Significance
Parkinson’s disease (PD) is the second most common neurodegenerative disorder. It is characterized by progressive deterioration of motor function caused by loss of dopamine-producing neurons. Currently, there is no treatment that could stop the progress of the disease. Loss-of-function mutations in Parkin account for ∼50% of early onset PD. Parkin functions as an E3 ubiquitin ligase that exhibits multiple protective roles especially in dopaminergic neurons. Here, we demonstrate that PICK1 directly binds to Parkin. PICK1 is a potent endogenous inhibitor of Parkin’s E3 ubiquitin ligase activity and blocks Parkin’s protective functions. Conversely, knockout of PICK1 enhances the neuroprotective effect of Parkin. Thus, the reduction of PICK1 may provide a therapeutic target for the treatment of PD.
Keywords: Parkinson’s disease, Parkin, E3 ligase, PICK1, BAR domain
Abstract
Parkin functions as a multipurpose E3 ubiquitin ligase, and Parkin loss of function is associated with both sporadic and familial Parkinson’s disease (PD). We report that the Bin/Amphiphysin/Rvs (BAR) domain of protein interacting with PRKCA1 (PICK1) bound to the really interesting new gene 1 (RING1) domain of Parkin and potently inhibited the E3 ligase activity of Parkin by disrupting its interaction with UbcH7. Parkin translocated to damaged mitochondria and led to their degradation in neurons, whereas PICK1 robustly inhibited this process. PICK1 also impaired the protective function of Parkin against stresses in SH-SY5Y cells and neurons. The protein levels of several Parkin substrates were reduced in young PICK1-knockout mice, and these mice were resistant to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-mediated toxicity. Taken together, the results indicate that PICK1 is a potent inhibitor of Parkin, and the reduction of PICK1 enhances the protective effect of Parkin.
Parkinson’s disease (PD) is a prevalent neurodegenerative disease characterized by dopaminergic neuron loss in the substantia nigra pars compacta (SNpc) and the formation of Lewy bodies (LBs). Several genes have been associated with the familial forms of PD (1), and Parkin mutations are the major cause of early onset PD (2). Structurally, Parkin is composed of an N-terminal ubiquitin-like (Ubl) domain, a linker region, a RING0 domain, a RING1 domain, an in-between RING (IBR) domain, and a RING2 domain at the C terminus. Parkin functions as an E3 ubiquitin ligase (3), and its E3 ligase activity can be positively modulated through neddylation and sulfhydration or regulators, such as CHIP, HSel-10, SUMO-1, and PINK1 (4–6). The E3 ligase activity of Parkin can also be negatively modified. Thermal and oxidative stress, S-nitrosylation, and dopamine may inactivate Parkin (7–11). BAG5 and 14-3-3ŋ bind to Parkin and inhibit its E3 ligase activity (12, 13). In addition, Cdk5, casein kinase-1, protein kinase A, protein kinase C, and c-Abl may phosphorylate Parkin and lead to its inactivation (4). The inactivation of Parkin contributes to sporadic PD (14). Although these regulators have been shown to inhibit the E3 ligase activity of Parkin, few reports have demonstrated their endogenous roles in vivo. In addition, no report has indicated that the function of these inhibitors is dependent on Parkin in vivo.
Parkin exhibits multivalent protective functions when overexpressed in various toxic conditions (15). However, the role of endogenous Parkin is poorly understood. Dopaminergic neuron loss has not been identified in the Parkin knockout (KO) mouse models that have been generated (16–19). In addition, dopaminergic neuron loss is not accelerated in Parkin KO mice administered 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) or 6-hydroxydopamine (20, 21). These findings have led to uncertainty regarding the role of endogenous Parkin. Although endogenous Parkin protects dopaminergic neurons from the mitochondrial DNA mutagenic stress (22), and acute KO of Parkin induces dopaminergic neuron loss and mitochondrial defects (23, 24), the potential protective function of endogenous Parkin requires more comprehensive studies.
PICK1 is a BAR domain-containing protein that is highly expressed in the brain (25). It has been reported that PICK1 could bind to Parkin and be monoubiquitinated (26). Monoubiquitination of PICK1 abolishes PICK1-dependent potentiation of ASIC2a. Surprisingly, we found that PICK1 potently inhibited the E3 ligase activity of Parkin. As a result, PICK1 compromised Parkin-mediated mitophagy and the protection against cell death. More importantly, several substrates of Parkin were reduced in young PICK1 KO mice, and these mice were resistant to MPTP-induced toxicity. These phenotypes were not identified in PICK1/Parkin double-KO (DKO) mice. Taken together, our findings suggest that PICK1 impairs the protective function of Parkin in vitro and in vivo via inhibition of the E3 ligase activity of Parkin.
Results
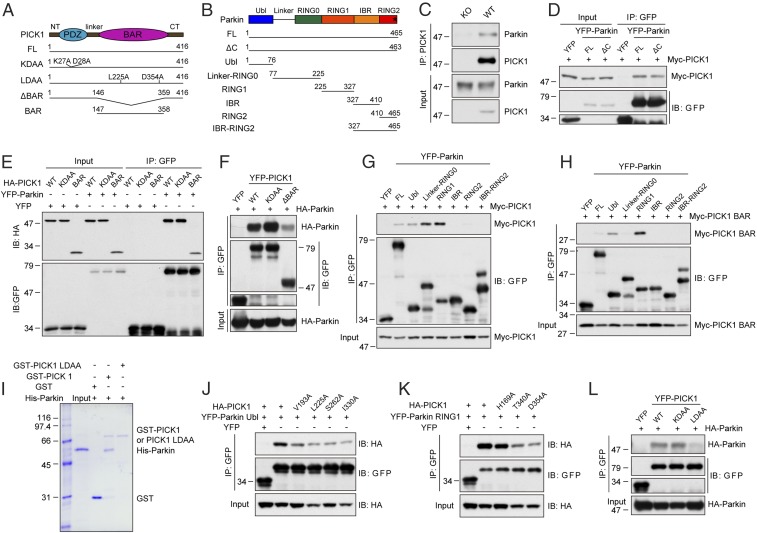
PICK1 Binds to Parkin Through the BAR Domain.
PICK1 contains a PDZ domain and has been reported to interact with the C-terminal PDZ-binding motif of Parkin in vitro using a heterologous expression system (Fig. 1 A and B) (26). To investigate the physiological relevance of the PICK1–Parkin interaction, we first performed in vivo coimmunoprecipitation in mouse brain homogenates and determined that Parkin coimmunoprecipitated with PICK1 (Fig. 1C). In HEK293T cells, PICK1 also interacted with Parkin (Fig. 1 D–G and L), consistent with the previous finding (26). However, Parkin (1-463) lacking the PDZ-binding motif retained the ability to bind to PICK1 (Fig. 1D). In addition, PICK1 KDAA, a mutant without PDZ-binding ability (27, 28), also interacted with Parkin (Fig. 1 E, F, and L). These results contradicted the previous report (26). A careful examination of the crystal structure of Parkin revealed that the C-terminal PDZ-binding motif of Parkin is not exposed (SI Appendix, Fig. S1A) (29). This finding provides further evidence that the interaction between PICK1 and Parkin is unlikely dependent on the PDZ domain. Instead, we found that the PICK1 BAR domain (subsequently referred to as PICK1 BAR) bound to Parkin (Fig. 1 E and H). Deletion of the BAR domain (PICK1 ΔBAR) reduced the interaction with Parkin (Fig. 1F), demonstrating that the interaction between PICK1 and Parkin is mainly dependent on the BAR domain. To determine which domain of Parkin binds to PICK1, serial deletion mutants of Parkin were generated (Fig. 1B). Both the full-length and the BAR domain of PICK1 potently bound to the Parkin RING1 domain and interacted with the Parkin Ubl domain to a lesser extent (Fig. 1 G and H). The Parkin linker-RING0 region might bind to other parts of PICK1 because it bound to the full-length PICK1, but not the BAR domain. To determine whether the interaction of PICK1 with Parkin is direct or indirect, we performed in vitro pull-down experiment with only purified proteins to eliminate the possibility of association through intermediate proteins. GST-PICK1 was found to associate with His-Parkin, as detected by Coomassie Blue staining (Fig. 1I). This indicates that PICK1 directly interacts with Parkin.
Fig. 1.
PICK1 binds to Parkin through the BAR domain. (A) Schematic representations of the structure of PICK1 and PICK1 mutants. CT, C-terminal; FL, full-length; NT, N-terminal. (B) Schematic representations of the structure of Parkin and Parkin mutants. The asterisk indicates the C-terminal PDZ-binding motif-FDV. (C) Coimmunoprecipitation (IP) of endogenous PICK1 and Parkin in mouse brain homogenates from WT and PICK1 KO mice. (D–H and J–L) Coimmunoprecipitation of PICK1 or PICK1 mutants with Parkin or Parkin mutants in whole-cell lysates from HEK293T cells expressing the indicated constructs. (I) In vitro binding between Parkin with GST, GST-PICK1, and GST-PICK1 LDAA. Coomassie Blue staining was used to stain proteins.
We subsequently aimed to determine the residues on the BAR domain of PICK1 that are critical for the interaction with Parkin. Systematic mutations in the BAR domain of PICK1 were generated and tested for their interactions with the Ubl and RING1 domains of Parkin. We identified that point mutations of V193, L225, S262, or I330 of PICK1 to alanine reduced the interaction between PICK1 and the Parkin Ubl domain (Fig. 1J). The PICK1 mutants H169A, T340A, or D354A reduced the interaction between PICK1 and the Parkin RING1 domain (Fig. 1K). The double mutations of L225 and D354 to alanine (PICK1 LDAA) almost completely abolished the PICK1–Parkin interaction (Fig. 1L). In addition, purified GST-PICK1 LDAA did not bind to purified His-Parkin (Fig. 1I). PICK1 LDAA still bound to ICA69 and GluA2, the known binding partners of the PICK1 BAR (25, 30) and the PDZ domain (28), respectively, which rules out the possibility that the reduced interaction with Parkin is a result of an altered structure of PICK1 (SI Appendix, Fig. S1 B and C).
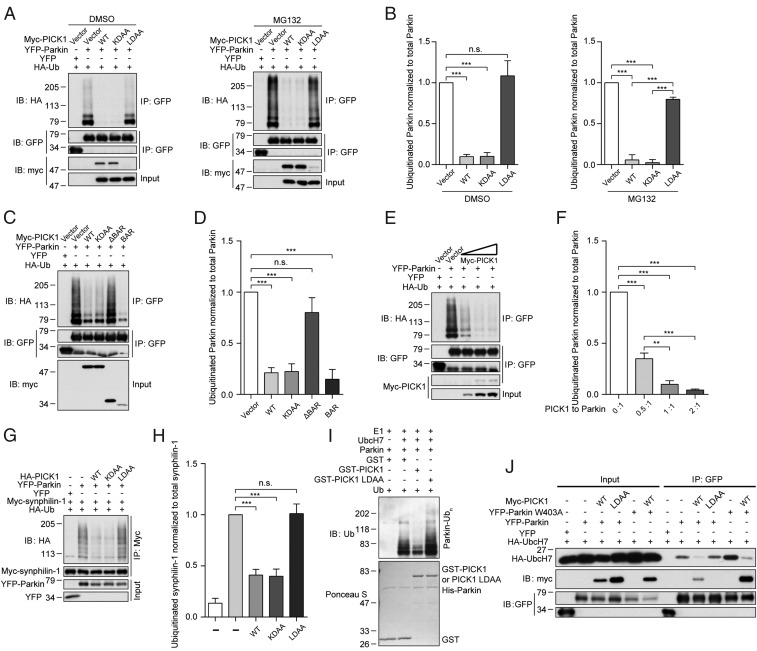
PICK1 Inhibits the E3 Ligase Activity of Parkin.
Parkin is capable of autoubiquitination. The autoubiquitination level represents the E3 ligase activity level of Parkin (3, 8). We surprisingly found that, in the presence of PICK1 or PICK1 KDAA, the autoubiquitination of Parkin was substantially reduced (Fig. 2 A and B). On the contrary, PICK1 LDAA did not influence the autoubiquitination of Parkin. In addition, after proteasome inhibition by MG132, Parkin autoubiquitination was still reduced, indicating that PICK1 does not accelerate the degradation of Parkin and the reduced autoubiquitination is likely a result of the inhibition of Parkin E3 ligase activity (Fig. 2 A and B). PICK1 and PICK1 KDAA interacted with Parkin, whereas PICK1 LDAA did not, indicating that the inhibitory effect is based on their interaction. In addition, the BAR domain of PICK1 also potently inhibited the autoubiquitination of Parkin, and the deletion of PICK1 BAR abolished the inhibition effect (Fig. 2 C and D). Moreover, the autoubiquitination of Parkin was decreased with an increased expression of PICK1, and their interaction was consistently enhanced (Fig. 2 E and F).
Fig. 2.
PICK1 inhibits the E3 ligase activity of Parkin. (A) Western blot analysis of the effects of PICK1, PICK1 KDAA, and PICK1 LDAA on autoubiquitination of Parkin in whole-cell lysates from HEK293T cells treated with DMSO or 10 µM MG132 for 6 h. GFP antibodies were used to immunoprecipitate YFP-Parkin. HA antibodies were used to detect polyubiquitinated Parkin. (B) Quantification of ubiquitinated Parkin normalized to total Parkin in A (n = 3). (C) Western blot analysis of the effects of PICK1 and PICK1 mutants on the autoubiquitination of Parkin. (D) Quantification of ubiquitinated Parkin normalized to total Parkin in C (n = 4). (E) Western blot analysis of the effect of increasing PICK1 expression on the autoubiquitination of Parkin. The cells were transiently transfected with different ratios of PICK1 to Parkin (0:1, 0.5:1, 1:1, and 2:1). (F) Quantification of ubiquitinated Parkin normalized to total Parkin in E (n = 4). (G) Western blot analysis of the effects of PICK1 and PICK1 mutants on the ubiquitination of synphilin-1 by Parkin. Myc antibodies were used to immunoprecipitate myc-synphilin-1. HA antibodies were used to detect polyubiquitinated synphilin-1. (H) Quantification of ubiquitinated synphilin-1 normalized to total synphilin-1 in G (n = 3). (I) In vitro ubiquitination of Parkin with or without PICK and PICK1 LDAA. Membrane was immunoblotted with ubiquitin antibodies in Upper, and Ponceau S staining was used in Lower. (J) Coimmunoprecipitation of Parkin and Parkin W403A with UbcH7, PICK1, and PICK1 LDAA. Error bars indicate mean ± SEM. One-way ANOVA with post hoc Bonferroni test. **P < 0.01; ***P < 0.001; n.s., not significant.
To investigate whether PICK1 regulates the ubiquitination of Parkin substrates, we examined the ubiquitination of synphilin-1. Consistent with a previous report (8), synphilin-1 was polyubiquitinated by Parkin (Fig. 2 G and H). However, in the presence of PICK1 or PICK1 KDAA, the ubiquitination of synphilin-1 was reduced, whereas there was no effect of PICK1 LDAA. Our results indicate that PICK1 binds to Parkin and inhibits its E3 ligase activity through the BAR domain. To investigate the direct inhibitory function of PICK1, we performed in vitro ubiquitination assay using purified recombinant proteins (Fig. 2I). The upper smear bands represented the autoubiquitination of Parkin. In the presence of PICK1, the autoubiquitination of Parkin was significantly reduced (Fig. 2I). As a control, PICK1 LDAA, which lacks the ability to interact with Parkin, also lost its ability to inhibit Parkin autoubiquitination.
We subsequently determined whether other BAR domain-containing proteins could also inhibit the E3 ligase activity of Parkin. The BAR domains of human arfaptin 1 and arfaptin 2 share 47% and 46% similarity, respectively, with the BAR domain of human PICK1. We found that the full-length arfaptin 1 and arfaptin 2 and their BAR domains had no inhibitory effect (SI Appendix, Fig. S2 A–D). In contrast, PICK1 and PICK1 BAR potently inhibited Parkin E3 ligase activity, suggesting a specific inhibitory role for PICK1 BAR (SI Appendix, Fig. S2 A–D). Consistently, PICK1 and PICK1 BAR interacted with Parkin in contrast to arfaptin 1 and arfaptin 2 and their BAR domains (SI Appendix, Fig. S2 A and C). In addition, to rule out the possibility that the ubiquitination of Parkin was caused by other E3 ligases, the catalytically inactive mutant Parkin C431F was employed (29). The ubiquitination level of Parkin was reduced by ∼81%, suggesting that most ubiquitination occurs through autoubiquitination (SI Appendix, Fig. S2 E and F). In addition, the ubiquitination level of Parkin was reduced by PICK1 to a similar level as Parkin C431F in cells.
We next investigated how PICK1 inhibits the E3 ligase activity of Parkin. UbcH7 is the ubiquitin-conjugating enzyme E2 for Parkin (3). UbcH7 binds to the RING1 domain of Parkin during the ubiquitination process (3, 29). Therefore, PICK1 may compete with UbcH7 to bind to the RING1 domain. Indeed, we found that, in the presence of PICK1, the interaction between UbcH7 and Parkin was nearly abolished (Fig. 2J). PICK1 LDAA, which did not bind to Parkin, had no effect on the Parkin–UbcH7 interaction.
Parkin is in an autoinhibited state, and a repressor element of Parkin blocks the binding region for UbcH7 (29). The Parkin W403A mutant relieves the repression effect of this element. Thus, the interaction between Parkin W403A and UbcH7 was subsequently enhanced (Fig. 2J). Interestingly, the interaction between Parkin W403A and UbcH7 was still reduced by PICK1. PICK1 also bound to Parkin W403A more potently compared with WT Parkin. Thus, PICK1 and UbcH7 may share a similar binding region on the RING1 domain; however, PICK1 has a higher affinity and blocks the access of UbcH7 to Parkin. Taken together, these results show that PICK1 inhibits the E3 ligase activity of Parkin by disrupting the interaction between Parkin and UbcH7.
PICK1 Promotes Parkin Aggresome Formation.
Parkin forms aggresomes following proteasome inhibition (31). In HEK293T cells treated with the proteasome inhibitor MG132, Parkin formed aggresomes as previously reported (SI Appendix, Fig. S3A). Surprisingly, PICK1 and PICK1 BAR led to Parkin aggresome formation in the absence of proteasome inhibition, whereas PICK1 LDAA or deletion of PICK1 BAR did not (SI Appendix, Fig. S3 A and B). To investigate whether Parkin aggresome formation was caused by a secondary effect of proteasome inhibition, transfected cells were treated with 5 µM MG132 for 16 h. The percentage of Parkin aggresomes in cells coexpressing PICK1 was significantly different from that in cells coexpressing vector control, PICK1 ΔBAR, or PICK1 LDAA, suggesting that Parkin aggresome formation induced by PICK1 could not be blocked by proteasome inhibition (SI Appendix, Fig. S3 A and B). In addition, the finding that PICK1 and PICK1 BAR rather than the mutants defective in Parkin binding lead to Parkin aggresome formation suggests that PICK1-induced Parkin aggresome formation is most likely a direct effect.
In HEK293T cells expressing synphilin-1, Parkin, and PICK1, synphilin-1 was also present in the Parkin aggresomes (SI Appendix, Fig. S3C). The spontaneous Parkin aggresomes in cells were ubiquitin positive as previously reported (31) (SI Appendix, Fig. S3D). The Parkin aggresomes induced by PICK1 or PICK1 BAR were also ubiquitin positive. In addition, PICK1-induced Parkin aggresomes were also strongly labeled with endogenous p62 (SI Appendix, Fig. S3E), which is an autophagy adaptor protein found in protein inclusion bodies (32). Interestingly, we found that, in these aggresomes, the Parkin signals appeared to be surrounded by PICK1 signals (SI Appendix, Fig. S3F).
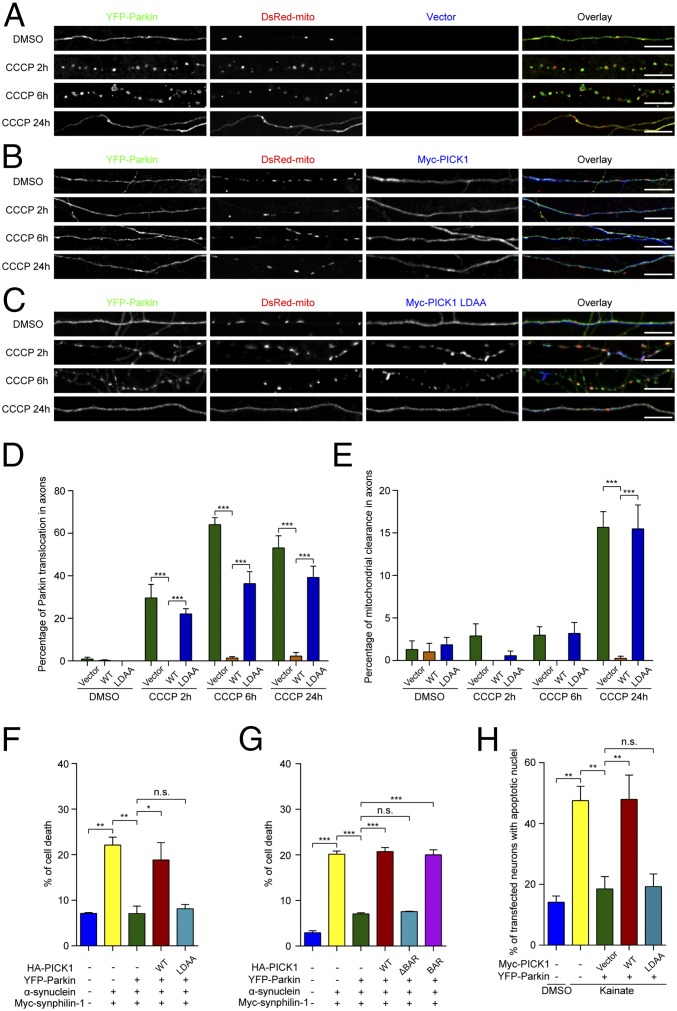
PICK1 Blocks Parkin’s Functions in Neurons.
PICK1 directly binds to Parkin and inhibits its E3 ligase activity. The E3 ligase activity is crucial for the protective role of Parkin (15). Therefore, we subsequently investigated whether PICK1 blocks Parkin’s functions in cells. Parkin is recruited to damaged mitochondria and promotes mitophagy (33, 34). Carbonyl cyanide m-chlorophenylhydrazone (CCCP) is a drug that uncouples the mitochondrial membrane potential, which damages mitochondria. Under basal conditions, Parkin was diffuse in axons when coexpressed with vector control, PICK1, or PICK1 LDAA (Fig. 3 A–C). After the neurons were treated with CCCP for 2, 6, or 24 h, Parkin had translocated to the axonal mitochondria marked by DsRed-mito (representative image for 24 h was not shown) (Fig. 3 A and D). After 24 h of CCCP treatment, ∼16% of axons showed dispersed DsRed-mito signals, demonstrating that the dysfunctional mitochondria have been cleared (Fig. 3 A and E). Parkin was also diffuse in the axons with mitochondrial clearance (Fig. 3A). Interestingly, the addition of PICK1 prevented the translocation of Parkin to damaged axonal mitochondria for up to 24 h of CCCP treatment, and mitochondrial clearance was also blocked (Fig. 3 B, D, and E). This is consistent with reports that inactive Parkin is incapable of mitochondrial translocation and clearance (29, 35). In contrast, PICK1 LDAA did not prevent the translocation of Parkin and the subsequent degradation of damaged mitochondria (Fig. 3 C–E).
Fig. 3.
PICK1 inhibits Parkin-mediated mitophagy and the protective function of Parkin against stresses in SH-SY5Y cells and neurons. (A–C) Cultured hippocampal neurons were transiently transfected with YFP-Parkin (green) and DsRed-mito (red) in combination with vector control (A, blue) or myc-PICK1 (B, blue) or myc-PICK1 LDAA (C, blue). The neurons were treated with DMSO or 13.5 µM CCCP for the indicated times. (Scale bars, 10 µm.) (D) Quantification of Parkin translocation in axons (n = 4). (E) Quantification of mitochondrial clearance in axons (n = 4). Error bars indicate mean ± SEM. Two-way ANOVA with post hoc Bonferroni test. ***P < 0.001. (F and G) Quantification of cell death in SH-SY5Y cells transfected with indicated constructs and treated with 10 µM MG132 for 18 h (n = 3 for both F and G). (H) Cultured hippocampal neurons were treated with DMSO or 25 µM kainate. Quantification of the percentage of transfected neurons with apoptotic nuclei (n = 5). Error bars indicate mean ± SEM. One-way ANOVA with post hoc Bonferroni test. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant.
Parkin was also diffuse in the cell bodies and dendrites when coexpressed with vector control, PICK1, or PICK1 LDAA under basal conditions (SI Appendix, Fig. S4A). After the neurons were treated with CCCP for 2, 6, or 24 h, Parkin had translocated to mitochondria in the cell bodies and dendrites (SI Appendix, Fig. S4 B–E). Similarly, the addition of PICK1 prevented the translocation of Parkin to mitochondria in these regions for up to 24 h of CCCP treatment, whereas PICK1 LDAA did not.
Next, whether PICK1 impairs Parkin-mediated protection against cell death was investigated. It has been shown that the coexpression of synphilin-1 and α-synuclein in SH-SY5Y cells with the addition of proteasome inhibitors induces an increase of cell death, which is indicated by an increased ratio of propidium iodide/Hoechst (8, 12). Parkin suppressed the toxicity of synphilin-1 and α-synuclein overexpression, which was characterized by a reduced ratio of propidium iodide/Hoechst (Fig. 3 F and G). However, the Parkin-mediated enhancement of cell survival was significantly reduced by PICK1 and PICK1 BAR, but not by PICK1 LDAA or PICK1 ΔBAR (Fig. 3 F and G).
Kainate causes excitotoxicity and induces apoptosis in neurons. Parkin can attenuate this toxicity (36). Whether PICK1 affects the Parkin-mediated attenuation of kainate toxicity was also investigated. Similar to the previous report (36), kainate induced apoptosis in both hippocampal and ventral midbrain dopaminergic neurons, as characterized by condensed nuclei (Fig. 3H and SI Appendix, Figs. S5 and S6). Parkin overexpression reduced the number of neurons that contained apoptotic nuclei. This Parkin-mediated protective effect was largely blocked by the expression of PICK1, but not by PICK1 LDAA (Fig. 3H and SI Appendix, Figs. S5 and S6). Taken together, PICK1 suppresses the protective functions of Parkin.
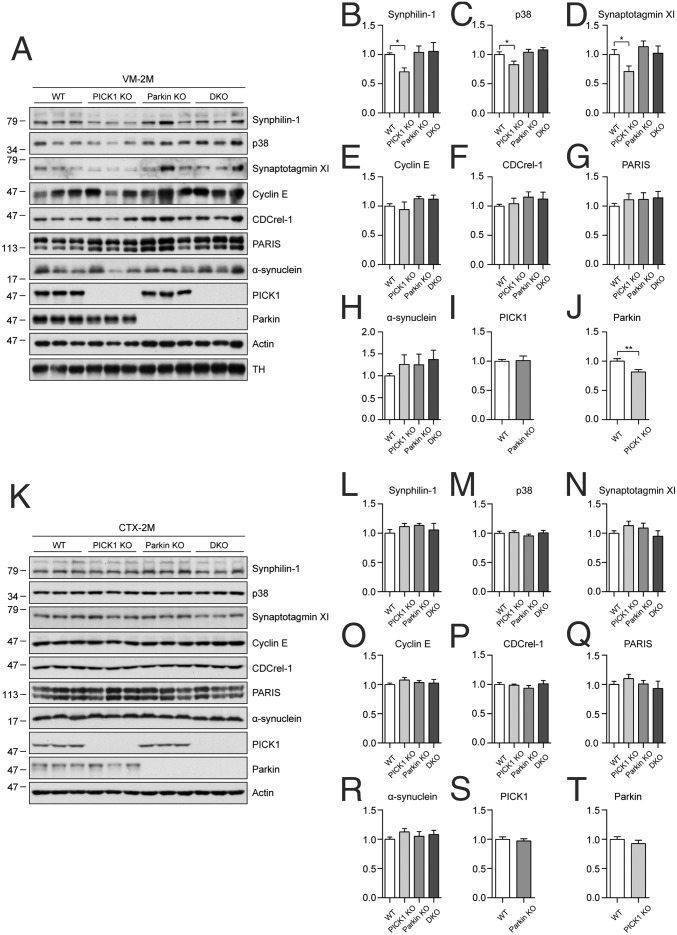
Several Parkin Substrates Are Reduced in the Ventral Midbrain of Young PICK1 KO Mice.
The loss of dopaminergic neurons is one hallmark of PD. Therefore, we subsequently examined whether PICK1 is present in dopaminergic neurons. We found that PICK1 was highly enriched in tyrosine hydroxylase (TH)-labeled dopaminergic neurons in the SNpc of WT mice, whereas no signal was detected in PICK1 KO mice (SI Appendix, Fig. S7A). Western blotting analysis of microdissected brain regions also confirmed that both PICK1 and Parkin were expressed in the ventral midbrain (VM), where the SNpc is located (SI Appendix, Fig. S7B). We also examined the protein expression profile in different tissues. Parkin was present in all of the tissues tested with relatively high expression in the testes, brain, stomach, and kidney (SI Appendix, Fig. S7C). This expression pattern was similar to PICK1, which was highly expressed in the brain, testes, and pancreas, consistent with previous findings (25).
PICK1 inhibits the E3 ligase activity of Parkin. This finding raises the possibility that, in PICK1 KO mice, Parkin would have higher E3 ligase activity without PICK1 inhibition. This may result in more degradation of its substrates. To test this hypothesis, the protein expression levels of Parkin substrates were determined in the VM of WT, PICK1 KO, Parkin KO, and PICK1/Parkin DKO mice. Three substrates, namely synphilin-1, p38, and synaptotagmin XI, were reduced in the VM of young PICK1 KO mice (2 mo of age) (Fig. 4 A–D). These substrates were not significantly changed in the VM of young Parkin KO mice. Interestingly, synphilin-1, p38, and synaptotagmin XI were not reduced in the VM of young PICK1/Parkin DKO mice, indicating that the reduction of these proteins in PICK1 KO mice is dependent on the function of Parkin. Moreover, Parkin was also reduced in the VM of PICK1 KO mice (Fig. 4 A and J), which further suggests that Parkin has higher E3 ligase activity without PICK1 and Parkin is subsequently autoubiquitinated and degraded. The other substrates tested, namely cyclin E, CDCrel-1, PARIS, and α-synuclein, were not significantly changed (Fig. 4 A and E–H). PICK1 was not significantly changed in the VM of Parkin KO mice (Fig. 4 A and I). The expression levels of Parkin substrates were also determined in the cortex of young mice. There were no significant changes in the substrates tested in the cortex (Fig. 4 K–R). In addition, Parkin and PICK1 were not significantly changed in the cortex of PICK1 KO and Parkin KO mice, respectively (Fig. 4 K, S, and T).
Fig. 4.
Several Parkin substrates are reduced in the ventral midbrain of young PICK1 KO mice. (A–T) Immunoblots (A and K) and quantification of immunoblots (B–J and L–T) for the indicated proteins to determine their expressions in the ventral midbrain (VM) (B–J, n = 12 per genotype) and cortex (CTX) (L–T, n = 9 per genotype) of 2-mo-old WT, PICK1 KO, Parkin KO, and PICK1/Parkin DKO mice. VM samples were normalized to TH, and CTX samples were normalized to actin. Error bars indicate mean ± SEM. One-way ANOVA with post hoc Bonferroni test for B–D; Student’s t test for J. *P < 0.05; ***P < 0.001.
The expression levels of Parkin substrates were also determined in the VM of old mice (12 mo of age). Interestingly, in old PICK1 KO mice, all substrates tested as well as Parkin were not significantly changed in the VM (SI Appendix, Fig. S8 A–H and J). In addition, we did not identify significant changes in the substrates or PICK1 in the VM of old Parkin KO or PICK1/Parkin DKO mice (SI Appendix, Fig. S8 A–I). Furthermore, no significant changes were identified in the cortex of PICK1 KO, Parkin KO, and PICK1/Parkin DKO mice with the exception of PARIS, which was reduced in PICK1/Parkin DKO mice (SI Appendix, Fig. S8 K–T).
Young PICK1 KO Mice Are Resistant to MPTP Toxicity.
Our current data indicate that PICK1 inhibits the E3 ligase activity of Parkin and reduces its protective function in cells. The reduced expressions of Parkin substrates in young PICK1 KO mice suggest increased Parkin E3 ligase activity, which may enhance the protective function of Parkin. We administered MPTP, which is widely used to induce dopaminergic neuron loss in rodents and primates (37), to investigate whether dopaminergic neurons in PICK1 KO mice are resistant to toxicity.
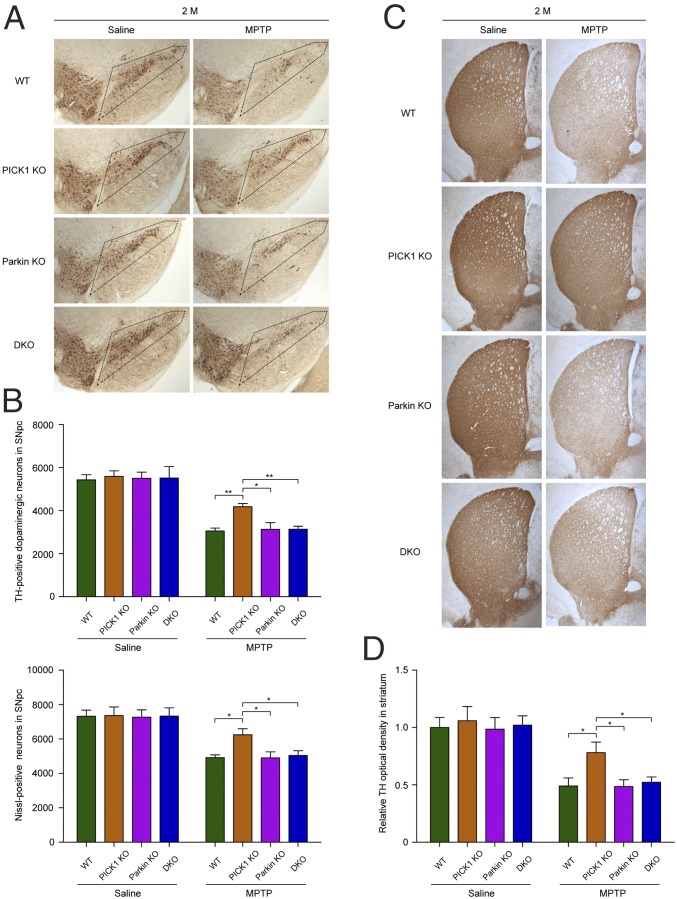
Both young (2-mo-old) and old (12-mo-old) WT, PICK1 KO, Parkin KO, and PICK1/Parkin DKO mice were administered saline or MPTP, and the number of TH-positive dopaminergic neurons within the SNpc was quantified. The numbers of dopaminergic neurons were similar among young WT, PICK1 KO, Parkin KO, and PICK1/Parkin DKO mice administered saline (Fig. 5 A and B). Following the induction of dopaminergic neuron loss in the SNpc with MPTP, the number of dopaminergic neurons in young PICK1 KO mice was significantly higher than the numbers in young WT, Parkin KO, and PICK1/Parkin DKO mice (Fig. 5 A and B). To rule out the possibility that the loss of TH signal is due to down-regulation of the TH marker, Nissl staining was performed, which confirmed that there was an actual neuronal loss in SNpc (Fig. 5B and SI Appendix, Fig. S9A). These results indicate that the loss of PICK1 renders a protective effect for dopaminergic neurons. In contrast, the number of dopaminergic neurons in Parkin KO mice did not differ significantly from the number in WT mice, consistent with a previous report (21). More importantly, the number of dopaminergic neurons in PICK1/Parkin DKO mice was also not significantly different from the number in WT mice, which further demonstrates that the function of PICK1 depends on Parkin, and when Parkin is absent, the loss of PICK1 has no effect.
Fig. 5.
Young PICK1 KO mice are resistant to MPTP-induced toxicity. (A) Immunohistochemical analysis of dopaminergic neurons from young (2-mo-old) mice administered saline or MPTP. The boxed regions indicate the location of the SNpc. (B) Quantification of TH- and Nissl-positive neurons in the SNpc. For TH staining: saline: WT, n = 9; PICK1 KO, n = 7; Parkin KO, n = 10; DKO, n = 6. MPTP: WT, n = 8; PICK1 KO, n = 9; Parkin KO, n = 8; DKO, n = 10. n = 6 per genotype for Nissl staining. (C) TH immunohistochemistry of the striatum. (D) Quantification of OD of TH in striatum. Saline: WT, n = 9; PICK1 KO, n = 7; Parkin KO, n = 10; DKO, n = 6. MPTP: WT, n = 8; PICK1 KO, n = 9; Parkin KO, n = 8; DKO, n = 10. The mice were killed 5 d after last dosage of MPTP administration. Error bars indicate mean ± SEM. Two-way ANOVA with post hoc Bonferroni test. *P < 0.05; **P < 0.01.
We also examined the TH-positive fiber density in striatum. Intensities of dopaminergic nerve terminals were clearly reduced following MPTP treatment, compared with saline-treated controls in WT mice (Fig. 5 C and D), indicating a loss of TH-positive fibers. PICK1 deficiency significantly rescued the loss of TH-positive fibers in striatum. No significant changes could be observed among other genotypes. Striatal dopamine levels also showed a similar pattern to TH-positive dopaminergic neuron number in SNpc and TH-positive fiber density in striatum (SI Appendix, Fig. S9B). This provided further support to the notion that loss of PICK1 renders a protective effect to MPTP toxicity.
Monoamine oxidase B (MAO-B) converts MPTP to MPP+ to elicit neurotoxicity (37). To ensure that the protection by PICK1 gene deletion is not caused by alteration in MPP+ metabolism, MAO-B activity and MPP+ levels in striatum were measured. The activity of MAO-B was not significantly changed among the four genotypes (SI Appendix, Fig. S9C) and MPP+ levels were also not significantly different among the four genotypes of MPTP administered mice (SI Appendix, Fig. S9D).
In contrast to the young PICK1 KO mice, in the old PICK1 KO mice administered MPTP, the number of dopaminergic neurons was not significantly different compared with the numbers in WT, Parkin KO, and PICK1/Parkin DKO mice administered MPTP (SI Appendix, Fig. S10 A and B). In addition, the numbers of dopaminergic neurons in Parkin KO and PICK/Parkin DKO mice were not significantly altered compared with the number in WT mice.
Subacute MPTP injection (25 mg/kg MPTP) causes progressive neuronal lesion and stabilizes by day 21 (38–40). To differentiate between neuronal protection versus delay of cell death, another batch of young mice were administered MPTP and killed 21 d after last dosage of MPTP. Deletion of PICK still showed a protective effect in 21 d after MPTP injection, similar to 5 d post-MPTP injection (SI Appendix, Fig. S11 A and B).
Discussion
Our study demonstrates that PICK1 is a potent endogenous inhibitor of Parkin. The BAR domain of PICK1 bound to the RING1 domain of Parkin and blocked its access to UbcH7, which rationally explains how PICK1 inhibits the E3 ligase activity of Parkin. As a consequence of this inhibition, PICK1 impaired Parkin-mediated mitophagy in neurons and diminished Parkin-mediated cell survival in vitro. More importantly, the endogenous role of PICK1 was verified in vivo. In young PICK1 KO mice, the loss of PICK1 boosted the activity of Parkin and enhanced its neuronal protective effect, as demonstrated by the reduction of several Parkin substrates in the VM, and these mice became resistant to MPTP-mediated toxicity. Furthermore, the endogenous inhibitory effect of PICK1 was dependent on Parkin because these effects were abolished in PICK1/Parkin DKO mice.
Selective inactivation of Parkin in the SNpc through nitrosative and dopaminergic stress and c-Abl phosphorylation have been reported in sporadic PD (8, 10, 41). Similarly, PARIS, a Parkin substrate, is selectively accumulated in the SNpc, but not in the cortex, of sporadic PD patients (23). The finding that Parkin substrates were only reduced in the VM but not in the cortex of young PICK1 KO mice suggests that Parkin may be selectively inhibited by PICK1 in the SNpc. However, the extent to which PICK1 contributes to the inactivation of Parkin and the pathogenesis of PD remains to be determined. Although we demonstrated that PICK1 elimination was initially beneficial to dopaminergic neurons through the enhancement of the E3 ligase activity of Parkin, Parkin could be inhibited by other compensatory mechanisms in the absence of PICK1 during aging, because its substrates were restored in the VM of old PICK1 KO mice, and these mice were no more tolerant of MPTP-induced toxicity. Thus, it would be interesting to figure out which inhibitors compensated for the inhibitory effect of PICK1 in old mice; moreover, the elimination of these inhibitors may provide a new perspective for the treatment of PD.
Many loss-of-function mutations in Parkin have been associated with familial PD. However, the potential endogenous role of Parkin remains poorly understood. The major difficulty originates from the finding that there are no or mild phenotypic defects in Parkin KO mice (16–19). A compensatory effect is one potential reason for no obvious dopaminergic neuron loss in Parkin KO mice. Consistently, to avoid the potential developmental compensation, the acute KO of Parkin in the SNpc induces dopaminergic neuron loss by 40% (23). The deposition of toxic substrates, such as PARIS, through the acute loss of Parkin may mediate the degeneration of dopaminergic neurons (23). Consistent with this notion, we found that the reduction of substrates in the VM of young PICK1 KO mice rendered these mice resistant to MPTP-mediated toxicity. This finding suggests that endogenous Parkin plays an important protective role, at least in part, by reducing toxic substrates. The results that the other tested substrates were not changed are likely because the subcellular localizations of these substrates may be different or the modulation of their expression by Parkin is activity dependent. In addition, normal protein levels in the VM of old PICK1 KO mice render these mice not more tolerant of MPTP-induced toxicity, which further supports the notion that the accumulation of toxic Parkin substrates induces dopaminergic neuron loss in PD. We did not identify significant differences for p38 and PARIS in the VM of Parkin KO mice, which is different from previous reports (23, 42). This result may be explained by the different targeting methods used to generate the Parkin KO mice (19) or age differences in the mice (23). Notably, PARIS was reduced in the cortex of 12-mo-old PICK1/Parkin DKO mice. However, the reason for this result is unclear.
Several reports have demonstrated that native Parkin is in an autoinhibited state (6, 29). However, YFP-tagged Parkin is constitutively active because this big tag force opens the Ubl domain of Parkin and exposes the RING1 domain. This may be different from the “physiological situation.” However, YFP- or GFP-tagged Parkin have been widely used to study Parkin functions and inhibitors (6, 12, 29). Thus, this cellular system can be readily employed to investigate the inhibitory function of PICK1. In addition, using in vitro ubiquitination assay, we explicitly demonstrated that PICK1 also inhibits the “weak” activity of native Parkin. Native Parkin is activated in a systematic manner (6, 43). As for when PICK1 steps in to inhibit Parkin during cellular process, it is not of our primary concern. Our focus in this study is to demonstrate that PICK1 is a potent inhibitor of Parkin both in vitro and in vivo.
Parkin aggresomes are proteinaceous inclusions that are formed following exposure to proteasome inhibitors (31) or various parkinsonian neurotoxins, including MPP+, paraquat, and rotenone (11, 44). Proteasome inhibition promotes autophagic degradation of protein inclusions/aggresomes (32). PICK1 inhibits the E3 ligase activity of Parkin; thus, it is possible that Parkin cannot promote self-degradation through the ubiquitin–proteasome system, and misfolded Parkin forms aggresomes for autophagic degradation. Consistent with this, the aggresomes are positive for p62, which is an autophagy adaptor protein (32). Therefore, Parkin aggresome formation may be a secondary effect of the inhibition of Parkin E3 ligase activity. Consistent with this notion, the inhibition of Parkin E3 ligase activity by oxidation also contributes to protein aggregation (11). Parkin aggresomes were surrounded by PICK1 and, similar to LBs, were synphilin-1 and ubiquitin positive. It is tempting to speculate that PICK1 is present in LBs and may play a role in LB formation.
Materials and Methods
Mice.
PICK1 KO mice have been previously described (45). Parkin KO mice were obtained from The Jackson Laboratory (stock number 006582). All animal procedures were approved by the Animal Ethics Committee of the Hong Kong University of Science and Technology (SI Appendix, SI Materials and Methods).
Cell Culture, Transfection, and Staining.
Full details are provided in SI Appendix, SI Materials and Methods.
In Vitro Binding.
In brief, 2 µg of GST or GST fusion proteins were bound to glutathione-Sepharose beads (Life Technologies) in binding buffer (50 mM Tris⋅HCl at pH 7.5, 140 mM NaCl, 0.1% Triton X-100 with protease inhibitors) at 4 °C for 1 h. The beads coupled with GST fusion protein were added to 2 µg of His-Parkin (Millipore) and incubated for 3 h at 4 °C. The beads were washed three times with TBS.
In Vitro Ubiquitination Assay.
In brief, 2 μg of purified GST or GST fusion proteins were incubated at 37 °C in 50 μL of reaction buffer (50 mM Tris⋅HCl, pH 7.5, 5 mM MgCl2, 2 mM DTT, and 2 mM ATP) containing 100 ng of E1 (Boston Biochem), 200 ng of E2 (UbcH7; Boston Biochem), 1 μg of His-tagged Parkin (Millipore), and 10 μg of ubiquitin (Boston Biochem). After incubation for 3 h, the reaction was stopped by addition of loading buffer.
Quantification of Parkin Aggresomes.
Transfected HEK293T cells were fixed and immunostained. Aggregates larger than 2.6 μm in diameter were designated Parkin aggresomes. The percentage of Parkin aggresomes was quantified as the ratio of the number of cells that contained one or more Parkin aggresomes to the number of total transfected cells.
Quantification of the Percentage of Axons or Cell Bodies and Dendrites with Parkin Translocation to Mitochondria and the Percentage of Mitochondrial Clearance in Axons.
The percentage of axons with Parkin translocation to mitochondria was calculated as the ratio of the number of axons with Parkin translocation to mitochondria to the number of total transfected axons. The percentage of cell bodies and dendrites with Parkin translocation to mitochondria was calculated as the ratio of the number of cells with Parkin translocation to mitochondria in the cell bodies and dendrites to the number of total transfected cells. The percentage of mitochondrial clearance was calculated as the ratio of the number of axons that exhibited diffuse DsRed signals to the number of total transfected axons.
Cell Death Assay.
SH-SY5Y cells were transfected by Lipofectamine 2000. Twenty-four hours after transfection, the cells were treated with 10 μM MG132 for 18 h. Hoechst (Molecular Probes) and propidium iodide (Molecular Probes) were stained to quantify the percentage of cell death. Transfected hippocampal neurons were treated with DMSO or 25 µM kainate for 2 h. Ventral midbrain neurons were infected with AAV-DJ expressing the respective proteins and treated with DMSO or 100 µM kainate for 10 h. DAPI was used to label the nuclei. Image J (NIH) was used to analyze the data, and the area of the nuclei determined the percentage of cell death.
MPTP Administration.
Young (2-mo-old) and old (12-mo-old) male WT, PICK1 KO, Parkin KO, and PICK1/Parkin DKO mice were administered MPTP or saline. The mice were s.c. administered a single low dose of MPTP HCl (25 mg per kg of body weight, calculated as a conjugated salt; Sigma-Aldrich) daily for 5 consecutive days. The control young and old male mice received an equivalent volume of 0.9% saline. The mice were killed 90 min, 5 d, or 21 d after the last injection, and the brains were dissected for further analysis.
Quantification of TH- and Nissl-Positive Neurons.
Every fourth floating 40-µm-thick cryosection that spanned the SNpc was collected for immunohistochemistry. Sections for Nissl staining were adjacent to the ones for TH staining. Unbiased counts of the total numbers of the TH- and Nissl-positive neurons within the SNpc of one hemisphere were performed using the optical fractionator workflow under Nikon Ni-U upright microscope equipped with MBF Stereo Investigator software. Regions of interest were outlined at low magnification (4× objective) and sampled at high magnification (100× oil objective). The following parameters were used in our study: sampling frame, 80 µm × 80 µm; grid area, 200 µm × 200 µm; guard region, 1.5 µm; sampling depth, 10 µm. Coefficient of error values for TH neurons and nuclei were both under 0.1 (SI Appendix, SI Materials and Methods).
Quantification of TH OD in Striatum.
All images were obtained with the Nikon Eclipse Ti inverted microscope with SPOT advanced software at the same magnification to allow the visualization of the semistriatum in a single field. To quantify the TH fibers, the OD was measured using ImageJ software. The OD of the corpus callosum was used as background and was subtracted from every measurement.
Statistical Analyses.
Data are represented as mean ± SEM. For the comparisons between two groups, unpaired two-tailed Student’s t tests were used to analyze the data. For the comparisons across more than two groups, the data were analyzed using one-way ANOVAs and adjusted with the Bonferroni’s correction. For data with more than one independent variable, two-way ANOVAs with Bonferroni’s post hoc analysis were used. A value of P < 0.05 was considered statistically significant and is indicated by asterisks as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. Yuan Shang for analyzing the structure of Parkin. We are also grateful to Dr. Zhuoyi Liang for providing guidance for the statistical analyses. Special thanks are given to Miss Shui Wa Yun for assistance in taking care of animals and genotyping. This work was supported, in part, by the Research Grants Council of the Hong Kong Special Administrative Region, China: Grants 16146516, 16102914, 663613, N_HKUST625/15, HKUST10/CRF/12R, C4011-14R, T13-607/12R, AoE/M-05/12, and AoE/M-604/16.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716506115/-/DCSupplemental.
References
- 1.Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson’s disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lücking CB, et al. French Parkinson’s Disease Genetics Study Group European Consortium on Genetic Susceptibility in Parkinson’s Disease Association between early-onset Parkinson’s disease and mutations in the parkin gene. N Engl J Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 3.Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 4.Walden H, Martinez-Torres RJ. Regulation of Parkin E3 ubiquitin ligase activity. Cell Mol Life Sci. 2012;69:3053–3067. doi: 10.1007/s00018-012-0978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandiver MS, et al. Sulfhydration mediates neuroprotective actions of parkin. Nat Commun. 2013;4:1626. doi: 10.1038/ncomms2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–374. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winklhofer KF, Henn IH, Kay-Jackson PC, Heller U, Tatzelt J. Inactivation of parkin by oxidative stress and C-terminal truncations: A protective role of molecular chaperones. J Biol Chem. 2003;278:47199–47208. doi: 10.1074/jbc.M306769200. [DOI] [PubMed] [Google Scholar]
- 8.Chung KKK, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 9.Yao D, et al. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11:1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- 11.Meng F, et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol Neurodegener. 2011;6:34. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalia SK, et al. BAG5 inhibits parkin and enhances dopaminergic neuron degeneration. Neuron. 2004;44:931–945. doi: 10.1016/j.neuron.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 13.Sato S, et al. 14-3-3eta is a novel regulator of parkin ubiquitin ligase. EMBO J. 2006;25:211–221. doi: 10.1038/sj.emboj.7600774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson TM, Dawson VL. Parkin plays a role in sporadic Parkinson’s disease. Neurodegener Dis. 2014;13:69–71. doi: 10.1159/000354307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson TM, Dawson VL. The role of parkin in familial and sporadic Parkinson’s disease. Mov Disord. 2010;25:S32–S39. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg MS, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 17.Itier JM, et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12:2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- 18.Von Coelln R, et al. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci USA. 2004;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci USA. 2005;102:2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez FA, Curtis WR, Palmiter RD. Parkin-deficient mice are not more sensitive to 6-hydroxydopamine or methamphetamine neurotoxicity. BMC Neurosci. 2005;6:71. doi: 10.1186/1471-2202-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas B, et al. MPTP and DSP-4 susceptibility of substantia nigra and locus coeruleus catecholaminergic neurons in mice is independent of parkin activity. Neurobiol Dis. 2007;26:312–322. doi: 10.1016/j.nbd.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickrell AM, et al. Endogenous parkin preserves dopaminergic substantia nigral neurons following mitochondrial DNA mutagenic stress. Neuron. 2015;87:371–381. doi: 10.1016/j.neuron.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin JH, et al. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens DA, et al. Parkin loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc Natl Acad Sci USA. 2015;112:11696–11701. doi: 10.1073/pnas.1500624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao M, et al. PICK1-ICA69 heteromeric BAR domain complex regulates synaptic targeting and surface expression of AMPA receptors. J Neurosci. 2007;27:12945–12956. doi: 10.1523/JNEUROSCI.2040-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joch M, et al. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol Biol Cell. 2007;18:3105–3118. doi: 10.1091/mbc.E05-11-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staudinger J, Lu J, Olson EN. Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-alpha. J Biol Chem. 1997;272:32019–32024. doi: 10.1074/jbc.272.51.32019. [DOI] [PubMed] [Google Scholar]
- 28.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 29.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 30.He J, Xia M, Tsang WH, Chow KL, Xia J. ICA1L forms BAR-domain complexes with PICK1 and is crucial for acrosome formation in spermiogenesis. J Cell Sci. 2015;128:3822–3836. doi: 10.1242/jcs.173534. [DOI] [PubMed] [Google Scholar]
- 31.Junn E, Lee SS, Suhr UT, Mouradian MM. Parkin accumulation in aggresomes due to proteasome impairment. J Biol Chem. 2002;277:47870–47877. doi: 10.1074/jbc.M203159200. [DOI] [PubMed] [Google Scholar]
- 32.Kraft C, Peter M, Hofmann K. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 33.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarou M, et al. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staropoli JF, et al. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- 37.Dauer W, Przedborski S. Parkinson’s disease: Mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 38.Tatton NA, Kish SJ. In situ detection of apoptotic nuclei in the substantia nigra compacta of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-treated mice using terminal deoxynucleotidyl transferase labelling and acridine orange staining. Neuroscience. 1997;77:1037–1048. doi: 10.1016/s0306-4522(96)00545-3. [DOI] [PubMed] [Google Scholar]
- 39.Przedborski S, Vila M. MPTP: A review of its mechanisms of neurotoxicity. Clin Neurosci Res. 2001;1:407–418. [Google Scholar]
- 40.Huang D, et al. Dynamic changes in the nigrostriatal pathway in the MPTP mouse model of Parkinson’s disease. Parkinsons Dis. 2017;2017:9349487. doi: 10.1155/2017/9349487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko HS, et al. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin’s ubiquitination and protective function. Proc Natl Acad Sci USA. 2010;107:16691–16696. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ko HS, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pao KC, et al. Probes of ubiquitin E3 ligases enable systematic dissection of parkin activation. Nat Chem Biol. 2016;12:324–331. doi: 10.1038/nchembio.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, et al. Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum Mol Genet. 2005;14:3885–3897. doi: 10.1093/hmg/ddi413. [DOI] [PubMed] [Google Scholar]
- 45.Gardner SM, et al. Calcium-permeable AMPA receptor plasticity is mediated by subunit-specific interactions with PICK1 and NSF. Neuron. 2005;45:903–915. doi: 10.1016/j.neuron.2005.02.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.