Abstract
One bead one compound (OBOC) libraries can be screened against serum samples to identify ligands to antibodies in this mixture. In this protocol, hit beads are identified by staining with a fluorescent labeled secondary antibody. When screens are conducted against two different sets of serum, antibodies, and ligands to them, can be discovered that distinguish the two populations. The application of DNA-encoding technology to OBOC libraries has allowed the use of 10 μm beads for library preparation and screening, which pass through a standard flow cytometer, allowing the fluorescent hit beads to be separated from beads displaying non-ligands easily. An important issue in using this approach for the discovery of antibody biomarkers is its analytical sensitivity. In other words, how abundant must an IgG be to allow it to be pulled out of serum in an unbiased screen using a flow cytometer? We report here a model study in which monoclonal antibodies with known ligands of varying affinities are doped into serum. We find that for antibody ligands typical of what one isolates from an unbiased combinatorial library, the target antibody must be present at 10-50 nM. True antigens, which bind with significantly higher affinity, can detect much less abundant serum antibodies.
Keywords: Combinatorial libraries, Peptidomimetic ligand, Antibodies, Biomarkers, Flow cytometry
Graphical abstract
A seminal problem in biomedicine today is the discovery of serum biomarkers for the diagnosis of disease, prediction of drug efficacy and a variety of other purposes. The adaptive immune system is an attractive source of such biomarkers,1 since it is likely that many disease states induce the production of antibodies against disease-specific antigens. We have developed one approach to the discovery of these antibodies called epitope surrogate technology.2-4 In this process a one bead one compound (OBOC) combinatorial library,5 created by split and pool synthesis,5, 6 is incubated with a pool of control serum samples and, after washing, the beads that display antibody ligands are visualized by staining with a fluorescently labeled secondary antibody. These are discarded and the remainder of the library is screened against a pool of serum samples of interest (the “case” population). Beads that “light up” in this screen display candidate ligands for potential antibody biomarkers that distinguish the case from the control population. The compounds mined from the OBOC library can, if validated as bona fide ligands,7 be used as an affinity reagent to enrich the putative disease-specific antibodies to which they bind. These antibodies, in turn, can be used in immunoprecipitation or similar experiments to identify their native antigens in a suitable tissue extract.8, 9
Until very recently, these screens employed relatively large (90 μm) TentaGel beads because of the requirement for enough compound to identify the structure of a hit by tandem mass spectrometry.10 Hits were separated from non-hits by a tedious procedure involving visual inspection of the entire library under a low power fluorescence microscope and manual picking of the fluorescent beads using a Pipetteman.11-13 However, the adaption of DNA encoding technology14-16 to OBOC libraries17 has allowed the use of much smaller 10 μm TentaGel beads for library synthesis and screening, since the identity of the bead-displayed compound can be determined much more sensitively through PCR amplification of the encoding tag and sequencing of the amplicon. 10 μm beads are about the size of a small mammalian cell, thus a flow cytometer can be employed to separate hits from non-hits.7, 16, 18 This process is much faster and simpler than manual bead collection, allowing the use of larger libraries. The flow cytometer also facilitates two-color screening experiments.19 For antibody biomarker discovery, the case and control antibody populations are labeled with green- and red-labeled secondary Fab antibodies,20 respectively, then mixed together and exposed to the bead-displayed library. The flow cytometer can be gated to collect beads that bind only red-stained (case) antibodies and not green-stained (control) IgGs.
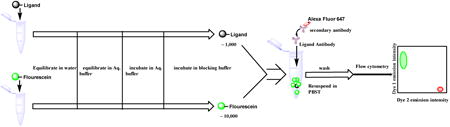
An important question is how abundant must a potential biomarker IgG be in order for it to be captured by a bead-displayed ligand in a screen using this new technology? In other words, how deeply can the immunoproteome be screened in the search for biomarkers? To address the sensitivity of ligand identification in this screening format, we started with the well-characterized anti-Flag antibody-Flag peptide (DYKDDDDK) interaction, which is of very high affinity (sub-nM apparent KD12 under the conditions used here). Altering the peptide sequence of Flag to (DYKHNNYN) (FLAG-D4H)21 increases the KD to 130 nM.12 The FLAG peptide or FLAG-D4H was synthesized on 10 μm TentaGel beads following a linker sequence. Approximately 1,000 of the peptide-displaying beads were doped into about 10,000 blank control beads (no peptide displayed). To distinguish the control beads from those displaying a peptide, the former were labeled in the protein-inaccessible, hydrophobic internal domain of the TentaGel matrix22 with carboxyfluorescein, whereas the peptide- displaying beads were unlabeled (Fig. 1).
Fig. 1. Schematic of the mock screening protocol employed in this study.
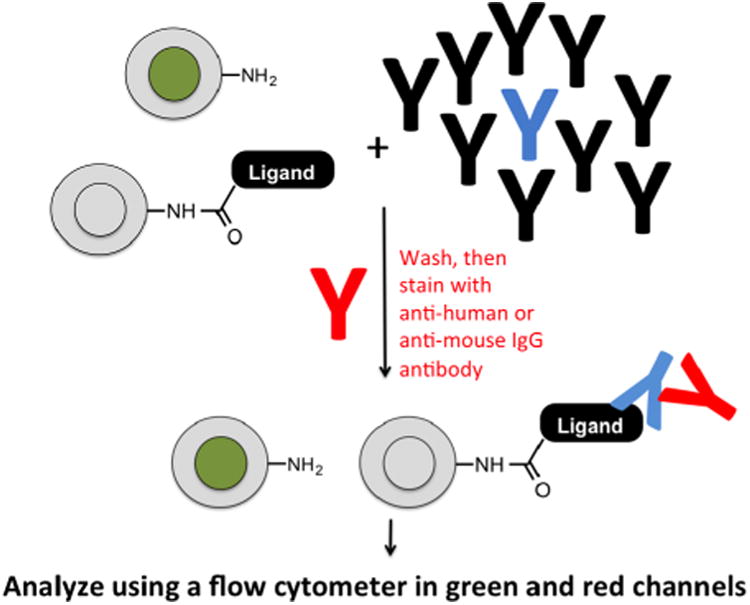
Cartoon of the mock screening strategy. “Blank beads” labeled internally with carboxyfluorescein were mixed with unlabeled beads displaying a known ligand to the antibody of interest at a ratio of 10:1. The monoclonal antibody targeted by the ligand (blue) was then doped into serum containing a multitude of IgG antibodies (black) at a known concentration. The beads and serum were mixed, washed, then stained with an Alexa Fluor 647 (A647)-conjugated secondary antibody. After a final wash the beads were analyzed by flow cytometry.
The bead mixture was incubated with mouse serum (45 mg/mL total protein, 18.1 mg/mL (1.2 × 10-4 M) IgG antibodies diluted 250-fold into PBST buffer) into which a known amount of mouse anti-FLAG antibody had been doped. Mouse serum does not contain antibodies to FLAG peptide, so this protocol allows us to control the amount of the target antibody in the serum precisely. After thorough washing, the beads were incubated with Alexafluor 647-labeled chicken anti-mouse secondary antibody, washed again, then analyzed using a flow cytometer, monitoring at 448 nm and 647 nm (green and red channels, respectively). The concentration of the anti-FLAG antibody was varied from 0 to 100 nM, with 1 pM being the lowest analyzed. The primary data, in the form of flow cytometry dot plots, are shown in Fig. 2.
Fig. 2. Binding of bead-displayed FLAG peptide to anti-FLAG antibody in mouse serum.
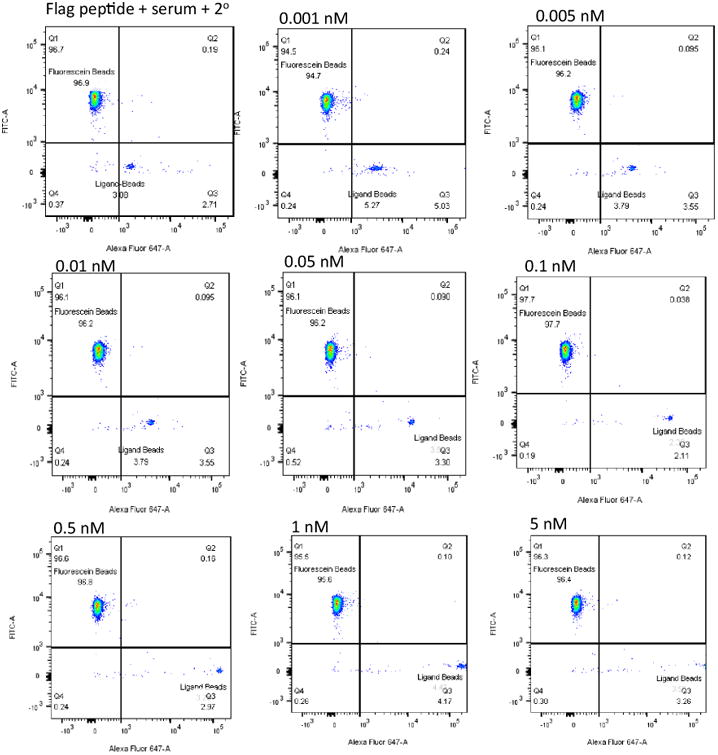
Mouse anti-FLAG antibody was added to mouse serum to provide the final concentrations indicated. The solution was then mixed with beads. Most of the beads did not display FLAG peptide, but were modified with fluorescein (see Fig. 1). A small percentage displayed the FLAG peptide, but lacked the fluorescein tag. The beads were incubated with the serum, washed, stained with A647-labeled anti-mouse IgG secondary antibody, washed again and analyzed by flow cytometry. The FLAG peptide-displaying, unlabeled beads shift to the right (indicating retention of the A647 label) in an anti-FLAG antibody dose-dependent fashion. The fluorescein-labeled beads lacking the FLAG peptide do not.
In the absence of added anti-FLAG antibody, the FLAG peptide displaying beads showed a slightly higher level of red fluorescence than the blank beads (top left of Fig. 2), reflecting some non-specific binding of FLAG peptide to mouse serum antibodies and/or the labeled secondary antibody. As anti-FLAG antibody was titrated into the serum, the red fluorescence exhibited by the FLAG peptide-displaying beads increased in a dose-dependent fashion, while the blank beads were unaffected, as expected. The fluorescence of the FLAG peptide-displaying beads in the red channel was cleanly separated from the background (defined by the top left dot plot in Fig. 2) at 0.01 nM anti-FLAG antibody and exhibited an intense signal (100-fold above background) at 0.5 nM anti-FLAG antibody (Figs. 2 and 3). Clearly, had this been a real screen, even low abundance antibodies could have been targeted for ligand discovery. These data, as well as that obtained from a variety of control experiments are shown in bar graph format in Fig. 3A.
Fig. 3. Level of fluorescence in the 647 channel displayed by the indicated beads in the presence or absence of mouse serum with the indicated amount of anti-FLAG antibody doped into the serum.
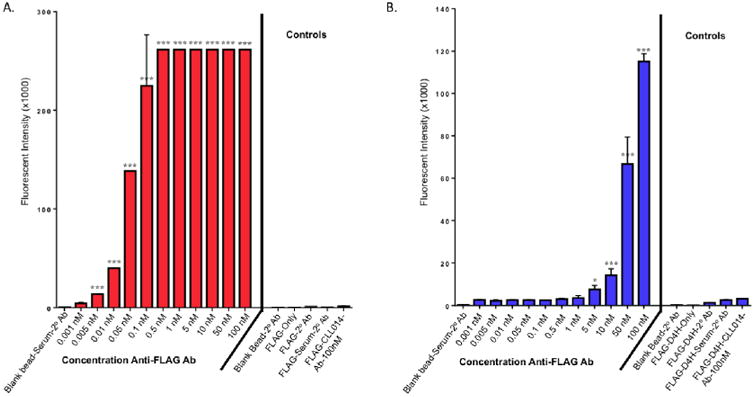
A. Beads displaying the FLAG peptide. B. Beads displaying the FLAGD4H peptide. The same controls are included on both graphs. Note the different scales in A and B.
Of course, the FLAG peptide has an affinity for anti-FLAG antibody that is much greater than a typical screening hit would have for a target antibody. Therefore, we repeated the same experiment using the more modest affinity FLAG-D4H peptide (apparent KD ≈ 133 nM).17 As shown in Fig. 3B (see SI Fig. 1 for the dot plot data) much higher levels of anti-FLAG antibody were necessary in order for the ligand-displaying beads to capture above background levels of anti-FLAG antibody. At 10 nM anti-FLAG antibody, the signal was barely above background, whereas at 50 nM anti-FLAG antibody robust binding was observed.
Finally, a similar experiment was repeated with KMS31, a ligand for the antigen-binding site of a B cell receptor isolated from a patient with chronic lymphocytic leukemia (CLL014). KMS31 is an actual screening hit derived from an OBOC library in which the target was a soluble, IgG version of the CLL014 BCR.23 When immobilized on a surface, KMS31 was shown to bind (CLL014) monoclonal antibody with an apparent KD of 67 nM under conditions similar to those used in the assays described here.23 KMS31 was synthesized (see SI Fig. 4) on 10 μM TentaGel beads and approximately 1,000 of these beads were doped into about 10,000 of the fluorescein-labeled beads. The beads were incubated with human serum (59 mg/mL total protein, 3.9 mg/mL total IgG (2.6 × 10-5 M) diluted 250-fold into PBST buffer) to which a known amount of purified CLL014 antibody had been added, followed by staining with Alexa Fluor 647-labeled goat anti-human IgG. After thorough washing, the beads were passed through the flow cytometer for analysis. To serve as controls, we used blank beads, KMS31 beads in serum without CLL014 Ab, KMS31 beads only (without CLL014 and secondary antibody), and KMS31 pre-incubated with Anti-FLAG Ab, then chicken anti-mouse IgG conjugated to Alexa Fluor 647. As shown in figure 4, retention of the CLL014 Ab from human serum could barely be detected above background when the antibody concentration reached 5-10 nM, but the beads were well separated from the background at 50 nM CLL014.
Fig. 4. Interaction of immobilized KMS31 with CLL014 antibody in human serum. 10 μM bead-displayed.
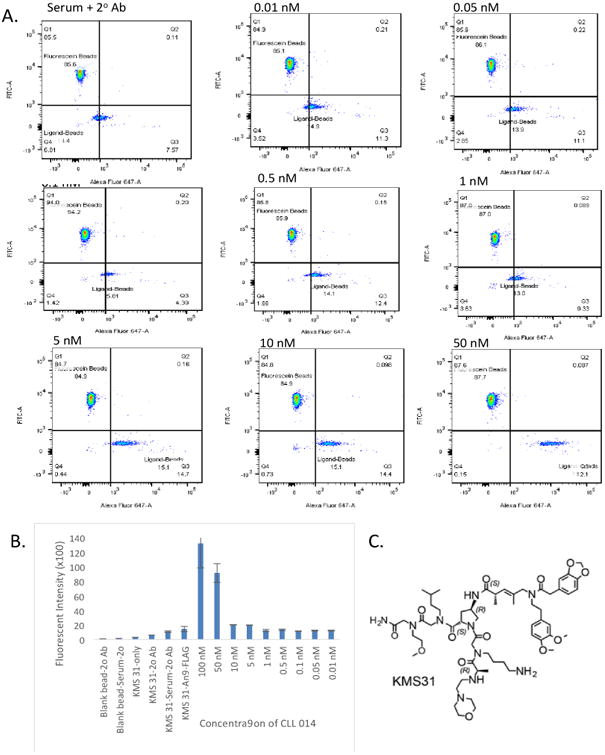
A. KMS31 was doped into fluorescein tagged beads, after equilibration in aqueous and in blocking buffer, the beads were incubated with CLL014 Antibody (titratedfrom 0.01 to 100 nM in human serum), followed by goat anti-human secondary antibody conjugated to Alexa Fluor 647. After thorough washing, the fluorescence signal due to the secondary antibody was measured using a flow cytometer. B. Bar graph representation of the data. C. Structure ofKMS31 (linkage to bead at the C-terminus not shown).
These mock screening experiments have shown that, as expected, higher affinity molecules can extract lower abundance antibodies from serum. The edge of detection of binding of the target antibodies to the bead displayed ligands studied here correlated roughly to the apparent KD of the complex. However, there was also a contribution of the selectivity of binding as well. For example, the background level of binding of human serum antibodies and/or the labeled secondary antibody to KMS31 was somewhat higher than the equivalent level of non-specific binding of mouse antibodies and/or the labeled anti-mouse IgG secondary to the peptides. This lower background allowed a weaker signal to be detected above background for the mouse antibodies (compare Figs. 2, 3 and 4b). The absolute numbers are instructive. KMS31 is typical of the type of ligand that is likely to be obtained as a screening hit using the type of libraries currently available.23-26 Therefore, it is likely that screens designed to identify ligands to antibodies that distinguish two sets of serum samples (for example, healthy controls vs. patients with a particular disease) will be able to identify ligands to antibodies that are present at a concentration ≥ 50 nM (≈ 7.5 μg/ml for an IgG). While it is remarkably difficult to find data on the absolute concentration of individual antibodies for many disease states, these data are available for antibodies induced by immunization and the post-vaccination levels of induced antibodies are generally in the 60-750 nM range.27-29 so the sensitivity of this technique is likely sufficient to discover many interesting new biomarkers, consistent with our studies published to date.2, 8, 9, 18, 30 However, as exemplified by the FLAG peptide, higher affinity molecules could dig deeper into the “immunoproteome”. In general, it is reasonable to anticipate that the chances of identifying higher affinity ligands for a given target will increase along with library size, assuming that the library is comprised of conformationally constrained and chemically diverse molecules. Thus, these studies highlight the importance of further development of combinatorial library design and synthesis to take full advantage of the potentially rich source of biomarkers represented by circulating antibodies.
Data Analysis
Data was analyzed using FlowJo_V10 software. Data represent mean ± SD of triplicate experiments. GraphPad Prism V 6.05 used to assess significance value. One-way ANOVA analysis comparing background (compound bead with serum and detection reagent) to each sample was used. Dunnett's test was used as correction for multiple comparisons. P<0.001=***, P<0.05=*
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (AG0528113). O.E. was supported by a Fulbright Foreign Student Program (PS00217127) sponsored by the Institute of International Education.
Footnotes
Author Contributions: O. E. synthesized the ligands, performed the experiments and wrote the first draft of the paper. S.S. assisted in the expression and purification of the CLL BCR IgG, measured total IgG in thehuman and mouse serum samples and provided general technical support and guidance to O.E.P. J. M. Synthesized the fluorescein-tagged beads, assisted in the Flow cytometric analysis andedited the paper. T. K. designed the study, oversaw the project, and edited the paper.
Notes: T.K. is a major shareholder in Deluge Biotechnologies, a biotechnology firm focused on the commercial development of epitope surrogate technology. Deluge provided no funding for this study, had no input, nor were any of its personnel involved.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson KS, LaBaer J. The sentinel within: Exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4:1123–1133. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doran TM, Sarkar M, Kodadek T. Chemical Tools To Monitor and Manipulate Adaptive Immune Responses. J Amer Chem Soc. 2016;138(19):6076–6094. doi: 10.1021/jacs.6b02954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy MM, Wilson R, Wilson J, et al. Identification of candidate IgG biomarkers for Alzheimer's Disease via combinatorial library screening. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raveendra B, Hao W, Baccala R, et al. Discovery of peptoid ligands for anti-Aquaporin 4 antibodies. Chem & Biol. 2013;20:350–359. doi: 10.1016/j.chembiol.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 6.Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991;354(6348):84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 7.Validation involves testing the markers on serum samples different than those used in the discovery effort. For a discussion of this point, see: Ransohoff, DF. Lessons from controvery: Ovarian cancer screening and serum proteomics. J of the NCI. 2005;97:315–319. doi: 10.1093/jnci/dji054. [DOI] [PubMed] [Google Scholar]
- 8.Doran TM, Morimoto J, Simanski S, et al. Discovery of Phosphorylated Peripherin as a Major Humoral Autoantigen in Type 1 Diabetes Mellitus. Cell Chem Biol. 2016;23(5):618–628. doi: 10.1016/j.chembiol.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doran TM, Simanski S, Kodadek T. Discovery of native autoantigens via antigen surrogate technology: Application to Type I diabetes. ACS Chem Biol. 2015;10:401–412. doi: 10.1021/cb5007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulick MG, Hart KM, Brinner KM, Tjandra M, Charych DH, Zuckermann RN. Cleavable hydrophilic linker for one-bead-one-compound sequencing of oligomer libraries by tandem mass spectrometry. J Comb Chem. 2006;8(3):417–426. doi: 10.1021/cc0501460. [DOI] [PubMed] [Google Scholar]
- 11.Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ, Kodadek T. Isolation of protein ligands from large peptoid libraries. J Amer Chem Soc. 2003;125(46):13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 12.Mendes K, Ndungu JM, Clark L, Kodadek T. Optimization of the Magnetic Recovery of Hits from One Bead One Compound Library Screens. ACS Comb Sci. 2015;17:506–517. doi: 10.1021/acscombsci.5b00090. [DOI] [PubMed] [Google Scholar]
- 13.Olivos HJ, Baccawat-Sikder K, Kodadek T. Quantum dots as a visual aid for screening bead-bound combinatorial libraries. ChemBiochem. 2003;4:1242–1245. doi: 10.1002/cbic.200300712. [DOI] [PubMed] [Google Scholar]
- 14.Brenner S, Lerner RA. Encoded combinatorial chemistry. Proc Natl Acad Sci U S A. 1992;89(12):5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark MA, Acharya RA, Arico-Muendel CC, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nature Chem Biol. 2009;5(9):647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 16.Needels MC, Jones DG, Tate EH, et al. Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proc Natl Acad Sci USA. 1993;90:10700–10704. doi: 10.1073/pnas.90.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacConnell AB, McEnaney PJ, Cavett VJ, Paegel BM. DNA-Encoded Solid-Phase Synthesis: Encoding Language Design and Complex Oligomer Library Synthesis. ACS Comb Sci. 2015;17:518–534. doi: 10.1021/acscombsci.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendes K, Malone ML, Ndungu JM, et al. High-throughput identification of DNA-encoded IgG ligands that distinguish active and latent mycobacterium tuberculosis infections. ACS Chem Biol. 2017;12:234–243. doi: 10.1021/acschembio.6b00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. A peptoid “antibody surrogate” that antagonizes VEGF receptor 2 activity. J Amer Chem Soc. 2008;130:5744–5752. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
- 20.Kattah MG, Alemi GR, Thibault DL, Balboni I, Utz PJ. A new two-color Fab labeling method for autoantigen protein microarrays. Nat Methods. 2006;3(9):745–751. doi: 10.1038/nmeth910. [DOI] [PubMed] [Google Scholar]
- 21.The name given this peptide stems from the fact that the change of the negatively charged D to the positively charged H has the greatest effect on reducing the affinity for the antibody
- 22.Liu R, Shih TC, Deng X, et al. Design, synthesis, and application of OB2C combinatorial peptide and peptidomimetic libraries. Methods Mol Biol. 2015;1248:3–22. doi: 10.1007/978-1-4939-2020-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarkar M, Liu Y, Qi J, et al. Targeting stereotyped B cell receptors from chronic lymphocytic leukemia patients with synthetic antigen surrogates. The Journal of biological chemistry. 2016;291:7558. doi: 10.1074/jbc.M115.701656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doran TM, Gao Y, Simanski S, McEnaney P, Kodadek T. High affinity binding of conformationally constrained synthetic oligomers to an antigen-specific antibody: Discovery of a diagnostically useful synthetic ligand for murine Type 1 Diabetes autoantibodies. Bioorg Med Chem Lett. 2015;25:4910–4917. doi: 10.1016/j.bmcl.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar M, Liu Y, Morimoto J, et al. Recognition of antigen-specific B cell receptors chronic lymphocytic leukemia patients by synthetic “antigen surrogates”. Chem & Biol. 2014;111:1670–1679. doi: 10.1016/j.chembiol.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kodadek T, McEnaney PJ. Towards vast libraries of scaffold-diverse, conformationally constrained oligomers. Chemical communications (Cambridge, England) 2016;52(36):6038–6059. doi: 10.1039/c6cc00617e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickering JW, Martins TB, Schroder MC, Hill HR. Comparison of a multiplex flow cytometric assay with enzyme-linked immunosorbent assay for auantitation of antibodies to tetanus, diphtheria, and Haemophilus influenzae Type b. Clin Diagn Lab Immunol. 2002;9(4):872–876. doi: 10.1128/CDLI.9.4.872-876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anttila M, Voutilainen M, Jantti V, Eskola J, Kayhty H. Contribution of serotype-specific IgG concentration, IgG subclasses and relative antibody avidity to opsonophagocytic activity against Streptococcus pneumoniae. Clinical and experimental immunology. 1999;118(3):402–407. doi: 10.1046/j.1365-2249.1999.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krudysz-Amblo J, Parhami-Seren B, Butenas S, et al. Quantitation of anti-factor VIII antibodies in human plasma. Blood. 2009;113(11):2587–2594. doi: 10.1182/blood-2008-08-174987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doran TM, Morimoto J, Simanski S, McEnaney PJ, Kodadek T. Reliable diagnosis of murine type 1 diabetes using a panel of autoantigens and “antigen surrogates” mounted onto a liquid array. Molecular bioSystems. 2015;11(11):3156–3163. doi: 10.1039/c5mb00521c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


