Significance
Lipoic acid is an enzyme cofactor found throughout the biological world that is required for key steps in central metabolism. In humans, defective lipoic acid synthesis results in defective energy production, accumulation of toxic levels of certain amino acids, and early death. The different pathways for lipoic acid synthesis put forth have not been validated by direct analysis of the postulated enzyme reactions, excepting a protein called LIPT1. Unfortunately, the enzyme activity reported for LIPT1 is misleading and seems to be an evolutionary remnant. We report that LIPT1 has a second “moonlighting” enzyme activity that fully explains the physiology of individuals lacking LIPT1 activity. We also document the postulated activity of LIPT2, another essential enzyme of the pathway.
Keywords: lipoic acid, mitochondrial disorder, inborn errors, glycine cleavage system, 2-oxoacid dehydrogenases
Abstract
The lack of attachment of lipoic acid to its cognate enzyme proteins results in devastating human metabolic disorders. These mitochondrial disorders are evident soon after birth and generally result in early death. The mutations causing specific defects in lipoyl assembly map in three genes, LIAS, LIPT1, and LIPT2. Although physiological roles have been proposed for the encoded proteins, only the LIPT1 protein had been studied at the enzyme level. LIPT1 was reported to catalyze only the second partial reaction of the classical lipoate ligase mechanism. We report that the physiologically relevant LIPT1 enzyme activity is transfer of lipoyl moieties from the H protein of the glycine cleavage system to the E2 subunits of the 2-oxoacid dehydrogenases required for respiration (e.g., pyruvate dehydrogenase) and amino acid degradation. We also report that LIPT2 encodes an octanoyl transferase that initiates lipoyl group assembly. The human pathway is now biochemically defined.
Although lipoic acid was discovered over 60 y ago as a covalently bound enzyme cofactor required for aerobic metabolism (1–3), it is only in recent years that the mechanisms of its biosynthesis have become understood (4–6). The importance of protein lipoylation is illustrated by disorders of this mitochondrial pathway, which result in grave metabolic defects and early death.
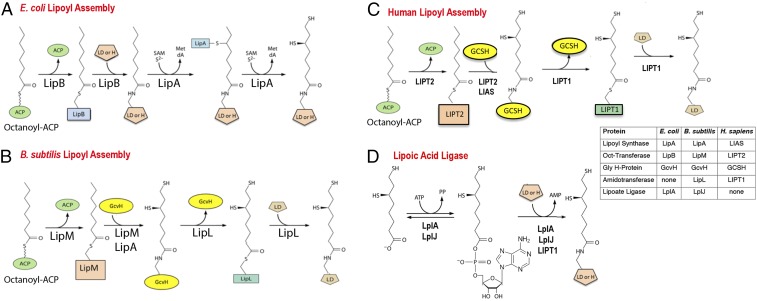
Lipoic acid biosynthesis is best described as an assembly process because lipoyl moieties are constructed on the enzyme subunits of the cognate enzymes via a markedly atypical pathway (7) (Fig. 1). Lipoic acid is an eight-carbon fatty acid in which sulfur atoms replace the hydrogen atoms of carbons 6 and 8 of the acyl chain (oxidation of the resulting disulfide gives lipoic acid). Genetic and biochemical studies in Escherichia coli showed that an octanoate moiety diverted from fatty acid synthesis by the LipB octanoyl transferase becomes attached to the ε-amino group of a specific lysine residue of the cognate enzyme proteins (4). The octanoylated proteins then become substrates for sulfur insertion by the S-adenosyl-l-methionine radical enzyme, LipA (Fig. 1). The lipoyl-modified proteins are the GcvH protein of glycine cleavage (8) and the small universally conserved protein domains located at the amino termini of the E2 subunits of the 2-oxoacid dehydrogenases required for aerobic metabolism and other reactions (4). The LipB–LipA pathway (Fig. 1A) is the simplest, but not the only lipoyl assembly pathway (4). Another bacterium, Bacillus subtilis, requires four proteins for lipoyl assembly rather than the two that accomplish the task in E. coli (4, 9, 10) (Fig. 1B). In contrast to E. coli where the lipoyl assembly pathway directly modifies each of the cognate proteins, B. subtilis assembles lipoyl moieties only on the H protein of the glycine cleavage system (4, 9, 10). The other lipoate-dependent enzymes obtain lipoyl moieties only upon transfer from the H protein. Essentially, the same pathway has recently been documented in Staphylococcus aureus (11). Thus, the small H protein (127 residues) has two functions in central metabolism: the glycine cleavage pathway of single carbon metabolism and lipoylation of the 2-oxoacid dehydrogenases required for aerobic metabolism and branched chain fatty acid synthesis (12). Indeed, B. subtilis strains that lack the H protein are unable to grow without lipoate (or supplements that bypass function of the key 2-oxoacid dehydrogenases) and cannot cleave glycine to serve as the sole nitrogen source (9, 12). The yeast, Saccharomyces cerevisiae, is thought to have a similar pathway (13), although little in vitro enzymology has been done due to the intractable nature of the yeast proteins.
Fig. 1.
Pathways of lipoyl assembly on cognate proteins. (A) The simplest assembly pathway is that of E. coli where only two enzymes are required (4). LipB transfers an octanoyl moiety from octanoyl-ACP to each of the cognate protein substrates. The LipA radical S-adenosyl-l-methionine enzyme then inserts two sulfur atoms to produce dihydrolipoyl moieties. (B) A more complex pathway is found in Firmicute bacteria such as Bacillus subtilis (10, 12, 68) and Staphyloccus aureus (11). In this pathway, lipoyl moieties are assembled on the GcvH protein of the glycine cleavage system and then transferred to the lipoyl domains (LDs) of the 2-oxoacid dehydrogenases. This pathway requires a lipoyl amidotransferase called LipL and a distinct octanoyl transferase called LipM. LipA catalyzes sulfur insertion as in A. (C) The pathway of lipoyl assembly in humans as elucidated in the present work. The pathway parallels the bacterial pathway of B. The differing nomenclatures of the human and bacterial lipoyl assembly proteins and enzymes are given in the Inset. Note that the LIPT1 acyl-enzyme intermediate is hypothetical. (D) The lipoate ligase reaction catalyzed by the lipoate salvage enzymes, E. coli LplA and B. subtilis LplJ (4). Acyl adenylate is a stably bound intermediate in the reaction. Human and bovine LIPT1s can catalyze only the second partial ligase reaction, transfer from the adenylate to the acceptor protein (17, 18). All three enzymes are active with octanoate in place of lipoate.
In mammals, all proteins involved in lipoyl assembly are located in the mitochondria. In humans, the first indicator of defective lipoyl assembly is generally the presence of abnormally elevated levels of lactate (derived by reduction of pyruvate accumulated due to pyruvate dehydrogenase deficiency) in urine and plasma. Subsequent measurements of glycine levels in body fluids allow these individuals to be divided into two groups (14, 15). Normal glycine levels indicate that the glycine cleavage system is functional, and thus the glycine cleavage H protein (GCSH) is lipoylated, whereas abnormally high glycine levels indicate a lack of GCSH lipoylation. Elevated brain glycine levels result in a host of neurological disorders, including neurodegeneration, encephalopathy, and neonatal-onset epilepsy (14, 15), whereas the lack of 2-oxoacid dehydrogenase lipoylation short-circuits function of the tricarboxylic acid cycle, resulting in severe respiratory deficiency and extreme muscle weakness (14, 15).
Human individuals having severely decreased levels of all lipoylated proteins have mutations in either LIAS, which encodes a lipoyl synthase known to functionally replace E. coli LipA (16), or LIPT2, which is proposed to encode an octanoyl transferase (14, 15). The patients who selectively retain GCSH lipoylation have mutations in a third gene, LIPT1 (14, 15). For decades, the reported LIPT1 enzymatic function has muddled interpretation of the human disorders. The reported activity for LIPT1 protein is transfer of a lipoyl moiety from lipoyl-adenylate to both GCSH and to the 2-oxoacid dehydrogenase E2 subunits (17, 18) (Fig. 1D). Modification of both acceptor proteins directly conflicts with the LIPT1 biochemical phenotype because individuals lacking LIPT1 activity should lack GCSH lipoylation whereas GCSH lipoylation (hence glycine cleavage) is normal in LIPT1 patients. A second argument against the physiological relevance of the reported LIPT1 “half-ligase” activity is that there seems to be no valid source of the lipoyl-adenylate required for the reaction. ACSM1, an extraordinarily promiscuous acyl-CoA synthetase (19), was reported to synthesize lipoyl-adenylate (20). However, LIPT1 utilizes both isomers of lipoate, whereas lipoylated proteins contain only the R isomer (20), which strongly argues against a role for ACSM1 in lipoate attachment. Despite these shortcomings, LIPT1 and ACSM1 have been ascribed roles in disorders of human lipoate metabolism, generally in uptake and attachment of dietary lipoic acid (21, 22). However, dietary lipoic acid supplementation has no effect on survival of mammals or tissue culture cells defective in lipoyl assembly (see below). The close analogy of the human lipoate metabolism defects to those of B. subtilis mutant strains defective in lipoylation led to the hypothesis that the relevant LIPT1 activity is transfer of lipoyl moieties from lipoyl-GCSH to the 2-oxoacid dehydrogenase subunits (4). In this scenario, LIPT1 would have lipoyl amidotransferase activity that parallels that demonstrated for B. subtilis LipL (9). It would follow that the lipoyl-adenylate activity would be an evolutionary remnant, as often seen in moonlighting proteins (23–25). It should be noted that recent models of the human disorders include a step in which LIPT1 somehow transfers lipoyl groups from GCSH to the 2-oxoacid dehydrogenase E2 subunits without invoking a mechanism for this transfer (15, 26, 27). We report biochemical evidence obtained by reconstruction of the human assembly pathway in E. coli plus direct biochemical assays with purified enzymes and acceptor proteins that demonstrate LIPT1 to be a lipoyl amidotransferase that catalyzes transfer of lipoyl moieties from GCSH to a 2-oxoacid dehydrogenase domain. We also demonstrate that purified LIPT2 is an octanoyl transferase and that LIPT2 functionally replaces the E. coli octanoyl transferase. These data define the biochemical mechanism of the human lipoyl assembly pathway.
Results
LipT1 Has Lipoyl Amidotransferase Activity.
In our first experiments to test LipT1 amidotransferase activity (Fig. 2), we constructed three compatible plasmids, each of which expresses one of the relevant human proteins: GCSH, LIPT1, or a hexahistidine-tagged lipoyl domain (LD) derived from the E2 subunit of pyruvate dehydrogenase. Expression of each codon-optimized protein was placed under a tightly controlled promoter. The plasmids were transformed into an E. coli ∆lipB strain to attempt construction of the human lipoyl transfer pathway in this bacterium (Fig. 2). We first induced GCSH expression from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter in the presence of lipoic acid to allow accumulation of lipoyl-GCSH (lipoate attachment was via the host LplA lipoate ligase). These cells were then washed free of lipoate and IPTG and suspended in fresh medium lacking lipoate and IPTG (supplementation with acetate and succinate allowed growth to proceed). Expression of LipT1 and the PDH inner E2 domain were then induced with arabinose to provide the enzyme and its putative substrate for lipoyl transfer from GCSH to the E2 domain. The E2 domain was then purified by Ni-chelate chromatography and analyzed by mass spectrometry. The apo-E2 domain (m/z of 13,814.3) was converted to lipoyl-E2 domain (m/z of 14,001.5) (Fig. 2E). The delta mass between the two forms was 187, whereas a lipoyl moiety is 188. The 14,001.5 species was not present when either the LIPT1 or GCSH-encoding plasmid was omitted (Fig. 2 C and D), indicating that the washing steps effectively removed lipoate and thereby rendered the host LplA lipoate ligase inactive.
Fig. 2.
(A) Flow chart of the reconstitution of LIPT1lipoyl transfer from lipoyl-GCSH to a human LD acceptor in the E. coli ∆lipB strain, XC.127. The cultures were grown in glycerol minimal medium with differing supplements as shown in the figure. Supplementation with succinate and acetate bypasses the need for 2-oxoacid dehydrogenase lipoylation (4). Strain XC.127 transformed with the plasmid encoding the His6-LD acceptor protein was additionally transformed with the GCSH plasmid plus the LIPT1 plasmid, the GCSH plasmid alone, or the LIPT1 plasmid alone. The resulting cultures were induced with IPTG in the presence of lipoate to allow the host LplA ligase to synthesize lipoyl-GCSH (if present). The cells of each culture were then collected by centrifugation and washed to remove lipoate and IPTG. After resuspension in glycerol minimal medium containing acetate and succinate, arabinose was added to the three cultures to induce expression of LIPT1 and His6-LD. The cultures were incubated to allow further growth and accumulation of the His6-LD. The cells were then collected and lysed, and the His6-LD of each culture was purified by Ni2+ chelate chromatography. The purified samples were then submitted for mass-spectrometric analysis and the proteins expressed in each sample are summarized in B, where + or − denotes expression. The electrospray mass-spectrometric scans for each culture are given in C–E. (C) Mass-spectrometric analysis of the His6-LD acceptor accumulated in the absence of the LIPT1 plasmid. The LD remained in the apo form (13,796.2 Da). Note that the apo LD mass was 18 Da less than that calculated (13,814.3 Da), consistent with either dehydration (−18 Da) or deamidation (−17 Da) during mass spectrometry (Protein Prospector, prospector.ucsf.edu). Dehydration seems more probable since almost a third of the protein is composed of residues (S, T, E, and D) known to undergo water loss. (D) Mass-spectrometric analysis of the His6-LD acceptor accumulated in the absence of the GCSH plasmid. The LD remained in the apo form (m/z 13,797.2 Da). (E) Mass-spectrometric scan of the His6-LD accumulated when both LIPT1 and GCSH were expressed. The mass of the lipoylated LD form (13,983.1 Da) agrees well with the calculated value (14,001.5 Da). The change in mass upon modification (calculated for lipoyl modification, 188 Da; observed, 187 Da) is within the accuracy of the instrument utilized. a.u., arbitrary units; Intens., intensity.
LipT1 Has Lipoyl Amidotransferase Activity but Lacks Octanoyl Amidotransferase Activity.
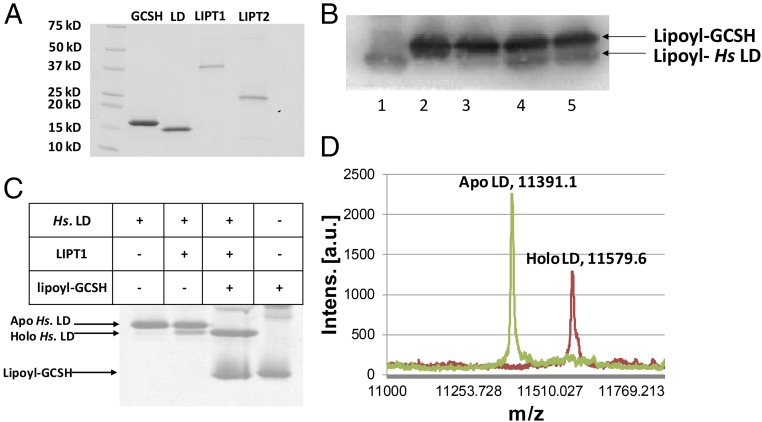
Given these encouraging results, we expressed each of the genes in E. coli and purified the proteins (Fig. 3A). The substrates needed to assay lipoyl amidotransferase activity were also prepared in pure form. These consisted of an acceptor domain consisting of the inner LD of the human pyruvate dehydrogenase E2 subunit and a lipoyl (or octanoyl) donor protein consisting of the modified human GCSH protein. Lipoyl-GCSH was synthesized from the apo protein using the B. subtilis LplJ lipoate ligase (Fig. 1D), ATP, and lipoic acid, whereas octanoyl-GCSH labeled with 14C in the octanoyl moiety was prepared with [1-14C]octanoic acid using the same enzymatic reaction.
Fig. 3.
LIPT1 catalyzes transfer of lipoyl moieties from GCSH to an LD derived from human pyruvate dehydrogenase. (A) Purification of the lipoate assembly proteins. The proteins were purified as described in Experimental Procedures and analyzed by SDS/PAGE on 4–20% polyacrylamide gels. The molecular weights of the Bio-Rad broad-range protein standards are indicated. (B) Western blot analysis of SDS/PAGE assay of LIPT1 lipoyl amidotransferase activity using anti-lipoyl antibody and B. subtilis LplJ-generated lipoyl-GCSH as the substrate. Lipoyl-GCSH was synthesized with LplJ plus ATP and lipoic acid and then purified using anion exchange chromatography to remove LplJ and residual ATP. Lanes: 1, standard lipoyl-LD prepared by LplJ modification of human LD (Hs apo-LD); 2, lipoyl-GCSH; 3, lipoyl-GCSH plus the human LD; 4 and 5, lipoyl-GCSH incubated with LIPT1 and LD. (C) LIPT1 lipoyl amidotransferase activity analyzed by urea gel electrophoresis. Loss of the positive charge of the modified lysine ϵ-amino group of the LD results in faster migration of the modified form on these gels. The gel was stained with Coomassie Blue. (D) Electrospray mass-spectrometric analysis of lipoylated human LD and the remaining apo LD from the reaction of gel (B, lane 4). The calculated difference in mass (delta mass) between the apo and lipoyl forms was 188, whereas the observed delta mass was 188.5. Note that, in A and B, trace levels of LD lipoylation were seen in the absence of GCSH, which is attributed to LIPT1-bound lipoyl-AMP that survives purification and crystallization (28). The traces of lipoylation that appears without LIPT1 in B seems likely to be due the lipoate assembly pathway of the wild-type E. coli strain used for protein production.
Lipoyl-GCSH purified free of ATP and LipJ was incubated with LIPT1 plus the LD acceptor protein. Three different assays were used (Fig. 3). Fig. 3C shows a mobility shift assay using gel electrophoresis in the presence of urea. In this assay, modification of the LD results in an increased rate of migration of the protein due to loss of the positive charge of the modified lysine (both LDs and glycine cleavage H subunits are unusually acidic proteins). In reactions that contained all of the reaction components, the LD was largely converted to a faster-migrating species, whereas no such shift was seen when a reaction component was omitted. A second assay evaluated transfer of lipoyl moieties from purified lipoyl-GCSH to the human LD. These reaction products were separated by SDS/PAGE followed by Western blotting with an anti-lipoate antibody (Fig. 3B). A species having the same mobility as the lipoyl-LD standard was formed in the complete reaction mixture but not when a component was omitted. Finally, a portion of the complete reaction mixture was analyzed by electrospray mass spectrometry. The two peaks observed were the remaining unmodified apo-LD substrate and lipoyl-LD. The difference in mass values of the two peaks was 188.5, whereas a lipoyl moiety has a mass of 188. Note that traces of LD modification were seen in the absence of lipoyl-GCSH (Fig. 3 B and C). Since prior workers demonstrated that LipT1 purified from E. coli contained bound lipoyl-adenylate (28) and can transfer the lipoyl-adenylate lipoyl moiety to LD domains (17), this low level of LD modification is attributed to bound lipoyl-adenylate that accompanied LIPT1 through purification of the protein.
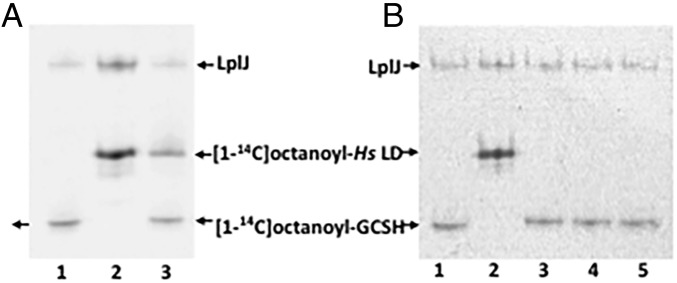
We also tested the ability of LIPT1 to transfer [1-14C]octanoyl moieties from octanoyl-GCSH to the human LD (Fig. 4) and found no detectable transfer. Note that the LD preparation was the same as that used in the lipoyl transfer experiments and was active with LplJ.
Fig. 4.
Ability of LIPT1 to transfer octanoyl moieties from GCSH to an LD derived from human pyruvate dehydrogenase. (A) Autoradiograms of urea–PAGE gels of assays testing LipT1-catalyzed octanoyl amidotransfer from purified [1-14C]octanoyl-GCSH to the lipoyl domain (LD). Lanes: 1, [1-14C]octanoyl-GCSH synthesized using B. subtilis LplJ plus ATP and [1-14C]octanoic acid; 2, [1-14C]octanoyl-LD standard synthesized with LD as in lane 1; 3, LD added to the [1-14C]octanoyl-GCSH synthesis reaction. The [1-14C]octanoyl-labeled LD band indicates that residual ATP remaining from the [1-14C]octanoyl-GCSH synthetic reaction was used by LplJ to modify the LD. (B) Octanoyl amidotransferase urea–PAGE gel assays performed in the presence of an ATP trap (hexokinase plus d-glucose) to prevent LplJ modification of the LD. Lanes: 1 and 2 are a repetition of the experiment of lanes 1 and 2 of the gel in A, respectively; 3, LD added to the [1-14C]octanoyl-GCSH synthesis reaction in the presence of the ATP trap (2 units of hexokinase and 10 mM d-glucose). The lack of LD labeling indicates that the trap eliminated the ATP remaining from the [1-14C]octanoyl-GCSH synthesis reaction; 4 and 5, [1-14C]octanoyl-GCSH with addition of LIPT1, the ATP trap, and the LD acceptor.
The Putative LIPT2 Octanoyl Transferase Catalyzes Transfer from Octanoyl-ACP to GCSH.
Human genetics investigators have postulated that LIPT2 is an octanoyl transferase (5, 15, 27) based on sequence alignments with the octanoyl transferases of E. coli (29, 30) and Mycobacterium tuberculosis (31) (Fig. 5A). However, sequence alignments within Pfam PF03099 protein family are not a trustworthy predictor of function and must be viewed with considerable caution (Fig. 5B). For example, the B. subtilis genome has been annotated as encoding three lipoate ligases (www.microbesonline.org). However, only one protein had ligase activity; the other two proteins catalyzed octanoyl transfer and lipoyl amidotransfer (9, 10). These considerations indicated that validation by direct assay of the postulated LIPT2 activity was required. The lack of such evidence became a more serious shortcoming when the first LIPT2 mutations were detected in human patients (27).
Fig. 5.
Alignment and phylogeny of LIPT2. (A) Alignment of LIPT2 with the enzymatically characterized LipB and LipM proteins. Unweighted sequence alignments were performed using T-Coffee (69) at the European Bioinformatics Institute website (https://www.ebi.ac.uk) using the default settings and displayed using Jalview. The sequence name indicates the enzyme type, the Uniprot code indicates the organism of origin, and the numbers indicate the amino acid residues displayed. Positions having 50% or greater identity are highlighted in blue. The catalytic cysteine residues of the LIPT2, LipB, and LipM are boxed and highlighted in black, as is the conserved lysine residue. The leucine-to-arginine mutations found in the human LIPT2 patients (27) are given in red. The edges of the alignment were trimmed using Jalview (70), so only the catalytic domain is shown. (B) Phylogeny of the LipB_LplA_LipM family (PF03099) was determined with sequences retrieved from the Pfam database (71). Multiple sequence alignments was done using ClustalW (72). The poorly conserved LplA N and C termini were removed. The phylogenetic tree was constructed using the maximum-likelihood method with the PhyML program (73, 74). PHYLIP Interleaved was used for alignment. Bootstrap analysis was set to 1,000 replicates.
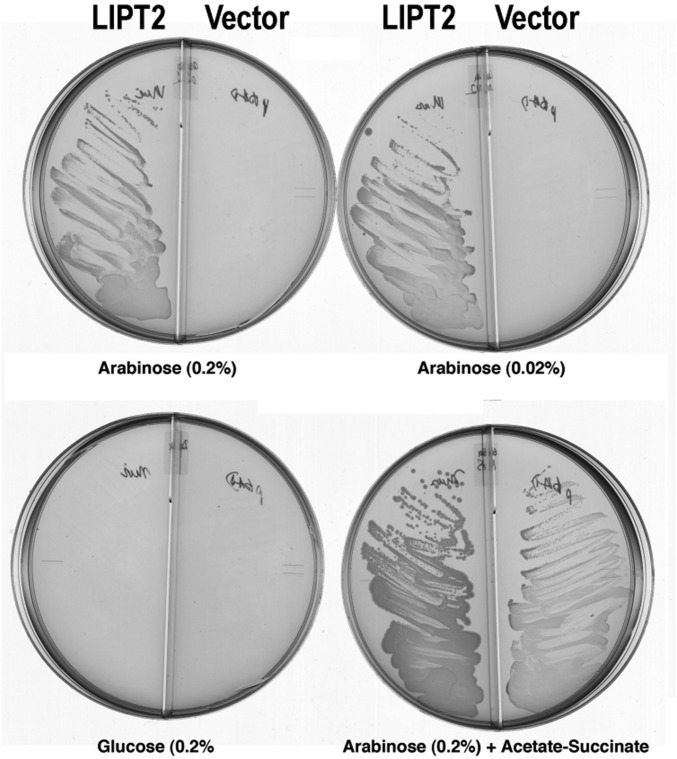
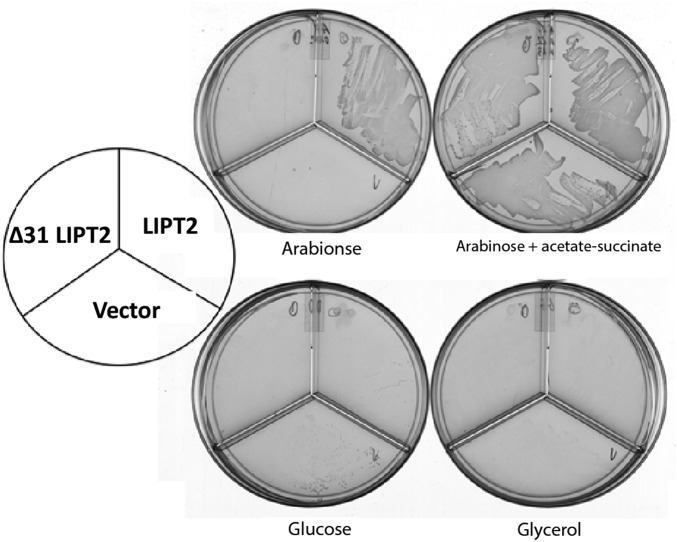
We began by asking whether LIPT2 could functionally replace the E. coli LipB octanoyl transferase, an enzyme essential for lipoyl assembly (29, 32). A synthetic LIPT2 gene with codons optimized for E. coli expression was inserted into the medium copy vector pBAD322A to be transcribed from the arabinose-inducible araBAD promoter and translated using the vector ribosome binding site. The mouse LIPT2 sequence was used because an unambiguous human sequence was not available when this work was initiated. The gene encoding the primary translation product was used because the E. coli and Arabidopsis LipBs are inactivated by small N-terminal truncations (29, 33). The LipT2 plasmid was transformed into an E. coli strain carrying deletions of the genes encoding both LipB and the LplA lipoate ligase. The latter mutation was included to avoid bypass of LipB by LplA mutations (34). The ∆lipB ∆liplA strain was grown on glycerol minimal medium containing acetate and succinate (which bypass the lack of lipoyl proteins) and then streaked on glycerol minimal medium plates containing various supplements. Growth proceeded only when the medium was supplemented with arabinose, the inducer of the araBAD promoter (Fig. 6). Slow growth was observed, which we attribute to the requirement that LIPT2 function with three bacterial protein substrates: ACP and the two 2-oxoacid dehydrogenase E2 subunits. These complementation results argued strongly that LIPT2 was indeed an octanoyl transferase and that E. coli octanoyl-ACP and E. coli E2 LDs should function in vitro as the octanoyl donor and acceptor, respectively.
Fig. 6.
Complementation of the lipB deletion mutation (∆lipB) of E. coli strain QC145 by expression of mouse LIPT2. A synthetic gene encoding the full-length mouse LIPT2 coding sequence was inserted into plasmid vector pBAD322A, which provided the transcription and translation sequences required for expression of the protein to give plasmid pCY754. Transcription was from the vector araBAD promoter, which is induced by arabinose and repressed by glucose. The plasmid was introduced into strain QC145 (∆lipB::cml ∆lplA::kan) (68), which carries a deletion of the lplA lipoate ligase gene in addition to the ∆lipB deletion. The lplA mutation was included because mutant LplA proteins can bypass the ∆lipB defect by scavenging cellular octanoic acid (34). The medium was M9 minimal salts with glycerol as carbon source (glycerol allows basal expression of the araBAD promoter) and 100 μg/mL sodium ampicillin to select for plasmid maintenance. Derivatives of strain QC145 carrying either the LIPT2 plasmid pCY754 or the empty vector were grown on glucose minimal salts containing 5 mM each of sodium acetate and sodium succinate, which bypasses the lipoylation requirement. These cultures were then streaked onto glycerol minimal salts plates supplemented with arabinose, glucose, or acetate plus succinate as given on the figure. The plates used are divided into sectors by plastic walls to prevent cross-feeding. The left sector of each plate contained the strain with the LIPT2 plasmid pCY754, whereas the right sector contained the empty pBAD322A vector. No growth was seen on plates that contained only glycerol (basal expression). The plates were incubated for 2 d at 37 °C.
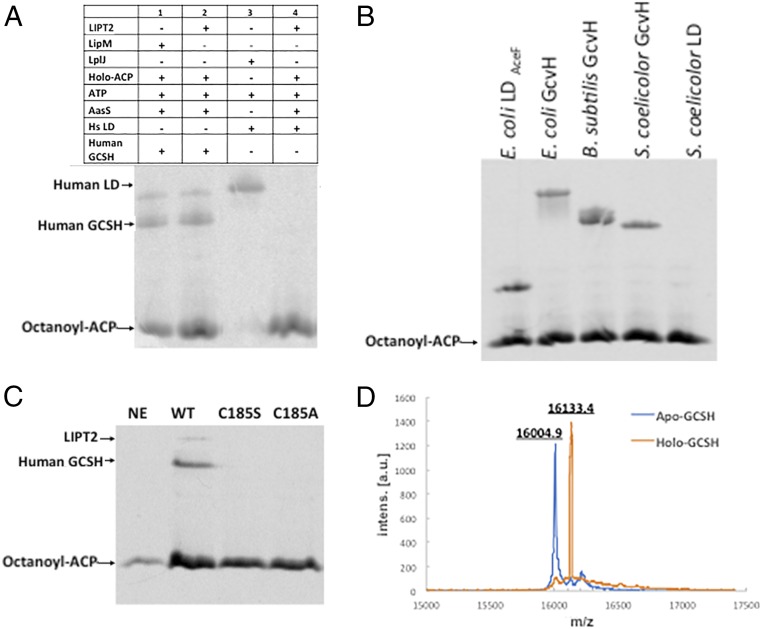
We then expressed a hexahistidine-tagged version of LIPT2 in E. coli and purified the enzyme to homogeneity (Fig. 3). LIPT2 readily transferred the octanoyl moiety from E. coli [14C]octanoyl-ACP to human GCSH but was unable to transfer an octanoyl moiety to the human LD (Fig. 7A). Note that the human LD domain was an excellent substrate for the B. subtilis LplJ ligase and as an acceptor in lipoyl amidotransfer assays, indicating that it had native structure. As expected from the observed functional replacement of E. coli LipB, LIPT2 modified an E. coli LD and also the GcvH proteins of E. coli and two other bacteria (Fig. 7B). In the LipB reaction, an octanoyl moiety is transferred from ACP to the LipB active-site cysteine thiol (30). This acyl-enzyme intermediate is then attacked by the ε-amino group of the target lysine reside to give the octanoylated acceptor protein (30, 35, 36). Based on the alignments of Fig. 5, Cys185 of LIPT2 was expected to be the site of acyl enzyme formation. To test this hypothesis, Cys185 was replaced with either alanine or serine. As expected from prior results with LipB (30), both mutant proteins lacked octanoyl transferase activity (Fig. 7C). Finally, the results obtained using the radioactive assay were validated by mass-spectroscopic analysis of LIPT2-catalyzed modification of human GCSH with a nonradioactive octanoyl moiety (Fig. 7D).
Fig. 7.
LIPT2 octanoyl transferase activity in vitro. The substrate used in these experiment was E. coli ACP acylated with [1-14C]octanoic acid by AasS acyl-ACP synthetase. (A) Lanes: 1, standard [1-14C]octanoyl-GCSH prepared using B. subtilis LipM; 2, LIPT2-catalyzed synthesis of [1-14C]octanoyl-GCSH; 3, standard [1-14C]octanoyl-LD prepared using B. subtilis LplJ; and 4, LIPT2 fails to catalyze transfer of [1-14C]octanoyl moiety from [1-14C]octanoyl-ACP to the human LD. (B) LIPT2 is active on a several bacterial GcvH and LD proteins. The bands migrating more slowly than octanoyl-ACP are the octanoylated acceptor proteins. The right-hand lanes are Streptomyces coelicolor acceptor proteins (the LD preparation tested was inactive). (C) Mutant LIPT2 proteins lacking C185 are inactive. Designations: NE, no LIPT2; WT, wild-type LIPT2; and C185S and C185A, respectively, denote proteins in which serine or alanine replaced cysteine 185. (D) Electrospray mass-spectrometric analysis of the unmodified (calculated mass of 15,997.6 Da) and octanoylated (calculated mass of 16,123.6 Da) forms of GCSH by LIPT2, respectively. Within the accuracy of the instrument, the protein masses agree well with the calculated values, as does the change in mass upon modification (calculated, 126 Da; observed, 128.5 Da). a.u., arbitrary units; Intens, intensity.
As first seen with the mouse LIPT2 (Fig. 6), a synthetic gene expressing the full-length human LIPT2 (84% identical to the mouse LIPT2) restored growth of the E. coli ∆lipB ∆lplA strain in the presence of arabinose (Fig. 8). However, the putative protease-processed mature form of human LIPT2 recently reported (26) (a deletion of residues 2–31) failed to complement (Fig. 8). Hence, although the cell death observed (26) was attributed to a lack of mitochondrial targeting, the LIPT2 construct also lacked activity. Indeed, LIPT2 is not processed. Several mass-spectral analyses of the human proteome report detection of LIPT2 peptides corresponding to the N-terminal seven residues plus residues 14–24 and 29–40 of the primary translation product (37–39) (the peptide data are collected at www.proteomicsdb.org), which would be lacking in the putative mature form (26). Note that, consistent with human LIPT2, loss of the 22 N-terminal E. coli LipB residues inactivated the protein (29). This might be explained by the similarities (underlined) seen in the N-terminal sequences of LIPT2 (residues 5–19, AVRLVRLGRVPYAEL, and LipB residues 5–14, LVRQLG—LPYEPI).
Fig. 8.
Complementation of the ∆lipB deletion of E. coli strain QC146 by expression of human LIPT2. Synthetic genes encoding either the full-length human LIPT2 or a derivative that lacked the first 31 residues (∆31, a methionine codon replaced residue 31 to permit translation) were inserted into vector pBAD322A as in Fig. 6 to give plasmids pCY1110 and pCY1108, respectively. The plasmids were introduced into strain QC146 (∆lipB ∆lplA) (75), and transformants were streaked onto plates containing 0.02% arabinose or another carbon source as given in Fig. 6. Note that, unlike the mouse LIPT2, 0.2% arabinose gave rapid growth, but growth soon halted, suggesting high-level expression of human LIPT2 is toxic.
Discussion
Our evidence that LIPT1 has lipoyl amidotransferase activity renders inoperative the models of the human disorders that include the partial ligase activity. This, plus the demonstration that LIPT2 is an octanoyl transferase, defines a straightforward pathway that fully explains each of the phenotypes of the human disorders. Individuals having mutations that result in loss of function of LIAS or LIPT2 are unable to assemble lipoyl moieties, and hence all cognate proteins remain in their unmodified and inactive apo forms. These individuals suffer high levels of body fluid glycine, lysine, and branched chain amino acids plus defective energy metabolism (14, 15). In contrast, although patients carrying LIPT1 mutations have normal lipoyl-GCSH and glycine cleavage levels, they suffer from defective energy metabolism plus high levels of lysine and branched chain amino acids (14, 15). Note that decreased 2-oxoacid dehydrogenase activities would additionally result in severely decreased levels of acetyl-CoA and succinyl-CoA, and thus modification of histones and other proteins could be compromised (40).
Our data plus prior work on mitochondrial fatty acid synthesis put the pathway (Fig. 1C) on a solid basis. Following demonstration that mammalian mitochondria contain soluble ACP in addition to the ACP molecules that become subunits of respiratory complex I (41), a complete type II fatty acid synthesis pathway in mammalian mitochondria was demonstrated by Smith and coworkers (42–44), who went on to show that this pathway provides the octanoyl-ACP required for lipoyl moiety synthesis (42–44). Moreover, these workers showed that blocking mitochondrial fatty acid synthesis in transgenic mice blocked protein lipoylation and resulted in a variety of serious physiological abnormalities (including early death) due to disruption of energy metabolism (45). A similar but less severe decrease in lipoyl protein assembly due to deficient mitochondrial fatty acid synthesis was recently reported in human patients (46). GCSH is the only gene of the pathway in which no human disorder maps, although mutations in the other genes specific to glycine cleavage (AMT and GLDC) are fairly abundant (47). This disparity seems due to the small size of the GCSH coding sequence (173 codons), a comparatively small mutational target relative to the coding sequences of AMT (403 codons) and GLDC (1020 codons). Mutations inactivating GCSH should have the same phenotype as mutations in LIAS or LIPT2 and thus would have more profound effects that those individuals having AMT or GLDC mutations, which retain normal energy metabolism and protein modification ability (47).
Our finding that LIPT1 has two mechanistically discrete enzyme activities fits the concept of enzyme evolution called “moonlighting” that has received strong support in recent years (23–25, 48–52), including its importance in diagnosis of inborn errors of metabolism (53, 54). It has been shown that a protein can acquire a second (moonlighting) function without concomitantly losing all or part of its original function. That is, mutations can enhance the moonlighting function without necessarily eliminating the ancestral function (23–25). The original ancestor of LIPT1 seems likely to have encoded a fully functional lipoate ligase analogous to E. coli LplA and B. subtilis LplJ (Fig. 1) rather than the present defective (half) ligase. In this scenario, evolution of the LIPT1 gene has resulted in a protein that has lost the ability to activate lipoate while acquiring amidotransferase activity and (temporarily?) retaining the lipoyl transfer activity of the defective ligase. Indeed, LIPT1 is a member of Pfam PF03099, a group of enzymes that are constructed on the same structural scaffold (4), albeit from diverse sequences and extra domains in the ligases. Surprisingly, despite their conserved structural scaffold, these enzymes perform chemically distinct reactions: They can be lipoate (or biotin) ligases, octanoyl transferases, or lipoyl amidotransferases (4). Even PF03099 enzymes that catalyze the same reaction via the same chemical mechanism can be divergent. The LipB and LipM octanoyl transferases share almost no sequence conservation, and their active-site cysteine residues are found on different loops of the common scaffold (35) (Fig. 5B). It would be interesting to produce a mutant LIPT1 that lacks the partial ligase activity while retaining its amidotransferase activity (assuming that the same lipoate binding site is used in both reactions). A straightforward approach would be to mutate the LIPT1 residues that are hydrogen bonded to the lipoyl adenylate adenosine moiety. However, this is problematical because those bonds are primarily formed with backbone atoms (28). It should be noted that, although both LIPT1 reactions involve transfer of a lipoyl moiety, the energetics of transfer are strikingly different. Lipoyl-adenylate contains a “high-energy” mixed anhydride linkage, and thus lipoyl transfer is extremely facile. Indeed, adenylates are known to readily modify protein amino groups without enzymatic assistance (55, 56). In contrast, the amide linking the lipoyl moiety to GCSH is among the lowest of “low-energy” linkages found in biology, and thus lipoyl transfer from this linkage is kinetically and chemically challenging.
Although modification of lipoyl proteins by incorporation of exogenously supplied lipoic acid has been invoked in models of human lipoate disorders (21, 22), there is a large body of evidence indicating that mammals are unable to use exogenous lipoic acid to bypass loss of the lipoyl assembly pathway. Dietary lipoate readily enters the bloodstream and tissues. Radioactive lipoic acid has been administered to mammals and its fate followed (57, 58). The labeled cofactor was quickly reduced and degraded by the β-oxidation pathway and no evidence for attachment of exogenously fed lipoate to proteins was reported. Moreover, studies of homozygous lipoyl synthase (LIAS) knockout mice (59), of LIAS, LIPT1, and LIPT2 patients (plus fibroblasts derived from patients) (14–16, 27, 60), and mammalian tissue culture cells blocked in synthesis of the lipoate backbone (42) invariably report that lipoic acid supplementation is without benefit. Moreover, lipoic acid supplementation did not significantly increase the levels of lipoyl-modified 2-oxoacid dehydrogenases or GCSH (14–16, 60).
Our finding that LIPT2 is unable to catalyze octanoyl transfer from octanoyl-GCSH to the human pyruvate dehydrogenase LD (Fig. 7A) is expected from the phenotype of the LIPT1 disorder. If octanoylation of the 2-oxoacid dehydrogenase E2 subunits did occur, this could provide a substrate for LIAS-catalyzed sulfur insertion as suggested (26). However, if this were the case, loss of LIPT1 activity would be bypassed and no LIPT1 metabolic disorder would exist.
Experimental Procedures
Chemicals and Growth Media.
The antibiotics and most chemicals used in this study were purchased from Millipore, Sigma, and Fisher, unless noted otherwise. American Radiolabeled Chemicals provided [1-14C]octanoic acid. DNA manipulation enzymes were from New England Biolabs). DNA sequencing was performed by AGCT. Invitrogen provided the Ni2+-agarose column. Growth media were as in prior publications.
Plasmids and Bacterial Strains (Table 1).
Table 1.
Bacterial strains and plasmids
| Strains/plasmids | Relevant genotype or description | Reference or derivation |
| E. coli strains | ||
| BL21(DE3) | E. coli B ompT hsdSB gal dcm (DE3) | Lab stock |
| DH5α | ∆(argF−lacZ)U169 Φ80 ∆(lacZ)M15 recA1 endA1 | Lab stock |
| MG1655 | Wild-type E. coli K-12 | Lab stock |
| BL21(Tuner) | ompT hsdSB lacY1 gal dcm (DE3 | Lab stock |
| QC145 | lipB::cml ∆lplA::kan of MG1655 | Ref. 68 |
| QC146 | ∆lipB ∆lplA of MG1655 | Ref. 68 |
| XC.080 | BL21 (DE3)/pXC.065 | This study |
| XC.083 | BL21 (Tuner)/pXC.044 | This study |
| XC.127 | ∆lipB of MG1655 | This study |
| XC.131 | XC.127/pXC.067, pXC.068, pXC.069 | This study |
| XC.139 | XC.127/pXC.067 and pXC.069 | This study |
| XC.184 | QC146/pXC.064 and pTARA | This study |
| XC.213 | DH5α/pXC.066 | This study |
| Plasmids | ||
| pET28b | T7 promoter expression vector, KanR | Novagen |
| pQE-2 | T5 promoter expression vector, AmpR | Qiagen |
| pTARA | T7 RNA polymerase expression | Ref. 76 |
| pEH1 | lacUV5 promoter expression vector, KanR | Ref. 77 |
| pBAD33 | araBAD promoter expression, p15 ori CmlR | Ref. 78 |
| pBAD1031G | araBAD promoter expression vector, p1031 ori GmR | Ref. 79 |
| pKD46 | Recombineering phage λred genes | Ref. 62 |
| pCP20 | Yeast Flp recombinase gene | Ref. 62 |
| pXC.004 | pET28b encoding native S. coelicolor GcvH | This study |
| pXC.044 | pET28b encoding N-terminal His6-human GCSH | This study |
| pXC.065 | pET28b encoding N-terminal His6-human LIPT1 | This study |
| pXC.066 | pQE-2 encoding N-terminal His6-mouse LIPT2 | This study |
| pXC.067 | pEH1 encoding native human GCSH | This study |
| pXC.068 | pBAD33 encoding human LIPT1 | This study |
| pXC.069 | pBAD1031G encoding N-terminal His6-human LD of pyruvate dehydrogenase E2 | This study |
| pCY754 | pBAD322A encoding full length mouse LIPT2 | This study |
| pCY1108 | pBAD322A encoding human LIPT2 lacking the 31 N-terminal residues. | This study |
| pCY1110 | pBAD322A encoding full-length human LIPT2 | This study |
Human genes synthesized with optimized E. coli codons encoding GCSH, the inner LD of the E2 component of the human pyruvate dehydrogenase complex (E2p), LIPT1, and human or mouse LIPT2 were from Epoch Life Science or Integrated DNA Technologies. All constructs were verified by sequencing. The human LD and LIPT1 genes were inserted into vector pET28b (Table 1) to generate plasmids pXC.064 and pXC.065 using restriction sites NdeI plus BamHI and NdeI plus HindIII, respectively. Mouse LIPT2 was amplified with primers oXC288/oXc289 and inserted into the NdeI plus SalI restriction sites of vector pQE-2 to give pXC.066. The human GCSH gene was amplified with primers oXC159/oXC160 (Table 2), which added BspHI and HindIII sites to allow ligation into NcoI plus HindIII-cut vector pEH1 downstream of an IPTG-inducible promoter to give pXC.067. Plasmid pXC.068 was generated by excising LIPT1 directly from pXC.065 with restriction enzymes XbaI plus HindIII and ligation into pBAD33 using the same restriction sites. Plasmid pXC.068 carries the pET28b ribosome binding site. Plasmid pXC.069 was generated by excision of the human His6-LD gene from pXC.064 via the NcoI and HindIII sites followed by ligated into the same sites of pBAD1031G (Table 1).
Table 2.
Oligonucleotides
| Oligonucleotide | Sequence, 5′–3′ |
| Mouse lipT2, forward (NdeI) | ATTCACATATGAGCCTGCCGGTGGTG |
| Mouse lipT2, reverse (SalI) | TATAGTCGACTTAGCTCGGGCTATCTTCGCTAATCAG |
| Human gcsH, forward (BspHI) | ATAATTCATGAGCGTGCGCAAATTCA |
| Human gcsH, reverse (HindIII) | TAGATAAGCTTTCACTCCTCAATGG |
| P1 priming site for E. coli lipB | TTTCCCCCACTTTTACTCATTCTCCACGGAGATGCCGTTTT |
| GTATCAGGATAAAATTCgtgtaggctggagctgcttc | |
| P2 priming site for E. coli lipB | AGATATTATGAGTAATGACCCAGTGTAAATTGGGCCA |
| TTGATGTATGGAAcatatgaatatcctcctta | |
| lipB 250 bp upstream, forward | AAGTGGTACAGCGCCATGCGC |
| lipB 250 bp downstream, reverse | TTGGAACGGAACGCTTTTGCTCACC |
Construction of the lipoate auxotroph strain XC127 (MG1655 ∆lipB) was performed as previously described using pKD3 (Table 1) as the template and P1–P2 as the primers (Table 2) (61, 62), and the chloramphenicol marker was excised by pCP20-encoded Flp recombinase encoded by pCP20 (Table 1) to yield XC.127. The ∆lipB construct was verified by sequencing a PCR product obtained using primers oXC134 and oXC135 (Table 2).
Protein Expression and Purification.
Hexahistidine-tagged versions of Homo sapiens GCSH and LIPT1 were expressed in E. coli BL21, whereas Mus musculus LIPT2 was expressed in DH5α. These strains were grown in 1 L of LB medium containing the antibiotics required for plasmid maintenance. Expression was induced by the addition of 25 µM IPTG at the start of the culture. Cells were harvested by centrifugation after incubation at 30 °C for 22 h. The proteins were purified by nickel affinity and anion exchange chromatographic steps as previously described (63). Protein concentrations were determined both by the Bradford assay (64) and at 280 nm using extinction coefficients calculated from the ProtParam program of the ExPASy tool website. Protein purity was monitored by SDS/PAGE.
To purify the hexahistidine-tagged human LD (E2p) in the purely apo form, plasmids pXC.064 and pTARA were cotransformed into the lipoic acid auxotrophic strain QC146 to yield strain XC.184. The strain was grown at 30 °C in M9 minimum medium with 0.8% glycerol, 5 mM acetate, and 5 mM succinate (pH 7.0), and 0.2% arabinose was added at culture initiation. IPTG was added to 100 µM when the culture reached an absorbance of 0.6 at 600 nm. The culture was incubated for another 6 h before the cells were frozen at −80 °C. The protein was purified by Ni2+ affinity chromatography followed by anion exchange chromatography as previously described (65). Bacillus subtilis LipM and GcvH, E. coli holo-acyl carrier protein (ACP) and LD, the Vibrio harveyi AasS, and the Streptomyces coelicolor GcvH and LD proteins were purified as described previously (12, 35, 65–67). Electrospray mass spectrometry was carried out as described previously (65).
Preparation of [1-14C]Octanoyl-GCSH and Assay of LIPT1-Catalyzed Octanoyl Transfer.
[1-14C]Octanoyl-labeled GCSH was prepared using B. subtilis lipoate protein ligase (LplJ) and purified by nickel affinity plus ion exchange chromatography as previously described (10). The 100-μL reaction mixture contained 50 mM sodium phosphate (pH 7.8), 2 mM MgCl2, 10 mM ATP, 0.5 mM sodium [1-14C]octanoate, 0.5 mM human GCSH, and 5 μM LplJ. The reaction was allowed to proceed for 4 h at 37 °C.
To test whether LIPT1 transfer from octanoyl-GCSH to apo LD, the substrate was [1-14C]octanoyl-GCSH (see above). Each reaction (30 µL) contained 20-µL [1-14C]octanoyl-GCSH mixtures (containing LplJ and residual ATP), 50 mM sodium chloride, 2 µM purified LipT1, 20 µM human LD, 2 units of hexokinase, and 10 mM d-glucose. Hexokinase plus d-glucose served as an ATP trap to remove any ATP remaining from the LplJ-catalyzed reaction. After incubation at 37 °C for 1 h, each reaction was loaded on a 15% native polyacrylamide gel containing 2 M urea, and separated by electrophoresis. The gel was stained with Coomassie R-250, soaked in Amplify (GE Healthcare), dried on filter paper, and exposed to preflashed Biomax XAR film (Kodak) at −80 °C for 24 h.
Preparation of Lipoyl-GCSH and Assay of LIPT1-Catalyzed Lipoyl Transfer.
To directly measure lipoyl amidotransfer by LIPT1, lipoyl-GCSH was synthesized as a substrate using LplJ. The reaction contained 50 mM sodium phosphate (pH 7.8), 10 mM ATP, 2 mM MgCl2, 1 mM sodium lipoate, and 10 µM LplJ, and was incubated at 37 °C for 4 h. The reaction was diluted 20-fold in 50 mM sodium phosphate buffer (pH 8.0) and purified by anion exchange chromatography using an AKTA Purifier10 (GE Healthcare) with a 5-mL POROS QE anion exchange column with a flow rate of 2.5 mL per min. Proteins were eluted with a 0–2 M NaCl gradient. Lipoyl-GcvH eluted at about 400 mM NaCl. Lipoyl amidotransfer to apo human LD protein in reactions (20 µL) containing 50 mM sodium phosphate (pH 7.8), 50 mM sodium chloride, 20 µM lipoyl-GCSH, 20 µM apo human LD, and 2 µM LIPT1. The reactions (20 µL) were incubated at 37 °C for 1 h, loaded on a 2 M urea–PAGE gel (15% polyacrylamide) for gel shift analysis, or loaded onto 15% SDS/PAGE gels for Western blot analysis.
Western Blot Analysis of LIPT1 Amidotransferase Activity.
Anti-lipoyl protein primary antibody was utilized to probe protein lipoylation, as described previously (65). Briefly, LIPT1-catalyzed amidotransfer reactions (20 µL) were loaded onto SDS/PAGE gel and transferred by electrophoresis to Immobilon-P membranes (Millipore) for 30 min at 60 V. The membranes were preblocked with TBS buffer (100 mM Tris base and 0.9% NaCl, pH 7.5) containing 0.1% Tween 20 and 5% nonfat milk powder. The membranes were probed for 1 h with an anti-lipoyl protein primary antibody (Calbiochem) diluted 1:10,000 in the above buffer. Following incubation with anti-rabbit secondary antibody (diluted 1:5,000; GE Healthcare Life Sciences), the labeled proteins (Human LD) were detected using Quantity One software.
Coupled AasS/LIPT2 Octanoyl Transfer Reaction.
The coupled reaction mixture (25 μL) contained 100 mM sodium phosphate (pH 7.2), 50 mM NaCl, 5 mM disodium ATP, 0.25 mM [1-14C]sodium octanoate, 50 μM holo-ACP, 2.5 μM of the V. harveyi AasS acyl-ACP synthetase, 2 μM LIPT2, and ∼20 μM human GCSH or another acceptor protein. The reaction was performed at 37 °C for 2 h. The products were electrophoresed on 2 M urea–PAGE (15% acrylamide), and then dried under vacuum at 65 °C for 2 h and exposed to preflashed Biomax XAR film (Kodak) at −80 °C for 24 h.
Acknowledgments
This work was supported by National Institutes of Health Grant AI15650 from the National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
References
- 1.Reed LJ. From lipoic acid to multi-enzyme complexes. Protein Sci. 1998;7:220–224. doi: 10.1002/pro.5560070125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed LJ. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J Biol Chem. 2001;276:38329–38336. doi: 10.1074/jbc.R100026200. [DOI] [PubMed] [Google Scholar]
- 3.Reed LJ, DeBUSK BG, Gunsalus IC, Hornberger CS., Jr Crystalline alpha-lipoic acid; a catalytic agent associated with pyruvate dehydrogenase. Science. 1951;114:93–94. doi: 10.1126/science.114.2952.93. [DOI] [PubMed] [Google Scholar]
- 4.Cronan JE. Assembly of lipoic acid on its cognate enzymes: An extraordinary and essential biosynthetic pathway. Microbiol Mol Biol Rev. 2016;80:429–450. doi: 10.1128/MMBR.00073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solmonson AD, DeBerardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. J Biol Chem. 2018;293:7522–7530. doi: 10.1074/jbc.TM117.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spalding MD, Prigge ST. Lipoic acid metabolism in microbial pathogens. Microbiol Mol Biol Rev. 2010;74:200–228. doi: 10.1128/MMBR.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X, Miller JR, Jiang Y, Marletta MA, Cronan JE. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem Biol. 2003;10:1293–1302. doi: 10.1016/j.chembiol.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Nesbitt NM, et al. Expression, purification, and physical characterization of Escherichia coli lipoyl(octanoyl)transferase. Protein Expr Purif. 2005;39:269–282. doi: 10.1016/j.pep.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 9.Christensen QH, Martin N, Mansilla MC, de Mendoza D, Cronan JE. A novel amidotransferase required for lipoic acid cofactor assembly in Bacillus subtilis. Mol Microbiol. 2011;80:350–363. doi: 10.1111/j.1365-2958.2011.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin N, Christensen QH, Mansilla MC, Cronan JE, de Mendoza D. A novel two-gene requirement for the octanoyltransfer reaction of Bacillus subtilis lipoic acid biosynthesis. Mol Microbiol. 2011;80:335–349. doi: 10.1111/j.1365-2958.2011.07597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zorzoli A, Grayczyk JP, Alonzo F., 3rd Staphylococcus aureus tissue infection during sepsis is supported by differential use of bacterial or host-derived lipoic acid. PLoS Pathog. 2016;12:e1005933. doi: 10.1371/journal.ppat.1005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao X, Hong Y, Zhu L, Hu Y, Cronan JE. Development and retention of a primordial moonlighting pathway of protein modification in the absence of selection presents a puzzle. Proc Natl Acad Sci USA. 2018;115:647–655. doi: 10.1073/pnas.1718653115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermes FA, Cronan JE. The role of the Saccharomyces cerevisiae lipoate protein ligase homologue, Lip3, in lipoic acid synthesis. Yeast. 2013;30:415–427. doi: 10.1002/yea.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayr JA, Feichtinger RG, Tort F, Ribes A, Sperl W. Lipoic acid biosynthesis defects. J Inherit Metab Dis. 2014;37:553–563. doi: 10.1007/s10545-014-9705-8. [DOI] [PubMed] [Google Scholar]
- 15.Tort F, Ferrer-Cortes X, Ribes A. Differential diagnosis of lipoic acid synthesis defects. J Inherit Metab Dis. 2016;39:781–793. doi: 10.1007/s10545-016-9975-4. [DOI] [PubMed] [Google Scholar]
- 16.Mayr JA, et al. Lipoic acid synthetase deficiency causes neonatal-onset epilepsy, defective mitochondrial energy metabolism, and glycine elevation. Am J Hum Genet. 2011;89:792–797. doi: 10.1016/j.ajhg.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara K, Okamura-Ikeda K, Motokawa Y. Lipoylation of acyltransferase components of alpha-ketoacid dehydrogenase complexes. J Biol Chem. 1996;271:12932–12936. doi: 10.1074/jbc.271.22.12932. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara K, Okamura-Ikeda K, Motokawa Y. Lipoate addition to acyltransferases of alpha-keto acid dehydrogenase complexes and H-protein of glycine cleavage system. Methods Enzymol. 1997;279:184–193. doi: 10.1016/s0076-6879(97)79022-0. [DOI] [PubMed] [Google Scholar]
- 19.Vessey DA, Lau E, Kelley M. Isolation and sequencing of cDNAs for the XL-I and XL-III forms of bovine liver xenobiotic-metabolizing medium-chain fatty acid:CoA ligase. J Biochem Mol Toxicol. 2000;14:11–19. doi: 10.1002/(sici)1099-0461(2000)14:1<11::aid-jbt2>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara K, Takeuchi S, Okamura-Ikeda K, Motokawa Y. Purification, characterization, and cDNA cloning of lipoate-activating enzyme from bovine liver. J Biol Chem. 2001;276:28819–28823. doi: 10.1074/jbc.M101748200. [DOI] [PubMed] [Google Scholar]
- 21.Baker PR, 2nd, et al. Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain. 2014;137:366–379. doi: 10.1093/brain/awt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soreze Y, et al. Mutations in human lipoyltransferase gene LIPT1 cause a Leigh disease with secondary deficiency for pyruvate and alpha-ketoglutarate dehydrogenase. Orphanet J Rare Dis. 2013;8:192. doi: 10.1186/1750-1172-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Zabad S, Liu H, Wang W, Jeffery C. MoonProt 2.0: An expansion and update of the moonlighting proteins database. Nucleic Acids Res. 2018;46:D640–D644. doi: 10.1093/nar/gkx1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Copley SD. Moonlighting is mainstream: Paradigm adjustment required. BioEssays. 2012;34:578–588. doi: 10.1002/bies.201100191. [DOI] [PubMed] [Google Scholar]
- 25.Copley SD. An evolutionary perspective on protein moonlighting. Biochem Soc Trans. 2014;42:1684–1691. doi: 10.1042/BST20140245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernardinelli E, et al. Mis-targeting of the mitochondrial protein LIPT2 leads to apoptotic cell death. PLoS One. 2017;12:e0179591. doi: 10.1371/journal.pone.0179591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habarou F, et al. Biallelic mutations in LIPT2 cause a mitochondrial lipoylation defect associated with severe neonatal encephalopathy. Am J Hum Genet. 2017;101:283–290. doi: 10.1016/j.ajhg.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujiwara K, et al. Crystal structure of bovine lipoyltransferase in complex with lipoyl-AMP. J Mol Biol. 2007;371:222–234. doi: 10.1016/j.jmb.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 29.Jordan SW, Cronan JE., Jr The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein:protein transferase. J Bacteriol. 2003;185:1582–1589. doi: 10.1128/JB.185.5.1582-1589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Miller JR, Cronan JE. The reaction of LipB, the octanoyl-[acyl carrier protein]:protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry. 2005;44:16737–16746. doi: 10.1021/bi051865y. [DOI] [PubMed] [Google Scholar]
- 31.Ma Q, et al. The Mycobacterium tuberculosis LipB enzyme functions as a cysteine/lysine dyad acyltransferase. Proc Natl Acad Sci USA. 2006;103:8662–8667. doi: 10.1073/pnas.0510436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris TW, Reed KE, Cronan JE., Jr Lipoic acid metabolism in Escherichia coli: The lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada M, Yasuno R, Jordan SW, Cronan JE, Jr, Wada H. Lipoic acid metabolism in Arabidopsis thaliana: Cloning and characterization of a cDNA encoding lipoyltransferase. Plant Cell Physiol. 2001;42:650–656. doi: 10.1093/pcp/pce081. [DOI] [PubMed] [Google Scholar]
- 34.Hermes FA, Cronan JE. Scavenging of cytosolic octanoic acid by mutant LplA lipoate ligases allows growth of Escherichia coli strains lacking the LipB octanoyltransferase of lipoic acid synthesis. J Bacteriol. 2009;191:6796–6803. doi: 10.1128/JB.00798-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen QH, Cronan JE. Lipoic acid synthesis: A new family of octanoyltransferases generally annotated as lipoate protein ligases. Biochemistry. 2010;49:10024–10036. doi: 10.1021/bi101215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassan BH, Cronan JE. Protein-protein interactions in assembly of lipoic acid on the 2-oxoacid dehydrogenases of aerobic metabolism. J Biol Chem. 2011;286:8263–8276. doi: 10.1074/jbc.M110.194191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11:M111.014050. doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maier SK, et al. Comprehensive identification of proteins from MALDI imaging. Mol Cell Proteomics. 2013;12:2901–2910. doi: 10.1074/mcp.M113.027599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 41.Cronan JE, Fearnley IM, Walker JE. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Lett. 2005;579:4892–4896. doi: 10.1016/j.febslet.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 42.Feng D, Witkowski A, Smith S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J Biol Chem. 2009;284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witkowski A, Joshi AK, Smith S. Coupling of the de novo fatty acid biosynthesis and lipoylation pathways in mammalian mitochondria. J Biol Chem. 2007;282:14178–14185. doi: 10.1074/jbc.M701486200. [DOI] [PubMed] [Google Scholar]
- 44.Witkowski A, Thweatt J, Smith S. Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. J Biol Chem. 2011;286:33729–33736. doi: 10.1074/jbc.M111.291591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith S, et al. Compromised mitochondrial fatty acid synthesis in transgenic mice results in defective protein lipoylation and energy disequilibrium. PLoS One. 2012;7:e47196. doi: 10.1371/journal.pone.0047196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heimer G, et al. University of Washington Center for Mendelian Genomics MECR mutations cause childhood-onset dystonia and optic atrophy, a mitochondrial fatty acid synthesis disorder. Am J Hum Genet. 2016;99:1229–1244. doi: 10.1016/j.ajhg.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azize NA, et al. Mutation analysis of glycine decarboxylase, aminomethyltransferase and glycine cleavage system protein-H genes in 13 unrelated families with glycine encephalopathy. J Hum Genet. 2014;59:593–597. doi: 10.1038/jhg.2014.69. [DOI] [PubMed] [Google Scholar]
- 48.Espinosa-Cantú A, Ascencio D, Barona-Gómez F, DeLuna A. Gene duplication and the evolution of moonlighting proteins. Front Genet. 2015;6:227. doi: 10.3389/fgene.2015.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeffery CJ. Why study moonlighting proteins? Front Genet. 2015;6:211. doi: 10.3389/fgene.2015.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo M, Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat Chem Biol. 2013;9:145–153. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 52.Copley SD. Toward a systems biology perspective on enzyme evolution. J Biol Chem. 2012;287:3–10. doi: 10.1074/jbc.R111.254714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Houten SM. Protein moonlighting in inborn errors of metabolism: The case of the mitochondrial acylglycerol kinase. J Inherit Metab Dis. 2017;40:755–756. doi: 10.1007/s10545-017-0090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sriram G, Martinez JA, McCabe ER, Liao JC, Dipple KM. Single-gene disorders: What role could moonlighting enzymes play? Am J Hum Genet. 2005;76:911–924. doi: 10.1086/430799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi-Rhee E, Schulman H, Cronan JE. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 2004;13:3043–3050. doi: 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Streaker ED, Beckett D. Nonenzymatic biotinylation of a biotin carboxyl carrier protein: Unusual reactivity of the physiological target lysine. Protein Sci. 2006;15:1928–1935. doi: 10.1110/ps.062187306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bustamante J, et al. Alpha-lipoic acid in liver metabolism and disease. Free Radic Biol Med. 1998;24:1023–1039. doi: 10.1016/s0891-5849(97)00371-7. [DOI] [PubMed] [Google Scholar]
- 58.Roy S, Packer L. Redox regulation of cell functions by alpha-lipoate: Biochemical and molecular aspects. Biofactors. 1998;7:263–267. doi: 10.1002/biof.5520070324. [DOI] [PubMed] [Google Scholar]
- 59.Yi X, Maeda N. Endogenous production of lipoic acid is essential for mouse development. Mol Cell Biol. 2005;25:8387–8392. doi: 10.1128/MCB.25.18.8387-8392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tort F, et al. Mutations in the lipoyltransferase LIPT1 gene cause a fatal disease associated with a specific lipoylation defect of the 2-ketoacid dehydrogenase complexes. Hum Mol Genet. 2014;23:1907–1915. doi: 10.1093/hmg/ddt585. [DOI] [PubMed] [Google Scholar]
- 61.Cao X, Zhu L, Hu Z, Cronan JE. Expression and activity of the BioH esterase of biotin synthesis is independent of genome context. Sci Rep. 2017;7:2141. doi: 10.1038/s41598-017-01490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi J, Cao X, Chen Y, Cronan JE, Guo Z. An atypical alpha/beta-hydrolase fold revealed in the crystal structure of pimeloyl-acyl carrier protein methyl esterase BioG from Haemophilus influenzae. Biochemistry. 2016;55:6705–6717. doi: 10.1021/acs.biochem.6b00818. [DOI] [PubMed] [Google Scholar]
- 64.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 65.Cao X, Cronan JE. The Streptomyces coelicolor lipoate-protein ligase is a circularly permuted version of the Escherichia coli enzyme composed of discrete interacting domains. J Biol Chem. 2015;290:7280–7290. doi: 10.1074/jbc.M114.626879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Lay NR, Cronan JE. In vivo functional analyses of the type II acyl carrier proteins of fatty acid biosynthesis. J Biol Chem. 2007;282:20319–20328. doi: 10.1074/jbc.M703789200. [DOI] [PubMed] [Google Scholar]
- 67.Jiang Y, Chan CH, Cronan JE. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry. 2006;45:10008–10019. doi: 10.1021/bi060842w. [DOI] [PubMed] [Google Scholar]
- 68.Christensen QH, Hagar JA, O’Riordan MX, Cronan JE. A complex lipoate utilization pathway in Listeria monocytogenes. J Biol Chem. 2011;286:31447–31456. doi: 10.1074/jbc.M111.273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 70.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview, version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McWilliam H, et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 75.Christensen QH, Cronan JE. The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. J Biol Chem. 2009;284:21317–21326. doi: 10.1074/jbc.M109.015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wycuff DR, Matthews KS. Generation of an AraC-araBAD promoter-regulated T7 expression system. Anal Biochem. 2000;277:67–73. doi: 10.1006/abio.1999.4385. [DOI] [PubMed] [Google Scholar]
- 77.Hashemzadeh-Bonehi L, et al. Importance of using lac rather than ara promoter vectors for modulating the levels of toxic gene products in Escherichia coli. Mol Microbiol. 1998;30:676–678. doi: 10.1046/j.1365-2958.1998.01116.x. [DOI] [PubMed] [Google Scholar]
- 78.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chakravartty V, Cronan JE. A series of medium and high copy number arabinose-inducible Escherichia coli expression vectors compatible with pBR322 and pACYC184. Plasmid. 2015;81:21–26. doi: 10.1016/j.plasmid.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]