Abstract
Functional characterization of adenylyl cyclase (AC) isoforms has proven challenging in mammalian cells because of the endogenous expression of multiple AC isoforms and the high background cAMP levels induced by nonselective AC activators. To simplify the characterization of individual transmembrane AC (mAC) isoforms, we generated a human embryonic kidney cell line 293 (HEK293) with low cAMP levels by knocking out two highly expressed ACs, AC3 and AC6, using CRISPR/Cas9 technology. Stable HEK293 cell lines lacking either AC6 (HEK-ACΔ6) or both AC3 and AC6 (HEK-ACΔ3/6) were generated. Knockout was confirmed genetically and by comparing cAMP responses of the knockout cells to the parental cell line. HEK-ACΔ6 and HEK-ACΔ3/6 cells revealed an 85% and 95% reduction in the forskolin-stimulated cAMP response, respectively. Forskolin- and Gαs-coupled receptor-induced activation was examined for the nine recombinant mAC isoforms in the HEK-ACΔ3/6 cells. Forskolin-mediated cAMP accumulation for AC1–6 and AC8 revealed 10- to 250-fold increases over the basal cAMP levels. All nine mAC isoforms, except AC8, also exhibited significantly higher cAMP levels than the control cells after Gαs-coupled receptor activation. Isoform-specific AC regulation by protein kinases and Ca2+/calmodulin was also recapitulated in the knockout cells. Furthermore, the utility of the HEK-ACΔ3/6 cell line was demonstrated by characterizing the activity of novel AC1 forskolin binding-site mutants. Hence, we have developed a HEK293 cell line deficient of endogenous AC3 and AC6 with low cAMP background levels for studies of cAMP signaling and AC isoform regulation.
Introduction
Adenylyl cyclases (ACs) are key transduction mediators of G protein–coupled receptor (GPCR) signaling by catalyzing the conversion of ATP into the secondary messenger, cyclic AMP (cAMP). Nine mammalian mACs (AC1–AC9) have been cloned, and one soluble AC (sAC) has also been characterized. Although mAC isoforms exhibit high structural homology, they are differentially regulated by G-protein subunits, Ca2+/calmodulin and protein kinases; depending on these regulatory properties, mACs are classified into four groups (reviewed in Dessauer et al., 2017). Group 1, comprising AC1, AC3, and AC8, is regulated by Ca2+/calmodulin. Group 2, which includes AC2, AC4, and AC7, is conditionally stimulated by Gβγ subunits. AC5 and AC6 belong to group 3, and both are inhibited by Ca2+. Lastly, AC9, the only mAC that is insensitive to the diterpene forskolin, is the sole member of group 4. These differential regulatory mechanisms of mAC isoforms, coupled with their tissue-specific expression patterns, correlate with knockout and overexpression studies that have implicated certain mACs with various physiologic processes (Sadana and Dessauer, 2009). Consequently, AC signaling dysfunction has emerged as a potential therapeutic target, and considerable efforts have been made to identify potent and isoform selective AC modulators (Pierre et al., 2009; Seifert et al., 2012; Dessauer et al., 2017).
Overexpression of mAC isoforms in various cellular models such as HEK293, COS-7, and Chinese hamster ovary cells, has allowed in vitro and in vivo characterization of AC activity (Mann et al., 2009; Brust et al., 2017). HEK293 is a human expression system that is widely used to study recombinant proteins because of the cells’ easy maintenance, rapid reproduction, and high transfection efficiency; however, mammalian cells usually express multiple AC isoforms, resulting in high background cAMP responses arising from these endogenous ACs. Therefore, cautious interpretation is required when evaluating Gαs-coupled receptor- and forskolin-mediated stimulation of individual mAC isoforms in these heterologous systems. Other approaches to examine mAC activity have used the Sf9 cell line from Spodoptera frugiperda, which expresses recombinant proteins in high yields and exhibits low cAMP levels (Schneider and Seifert, 2010). In vitro studies to assess AC activity for all nine membrane isoforms and to characterize mAC inhibitors’ activity and selectivity have been successfully performed in Sf9 cells (Pinto et al., 2008; Brand et al., 2013); however, Sf9 cells also express an endogenous AC that is stimulated by forskolin and can be inhibited by P-site AC inhibitors (Gille et al., 2004; Pinto et al., 2008). In addition, possible disadvantages of using these insect cells include the difficulty of monitoring in real-time the spatiotemporal dynamics of the cAMP pathway and the potential lack of expression of key upstream regulators or downstream effectors of AC isoforms.
To facilitate the selective characterization of AC isoforms’ activity and regulation, it was the objective of this study to generate an isolated mammalian cell model with low cAMP production after drug-stimulated conditions. The CRISPR/Cas 9 technology was used to knock out two abundant endogenous ACs (AC3 and AC6) in HEK293 cells. Our HEK-ACΔ3/6 cell line showed a markedly reduced response to forskolin and Gαs-coupled receptor agonists and demonstrated to be a suitable cellular model to evaluate the individual responses of the nine mammalian mAC isoforms to forskolin, GPCRs, and other regulators, such as Ca2+ and protein kinases. The HEK-ACΔ3/6 cells were also used to stably express representative AC isoforms for membrane-based in vitro assays. This new cellular model proved to be a valuable tool to functionally characterize the activity profiles of a series of AC1 forskolin-binding site mutants that were difficult to interpret in the parental HEK293 cells owing to the high cAMP background. This cellular model offers advantages for future structure-function and structural biology studies, as well as high- throughput screening endeavors to identify AC isoform-selective regulators.
Materials and Methods
Reagents.
The sgRNA oligos and primers for plasmid construction or qPCR analysis were synthesized by Integrated DNA Technologies (IDT) (Coralville, IA). Dulbecco’s modified Eagle’s medium, penicillin-streptomycin-amphotericin B solution (antibiotic-antimycotic), cell dissociation buffer, phosphate-buffered saline, and OptiMEM were purchased from Life Technologies (Carlsbad, CA). The HEK293 cell line was obtained from ATCC (Manassas, VA). Bovine calf serum, Fetal Clone I, and HEPES were obtained from Hyclone (GE Healthcare, Pittsburgh, PA). Lipofectamine 2000 was purchased from Invitrogen (Carlsbad, CA), and XtremeGENE HP was obtained from Roche (Basel, Switzerland). A23187, adenosine 5′-triphosphate (ATP) disodium salt hydrate, bovine serum albumin, 3-isobutyl-1-methylxanthine (IBMX), (−)-isoproterenol (+)-bitartrate salt, magnesium chloride (MgCl2) anhydrous, puromycin, and Tween 20 were purchased from Sigma-Aldrich (St. Louis, MO). The Pierce BCA protein assay kit was obtained from Thermo Fisher Scientific (Waltham, MA), and forskolin, phorbol 12-myristate 13-acetate (PMA), and prostaglandin E2 (PGE2) were from Tocris (Bio-Techne, Minneapolis, MN). The 384-well white opaque plates (cat. no. 6007680) were from PerkinElmer (Waltham, MA), and the cAMP detection reagents, HTRF cAMP Dynamic 2, were obtained from Cisbio (Bedford, MA). The QuickExtract DNA extraction solution was purchased from Epicentre Biotechnologies (Illumina, Madison, WI), and the BbsI restriction enzyme and Phusion high-fidelity DNA polymerase were from New England Biolabs (Ipswich, MA). The RNeasy Mini kit was manufactured by QIAGEN (Hilden, Germany), and the iScript cDNA synthesis kit and iTaq Universal SYBR Green Supermix were from Bio-Rad Laboratories (Hercules, CA).
Plasmids.
The CRISPR/Cas9 vector, pSpCas9(BB)-2A-Puro (PX459), was a gift from Feng Zhang (plasmid no. 62988; Addgene, Cambridge, MA). The plasmids encoding for human AC1, AC3, AC4, and AC7 genes were synthesized, codon optimized for mammalian expression, and cloned into a pcDNA3.1+ expression vector by Genscript Biotech (Piscataway, NJ). The pcDNA3.1+/hAC5, pcDNA3.1+/hAC6, pcDNA3.1+/hAC9, and pQE60/Gαs-His vectors were gifts from Carmen Dessauer. The pReceiver/hAC8 vector was purchased from GeneCopoeia (Rockville, MD), and the pcDNA3.1+/hAC2 plasmid was obtained from Genscript Biotech. The corresponding DNA mutations of the mutant AC1 constructs (pcDNA3.1+/AC1-W419A, pcDNA3.1+/AC1-V423G, pcDNA3.1+/AC1-N878A, and pcDNA3.1+/AC1-S924P) were incorporated into the pcDNA3.1+/hAC1 plasmid by Genscript Biotech. All the adenylyl cyclase genes encoded for untagged proteins with expression driven by a cytomegalovirus promoter.
Design and Construction of CRISPR/Cas9 Plasmids.
Two abundant ACs in HEK293 cells are AC3 and AC6 encoded by the ADCY3 and ADCY6 genes, respectively (Dessauer et al., 2017). To generate the HEK-ACΔ6 and HEK-ACΔ3/6 knockout cell lines, we used the CRISPR/Cas9 system developed by the Zhang laboratory in which the pSpCas9(BB)-2A-Puro (PX459) vector coexpresses the single guide RNA (sgRNA), the human-codon optimized Cas9 nuclease, and a puromycin resistance gene (Ran et al., 2013). The specific sgRNAs for each gene were selected from the CRISPR/Cas9 screening library designed by Wang et al. (2014), and the following were the targeting sequences for the ADCY3 and ADCY6 genes: 5′ GGGAGAAGACCAAGACTGGG 3′ and 5′ TGGGTGGCTCTGCATCCCGG 3′, respectively. To insert the customized sgRNAs into the PX459 vector, two oligos per sgRNA (ADCY3: 5′-CACC-GGGAGAAGACCAAGACTGGGG-3′ and 5′-AAAC-CCCCAGTCTTGGTCTTCTCCC-3′, ADCY6: 5′-CACC-GTGGGTGGCTCTGCATCCCGG-3′ and 5′-AAAC-CCGGGATGCAGAGCCACCCAC-3′) were synthesized by IDT, annealed according to the protocol by Ran et al. (2013), and cloned into the BbsI site of the PX459 backbone vector. Correct insertion of the sgRNAs was confirmed by Sanger sequencing.
Stable Knockout Cell Lines: Generation and Validation.
HEK293 cells were transfected with either the sgRNA for AC6 (HEK-ACΔ6) or both AC3 and AC6 sgRNAs (HEK-ACΔ3/6) using Lipofectamine 2000. After 24-hour transfection, the media were replaced with puromycin (2 mg/ml) containing media to select for sgRNA- and Cas9-expressing cells. Transfected cells were then seeded sparsely into 10-cm dishes after being exposed to puromycin for 72 hours. Cell colonies were allowed to form for 2–3 weeks in regular HEK293 media consisting of Dulbecco’s modified Eagle’s medium supplemented with 5% bovine calf serum, 5% fetal clone I, and 1% penicillin-streptomycin-amphotericin B solution. HEK-ACΔ6 and HEK-ACΔ3/6 cell clones were screened for loss of function of AC3 and AC6 genetically and by comparing cAMP responses of the knockout cells and the parental HEK293 cell line. For genotypic characterization of the clones, genomic DNA was extracted from the HEK-ACΔ6 and HEKAC3/AC6 KO clones with QuickExtract DNA extraction solution according to the manufacturer’s instructions. A portion of the ADCY3 and ADCY6 genes around the sgRNA site was amplified by polymerase chain reaction (PCR) from the genomic DNA with the following AC3 and AC6 set of primers (ADCY3: 5′-GTTAAAGCCCGTCTAGTATTG-3′ and 5′-CATCAGTCGACCACACGTCG-3′, ADCY6: 5′-ATGTCATGGTTTAGTGGCCTCC-3′ and 5′-CGTGTAGGCGATGTAGACAA-3′). The PCR conditions were as follows: after initial denaturation at 98°C for 30 seconds, 30 cycles were performed of denaturation at 98°C for 10 seconds, annealing at 52°C for 30 seconds, and extension at 72°C for 30 seconds, followed by a final extension at 72°C for 10 minutes. The PCR products were analyzed by agarose gel electrophoresis and by Sanger sequencing. The resulting gene mutation for the isolated HEK-ACΔ6 cell line was a single base pair insertion in both alleles of the ADCY6 gene that caused a frame shift and subsequent disruption of ADCY6 protein expression (Supplemental Fig. 2b). For the isolated HEK-ACΔ3/6 cell line, ADCY3 expression was disrupted by a single base pair deletion for one allele and 2-bp insertion for the other allele, whereas loss of function of ADCY6 was a result of a single base pair deletion in both alleles of the gene (Supplemental Fig. 2c). Phenotypic changes in the HEK-ACΔ6 and HEK-ACΔ3/6 cell lines were assessed by comparing the forskolin- and Gαs-coupled receptor-stimulated cAMP responses in the knockout cells and the parental HEK293 cell line.
RT-qPCR.
Total RNA of the HEK293, HEK-ACΔ6 or HEK-ACΔ3/6 cells was isolated using the RNeasy Mini kit according to the manufacturer’s protocol, and the first-strand cDNA was synthesized from 500 ng of total RNA using the iScript cDNA synthesis kit. The RT-qPCR reactions were performed in 384-well plates using iTaq Universal SYBR Green Supermix detection reagent and 10 ng of cDNA template. Primers for the nine mACs were designed using PrimerQUEST (IDT), and their specificity was checked with PrimerBLAST (Ye et al., 2012). The primer sequences for the three housekeeping genes (Actin, GAPDH, and 18S_rRNA) were selected from previously described reports in the literature. The forward and reverse primer sequences used for each corresponding gene are listed in Supplemental Table 1. Three biologic samples were analyzed per cell line, and each primer set was run in duplicate. The RT-qPCR protocol was as follows: after initial activation at 95°C for 10 minutes, 40 cycles were completed of denaturation at 95°C for 15 seconds, primer annealing at 60°C for 1 minute, and extension at 72°C for 1 minute, followed by a final melting curve step. Ct values of target genes were normalized to the Ct value of the housekeeping gene 18S_rRNA (ΔCt: Ct [Target gene] – Ct [18S_rRNA]), and fold change in mRNA levels relative to the parental cell line was calculated for each target gene as follows: 2^-(ΔCt [knockout cell line] – ΔCt [HEK293]). The qPCR products from the total RNA isolated from the HEK293 cells were run in a 2% agarose gel.
cAMP Assays in Cells.
Parental HEK293, HEK-ACΔ6, and HEK-ACΔ3/6 cells were washed with phosphate-buffered saline, dissociated from the culture plate with cell dissociation buffer, and centrifuged twice at 150g for 5 minutes. The cell pellet was resuspended in Opti-MEM, and cells were counted and seeded in a white opaque 384-well plate at a cell density of 15,000 cells/well for the functional characterization of the knockout cells and 10,000 cells/well for the experiments with the transient transfected HEK-ACΔ3/6 cells. Cells were incubated for 2 hours at 37°C with 5% CO2 before the drug treatments to ensure that cells had adhered to the plate. HEK293, HEK-ACΔ6, HEK-ACΔ3/6, or transiently transfected HEK293 and HEK-ACΔ3/6 cells were treated with forskolin for 1 hour; the EP2R agonist, PGE2, for 15 minutes; or the β2AR agonist, isoproterenol, for 5 minutes. For Ca2+- and protein kinase C (PKC)-mediated AC activation and the synergistic experiments, cells were stimulated for 1 hour with 40 μM (4×) A23187 or 4 μM (4×) PMA in the absence or presence of 40 μM (4×) PGE2 or 200 μM (4×) forskolin respectively. To evaluate Ca2+ inhibition of AC5 and AC6 activity, transfected cells were treated with 40 μM (4×) forskolin, followed by vehicle or 40 μM (4×) A23187 for 1-hour incubation. AC activity was triggered in HEK293 and HEK-ACΔ3/6 cells transiently expressing wild-type AC1 (AC1-WT) or mutant AC1 (AC1-W419A, AC1-V423G, AC1-N878A, and AC1-S924P) cells with increasing concentrations of forskolin in the presence or absence of 12 μM (4×) A23187 for a period of 1 hour. All the drug treatments were performed in the presence of the phosphodiesterase inhibitor, IBMX, at a final concentration of 500 μM. After the corresponding incubation periods, Cisbio HTRF cAMP detection reagents, d2-labeled cAMP, and anti-cAMP cryptate were added to all wells, and time-resolved fluorescence was measured at dual-emission wavelengths (620 and 665 nm) after 1-hour incubation at room temperature. Cyclic AMP accumulation per well was calculated using GraphPad Prism (San Diego, CA) by interpolating the 620/665 signal ratios from a cAMP standard curve ran in parallel.
ACs Transient Transfections.
HEK-ACΔ3/6 cells were plated overnight before transfection in a 12-well plate. The cells were then transiently transfected with Lipofectamine 2000 in a 1:2 (DNA:Lipofectamine) ratio or with XtremeGENE HP in a 1:1.5 or a 1:3 (DNA:XtremeGENE HP) ratio according to the manufacturer’s protocol. Lipofectamine 2000 was used to transfect AC1-AC6 and AC8 for the forskolin, PKC, and Ca2+ inhibition experiments. XtremeGENE HP was used in a 1:1.5 ratio to transfect all mAC isoforms for the Ca2+- and Gαs-mediated AC activation assays, and it was used in a 1:3 ratio to transfect AC7 and AC9 in the forskolin experiments. After transfection, cells were cultured for 40 hours before cAMP responses were measured as described already herein.
AC1, AC2, AC5, and AC8 Stable Cell Pool Generation.
HEK-ACΔ3/6 cells were seeded overnight in a six-well plate to ensure that at the time of transfection, the cells had recovered and had reached 70% confluency. XtremeGENE was used in a 1:3 ratio (DNA:XtremeGENE) to transfect the pcDNA3.1+/hAC1, pcDNA3.1+/hAC2, pcDNA3.1+/hAC5, and pReceiver/hAC8 constructs. Media was replaced after 48-hour transfection to G418 (600 μg/ml) or puromycin (4 μg/ml) containing media for the cells transfected with the pcDNA3.1 or the pReceiver expression vectors, respectively. The HEK-ACΔ3/6-AC1, HEK-ACΔ3/6-AC2, and HEK-ACΔ3/6-AC5 pooled cell lines were maintained and proliferated for cellular membrane assays in media containing G418 (300 μg/ml), and the HEK-ACΔ3/6-AC8 pooled cell line was maintained in media containing puromycin (2 μg/ml).
Isolation of Cellular Membranes.
HEK-ACΔ3/6, HEK-ACΔ3/6-AC1, HEK-ACΔ3/6-AC2, HEK-ACΔ3/6-AC5, and HEK-ACΔ3/6-AC8 cells were proliferated in 15-cm cell culture dishes to a 90% confluency. Media were decanted and replaced with 10 ml of ice-cold lysis buffer containing 1 mM HEPES, 2 mM EDTA, and 1 mM EGTA at pH 7.4. The cell culture dish was incubated for 10 minutes with the lysis buffer on ice, and cells were scraped off using a disposable cell lifter. The cell suspension was transferred to a high-speed centrifuge tube and centrifuged at 30,000g for 20 minutes at 4°C. Supernatant was discarded, and the cell pellet was resuspended in binding buffer containing 4 mM MgCl2 and 50 mM Tris at pH 7.4. After the cell suspension was homogenized for 5 seconds using a Kinematica (Lucerne, Switzerland) homogenizer, it was divided into 1 ml-aliquots that were centrifuged at 10,000g for 10 minutes at 4°C. Supernatant was discarded, and membrane pellets were frozen and stored at −80°C until the day of the assay.
Gαs Purification and Activation.
Recombinant Gαs-His protein was expressed and purified using immobilized metal affinity and ion-exchange chromatography as described previously (Lee et al., 1994) and stored at −80°C until it was activated for membrane assays. The purified Gαs protein was activated by incubation on ice in 50 mM HEPES, 2 mM DTT, 250 μM GTPγS, and 1 mM MgCl2 for 20 minutes, followed by a 30-minute incubation at 30°C (Chen-Goodspeed et al., 2005).
cAMP Assays in Membranes.
Cells membranes were thawed on ice and resuspended in washing buffer containing 2 mM Tris, 1 mM EGTA, and 1 mg/ml bovine serum albumin. Membrane suspension was centrifuged at 10,000g for 10 minutes at 4°C; supernatant was aspirated, and the washing step was repeated two more times. After the final wash, the membrane pellet was resuspended in membrane buffer containing 33 mM HEPES, 0.1% Tween 20, and 1 mM EGTA. Protein concentration of the membrane suspension was measured using the Pierce BCA Protein Assay kit, and the protein concentration was adjusted to 250 μg/ml. Membrane suspensions were plated in a white opaque 384-well plate at 10 μl/well followed by the addition of increasing concentrations of Gαs or forskolin—prepared in membrane buffer without EGTA—and stimulation buffer containing 33 mM HEPES, 0.1% Tween 20, 2.5 mM MgCl2, 250 μM ATP, and 500 μM IBMX at pH 7.4. Cellular membranes were incubated with the stimulants for 45 minutes at room temperature, and cAMP accumulation was measured using the Cisbio HTRF cAMP detection kit as described already herein for the cAMP assays in cells.
Results
CRISPR/Cas9-Based HEK-ACΔ6 and HEK-ACΔ3/6 Cell Lines Display Reduced cAMP Levels in Response to Forskolin or Gαs Agonists.
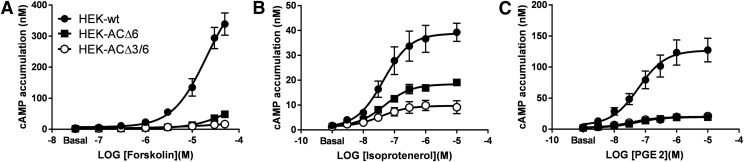
HEK293 cells express multiple AC isoforms that, when stimulated by forskolin and/or endogenous Gαs-coupled receptors, promote high levels of cAMP. It has been reported, based on qPCR studies, along with our own qPCR results (Supplemental Fig. 1a), that HEK293 cells express AC1, AC2, AC3, AC5, AC6, AC7, and AC9 mRNA (Ludwig and Seuwen, 2002; Atwood et al., 2011). Preliminary and published functional studies further suggest that AC3 and AC6 are important for drug-stimulated cAMP accumulation in HEK293 cells (Yu et al., 2014). Therefore, to decrease cAMP levels in HEK293 cells, we cloned sgRNA sequences that targeted two abundant endogenous ACs (AC3 and AC6) into a CRISPR/Cas9 expression system. HEK-ACΔ6 and HEK-ACΔ3/6 cell lines were generated lacking either AC6, or both AC3 and AC6, respectively. We characterized the genotype of the knockout cells (Supplemental Fig. 2) and determined loss of function of the ADCY3(AC3) and ADCY6(AC6) genes by comparing the drug-induced cAMP responses of the knockout cells with the parental cell line. The activity of the endogenous ACs was stimulated by two common mechanisms: direct AC activation with forskolin (Fig. 1A) or Gαs subunit–mediated activation elicited by the β2AR agonist, isoproterenol (Fig. 1B), or the EP2R agonist, PGE2 (Fig. 1C). Knocking out the ADCY6 gene in the HEK-ACΔ6 cell line reduced the cAMP response to forskolin by nearly 85% and by an additional 10% when AC3 expression was also disrupted, resulting in an overall reduction of cAMP accumulation by 95% in the HEK-ACΔ3/6 cells compared with the parental cell line (Fig. 1A). Likewise, HEK-ACΔ6 cells exhibited 50% of the maximal response to isoproterenol (Fig. 1B), whereas the HEK-ACΔ3/6 cells displayed >75% lower cAMP accumulation than the HEK293 parental cell line. In the case of the EP2R-mediated activation, HEK-ACΔ6 cells showed >85% reduction of the maximal response to PGE2, but no significant differences in PGE2-stimulated cAMP levels were noted between the HEK-ACΔ6 and HEK-ACΔ3/6 cells (Fig. 1C). No major changes in the relative mRNA levels of the mACs were observed in the HEK-ACΔ6 and HEK-ACΔ3/6 cell lines compared with parental HEK293 cells, except for a reduction of AC2 mRNA levels in both knockout cell lines (Supplemental Fig. 1b).
Fig. 1.
Functional characterization of HEK-ACΔ6 and HEK-ACΔ3/6 cell lines. Parental HEK293 cells and the CRISPR/Cas9 based HEK-ACΔ6 and HEK-ACΔ3/6 cells were incubated at room temperature with increasing concentrations of the direct AC activator, forskolin (A), or the Gαs-coupled receptors agonists isoproterenol (B) or PGE2 (C), and cAMP accumulation was measured. *HEK-ACΔ6 and HEK-ACΔ3/6 responses to PGE2 overlap in (C). Data represent the mean and S.E.M. of three to four independent experiments conducted in duplicate.
Forskolin-Mediated Responses of mAC Isoforms in Knockout Cells.
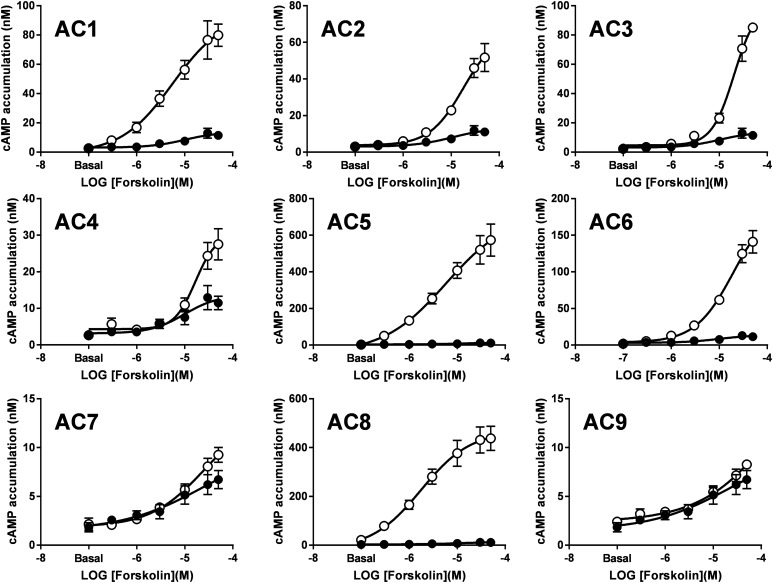
Forskolin directly binds to a hydrophobic pocket at the interface between the AC catalytic domains, and it elicits stimulatory effects on ACs 1 to 8 by promoting a conformational change that facilitates cAMP synthesis (Tesmer et al., 1997). Hence, we used our CRISPR/Cas9-based cell line to selectively assess forskolin-mediated responses in the nine mAC isoforms. Cyclic AMP levels were evaluated at increasing concentrations of forskolin in the HEK-ACΔ3/6 cell line transfected with control vector (Venus) or the individual mAC isoforms (Fig. 2). Forskolin induced cAMP production at a wide range of efficacies in the knockout cells expressing ACs, except for those transfected with AC7 and AC9. These forskolin-mediated responses ranged from 10- to a 250-fold increase over the basal cAMP levels for cells expressing AC1–AC6 and AC8 compared with a 4-fold increase over basal of the Venus-transfected cells. AC5- and AC8-expressing cells displayed the most robust cAMP response to forskolin relative to the other isoforms, whereas cells expressing AC9 were unresponsive to forskolin, as previously described (Yan et al., 1998). Lack of selectivity of commercially available mAC antibodies or epitope tags on the constructs used here prevented the assessment of protein expression for a more rigorous comparison of forskolin-mediated cAMP responses across the other AC isoforms.
Fig. 2.
Forskolin-stimulated cAMP responses for the nine mACs in the HEK-ACΔ3/6 cell line. HEK-ACΔ3/6 cells were transiently transfected with Venus (solid circles) or the individual mAC isoforms (open circles) as indicated. After 40 hours, cAMP accumulation was stimulated with increasing concentrations of forskolin at room temperature. Data represent the mean and S.E.M. of three to four independent experiments conducted in duplicate.
Gαs-Coupled Receptor Signaling through mACs.
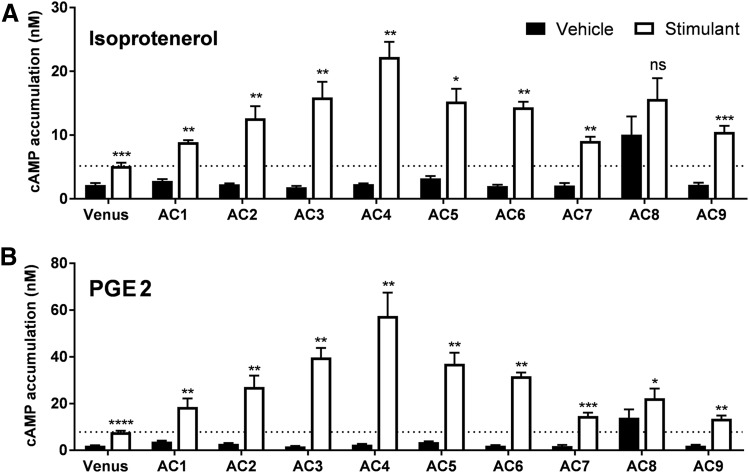
Activation of GPCRs linked to Gαs is another major mechanism to activate mACs. Therefore, we evaluated the specific mAC isoform responses after activation of the endogenously expressed β2-adrenergic receptor (β2AR) and prostaglandin EP2 receptor (EP2R) (Andressen et al., 2006; Atwood et al., 2011). HEK-ACΔ3/6 cells transiently transfected with the different mACs were treated with a saturating concentration of isoproterenol (Fig. 3A) or PGE2 (Fig. 3B). Activation of both Gαs-coupled receptors triggered a significant increase in cAMP levels above the basal response in all AC isoforms, except for AC8. AC8-expressing cells displayed high basal and forskolin-mediated activity, but cAMP levels did not increase significantly upon Gαs stimulation. Surprisingly, cells expressing AC4 showed the highest cAMP response to Gαs-coupled receptor activation, especially considering that this isoform was poorly activated by forskolin (Fig. 2). Knockout cells transfected with AC7 and AC9 also showed a 2-fold increase over Venus-transfected cells when stimulated by Gαs-coupled receptors. This finding suggests that both isoforms are being expressed, and the lack of response of these isoforms to forskolin activation may reflect their unique regulatory properties (Hacker et al., 1998; Yan et al., 2001).
Fig. 3.
Gαs-coupled receptor-mediated activation of the nine mACs in the HEK-ACΔ3/6 cell line. HEK-ACΔ3/6 cells were transiently transfected with Venus or the individual mAC isoforms as indicated. After 40 hours, cAMP accumulation was stimulated with 10 μM isoproterenol (A) or 10 μM PGE2 (B). Data represent the mean and S.E.M. of three to four independent experiments conducted in duplicate. Statistical analysis was performed using paired t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 compared with the cAMP levels of the vehicle-treated cells.
Selective Regulation of mAC Isoforms in the CRISPR/Cas9-Based Cell Line.
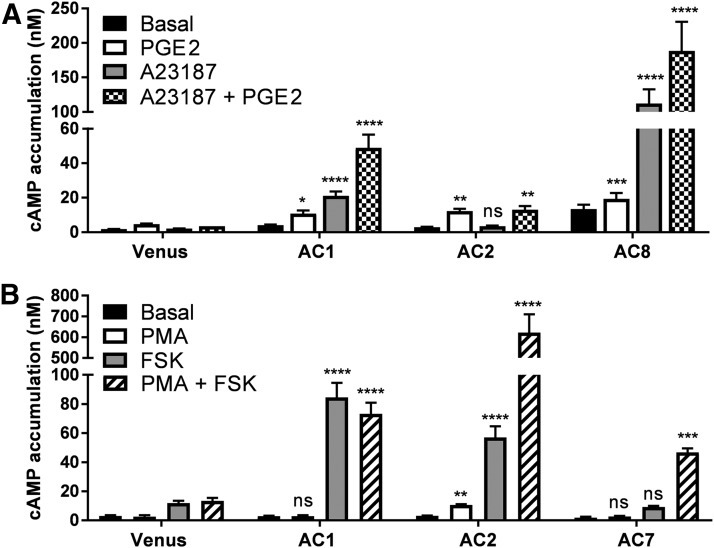
In addition to activation by forskolin and Gαs-proteins, mAC isoforms are selectively regulated by Ca2+ or protein kinases. Thus, we further explored AC1/AC8- and AC5/AC6- selective regulation by Ca2+, as well as, activation of AC2/AC7 by PKC in the HEK-ACΔ3/6 cells (Fig. 4; Supplemental Fig. 3). First, we examined the response of Ca2+/calmodulin-stimulated cyclases, AC1 and AC8, in the HEK-ACΔ3/6 cell line (Fig. 4A). An increase in intracellular Ca2+ with the calcium ionophore, A23187, elevated cAMP levels 5-fold over basal levels in AC1-expressing cells and by 8-fold in AC8-expressing cells, whereas Venus (mock) or AC2 (negative control) transfected cells failed to respond to Ca+2 influx. Additionally, we assessed whether Ca2+-induced AC1 and AC8 activation was differentially modulated by simultaneous activation with Gαs. Stimulation with A23187 and PGE2 (Gαs) elicited an apparent synergistic effect on HEK-ACΔ3/6 cells expressing AC1 since the cAMP response was greater for the combination of Ca2+ and Gαs stimulation than to the additive response to both drugs. Instead, AC8-expressing knockout cells were not robustly stimulated by Gαs, and no synergistic effect was observed when intracellular Ca2+ was elevated after Gαs activation. In contrast to AC1 and AC8, cAMP accumulation in response to forskolin of AC5- and AC6-expressing cells was decreased by 34% and 24%, respectively, when cells were cotreated with A23187 (Supplemental Fig. 3).
Fig. 4.
Ca+2 and PKC regulation of AC isoforms in the HEK-ACΔ3/6 cell line. (A) HEK-ACΔ3/6 cells were transiently transfected with the Venus control plasmid, AC1, AC2, or AC8. Cells were incubated with 10 μM A23187, 10 μM PGE2, or a combination of both, as indicated, and cAMP accumulation was measured. (B) Venus, AC1, AC2, or AC7-transfected HEK-ACΔ3/6 cells were incubated with 1 μM PMA, 50 μM forskolin, or a combination of both, as indicated, and cAMP accumulation was measured. Data represent the mean and S.E.M. of three to four independent experiments conducted in duplicate. Statistical analysis was performed using one-way ANOVA, followed by a Dunnett’s comparison. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 compared AC-transfected cAMP responses to the responses of the Venus-transfected cells for each stimulation condition.
Protein kinases are another set of important regulators of mACs. Particularly, activation of PKC with the phorbol ester PMA directly stimulates AC2 and AC7 activity (Jacobowitz and Iyengar, 1994; Harry et al., 1997). To determine whether AC2 and AC7 expression will give rise to PKC-stimulated cAMP accumulation in the HEK-ACΔ3/6 cells, transiently transfected cells were incubated with PMA (Fig. 4B). In the presence of the phorbol ester, AC2-expressing cells showed a 3-fold increase over basal levels. In contrast, cells expressing AC7 (or AC1) showed no significant response to PMA; however, when AC2 or AC7 activity was stimulated with PMA and forskolin, both isoforms appeared to exhibit a robust synergistic response. In the case of AC7, the PMA and forskolin combination was able to induce a cAMP signal that was significantly greater than control cells, despite an absent response to forskolin or PMA alone.
AC Activity in Cellular Membranes.
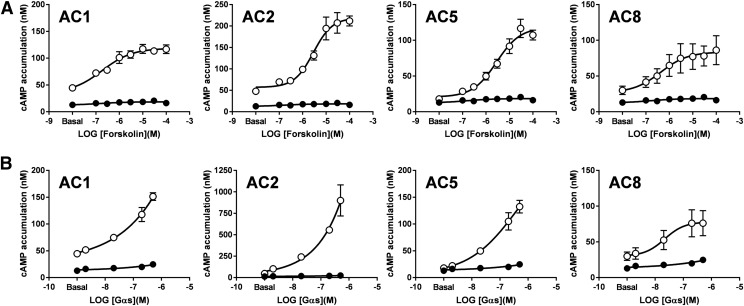
In vitro studies of native or overexpressed mACs in membrane fractions of insect or mammalian cells have enabled the study of AC activity in a controlled environment, providing valuable information about their pharmacology, regulatory mechanisms, and kinetic properties. Consequently, HEK-ACΔ3/6 pooled cell lines were generated that stably express AC1, AC2, AC5, or AC8 to isolate cellular membranes for in vitro studies. Forskolin- and Gαs-mediated cAMP accumulation was examined on membrane preparations in the presence of the metal ion cofactor Mg2+ and the substrate ATP (Fig. 5). An increase over basal catalytic activity was observed on cellular membranes overexpressing mACs with increasing concentrations of forskolin or purified Gαs, and these cAMP responses were significantly greater than the cAMP levels observed on the isolated membranes from untransfected HEK-ACΔ3/6 cells. Membranes isolated from HEK-ACΔ3/6-AC5 cells exhibited a markedly increased cAMP response to forskolin. Purified Gαs appeared to be more efficacious than forskolin in stimulating AC2 activity.
Fig. 5.
Forskolin and Gαs-induced cAMP responses of isolated membranes from HEK-ACΔ3/6 cells overexpressing mACs. Membranes isolated from HEK-ACΔ3/6 cells (solid circles) or HEK-ACΔ3/6 cells stably expressing AC1, AC2, AC5, or AC8 (open circles) were stimulated with increasing concentrations of (A) forskolin or (B) purified Gαs, and cAMP accumulation was measured. Data represent the mean and S.E.M. of three independent experiments conducted in duplicate.
Mutant AC1 Constructs Show Different AC Responses in HEK293 and HEK-ACΔ3/6 Cells.
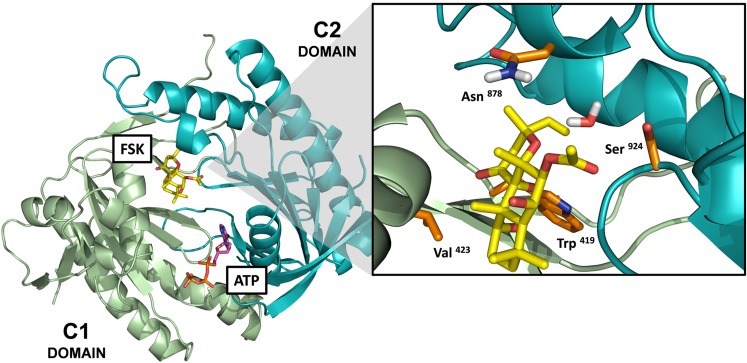
To further demonstrate that our HEK-ACΔ3/6 cell line is an improved cellular model to study ACs, the cAMP responses of four mutant AC1 constructs (AC1-W419A, AC1-V423G, AC1-N878A, and AC1-S924P) were examined in the HEK-ACΔ3/6 and HEK293 cells. Based on the crystal structure of the C1(AC5) and C2(AC2) catalytic domains in complex with Gαs and forskolin (Tesmer et al., 1997), the AC1 constructs were designed to incorporate single amino acid mutations at the forskolin-binding site. The mutations introduced in the AC1-W419A, AC1-V423G, and AC1-S924P constructs were intended to perturb the forskolin-binding pocket and impair the response to forskolin, taking into consideration that all three residues are conserved across forskolin-sensitive isoforms, and Trp419 and Val423 (AC1 numbering) in the C1 domain directly interact with the diterpene, and Ser924 (AC1 numbering) in the C2 domain creates a bridge with forskolin and a nearby water molecule (Tesmer et al., 1997). Instead, the Asn878 residue is located within the forskolin binding site, but it is not conserved, and the mutation incorporated into the AC1-N878A construct was designed to resemble the forskolin-binding pocket of the other Ca2+/calmodulin-stimulated cyclase, AC8. Figure 6 shows a structural model of the AC1 catalytic domains with the location of the mutated residues within the forskolin-binding site.
Fig. 6.
Location of mutated residues on AC1 constructs within the forskolin-binding site. Model of the catalytic domains of AC1 based on the crystal structure of the C1(AC5)-C2(AC2) catalytic domains (PDB: 1AZS) (Tesmer et al., 1997). The model includes the ATP and forskolin (FSK) molecules bound at their pseudosymmetric binding sites within the interface of the C1-C2 domains. The enlarge panel corresponds to the forskolin-binding site, and the side chains of the mutated amino acids are represented in orange sticks. The amino acid numbering corresponds to the numbering of the full amino acid sequence for human AC1. *In the 1AZS crystal structure, the Asn878 residue corresponds to Lys897 on the C2 domain of AC2.
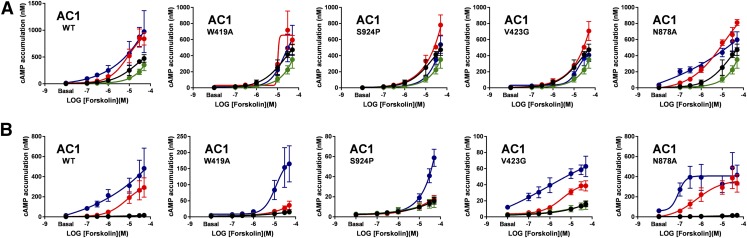
AC activity triggered by forskolin, A23187, or a combination of both was examined on transiently transfected HEK293 and HEK-ACΔ3/6 cells expressing Venus, AC1-WT, and the mutant AC1 constructs (Fig. 7). As expected, Venus-transfected HEK293 cells displayed greater cAMP levels to forskolin-induced activation than the HEK-ACΔ3/6 cells. In the presence of the calcium ionophore A23187 the forskolin response of the HEK293 cells was reduced compared with the responses mediated by forskolin alone, but no difference was observed on forskolin activation of Venus-transfected HEK-ACΔ3/6 cells in the presence of A23187. At the highest concentration of forskolin (50 μM), AC1-WT and Venus-transfected HEK293 cells showed a 3-fold difference in cAMP levels compared with a 20-fold difference between HEK-ACΔ3/6 cells expressing AC1-WT and Venus. Although both cell models revealed A23187 potentiation of forskolin-stimulated AC1 activity at low forskolin concentrations, the A23187-mediated potentiation in the AC1-expressing HEK-ACΔ3/6 cells was evident for all forskolin concentrations.
Fig. 7.
Cyclic AMP responses to forskolin and A23187 of AC1 mutants expressed in HEK293 and HEK-ACΔ3/6 cells. HEK293 (A) or HEK-ACΔ3/6 cells (B) were transiently transfected with Venus, AC1-wt, or an AC1 mutant construct, as indicated. After 40 hours, cAMP accumulation was stimulated with increasing concentrations of forskolin in the absence (red) or presence (blue) of 3 μM A23187. Black and green circles represent the forskolin response of Venus-transfected cells in the presence or absence of A23187, respectively. Data represent the mean and S.E.M. of three independent experiments conducted in duplicate.
In the HEK293 cells, AC1-W419A and AC1-S924P mutants appeared to be functionally inactive because neither forskolin nor A23187, or a combination of both, appeared to have an effect on cAMP accumulation. The AC1-V423G mutant seemed to be insensitive to forskolin but was responsive to A23187 (Supplemental Fig. 4). Surprisingly, the effects of forskolin and A23187 on the AC1-N878A mutant were substantially greater than those observed for AC1-WT in the HEK293 cells. As anticipated, expression of the AC1 mutants in the HEK-ACΔ3/6 cells provided more distinct and observable pharmacologic profiles in response to the stimulants. The AC1-W419A and AC1-S924P mutants showed little to no response when stimulated with forskolin or A23187, but the combination of the two reagents was able to induce a distinguishable cAMP signal that was significantly higher than the control cells. In the case of the AC1-V423G mutant, expression in the HEK-ACΔ3/6 cellular model revealed that this mutant was responsive to forskolin, A23187, and a combination of both agents, similar to AC1-WT. The extent of AC activation in response to forskolin, however, was vastly reduced in the AC1-V423G mutant in comparison with AC1-WT. Although a stimulant-induced cAMP response was observed for the AC1-N878A mutant in the HEK293 cells, the forskolin dose-response curves in the presence or absence of A23187 for AC1-N878A were shifted to the left in the HEK-ACΔ3/6 cells.
Different cAMP responses were observed for AC1-WT and the AC1 forskolin-binding site mutants in response to A23187, but these distinct responses were the same when the AC1 constructs were expressed in either the parental HEK293 or the HEK-ACΔ3/6 cells (Supplemental Fig. 4).
Discussion
We used CRISPR/Cas9 technology to disrupt the endogenous expression of AC3 and AC6 to develop a HEK293 cell line with a minimal background level of drug-stimulated cAMP. Functional characterization of the HEK-ACΔ6 and HEK-ACΔ3/6 cell lines revealed that the cAMP responses to forskolin and Gαs-coupled receptor activation were mediated predominantly by AC6 in HEK293 cells. Differences in the cAMP responses elicited by β2AR and EP2R activation of these knockout cells also suggested that endogenous AC3 and AC6 contribute to a different extent to the overall cAMP accumulation in HEK293 cells. Work with recombinant D1 and D5 dopamine receptors using AC-directed siRNAs also suggested that multiple ACs (i.e., AC3, AC5, AC6, AC7, and AC9) contribute differentially to dopamine agonist–stimulated cAMP accumulation in HEK293 cells (Yu et al., 2014). Although it has been determined in vitro that Gαs activates all mAC isoforms (reviewed in Simonds, 1999), cellular localization of the receptor and/or the AC presumably enable specific GPCR-AC coupling (Yu et al., 2014; Eichel and von Zastrow, 2018; Johnstone et al., 2018).
Forskolin has been a valuable tool for studying the functional roles of ACs and cAMP in various biologic processes. Despite the lack of tools to quantitate AC expression in cell models accurately, a few general conclusions can be made. The increased potency of forskolin observed in the AC1-expressing HEK-ACΔ3/6 membranes relative to AC2, AC5, and AC8 is consistent with previous studies of forskolin and forskolin analogs on Sf9 membranes (Pinto et al., 2008). Furthermore, no major differences in the efficacy of forskolin to activate AC1, AC2, and AC5 were observed in the Sf9 membranes (Pinto et al., 2008) or our knockout cell membranes, suggesting that the increased potency on AC1 agrees with the higher binding affinity of forskolin for AC1, as demonstrated by photoaffinity assays with an iodinated forskolin derivative in Sf9 membranes (Sutkowski et al., 1994). Also, the overall fold-change response to forskolin over the basal cAMP levels was significantly lower in the isolated membranes than in cells, supporting the proposed contribution of endogenous Gαs activity on forskolin-mediated AC activation (Insel and Ostrom, 2003). In addition, the synergistic effects of forskolin and Gαs and the observation that the forskolin-binding affinity for AC2, AC5, and AC6 increases in the presence of Gαs also support the larger responses observed in intact cells (Sutkowski et al., 1994; Alewijnse et al., 1997).
Isolated membrane fractions from cells overexpressing AC isoforms have also been a key tool in the analysis of AC function and regulation. These in vitro assays are a more direct approach to discern the effects of forskolin, Gαs, and other modulators on AC activity, particularly when evaluating the AC responses to Gαs, given that Gαs-stimulation in cell-based assays is contingent on GPCR expression levels, agonist sensitivity, and GPCR-AC subcellular localization (Ostrom and Insel, 2004). For example, all membrane-bound AC isoforms except AC8 were stimulated by Gαs-coupled receptor activation in HEK-ACΔ3/6 cells. The absence of an AC8 response in HEK293 cells is consistent with previous studies in intact cells (Nielsen et al., 1996); however, this result is not readily explained by the inability of Gαs to stimulate AC8 because in vitro studies with AC8-expressing HEK-ACΔ3/6 membrane preparations showed a robust response. AC2-expressing HEK-ACΔ3/6 membranes showed a significantly greater response to purified Gαs than to forskolin, as it was previously reported in Sf9 membranes (Sutkowski et al., 1994), whereas in the HEK-ACΔ3/6 cells overexpressing AC2, the fold-change over the basal response to forskolin stimulation was greater than to Gαs-coupled receptor activation. In conclusion, the ability to use the HEK-ACΔ3/6 cell line for both in vitro and cell-based assays provided insightful information about distinct AC isoform responses to general AC activators and demonstrated that our knockout cell line is an adequate system for studying mACs.
Because our newly developed HEK-ACΔ3/6 cell line is derived from mammalian cells, it also offers improvements over previously used Sf9 models. Significant differences in cholesterol content and glycosylation patterns are observed between mammalian and insect cells. The cholesterol:phospholipid ratio is more than an order of magnitude lower in membranes isolated from Sf9 compared with mammalian cells (Gimpl et al., 1995). Importantly, AC isoforms 1, 3, 5, 6, and 8 are found in cholesterol-rich lipid rafts, and the regulation of several isoforms is directly affected by localization into these domains (Bogard et al., 2011). For example, removal of membrane cholesterol with methyl-β-cyclodextrin relieves inhibition of AC6 by Ca2+ entry (Fagan et al., 1996). Furthermore, cholesterol depletion disrupts Ca2+-mediated stimulation of AC8 in HEK293 cells (Smith et al., 2002). Glycosylation patterns arising from proteins produced in mammalian cells are often quite different from those observed in insect cells (reviewed in Betenbaugh et al., 2004). Glycosylation has been shown to regulate the activity of AC6 and AC8. Mutation of glycosylation sites N805 and N890 in AC6 resulted in decreased stimulation by forskolin and decreased inhibition by PKC and D2 receptor signaling without affecting plasma-membrane localization (Wu et al., 2001). Additionally, glycosylation of AC8 is required for proper localization into cholesterol-rich membrane regions (Pagano et al., 2009). Taken together, membranes harvested from our newly developed cell line have advantages over Sf9 membranes regarding cholesterol content and glycosylation, both of which can regulate AC function in vitro. Furthermore, mammalian expression systems have become attractive for production of recombinant proteins (reviewed in Bandaranayake and Almo, 2014); thus, this cell line can offer a platform to screen recombinant AC expression and activity rapidly using forskolin-stimulated cAMP production.
Selective regulatory properties of AC isoforms were also recapitulated in the HEK-ACΔ3/6 cells. Consistent with previous reports in the literature, Ca2+ influx stimulated AC1 and AC8 and inhibited forskolin-stimulated activity of AC5 and AC6 (Tang et al., 1991; Katsushika et al., 1992; Cali et al., 1994). Likewise, our results with the AC1- and AC8-expressing cells for cotreatments with the calcium ionophore, A23187, and PGE2 were also in agreement with previous reports in intact cells, where only AC1 displayed an apparent synergistic response to Ca2+ stimulation in the presence of activated Gαs (Nielsen et al., 1996; Cumbay and Watts, 2001). By taking advantage of the unique regulatory properties of each mAC isoform, we ultimately demonstrated specific mechanisms to activate selectively all nine mACs with an improved signal window. We were also able to unmask subtle cAMP responses for AC isoforms, such as AC7 or AC9, that failed to display a robust cAMP response to various stimulatory mechanisms in the HEK-ACΔ3/6 cell model.
By disrupting the expression of AC3 and AC6 in HEK293 cells, enhancement of the signal window upon stimulation of AC activity in the knockout cells enabled AC responses that previously were masked by the high background cAMP levels. The cAMP responses detected for the previously uncharacterized AC1 constructs containing mutations within the forskolin-binding pocket were significantly different when these constructs were expressed in HEK293 or HEK-ACΔ3/6 cells. Mutation of the Trp419 or Ser924 residues were predicted to disrupt forskolin activity based on the binding interactions revealed by the crystal structure of the mAC catalytic domains (Tesmer et al., 1997). This appeared to be true in HEK293 cells, but in the HEK-ACΔ3/6 cells forskolin in combination with A23187 induced a distinct increase of cAMP, demonstrating only a partial loss of function. The analogous mutation on Ser942 of AC2 (AC2-S942P) is forskolin-insensitive in the absence or presence of Gαs, despite showing similar Gαs-mediated stimulation to AC2-WT (Brand et al., 2013), indicating that analogous mutations of conserved residues within the forskolin binding site of AC isoforms may differentially affect the cAMP responses to distinct stimulatory agents. The crystal structure of the C1-C2 heterodimer also suggested that Val423 directly interacts with forskolin (Tesmer et al., 1997). Comparison studies with AC1-V423G and AC1-WT revealed similar potency but reduced efficacy for forskolin, suggesting that Val423 is important, but not essential, for forskolin binding. That there were no significant effects of the mutation on the cAMP response mediated by A23187 alone also suggests that the AC1-WT and AC1-V423G constructs were expressed at similar levels. The robust enhancement of cAMP accumulation detected for AC1-N878A in comparison with AC1-WT was unanticipated. All the forskolin-sensitive mACs have a charged or polar residue at this position on the forskolin-binding site, except AC3 and AC8 (Seifert et al., 2012). By mutating Asn878 in AC1 to the counterpart Ala residue in AC8, it appears that the responses of AC1-N878A are comparable to the robust cAMP responses elicited by forskolin and A23187 on AC8-expressing cells. The HEK-ACΔ3/6 cell line is a useful tool that has several advantages over the parental cell line for the study of mutant ACs with reduced catalytic activity and changes in forskolin sensitivity.
For future applications, the HEK-ACΔ3/6 cellular system provides a more controlled environment for high-throughput screening assays of selective AC modulators ultimately to lower false-positive rates (nonselective compounds) and simplify hit validation. This cell line could also be used as a mammalian expression platform for production of purified, recombinant AC protein, as analysis of expression and activity of constructs could be evaluated by simply using forskolin-stimulated activity. Our CRISPR/Cas-based cell line is a valuable human cellular system to study AC isoforms in an unbiased manner and is a tool that could simplify and aid future efforts to develop selective AC modulators.
Acknowledgments
We acknowledge the National Institutes of Health and the Department of Medicinal Chemistry and Molecular Pharmacology (MCMP), Purdue University, for the support of this research.
Abbreviations
- AC
adenylyl cyclase
- β2AR
β2-adrenergic receptor
- Cas
CRISPR-associated system
- CRISPR
clustered regularly interspaced short palindromic repeats
- EP2R
prostaglandin E2 receptor
- GPCR
G protein–coupled receptor
- HEK293
human embryonic kidney cell line 293
- IBMX
3-isobutyl-1-methylxanthine
- IDT
Integrated DNA Technologies
- mAC
transmembrane AC
- PCR
polymerase chain reaction
- PGE2
prostaglandin E2
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- RT-qPCR
real-time quantitative PCR
- sgRNA
single-guide RNA
Authorship Contributions
Participated in research design: Soto-Velasquez, Hayes, Alpsoy, Dykhuizen, Watts.
Conducted experiments: Soto-Velasquez.
Performed data analysis: Soto-Velasquez, Dykhuizen.
Wrote or contributed to the writing of the manuscript: Soto-Velasquez, Hayes, Watts.
Footnotes
This work was financially supported by the National Institutes of Health [MH101673] and the Medicinal Chemistry and Molecular Pharmacology (MCMP) Research Enhancement Award.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Alewijnse AE, Smit MJ, Rodriguez Pena MS, Verzijl D, Timmerman H, Leurs R. (1997) Modulation of forskolin-mediated adenylyl cyclase activation by constitutively active G(S)-coupled receptors. FEBS Lett 419:171–174. [DOI] [PubMed] [Google Scholar]
- Andressen KW, Norum JH, Levy FO, Krobert KA. (2006) Activation of adenylyl cyclase by endogenous G(s)-coupled receptors in human embryonic kidney 293 cells is attenuated by 5-HT(7) receptor expression. Mol Pharmacol 69:207–215. [DOI] [PubMed] [Google Scholar]
- Atwood BK, Lopez J, Wager-Miller J, Mackie K, Straiker A. (2011) Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics 12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaranayake AD, Almo SC. (2014) Recent advances in mammalian protein production. FEBS Lett 588:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betenbaugh MJ, Tomiya N, Narang S, Hsu JT, Lee YC. (2004) Biosynthesis of human-type N-glycans in heterologous systems. Curr Opin Struct Biol 14:601–606. [DOI] [PubMed] [Google Scholar]
- Bogard AS, Xu C, Ostrom RS. (2011) Human bronchial smooth muscle cells express adenylyl cyclase isoforms 2, 4, and 6 in distinct membrane microdomains. J Pharmacol Exp Ther 337:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand CS, Hocker HJ, Gorfe AA, Cavasotto CN, Dessauer CW. (2013) Isoform selectivity of adenylyl cyclase inhibitors: characterization of known and novel compounds. J Pharmacol Exp Ther 347:265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust TF, Alongkronrusmee D, Soto-Velasquez M, Baldwin TA, Ye Z, Dai M, Dessauer CW, van Rijn RM, Watts VJ. (2017) Identification of a selective small-molecule inhibitor of type 1 adenylyl cyclase activity with analgesic properties. Sci Signal 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali JJ, Zwaagstra JC, Mons N, Cooper DM, Krupinski J. (1994) Type VIII adenylyl cyclase. A Ca2+/calmodulin-stimulated enzyme expressed in discrete regions of rat brain. J Biol Chem 269:12190–12195. [PubMed] [Google Scholar]
- Chen-Goodspeed M, Lukan AN, Dessauer CW. (2005) Modeling of G alpha(s) and Galpha(i) regulation of human type V and VI adenylyl cyclase. J Biol Chem 280:1808–1816. [DOI] [PubMed] [Google Scholar]
- Cumbay MG, Watts VJ. (2001) Heterologous sensitization of recombinant adenylate cyclases by activation of D(2) dopamine receptors. J Pharmacol Exp Ther 297:1201–1209. [PubMed] [Google Scholar]
- Dessauer CW, Watts VJ, Ostrom RS, Conti M, Dove S, Seifert R. (2017) International Union of Basic and Clinical Pharmacology. CI. structures and small molecule modulators of mammalian adenylyl cyclases. Pharmacol Rev 69:93–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichel K, von Zastrow M. (2018) Subcellular organization of GPCR signaling. Trends Pharmacol Sci 39:200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan KA, Mahey R, Cooper DM. (1996) Functional co-localization of transfected Ca(2+)-stimulable adenylyl cyclases with capacitative Ca2+ entry sites. J Biol Chem 271:12438–12444. [DOI] [PubMed] [Google Scholar]
- Gille A, Lushington GH, Mou TC, Doughty MB, Johnson RA, Seifert R. (2004) Differential inhibition of adenylyl cyclase isoforms and soluble guanylyl cyclase by purine and pyrimidine nucleotides. J Biol Chem 279:19955–19969. [DOI] [PubMed] [Google Scholar]
- Gimpl G, Klein U, Reiländer H, Fahrenholz F. (1995) Expression of the human oxytocin receptor in baculovirus-infected insect cells: high-affinity binding is induced by a cholesterol-cyclodextrin complex. Biochemistry 34:13794–13801. [DOI] [PubMed] [Google Scholar]
- Hacker BM, Tomlinson JE, Wayman GA, Sultana R, Chan G, Villacres E, Disteche C, Storm DR. (1998) Cloning, chromosomal mapping, and regulatory properties of the human type 9 adenylyl cyclase (ADCY9). Genomics 50:97–104. [DOI] [PubMed] [Google Scholar]
- Harry A, Chen Y, Magnusson R, Iyengar R, Weng G. (1997) Differential regulation of adenylyl cyclases by Galphas. J Biol Chem 272:19017–19021. [DOI] [PubMed] [Google Scholar]
- Insel PA, Ostrom RS. (2003) Forskolin as a tool for examining adenylyl cyclase expression, regulation, and G protein signaling. Cell Mol Neurobiol 23:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobowitz O, Iyengar R. (1994) Phorbol ester-induced stimulation and phosphorylation of adenylyl cyclase 2. Proc Natl Acad Sci USA 91:10630–10634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TB, Agarwal SR, Harvey RD, Ostrom RS. (2018) cAMP signaling compartmentation: adenylyl cyclases as anchors of dynamic signaling complexes. Mol Pharmacol 93:270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsushika S, Chen L, Kawabe J, Nilakantan R, Halnon NJ, Homcy CJ, Ishikawa Y. (1992) Cloning and characterization of a sixth adenylyl cyclase isoform: types V and VI constitute a subgroup within the mammalian adenylyl cyclase family. Proc Natl Acad Sci USA 89:8774–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Linder ME, Gilman AG. (1994) Expression of G-protein alpha subunits in Escherichia coli. Methods Enzymol 237:146–164. [DOI] [PubMed] [Google Scholar]
- Ludwig MG, Seuwen K. (2002) Characterization of the human adenylyl cyclase gene family: cDNA, gene structure, and tissue distribution of the nine isoforms. J Recept Signal Transduct Res 22:79–110. [DOI] [PubMed] [Google Scholar]
- Mann L, Heldman E, Bersudsky Y, Vatner SF, Ishikawa Y, Almog O, Belmaker RH, Agam G. (2009) Inhibition of specific adenylyl cyclase isoforms by lithium and carbamazepine, but not valproate, may be related to their antidepressant effect. Bipolar Disord 11:885–896. [DOI] [PubMed] [Google Scholar]
- Nielsen MD, Chan GC, Poser SW, Storm DR. (1996) Differential regulation of type I and type VIII Ca2+-stimulated adenylyl cyclases by Gi-coupled receptors in vivo. J Biol Chem 271:33308–33316. [DOI] [PubMed] [Google Scholar]
- Ostrom RS, Insel PA. (2004) The evolving role of lipid rafts and caveolae in G protein-coupled receptor signaling: implications for molecular pharmacology. Br J Pharmacol 143:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M, Clynes MA, Masada N, Ciruela A, Ayling LJ, Wachten S, Cooper DM. (2009) Insights into the residence in lipid rafts of adenylyl cyclase AC8 and its regulation by capacitative calcium entry. Am J Physiol Cell Physiol 296:C607–C619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre S, Eschenhagen T, Geisslinger G, Scholich K. (2009) Capturing adenylyl cyclases as potential drug targets. Nat Rev Drug Discov 8:321–335. [DOI] [PubMed] [Google Scholar]
- Pinto C, Papa D, Hübner M, Mou TC, Lushington GH, Seifert R. (2008) Activation and inhibition of adenylyl cyclase isoforms by forskolin analogs. J Pharmacol Exp Ther 325:27–36. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadana R, Dessauer CW. (2009) Physiological roles for G protein-regulated adenylyl cyclase isoforms: insights from knockout and overexpression studies. Neurosignals 17:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider EH, Seifert R. (2010) Sf9 cells: a versatile model system to investigate the pharmacological properties of G protein-coupled receptors. Pharmacol Ther 128:387–418. [DOI] [PubMed] [Google Scholar]
- Seifert R, Lushington GH, Mou TC, Gille A, Sprang SR. (2012) Inhibitors of membranous adenylyl cyclases. Trends Pharmacol Sci 33:64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonds WF. (1999) G protein regulation of adenylate cyclase. Trends Pharmacol Sci 20:66–73. [DOI] [PubMed] [Google Scholar]
- Smith KE, Gu C, Fagan KA, Hu B, Cooper DM. (2002) Residence of adenylyl cyclase type 8 in caveolae is necessary but not sufficient for regulation by capacitative Ca(2+) entry. J Biol Chem 277:6025–6031. [DOI] [PubMed] [Google Scholar]
- Sutkowski EM, Tang WJ, Broome CW, Robbins JD, Seamon KB. (1994) Regulation of forskolin interactions with type I, II, V, and VI adenylyl cyclases by Gs alpha. Biochemistry 33:12852–12859. [DOI] [PubMed] [Google Scholar]
- Tang WJ, Krupinski J, Gilman AG. (1991) Expression and characterization of calmodulin-activated (type I) adenylylcyclase. J Biol Chem 266:8595–8603. [PubMed] [Google Scholar]
- Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. (1997) Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha.GTPgammaS. Science 278:1907–1916. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GC, Lai HL, Lin YW, Chu YT, Chern Y. (2001) N-glycosylation and residues Asn805 and Asn890 are involved in the functional properties of type VI adenylyl cyclase. J Biol Chem 276:35450–35457. [DOI] [PubMed] [Google Scholar]
- Yan SZ, Beeler JA, Chen Y, Shelton RK, Tang WJ. (2001) The regulation of type 7 adenylyl cyclase by its C1b region and Escherichia coli peptidylprolyl isomerase, SlyD. J Biol Chem 276:8500–8506. [DOI] [PubMed] [Google Scholar]
- Yan SZ, Huang ZH, Andrews RK, Tang WJ. (1998) Conversion of forskolin-insensitive to forskolin-sensitive (mouse-type IX) adenylyl cyclase. Mol Pharmacol 53:182–187. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Sun M, Villar VA, Zhang Y, Weinman EJ, Felder RA, Jose PA. (2014) Differential dopamine receptor subtype regulation of adenylyl cyclases in lipid rafts in human embryonic kidney and renal proximal tubule cells. Cell Signal 26:2521–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]