DOI] [PubMed] [Google Scholar]
- 57.Reese J, Paria BC, Brown N, Zhao X, Morrow JD, Dey SK. Coordinated regulation of fetal and maternal prostaglandins directs successful birth and postnatal adaptation in the mouse. Proc Natl Acad Sci U S A. 2000;97:9759–9764. doi: 10.1073/pnas.97.17.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loftin CD, Trivedi DB, Tiano HF, Clark JA, Lee CA, Epstein JA, Morham SG, Breyer MD, Nguyen M, Hawkins BM, Goulet JL, Smithies O, Koller BH, Langenbach R. Failure of ductus arteriosus closure and remodeling in neonatal mice deficient in cyclooxygenase-1 and cyclooxygenase-2. Proc Natl Acad Sci U S A. 2001;98:1059–1064. doi: 10.1073/pnas.031573498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reese J, Anderson JD, Brown N, Roman C, Clyman RI. Inhibition of cyclooxygenase isoforms in late- but not midgestation decreases contractility of the ductus arteriosus and prevents postnatal closure in mice. Am J Physiol Regul Integr Comp Physiol. 2006;291:R1717–1723. doi: 10.1152/ajpregu.00259.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reese J, O'Mara PW, Poole SD, Brown N, Tolentino C, Eckman DM, Aschner JL. Regulation of the fetal mouse ductus arteriosus is dependent on interaction of nitric oxide and COX enzymes in the ductal wall. Prostaglandins Other Lipid Mediat. 2009;88:89–96. doi: 10.1016/j.prostaglandins.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Chen D, Chen K, Jubran A, Ramirez A, Astrof S. Endothelium in the pharyngeal arches 3, 4 and 6 is derived from the second heart field. Dev Biol. 2017;421:108–117. doi: 10.1016/j.ydbio.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivey KN, Sutcliffe D, Richardson J, Clyman RI, Garcia JA, Srivastava D. Transcriptional regulation during development of the ductus arteriosus. Circ Res. 2008;103:388–395. doi: 10.1161/CIRCRESAHA.108.180661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coceani F. Control of the ductus arteriosus--a new function for cytochrome P450, endothelin and nitric oxide. Biochem Pharmacol. 1994;48:1315–1318. doi: 10.1016/0006-2952(94)90552-5. [DOI] [PubMed] [Google Scholar]
- 64.Momma K, Toyono M. The role of nitric oxide in dilating the fetal ductus arteriosus in rats. Pediatr Res. 1999;46:311–315. doi: 10.1203/00006450-199909000-00010. [DOI] [PubMed] [Google Scholar]
- 65.Richard C, Gao J, LaFleur B, Christman BW, Anderson J, Brown N, Reese J. Patency of the preterm fetal ductus arteriosus is regulated by endothelial nitric oxide synthase and is independent of vasa vasorum in the mouse. Am J Physiol Regul Integr Comp Physiol. 2004;287:R652–660. doi: 10.1152/ajpregu.00049.2004. [DOI] [PubMed] [Google Scholar]
- 66.Thebaud B, Michelakis E, Wu XC, Harry G, Hashimoto K, Archer SL. Sildenafil reverses O2 constriction of the rabbit ductus arteriosus by inhibiting type 5 phosphodiesterase and activating BK(Ca) channels. Pediatr Res. 2002;52:19–24. doi: 10.1203/00006450-200207000-00006. [DOI] [PubMed] [Google Scholar]
- 67.Momma K, Toyoshima K, Imamura S, Nakanishi T. In vivo dilation of fetal and neonatal ductus arteriosus by inhibition of phosphodiesterase-5 in rats. Pediatr Res. 2005;58:42–45. doi: 10.1203/01.PDR.0000156370.50874.3C. [DOI] [PubMed] [Google Scholar]
- 68.Toyoshima K, Momma K, Imamura S, Nakanishi T. In vivo dilatation of the fetal and postnatal ductus arteriosus by inhibition of phosphodiesterase 3 in rats. Biol Neonate. 2006;89:251–256. doi: 10.1159/000089954. [DOI] [PubMed] [Google Scholar]
- 69.Chan S, Yan C. PDE1 isozymes, key regulators of pathological vascular remodeling. Curr Opin Pharmacol. 2011;11:720–724. doi: 10.1016/j.coph.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai Y, Nagel DJ, Zhou Q, Cygnar KD, Zhao H, Li F, Pi X, Knight PA, Yan C. Role of cAMP-phosphodiesterase 1C signaling in regulating growth factor receptor stability, vascular smooth muscle cell growth, migration, and neointimal hyperplasia. Circ Res. 2015;116:1120–1132. doi: 10.1161/CIRCRESAHA.116.304408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goyal R, Clyman RI. Response to Coceani et al. Pediatr Res. 2017;82:175. doi: 10.1038/pr.2017.125. [DOI] [PubMed] [Google Scholar]
- 72.Coceani F, Scebba F, Angeloni D. Ductus arteriosus: gene profile for fetal maturation versus postnatal closure. Pediatr Res. 2017;82:174. doi: 10.1038/pr.2017.124. [DOI] [PubMed] [Google Scholar]
- 73.Cahan P, Ahmad AM, Burke H, Fu S, Lai Y, Florea L, Dharker N, Kobrinski T, Kale P, McCaffrey TA. List of lists-annotated (LOLA): a database for annotation and comparison of published microarray gene lists. Gene. 2005;360:78–82. doi: 10.1016/j.gene.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Cahan P, Rovegno F, Mooney D, Newman JC, St Laurent G, 3rd, McCaffrey TA. Meta-analysis of microarray results: challenges, opportunities, and recommendations for standardization. Gene. 2007;401:12–18. doi: 10.1016/j.gene.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res. 2012;40:3785–3799. doi: 10.1093/nar/gkr1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eidem HR, Ackerman WEt, McGary KL, Abbot P, Rokas A. Gestational tissue transcriptomics in term and preterm human pregnancies: a systematic review and meta-analysis. BMC Med Genomics. 2015;8:27. doi: 10.1186/s12920-015-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hughes DA, Kircher M, He Z, Guo S, Fairbrother GL, Moreno CS, Khaitovich P, Stoneking M. Evaluating intra- and inter-individual variation in the human placental transcriptome. Genome Biol. 2015;16:54. doi: 10.1186/s13059-015-0627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manoli T, Gretz N, Grone HJ, Kenzelmann M, Eils R, Brors B. Group testing for pathway analysis improves comparability of different microarray datasets. Bioinformatics. 2006;22:2500–2506. doi: 10.1093/bioinformatics/btl424. [DOI] [PubMed] [Google Scholar]
- 79.Levet S, Ouarne M, Ciais D, Coutton C, Subileau M, Mallet C, Ricard N, Bidart M, Debillon T, Faravelli F, Rooryck C, Feige JJ, Tillet E, Bailly S. BMP9 and BMP10 are necessary for proper closure of the ductus arteriosus. Proc Natl Acad Sci U S A. 2015;112:E3207–3215. doi: 10.1073/pnas.1508386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Staiculescu MC, Kim J, Mecham RP, Wagenseil JE. Mechanical behavior and matrisome gene expression in the aneurysm-prone thoracic aorta of newborn lysyl oxidase knockout mice. Am J Physiol Heart Circ Physiol. 2017;313:H446–H456. doi: 10.1152/ajpheart.00712.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.