Abstract
Background
In addition to endogenous opioids, a number of peptide sequences, derived from endogenous (hemorphins, alphaS1-casomorphin), and exogenous proteins (casomorphins, exorphins) have been reported, possessing opioid activity. In the present work, we report the identification of a new peptide, receptorphin (Tyr-Ile-Phe-Asn-Leu), derived from the sequence of the second transmembrane loop of the opioid receptor. This sequence is unique for the opioid receptor, and conserved in all species and receptor-types.
Results and Discussion
Receptorphin competes for opioid binding, presenting a kappa-receptor interaction, while it binds equally to delta- and mu- opioid and somatostatin-binding sites, and inhibits the cell proliferation of a number of human cancer cell lines, in a dose-dependent and reversible manner, at the picomolar or the nanomolar range. Receptorphin shows a preferential action on prostate cancer cells.
Conclusion
Our work identifies, for the first time a peptide, in a receptor sequence, possessing ligand-agonistic activities. A hypothesis, based on receptorphin liberation after cell death, is presented, which could tentatively explain the time-lag observed during opioid antiproliferative action.
Background
Endogenous opioids derive from three main precursor proteins, namely proenkephalin A, proenkephalin B or prodynorphin and proopiomelanocortin (POMC), through alternative post-translational processing. These three precursor proteins give rise to at least seven endogenous opioid peptides [1]. All these peptides usually have a very brief half-life, with the exception of beta-endorphin, and therefore it is believed to act locally. Opioid binding sites, characterized pharmacologically, belong to three main categories, namely delta, mu and kappa. Different subtypes at each opioid site were equally reported (δ1, δ2, μ1, μ2, κ1, κ2, κ3). A differential distribution of each opioid site was found in the central and the peripheral nervous system [2], while each endogenous opioid ligand binds with a different selectivity to each site [3,4]. The molecular characterization of opioid receptors was obtained in the '90s. The cDNA and the aminoacid sequence of the three main opioid receptors was reported, and found to follow the general scheme of the seven loops membrane receptors [5]. The homology among the reported opioid receptor sequences varies between 50 and 64%. It is interesting to note that transmembrane and intracellular domains, presents a greater homology (~69%) as compared to that of their extracellular part [5-7].
Opioids interfere with a number of physiological actions in the nervous system, including nociception, cognition, and release of hormones or neurotransmitters. In addition, a specific modulatory action of opioids was reported in the peripheral nervous system [8,9], and immune-related cells [10-12]. The biological actions of opioids necessitate the presence of opioid receptors, their ligand and the ligand-specific degradation system in the proximity of their site of action. This is true for the nervous and the immune system, while the hypothalamic-pituitary portal circulation provides opioids to the pituitary gland [13]. In addition, opioids act in other tissues, and recent reports indicate that these agents possess potent antiproliferative actions in tumor cells [14-22], inducing arrest of cell proliferation and apoptosis [23-25]. β-endorphin, an opioid peptide produced mainly by the pituitary and the immune system, with a sufficiently high biological half-life, and/or locally produced opioids were proposed as potential endogenous opioid mediators of this antiproliferative effect [15,17]. In addition to the endogenous classical opioid peptides, a number of other opioid agonists has been reported, derived from food (exorphins) [26,27], hemoglobin (hemorphins) [28-31] or milk-caseins (casomorphins) [16,32-39]. These peptides can also compete for opioid binding, and decrease cell proliferation in different human and animal cell systems, and could be additional sources of locally-acting opioids. Finally, a number of synthetic peptides, usually produced through combinatorial search of peptide libraries [40,41] have shown an increased affinity for one or multiple opioid binding sites.
In the present work, we report another peptide, derived from the conserved second transmembrane segment of the opioid receptor, with potent antiproliferative activity, in different human cancer cell lines. Its sequence is Tyr-Ile-Phe-Asn-Leu. We named it receptorphin, and we report that it further competes for opioid agonist binding.
Results
Opioid receptor binding selectivity of receptorphin
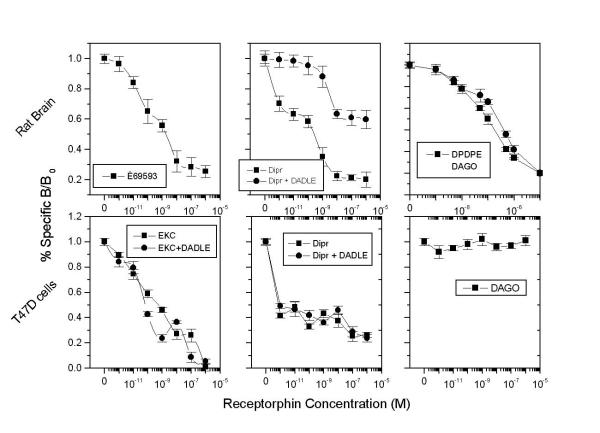
Figure 1 (upper panel) shows the competition of receptorphin for opioid ligand binding on different sites in the rat brain. As shown, the peptide displaces radiolabelled DPDPE and DAGO (selective ligands for the delta and mu opioid binding site) in a dose-dependent manner, with IC50s of 8.25 × 10-8 and 1.81 × 10-7 M respectively. In addition, [3H]U69-593 was equally displaced by receptorphin from kappa1 opioid sites. This peptide competes equally with kappa3 opioid sites, as [3H]diprenorphine binding, in the presence of micromolar concentrations of DADLE (which masks kappa2, delta and mu sites, on which the radioligand could equally bind [4]) does not modify the competition curves of receptorphin. The analysis of the binding curves is presented in Table 1.
Figure 1.
Displacement of different opioid ligands by receptorphin, in T47D cells and rat brain membranes. Upper panels: Rat brain membranes. Lower panels: T47D cells See text for conditions of binding. Mean ± SEM of three experiments in duplicate.
Table 1.
Inhibitory concentrations 50% (IC50) ± SE of receptorphin competition on different opioid sites, assayed by displacement binding of a variety of opioid ligands.
| Opioid Source | DPDPE or DSLET | DAGO | EKC | Diprenorphin | EKC +DADLE | Diprenorphin +DADLE |
| MCF7 | 3.25 ± 0.42 × 10-9 | 1.43 ± 0.20 × 10-10 | 6.65 ± 1.01 × 10-11 | 5.78 ± 0.64 × 10-13 | 1.59 ± 0.14 × 10-10 | >10-12 |
| T47D | ND | ND | 5.14 ± 0.74 × 10-10 | >10-12 | 5.41 ± 0.57 × 10-11 | >10-12 |
| DU145 | ND | 6.52 ± 0.58 × 10-11 | 2.34 ± 0.55 × 10-11 | ND | 2.87 ± 0.22 × 10-11 | ND |
| PC3 | ND | 4.30 ± 0.97 × 10-11 | >10-6 | ND | >10-6 | ND |
| Rat | 8.25 ± 0.81 × 10-8 | 1.81 ± 0.23 × 10-7 | *1.85 ± 0.11 × 10-10 | **5.77 ± 0.87 × 10-10 | - | 1.85 ± 0.43 × 10-10 |
| Brain | 2.65 ± 0.13 × 10-9 |
*[3H]U60-593 was used as a selective kappa1 opioid ligand **Displacement curves were best fitted with a two-site model. See text for details. IC50s (in Moles/L) were calculated by sigmoidal fitting of displacement experiments. ND = not detected. Mean ± SE of three different experiments performed in duplicate.
Previous results have shown that opioid binding sites could also been identified in a number of breast [17,19] and prostate cell lines [38]. Binding of opioid agonists on these sites induces a decrease of cell proliferation. Figure 1 (lower panel) shows the competition of receptorphin for opioid binding on T47D cells. The interaction of the peptide with the different subtypes of the kappa opioid site was obtained by the use of a combination of radiolabelled ligands (ethylketocyclazocine and diprenorphine) and effectors (DADLE), as described under Material and Methods. Receptorphin competes for binding to kappa opioid sites, with a very high affinity, presenting in this cell line too a main interaction with kappa1 and kappa3 subtypes, as microsomal concentrations of DADLE do not modify substantially opioid binding. The interaction of receptorphin with delta and mu sites in T47D cells was inexistant, due to the minimal number of these sites in T47D cells. In contrast, receptorphin displaces [3H]DPDPE and [3H]DAGO, in MCF7 cells, bearing delta and mu binding sites. A summary of the interaction of receptorphin with opioid sites in other cell lines is presented in Table 1.
Cell proliferation
The above results indicate that receptorphin competes for opioid binding on human breast and prostate cancer cells, bearing opioid receptors. A main effect of opioid ligands in these cancer cell lines is the decrease of cell proliferation [16,17,19,38]. We have therefore examined the antiproliferative action of receptorphin, in the same cell lines. Figure 2 and Table 2 present the effect of receptorphin on the proliferation of breast (MCF7 and T47D) and prostate cancer derived cell lines (PC3 and DU145). As shown, this peptide, at concentrations ranging from 10-12 to 10-6 M inhibits cell proliferation of breast (MCF7 and T47D) and prostate (DU145, PC3) cell lines by 41, 49, 59 and 27% respectively. IC50 was 0.15, 0.07, 0.44 and 6.98 nM respectively. The general opioid antagonist diprenorphin (10-6 M) shifted the effect of receptorphin by one log in breast cancer cell lines, while its action was much more pronounced in DU145 and PC3 cancer cell lines, indicating that opioid antagonists reverse, at least partially the antiproliferative effect of the peptide. In contrast, the selective kappa1 antagonist Nor-Binaltorphine (NorBNI) (10-6 M) produced only a partial inhibition of receptorphin binding. This result indicates that receptorphin might interact equally with other sites, different from classical opioid receptors.
Figure 2.
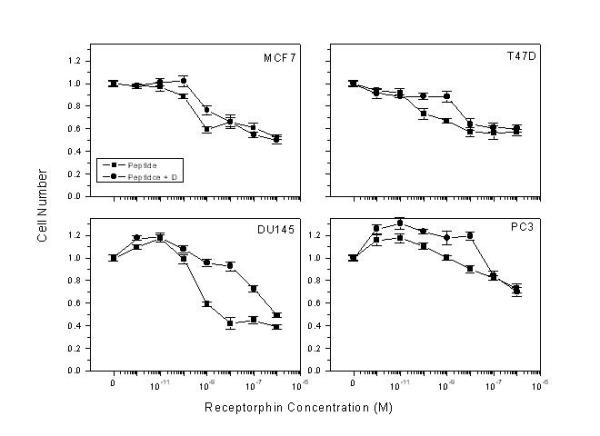
Effect of receptorphin on the proliferation of breast (MCF7, T47D) and prostate (DU145, PC3) human cancer cell lines. Cells were plated at an initial density of 20000 cells/well, and the effect of the peptide (changed every day) was assayed after two cell cycles (4th day). The number of cells therefore, in control conditions (in the absence of the peptide) was 80000/well. Cell viability was in all cases >95%. Diprenorphin was added simultaneously with receptorphin, at a concentration of 10-6 M. Mean ± SEM of three different experiments performed in triplicate.
Table 2.
Inhibitory concentrations 50% (IC50) and maximal inhibition of cell growth of different breast and prostate cancer cell lines by receptorphin.
| Receptorphin | Maximum Inhibition | Receptorphin+ Diprenorphin | Maximum Inhibition | |
| MCF7 | 1.51 ± 0.18 × 10-10 | 0.41 | 1.69 ± 0.10 × 10-9 | 0.49 |
| T47D | 7.01 ± 0.36 × 10-11 | 0.44 | 3.61 ± 0.13 × 10-9 | 0.48 |
| LNCaP | 8.70 ± 0.87 × 10-8 | 0.58 | 2.88 ± 0.15 × 10-9 | 0.61 |
| DU145 | 4.36 ± 0.19 × 10-10 | 0.59 | 3.44 ± 2.32 × 10-7 | 0.53 |
| PC3 | 6.98 ± 0.11 × 10-9 | 0.27 | 7.00 ± 0.14 × 10-7 | 0.31 |
Curves were obtained by sigmoidal fitting of data presented in Figure 2. Concentrations are expressed in Moles/L. Mean ± SE of three different experiments performed in triplicate.
Interaction of receptorphin with other membrane receptor systems
As presented in Figure 2, the addition of the general opioid antagonist diprenorphine in the culture medium decreases, but does not abolish the antiproliferative effect of receptorphin. This result could suggest a possible interaction of the peptide with other membrane sites, different from opioid binding sites. A similar result was equally onserved with a number of casomorphins, in the same cell lines [16,38]. Further analysis has revealed an interaction of casomorphins with somatostatin binding sites [16,38,42]. In order to identify a possible interaction of receptorphin with somatostatin sites, we have used the LNCaP prostate cancer cell line, which, according to our previous results does not express opioid binding sites [38]. Our results are shown in Figure 3. Receptorphin inhibited cell growth by 60%, with an IC50 of 87 nM, significantly higher than that observed for other cell lines. In this particular case, diprenorphine (10-6 M) enhanced rather than inhibited the antiproliferative effect by a factor of 30 (IC50 2.8 nM), indicating that other membrane receptor systems might be also implicated in the action of the peptide. Competition of receptorphin for [125I]Tyr11-somatostatin14 binding revealed that the peptide could decrease somatostatin binding with an IC50 of 18.9 ± 1.02 × 10-9 M, comparable with the antiproliferative effect of the peptide on LNCaP cells, indicating a significant interaction of receptorphin with somatostatin receptors.
Figure 3.
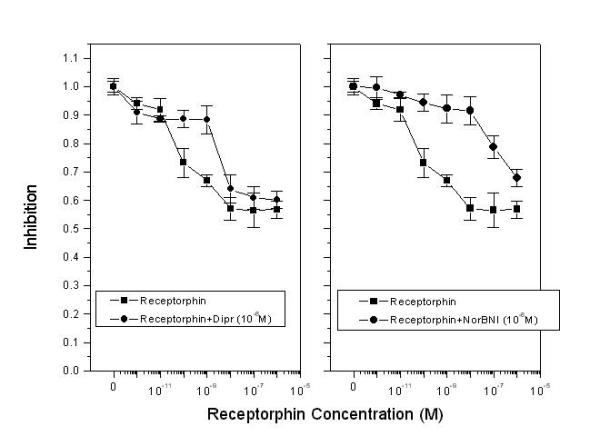
Competition of receptorphin action by general and selective opioid antagonists in T47D cells. T47D cells were plated at an initial density of 20000 cells/well, and the effect of the peptide (changed every day) was assayed after two cell cycles (4th day). The number of cells therefore, in control conditions (in the absence of the peptide) was 80000/well. Cell viability was in all cases >95%. Diprenorphin (A) or NorBNI (B) were added simultaneously with receptorphin, at a concentration of 10-6 M. Mean ± SEM of three different experiments performed in triplicate.
Discussion
In recent years, a number of endogenous and food-derived proteins have been identified as potential sources of opioid or opiomimetic peptides. They include gluten, hemoglobin and caseins [26,27,32-37,39]. By limited proteolysis of such proteins, a number of peptides have been identified, which presents features of opiomimetic action: binding to opioid receptors, competition for opioid ligands, opiomimetic effects, and reversion of their action by the addition of opioid antagonists. These opioid peptide sources could resolve the problem of opioid availability in organs, in which classical opioid precursor were not identified. Indeed, the blood flow disposition of endogenous opioid peptides is limited, due to the very short half-life of these peptides, with the exception of beta-endorphin.
The results of the present investigation identify another peptide, receptorphin, derived from the opioid receptor itself. Receptorphin sequence is comprised to the conserved structure of the second transmembrane segment of the opioid receptor (see Table 3 and Figure 5). Receptorphin exhibits some characteristics of an opioid ligand:
Table 3.
Position of receptorphin in different reported opioid receptor sequences.
| Receptor | Source | Length (a.a.) | Receptorphin position | Reference |
| Delta | Zebrafish | 373 | 90–94 | [51] |
| Delta | Human | 372 | 87–91 | [52] |
| Mu | Human | 400 | 98–102 | [53][54][55] |
| Mu variant | Human | 392 | 107–112 | [55] |
| Mu | Mouse | 367 | 86–90 | [56] |
| Mu | Rat | 398 | 69–100 | [57][58][59][60][61][62][63][64] |
| Mu | Bovine | 401 | 109–113 | [65] |
| Kappa | Rat | 380 | 97–101 | [66] |
| Kappa | Guinea Pig | 380 | 97–101 | [67] |
| Kappa | Mouse | 380 | 97–101 | [56] |
| Kappa | Human | 380 | 97–101 | [68][69] |
| Orphan | Human | 370 | 89–93 | [70][71] |
| Orphan | Rat | 357 | 86–90 | [72] |
| Orphan | Mouse | 367 | 87–92 | [56][73][74][75] |
| Orphan | Rat | 367 | 86–90 | [76][77][78][79][80][81] |
Figure 5.
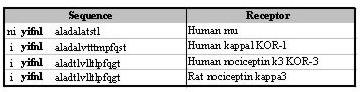
Alignment of the sequences of the second transmembrane loop of different opioid receptors. Sequences are derived from references in Table 3. The sequence of receptorphin is indicated in bold letters.
Figure 4.
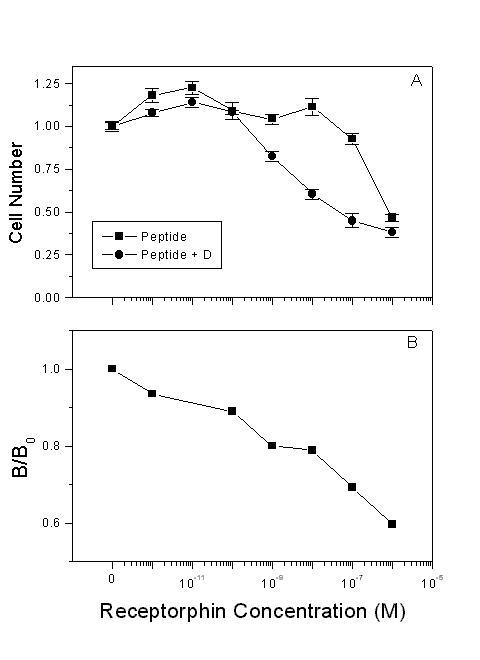
Interaction of receptorphin with somatostatin binding sites. A: Effect of receptorphin on the proliferation of LNCaP cells, not-presenting opioid binding sites [38]. Conditions of cell growth were similar with those presented in the legend of Figure 2, with the exception that cells were assayed at day 6, as the doubling time of LNCaP cells is longer (3.02d). Mean ± SEM of three experiments in triplicate. B. Competition of receptorphin for somatostatin binding. Displacement of radiolabelled [125I]Tyr11Somatostatin14 by varying concentrations of receptorphin. See text for details of binding.
1. It competes for opioid ligand binding on opioid sites, exhibiting principally selectivity for kappa opioid receptors (especially kappa1 and kappa3). Its affinity, in human breast and prostate cancer cell lines, is higher than that of the prototype ligands [14,17,19]. In contrast, in rat brain membranes, the affinity of receptorphin is lower than that of DAGO or DPDPE. It is not actually known whether this discrepancy is due to the difference of normal and neoplastic tissue, in which a possible alternative splicing of the opioid receptor could occur, to the conditions of binding (hypotonic medium in rat brain as compared to isotonic medium in whole cell binding) or to species differences. A similar discrepancy was also observed in the case of αS1-casomorphin and other casomorphin peptides [16,37,38]. In addition, receptorphin interacts with somatostatin binding, a result explaining the non-complete reversion of its effect by opioid antagonists. A similar interaction was equally observed with a number of other food-derived peptides, in the same system [16,38].
2. Receptorphin decreases cell proliferation in different breast and prostate cell lines, in a dose-dependent and reversible manner, as reported for other opioid alkaloids and casomorphin ligands [16,17,38]. Comparing the IC50 of receptorphin with that obtained in the same cell lines by a number of other opioid peptides [16,37,38] it is derived that this new opioid agonist is almost as potent as alphaS1-casomorphin in the prostate, while it is much less potent (at least by a factor of 20) in the breast. Comparable maximal inhibition of growth was obtained by receptorphin and other casomorphin peptides, both in the breast and the prostate [16,38], representing possibly, the maximum opioid-related effect on these cell lines. The physiological relevance of this prostate selectivity of receptorphin is not known.
The above results are, of course, not enough to characterize fully a possible new opioid peptide. Further work, implying more classical opioid effects in cells (GTP binding, cAMP inhibition) and organs or animals might be necessary for the complete identification of receptorphin as an opioid agonist. Nevertheless, the fact that receptorphin binds with a high affinity to opioid sites in different organs (brain and cancer cell lines) in which opioid receptors were identified, and its antiproliferative action in different cancer cell lines is inhibited by the general opioid antagonist diprenorphine apply for an opioid activity of this peptide.
The sequence of receptorphin (Tyr-Ile-Phe-Asn-Leu) is conserved in all reported opioid receptors, from a variety of species (Table 3). In contrast, this sequence is restricted to the opioid receptor, and is not detected in other seven-loop membrane receptors [43]. It is interesting that, at the same position (second transmembrane segment) of the different types of the somatostatin receptor (SSTR-1 to 5) which present the greater homology with opioid receptors [5], in different species, a peptide with the sequence Tyr-Ile(or-Leu)-Leu-Asn-Leu exists, presenting an homology in structure with receptorphin [44-48]. Furthermore, two or three amino acids of receptorphin (Phe-Asn-Leu) interfere with the agonist or antagonist binding, at least indirectly, participating in the formation of the ligand envelope [43]. This might be the reason of the conservation of the receptorphin sequence in all opioid receptor sequences (Table 3). Comparing receptorphin (Tyr-Ile-Phe-Asn-Leu) with other opioid peptides, it is observed that it shares Tyr at position 1 and Leu in position 5. In contrast, the presence of Ile and Asn at positions 2 and 4 and the non-classical Phe at position 3 are unique in receptorphin. This non-classical amino-acid composition may confer the described binding characteristics to this new peptide (Table 1), which bears the classical pharmacophoric groups of classical opioid agonists (Tyr, Phe, Leu).
Conclusions
In the present paper, we report the identification of a ligand-specific peptide, in the sequence of its cognitive receptor structure. Receptorphin (Tyr-Ile-Phe-Asn-Leu) has a unique sequence, conserved in all opioid receptor types and species. The peptide competes with a high affinity for opioid binding, showing a preferential interaction with kappa (κ1 and κ3), while it competes with delta, mu, and somatostatin sites with a lower affinity. It can therefore be considered as a non-selective opioid ligand. Although this finding is interesting per se, the current knowledge permits only speculations about a possible biological role of the peptide, if any. Receptorphin is flagged by Ile at position -1 and always by Ala at position 6 (see Figure 5), making it a putative target of peptidases, while its location, in the hydrophobic membrane environment, protects it from a possible hydrolytic-enzyme action. A possible hypothesis could be that receptorphin might act as an opioid peptide after cell death. In this case, after intra- and extracellular domains of opioid receptor destruction by different liberated proteolytic enzymes, and rupture of the membrane structure, the leading Ile-1 of receptorphin could be cleaved by exopeptidases, while the following Ala6 might be removed by basic endopeptidases, leading to the liberation of the active peptide. If such a mechanism occurs, opioid action could be potentiated after an opioid- and/or other inducers-related cell death. Indeed, it is tentative to assume that after an opioid mediated cellular death [23-25], receptorphin liberation might trigger a positive feedback loop, propagating the opioid effects to a number of adjacent opioid receptor-positive cells. This mechanism could possibly explain the time-lag found in opioid-related cell proliferation and apoptosis, reported in a number of malignant cell lines [15-17,37,38]. Nevertheless, this putative receptorphin implication in cell proliferation remains highly speculative, until its presence could be detected in cell cultures. In addition, it might be of interest to detect similar structures in the sequence of other members of the seven-loop superfamily, in order to investigate whether this finding is unique to the opioid receptor itself, or could be extended to its other members.
Methods
Peptide synthesis
Receptorphin (Tyr-Ile-Phe-Asn-Leu) was synthesized by conventional peptide chemistry methods. t-butoxycarboxyl (Boc) groups were used for protection of the a-amino-groups, while a t-butyl group was used for the protection of the phenolic hydroxy-group. The dichloro-carbodi-imiole/1-hydroxybenzo-triazo method was used for the coupling of the protected amino acids. Removal of the t-butyl and Boc groups was achieved by trifluoroacetic acid. Peptide purification was made by semipreparative HPLC on a reverse phase C-18 Nucleosil column, on a 30 min linear gradient of 15–85% methanol in 0.1% aqueous trifluoroacetic acid.
Cell lines and culture conditions
Two breast cancer (MCF7 and T47D), and tho prostate cell lines (PC3 and DU145), bearing opioid binding sites [16,17,19,38], were used, in order to assay the opioid activity of receptorphin. DU145 and MCF7 cell lines were purchased from DSMZ (Braunschweig, Germany), while T47D, and PC3 cells were from the European Collection of Cell Cultures (Salisbury, UK). T47D and DU145 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS). PC3 cells were cultured in DMEM medium with 10% FBS, while, for MCF7 cell culture, DMEM/F12 medium was used, supplemented with 5 μg/ml insulin (Sigma, St Louis, MI) and 10% FBS. All cell lines were maintained in a humidified atmosphere of 5% CO2 in air. All culture media were from Gibco BRL (Life Technologies, Paisley, UK). Medium, supplemented or not with receptorphin, was changed every day. Without addition of any drug, the proliferation time of all cell lines was two days. Receptorphin was dissolved in phosphate buffered saline shortly before use.
Cell proliferation
Cells were plated in 24-well plates, at an initial density of 2 × 104 cells, with 1.0 ml medium per well. All drugs were added to cultures one day after seeding (designated as day 0), in order to ensure uniform attachment of cells at the onset of the experiments. Cell growth was measured by the tetrazolium salt assay [49]. Cells were incubated for 4 h at 37°C with the tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide), and metabolically active cells reduced the dye to purple formazan. Dark blue crystals were dissolved with propanol, and the absorbance was measured at 570 nm and compared against a standard curve of known numbers of cells. All experiments were performed, at minimum, three times in triplicate.
Binding conditions
Cancer cell lines
Ligand binding assays on whole cells (106 cells/well) were performed as described in Hatzoglou et al [14,16,17], in a total volume of 0.5 ml phosphate buffered saline (10 mM phosphate, 150 mM NaCl, pH 7.4), containing radioactive opioids or somatostatin, without (total binding), with 10-5 M of the same unlabelled agent (non specific binding), or with varying concentrations of receptorphin, ranging from 10-12 to 10-6 M. Cells were incubated for 2 h at room temperature (18–22°C). At the end of the incubation period, cells were washed twice with cold buffer, in order to eliminate the unbound radioactivity. Cells were removed from plates with 0.4 ml 2N NaOH, mixed with 4 ml scintillation cocktail (SigmaFluor, Sigma, St Louis, MI) and counted in a scintillation counter (Tricarb, Series 4000, Packard), with a 60% efficiency for Tritium, for opioids and in a Tricarb (Packard) gamma counter with a 95% efficiency for [125I] (somatostatin binding). Binding was repeated three times (in duplicate).
A number of different opioid ligands were used in order to identify specific opioid binding sites. [3H] [D-Pen2, D-Pen5]enkephalin (DPDPE) was used as a selective delta ligand, and [3H] [N-me-Phe4, Gly5-ol]enkephalin (DAGO) as a selective mu ligand. [3H]ethylketocyclazocine (EKC) and [3H]diprenorphine were used as general opioid ligands, sharing nevertheless, at nanomolar concentrations, a different receptor recognition spectrum: ethylketocyclazocine recognizes delta, mu, and the kappa1 and kappa2 (or epsilon) subtypes of the kappa binding site, while diprenorphine recognizes delta, mu, kappa2 and kappa3 sites [3,4]. The addition of micromolar concentrations of DADLE masks the ligand interaction with delta, mu, and kappa2 opioid sites, permitting the differentiation of the effect of the tracer with the kappa1 and the kappa3 opiod site respectively [3,4]. Control experiments, in our laboratory have proven that by the use of these combinations we obtain similar results for the detection of kappa1 and kappa3 opioid sites, as with the use of the selective ligands U50488 and Met5-enkephalin-Arg6-Phe7.
Rat brain membranes
Rat brain membrane preparation and opioid binding was performed as described previously by Loukas et al. [39]. Briefly, binding was performed in Tris-HCl buffer (10 mM, pH 7.4), in a final volume of 1.0 ml. The protein concentration was 300 μg/assay. Binding was initiated by the addition of 2 nM of the selective ligand ([3H]DPDPE for δ or [3H]DAGO for μ sites, [3H]U69-593 for kappa1 site and [3H]diprenorphine as a general opioid ligand, as discussed above). Non specific binding was estimated in the presence of 10-5 M naloxone or diprenorphin. Peptide concentrations varied from 10-12 to 10-6 M. After the incubation, bound radioactivity was separated by filtration, under reduced pressure, through GF/D filters, previously soaked in Tris-HCl buffer, and rinsed twice with ice-cold buffer.
The results of binding assays were analyzed by the Origin (MicroCal, Northampton, MA.) V 5 package, using equations described by Munson and Rodbard [50].
Radiochemicals and chemicals
[3H]ethylketocyclazocine (S.A. 18 Ci/mmol), [3H] [D-Pen2, D-Pen5]enkephalin (DPDPE) (S.A. 37 Ci/mmol) and [3H]U69-593 (S.A. 39.7 Ci/mmol) were bought from New England Nuclear Co (Zaventum, Belgium). [3H]diprenorphine (S.A. 29 Ci/mmol), [3H] [N-me-Phe4, Gly5-ol]enkephalin (DAGO, S.A. 47.7 Ci/mmol) and [125I]Tyr11-somatostatin14 (S.A. 2000 Ci/mmol) were from Amersham (Buckinghabshire, UK). Ethylketocyclazocine was a gift from Sterling-Winthrop. Diprenorphine was from Reckit and Coleman Co. All other chemicals were either from Merck (Darmstad, Germany) or from Sigma (St Louis MO).
Acknowledgments
Acknowledgements
Work partially supported by the University of Crete, General Secretariat of Research and Technology and Varelas S.A. grants.
Contributor Information
Marilena Kampa, Email: kampa@med.uoc.gr.
Spyros Loukas, Email: spyro@mail.demokritos.gr.
Andreas Tsapis, Email: Andreas.Tsapis@ipsc.u-psud.fr.
Elias Castanas, Email: castanas@med.uoc.gr.
References
- Hollt V. Opioid peptide processing and receptor selectivity. Annu Rev Pharmacol Toxicol. 1986;26:59–77. doi: 10.1146/annurev.pa.26.040186.000423. [DOI] [PubMed] [Google Scholar]
- Castanas E, Blanc D, Bourhim N, Cupo A, Cantau P, P G. Reassessment of opioid binding sites in different areas of the rat brain. Neuropeptides. 1986;7:369–380. doi: 10.1016/0143-4179(86)90030-2. [DOI] [PubMed] [Google Scholar]
- Castanas E, Bourhim N, Giraud P, Boudouresque F, Cantau P, Oliver C. Interaction of opiates with opioid binding sites in the bovine adrenal medulla: I Interaction with delta and mu sites. J Neurochem. 1985;45:677–687. doi: 10.1111/j.1471-4159.1985.tb04046.x. [DOI] [PubMed] [Google Scholar]
- Castanas E, Bourhim N, Giraud P, Boudouresque F, Cantau P, Oliver C. Interaction of opiates with opioid binding sites in the bovine adrenal medulla: II Interaction with kappa sites. J Neurochem. 1985;45:688–699. doi: 10.1111/j.1471-4159.1985.tb04047.x. [DOI] [PubMed] [Google Scholar]
- Reisine T, Bell GI. Molecular biology of opioid receptors. Trends in Neurosci. 1993;16:506–510. doi: 10.1016/0166-2236(93)90194-Q. [DOI] [PubMed] [Google Scholar]
- Chen Y, Mestek A, Liu J, Hurley JL, Yu L. Molecular clonning and functional expression of a mu-opioid receptor from rat brain. Mol Pharmacol. 1993;44:8–12. [PubMed] [Google Scholar]
- Minami M, Toya T, Katao V, MacKawa K, Makamura S, Onegi T, Kaneko S, Satoh M. Clonning and expression of a cDNA for the rat k-opioid receptor. FEBS Letters. 1993;329:291–295. doi: 10.1016/0014-5793(93)80240-U. [DOI] [PubMed] [Google Scholar]
- Castanas E, Giraud P, Audigier Y, Drissi R, Boudouresque F, Conte-Devolx B, Oliver C. Opiate binding sites on bovine adrenal medulla and six human pheochromocytomas. Life Sci. 1983;33:295–298. doi: 10.1016/0024-3205(83)90501-5. [DOI] [PubMed] [Google Scholar]
- Giraud P, Castanas E, Conte-Devolx B, Boudouresque F, Taquet H, Orlando M, Jaquet P, Eiden LE, Cesselin F, Gunz G, Trigano M. Enkephalins in human pheochromocytoma: Biosynthesis and secretion. Europ Heart J. 1982;3:19–22. doi: 10.1093/eurheartj/3.suppl_c.19. [DOI] [PubMed] [Google Scholar]
- Roy S, Loh HH. Effects of opioids on the immune system. Neurochem Res. 1996;21:1375–1386. doi: 10.1007/BF02532379. [DOI] [PubMed] [Google Scholar]
- Scharrer B, Stefano GB. Neuropeptides and Immunoregulation. Neuropeptides and Immunoregulation: Springer-Verlag, Berlin; 1994.
- Stefano GB, Scharrer B, Smith EM, Hughes TK, Magazine HI, Bilfinger TV, Hartman AR, Fricchione GL, Liu Y, Makman MH. Opioid and Opiate Immunoregulation Processes. Critical Reviews in Immunology. 1996;16:109–144. doi: 10.1615/critrevimmunol.v16.i2.10. [DOI] [PubMed] [Google Scholar]
- Castanas E, Giraud P, Drissi R, Chabrier P-E, Conte-Devolx B, Boudouresque F, Cantau P, Cesselin F, Cupo A, Eiden LE, Oliver C. Characterization of enkephalins and related peptides in rat hypophyseal portal blood. Brain Res. 1984;310:1–6. doi: 10.1016/0006-8993(84)90003-9. [DOI] [PubMed] [Google Scholar]
- Hatzoglou A, Gravanis A, Margioris AN, Zoumakis E, Castanas E. Identification and characterization of opioid-binding sites present in the Ishikawa human endometrial adenocarcinoma cell line. J Clin Endocrinol Metab. 1995;80:418–423. doi: 10.1210/jc.80.2.418. [DOI] [PubMed] [Google Scholar]
- Hatzoglou A, Bakogeorgou E, Papakonstanti E, Stournaras C, Emmanouel DS, Castanas E. Identification and characterization of opioid and somatostatin binding sites in the opossum kidney (OK) cell line and their effect on growth. J Cell Biochem. 1996;63:410–421. doi: 10.1002/(SICI)1097-4644(19961215)63:4<410::AID-JCB3>3.3.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Hatzoglou A, Bakogeorgou E, Hatzoglou C, Martin PM, Castanas E. Antiproliferative and receptor binding properties of alpha- and beta-casomorphins in the T47D human breast cancer cell line. Eur J Pharmacol. 1996;310:217–223. doi: 10.1016/0014-2999(96)00339-1. [DOI] [PubMed] [Google Scholar]
- Hatzoglou A, Bakogeorgou E, Castanas E. The antiproliferative effect of opioid receptor agonists on the T47D human breast cancer cell line, is partially mediated through opioid receptors. Eur J Pharmacol. 1996;296:199–207. doi: 10.1016/0014-2999(95)00703-2. [DOI] [PubMed] [Google Scholar]
- Hytrek SD, McLaughlin PJ, Lang CM, Zagon IS. Inhibition of human colon cancer by intermittent opioid receptor blockade with naltrexone. Cancer Lett. 1996;101:159–164. doi: 10.1016/0304-3835(96)04119-5. [DOI] [PubMed] [Google Scholar]
- Maneckjee R, Biswas R, Vonderhaar BK. Binding of opioids to human MCF-7 breast cancer cells and their effects on growth. Cancer Res. 1990;50:2234–2238. [PubMed] [Google Scholar]
- Zagon IS, Hytrek SD, McLaughlin PJ. Opioid growth factor tonically inhibits human colon cancer cell proliferation in tissue culture. Am J Physiol. 1996;271:R511–R518. doi: 10.1152/ajpregu.1996.271.3.R511. [DOI] [PubMed] [Google Scholar]
- Zagon IS, McLaughlin PJ. Endogenous opioid systems regulate growth of neural tumor cells in culture. Brain Res. 1989;490:14–25. doi: 10.1016/0006-8993(89)90425-3. [DOI] [PubMed] [Google Scholar]
- Zagon IS, McLaughlin PJ, Goodman SR, Rhodes RE. Opioid receptors and endogenous opioids in diverse human and animal cancers. J natl Cancer Inst. 1987;79:1059–1065. [PubMed] [Google Scholar]
- Maneckjee R, Minna JD. Opioids induce while nicotine suppresses apoptosis in human lung cancer cells. Cell Growth Differ. 1994;5:1033–1040. [PubMed] [Google Scholar]
- Sueoka N, Sueoka E, Okabe S, Fujiki H. Anti-cancer effects of morphine through inhibition of tumour necrosis factor-alpha release and mRNA expression. Carcinogenesis. 1996;17:2337–2341. doi: 10.1093/carcin/17.11.2337. [DOI] [PubMed] [Google Scholar]
- Sueoka E, Sueoka N, Kai Y, Okabe S, Suganuma M, Kanematsu K, Yamamoto T, Fujiki H. Anticancer activity of morphine and its synthetic derivative, KT-90, mediated through apoptosis and inhibition of NF-kappaB activation. Biochem Biophys Res Commun. 1998;252:566–570. doi: 10.1006/bbrc.1998.9695. [DOI] [PubMed] [Google Scholar]
- Dohan FC. Hypothesis: Genes and neuroactive peptides from food as cause of schizophrenia. Neural peptides and neuronal communication. Edited by Costa E, Trabucchi M. pp. 535–548. New York: Raven Press; 1980. pp. 535–548. [PubMed]
- Zioudrou C, Streaty RA, Klee WA. Opioid peptides derived from food proteins. J Biol Chem. 1979;254:2446–2449. [PubMed] [Google Scholar]
- Moeller I, Chai SY, Smith I, Lew R, Mendelsohn FA. Haemorphin peptides may be endogenous ligands for brain angiotensin AT4 receptors. Clin Exp Pharmacol Physiol Suppl. 1998;25:S68–S71. doi: 10.1111/j.1440-1681.1998.tb02304.x. [DOI] [PubMed] [Google Scholar]
- Nyberg F, Sanderson K, Glamsta EL. The hemorphins: a new class of opioid peptides derived from the blood protein hemoglobin. Biopolymers. 1997;43:147–156. doi: 10.1002/(SICI)1097-0282(1997)43:2<147::AID-BIP8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Galoyan AA. Primary structure and biological activity of hemoglobin-related hypothalamic peptides. Biopolymers. 1997;43:135–137. doi: 10.1002/(SICI)1097-0282(1997)43:2<135::AID-BIP6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Garreau I, Sannier F, Piot JM. Opioid peptides derived from hemoglobin: hemorphins. Biopolymers. 1997;43:75–98. doi: 10.1002/(SICI)1097-0282(1997)43:2<75::AID-BIP2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Brantl V, Teschemacher H, Henschen A, Lottspeich F. Novel opioid peptides derived from casein (â-casomorphins): I. Isolation from bovine casein peptone. Hoppe-Seyler's Z Physiol Chem. 1979;360:1211–1216. doi: 10.1515/bchm2.1979.360.2.1211. [DOI] [PubMed] [Google Scholar]
- Meisel H, Frister H, Schlimme E. Biologically active peptides in milk proteins. Z Ernahrungswiss. 1989;28:267–278. doi: 10.1007/BF02019390. [DOI] [PubMed] [Google Scholar]
- Meisel H. Biochemical properties of regulatory peptides derived from milk proteins. Biopolymers. 1997;43:119–128. doi: 10.1002/(SICI)1097-0282(1997)43:2<119::AID-BIP4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Meisel H, Bockelmann W. Bioactive peptides encrypted in milk proteins: proteolytic activation and thropho-functional properties. Antonie Van Leeuwenhoek. 1999;76:207–215. doi: 10.1023/A:1002063805780. [DOI] [PubMed] [Google Scholar]
- Schlimme E, Meisel H. Bioactive peptides derived from milk proteins. Structural, physiological and analytical aspects. Nahrung. 1995;39:1–20. doi: 10.1002/food.19950390102. [DOI] [PubMed] [Google Scholar]
- Kampa M, Loukas S, Hatzoglou A, Martin P, Martin PM, Castanas E. Identification of a novel opioid peptide (Tyr-Val-Pro-Phe-Pro) derived from human alpha S1 casein (alpha S1-casomorphin, and alpha S1-casomorphin amide). Biochem J. 1996;319:903–908. doi: 10.1042/bj3190903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa M, Bakogeorgou E, Hatzoglou A, Damianaki A, Martin PM, Castanas E. Opioid alkaloids and casomorphin peptides decrease the proliferation of prostatic cancer cell lines (LNCaP, PC3 and DU145) through a partial interaction with opioid receptors. Eur J Pharmacol. 1997;335:255–265. doi: 10.1016/S0014-2999(97)01213-2. [DOI] [PubMed] [Google Scholar]
- Loukas S, Varoucha D, Zioudrou C, Streaty RA, Klee WA. Opioid activities and structures of á-casein derives exorphins. Biochemistry. 1983;22:4567–4573. doi: 10.1021/bi00288a034. [DOI] [PubMed] [Google Scholar]
- Becker J, Wallace A, Garzon A, Ingallinella P, Bianchi E, Cortese R, Simonin F, Kieffer BL, Pessi A. Ligands for kappa-opioid and ORL1 receptors, identified from a conformationally constrained peptide combinatorial library. J Biol Chem. 1999;274:27513–27522. doi: 10.1074/jbc.274.39.27513. [DOI] [PubMed] [Google Scholar]
- Balboni G, Salvadori S, Guerrini R, Bianchi C, Santagada V, Calliendo G, Bryant SD, Lazarus LH. Opioid pseudopeptides containing heteroaromatic or heteroaliphatic nuclei. Peptides. 2000;21:1663–1671. doi: 10.1016/S0196-9781(00)00315-6. [DOI] [PubMed] [Google Scholar]
- Hatzoglou A, Ouafik L, Bakogeorgou E, Thermos K, Castanas E. Morphine cross-reacts with somatostatin receptor SSTR2 in the T47D human breast cancer cell line and decreases cell growth. Cancer Res. 1995;55:5632–5636. [PubMed] [Google Scholar]
- Zhorov BS, Ananthanarayanan VS. Homology models of ì-opioid receptor with organic and inorganic cations at conserved aspartates in the second and third transmembrane domains. Arch Biochem Biophys. 2000;375:31–49. doi: 10.1006/abbi.1999.1529. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Post SR, Wang K, Tager HS, Bell GI. Seino, S. Cloning and functional characterization of a family of human and mouse somatostatin receptors expressed in brain, gastrointestinal tract, and kidney. Proc Natl Acad Sci USA. 1992;89:251–255. doi: 10.1073/pnas.89.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Stoffel M, Espinosa RI, Xiang KS, Seino M, Seino S, Le Beau MM, Bell GI. Human somatostatin receptor genes: localization to human chromosomes 14, 17, and 22 and identification of simple tandem repeat polymorphisms. Genomics. 1993;15:449–452. doi: 10.1006/geno.1993.1088. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Reisine T, Law SF, Ihara Y, Kubota A, Kagimoto S, Seino M, Seino Y, Bell GI, Seino S. Somatostatin receptors, an expanding gene family: cloning and functional characterization of human SSTR3, a protein coupled to adenylyl cyclase. Mol Endocrinol. 1992;6:2136–2142. doi: 10.1210/me.6.12.2136. [DOI] [PubMed] [Google Scholar]
- Rohrer L, Raulf F, Bruns C, Buettner R, Hofstaedter F, Schule R. Cloning and characterization of a fourth human somatostatin receptor. Proc Natl Acad Sci USA. 1993;90:4196–4200. doi: 10.1073/pnas.90.9.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Kagimoto S, Kubota A, Yasuda K, Masuda K, Someya Y, Ihara Y, Li Q, Imura H, Seino S, Seino Y. Cloning, functional expression and pharmacological characterization of a fourth (hSSTR4) and a fifth (hSSTR5) human somatostatin receptor subtype. Biochem Biophys Res Commun. 1993;195:844–852. doi: 10.1006/bbrc.1993.2122. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1973;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Munson PJ, Rodbard D. LIGAND: A versatile computerized approach to characterization of ligand-binding systems. Anal Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- Barrallo A, Gonzalez-Sarmiento R, Porteros A, Garcia-Isidoro M, Rodriguez RE. Cloning, molecular characterization, and distribution of a gene homologous to delta opioid receptor from zebrafish (Danio rerio). Biochem Biophys Res Commun. 1998;245:544–548. doi: 10.1006/bbrc.1998.8496. [DOI] [PubMed] [Google Scholar]
- Knapp RJ, Malatynska E, Fang L, Li X, Babin E, Nguyen M, Santoro G, Varga EV, Hruby VJ, Roeske WR, et al. Identification of a human delta opioid receptor: cloning and expression. Life Sci. 1994;54:PL463–PL469. doi: 10.1016/0024-3205(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Wang JB, Johnson PS, Persico AM, Hawkins AL, Griffin CA, Uhl GR. Human mu opiate receptor. cDNA and genomic clones, pharmacologic characterization and chromosomal assignment. FEBS Lett. 1994;338:217–222. doi: 10.1016/0014-5793(94)80368-4. [DOI] [PubMed] [Google Scholar]
- Mestek A, Hurley JH, Bye LS, Campbell AD, Chen Y, Tian M, Liu J, Schulman H, Yu L. The human mu opioid receptor: modulation of functional desensitization by calcium/calmodulin-dependent protein kinase and protein kinase C. J Neurosci. 1995;15:2396–2406. doi: 10.1523/JNEUROSCI.15-03-02396.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bare LA, Mansson E, Yang D. Expression of two variants of the human mu opioid receptor mRNA in SK-N-SH cells and human brain. FEBS Lett. 1994;354:213–216. doi: 10.1016/0014-5793(94)01129-X. [DOI] [PubMed] [Google Scholar]
- Nishi M, Takeshima H, Mori M, Nakagawara K, Takeuchi T. Structure and chromosomal mapping of genes for the mouse kappa-opioid receptor and an opioid receptor homologue (MOR-C). Biochem Biophys Res Commun. 1994;205:1353–1357. doi: 10.1006/bbrc.1994.2814. [DOI] [PubMed] [Google Scholar]
- Chen Y, Mestek A, Liu J, Hurley JA, Yu L. Molecular cloning and functional expression of a mu-opioid receptor from rat brain. Mol Pharmacol. 1993;44:8–12. [PubMed] [Google Scholar]
- Fukuda K, Kato S, Mori K, Nishi M, Takeshima H. Primary structures and expression from cDNAs of rat opioid receptor delta- and mu-subtypes. FEBS Lett. 1993;327:311–314. doi: 10.1016/0014-5793(93)81011-N. [DOI] [PubMed] [Google Scholar]
- Wang JB, Imai Y, Eppler CM, Gregor P, Spivak CE, Uhl GR. mu opiate receptor: cDNA cloning and expression. Proc Natl Acad Sci USA. 1993;90:10230–10234. doi: 10.1073/pnas.90.21.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RC, Mansour A, Akil H, Watson SJ. Cloning and pharmacological characterization of a rat mu opioid receptor. Neuron. 1993;11:903–913. doi: 10.1016/0896-6273(93)90120-G. [DOI] [PubMed] [Google Scholar]
- Eppler CM, Hulmes JD, Wang JB, Johnson B, Corbett M, Luthin DR, Uhl GR, Linden J. Purification and partial amino acid sequence of a mu opioid receptor from rat brain. J Biol Chem. 1993;268:26447–26451. [PubMed] [Google Scholar]
- Bunzow JR, Zhang G, Bouvier C, Saez C, Ronnekleiv OK, Kelly MJ, Grandy DK. Characterization and distribution of a cloned rat mu-opioid receptor. J Neurochem. 1995;64:14–24. doi: 10.1046/j.1471-4159.1995.64010014.x. [DOI] [PubMed] [Google Scholar]
- Sedqi M, Roy S, Ramakrishnan S, Elde R, Loh HH. Complementary DNA cloning of a mu-opioid receptor from rat peritoneal macrophages. Biochem Biophys Res Commun. 1995;209:563–574. doi: 10.1006/bbrc.1995.1538. [DOI] [PubMed] [Google Scholar]
- Zastawny RL, George SR, Nguyen T, Cheng R, Tsatsos J, Briones-Urbina R, O'Dowd BF. Cloning, characterization, and distribution of a mu-opioid receptor in rat brain. J Neurochem. 1994;62:2099–2105. doi: 10.1046/j.1471-4159.1994.62062099.x. [DOI] [PubMed] [Google Scholar]
- Onoprishvili I, Andria ML, Vilim FS, Hiller JM, Simon EJ. The bovine mu-opioid receptor: cloning of cDNA and pharmacological characterization of the receptor expressed in mammalian cells. Brain Res Mol Brain Res. 1999;73:129–137. doi: 10.1016/S0169-328X(99)00249-1. [DOI] [PubMed] [Google Scholar]
- Meng F, Xie G-X, Thompson RC, Mansour A, Goldstein A, Watson SJ, Akil H. Cloning and pharmacological characterization of a rat kappa opioid receptor. Proc Natl Acad Sci USA. 1993;90:9954–9958. doi: 10.1073/pnas.90.21.9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie GX, Meng F, Mansour A, Thompson RC, Hoversten MT, Goldstein A, Watson SJ, Akil H. Primary structure and functional expression of a guinea pig kappa opioid (dynorphin) receptor. Proc Natl Acad Sci USA. 1994;91:3779–3783. doi: 10.1073/pnas.91.9.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson E, Bare L, Yang D. Isolation of a human kappa opioid receptor cDNA from placenta. Biochem Biophys Res Commun. 1994;202:1431–1437. doi: 10.1006/bbrc.1994.2091. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen C, Xue J. Cloning of a human .kappa. opioid receptor from the brain. Life Sci. 1995;56:201–207. doi: 10.1016/0024-3205(94)00507-O. [DOI] [PubMed] [Google Scholar]
- Mollereau C, Parmentier M, Mailleux P, Butour JL, Moisand C, Chalon P, Caput D, Vassart G, Meunier JC. ORL1, a novel member of the opioid receptor family. Cloning, functional expression and localization. FEBS Lett. 1994;341:33–38. doi: 10.1016/0014-5793(94)80235-1. [DOI] [PubMed] [Google Scholar]
- Wick MJ, Minnerath SR, Roy S, Ramakrishnan S, Loh HH. Expression of alternate forms of brain opioid 'orphan' receptor mRNA in activated human peripheral blood lymphocytes and lymphocytic cell lines. Brain Res Mol Brain Res. 1995;32:342–347. doi: 10.1016/0169-328X(95)00096-B. [DOI] [PubMed] [Google Scholar]
- Halford WP, Gebhardt BM, Carr DJJ. Functional role and sequence analysis of a lymphocyte orphan opioid receptor. J Neuroimmunol. 1995;59:91–101. doi: 10.1016/0165-5728(95)00030-6. [DOI] [PubMed] [Google Scholar]
- Matthes H, Seward EP, Kieffer B, North RA. Functional selectivity of orphanin FQ for its receptor coexpressed with potassium channel subunits in Xenopus laevis oocytes. Mol Pharmacol. 1996;50:447–450. [PubMed] [Google Scholar]
- Pan YX, Cheng J, Xu J, Rossi G, Jacobson E, Ryan-Moro J, Brooks AI, Dean GE, Standifer KM, Pasternak GW. Cloning and functional characterization through antisense mapping of a kappa 3-related opioid receptor. Mol Pharmacol. 1995;47:1180–1188. [PubMed] [Google Scholar]
- Halford WP, Gebhardt BM, Carr DJ. Functional role and sequence analysis of a lymphocyte orphan opioid receptor. J Neuroimmunol. 1995;59:91–101. doi: 10.1016/0165-5728(95)00030-6. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Kato S, Mori K, Nishi M, Takeshima H, Iwabe N, Miyata T, Houtani T, Sugimoto T. cDNA cloning and regional distribution of a novel member of the opioid receptor family. FEBS Lett. 1994;343:42–46. doi: 10.1016/0014-5793(94)80603-9. [DOI] [PubMed] [Google Scholar]
- Bunzow JR, Saez C, Mortrud M, Bouvier C, Williams JT, Low M, Grandy DK. Molecular cloning and tissue distribution of a putative member of the rat opioid receptor gene family that is not a mu, delta or kappa opioid receptor type. FEBS Lett. 1994;347:284–288. doi: 10.1016/0014-5793(94)00561-3. [DOI] [PubMed] [Google Scholar]
- Chen Y, Fan Y, Liu J, Mestek A, Tian M, Kozak CA, Yu L. Molecular cloning, tissue distribution and chromosomal localization of a novel member of the opioid receptor gene family. FEBS Lett. 1994;347:279–283. doi: 10.1016/0014-5793(94)00560-5. [DOI] [PubMed] [Google Scholar]
- Lachowicz JE, Shen Y, Monsma FJJ, Sibley DR. Molecular cloning of a novel G protein-coupled receptor related to the opiate receptor family. J Neurochem. 1995;64:34–40. doi: 10.1046/j.1471-4159.1995.64010034.x. [DOI] [PubMed] [Google Scholar]
- Wang JB, Johnson PS, Imai Y, Persico AM, Ozenberger BA, Eppler CM, Uhl GR. cDNA cloning of an orphan opiate receptor gene family member and its splice variant. FEBS Lett. 1994;348:75–79. doi: 10.1016/0014-5793(94)00557-5. [DOI] [PubMed] [Google Scholar]
- Wick MJ, Minnerath SR, Lin X, Elde R, Law PY, Loh HH. Isolation of a novel cDNA encoding a putative membrane receptor with high homology to the cloned mu, delta, and kappa opioid receptors. Brain Res Mol Brain Res. 1994;27:37–44. doi: 10.1016/0169-328X(94)90181-3. [DOI] [PubMed] [Google Scholar]