Significance
Chromosomal inversions play an important role in local adaptation. Strong evidence exists of selection acting on inversions, but the genic targets inside them are largely unknown. Here we take a systems genetics approach, analyzing two inversion systems implicated in climatic adaption by Anopheles gambiae. We profiled physiology, behavior, and transcription in four different karyotypic backgrounds derived from a common parental colony. Acclimation to different climatic regimes resulted in pervasive inversion-driven phenotypic differences whose magnitude and direction depended upon gender, environment, and epistatic interactions between inversions. Inversion-affected loci were significantly enriched inside inversions, as predicted by local adaptation theory. Drug perturbation supported lipid homeostasis and energy balance as inversion-regulated functions, a finding supported by research on climatic adaptation in multiple systems.
Keywords: Anopheles gambiae, aridity, chromosomal inversion, climate adaptation, systems genetics
Abstract
Inversion polymorphisms in the African malaria vector Anopheles gambiae segregate along climatic gradients of aridity. Despite indirect evidence of their adaptive significance, little is known of the phenotypic targets of selection or the underlying genetic mechanisms. Here we adopt a systems genetics approach to explore the interaction of two inversions on opposite arms of chromosome 2 with gender, climatic conditions, and one another. We measure organismal traits and transcriptional profiles in 8-d-old adults of both sexes and four alternative homokaryotypic classes reared under two alternative climatic regimes. We show that karyotype strongly influences both organismal traits and transcriptional profiles but that the strength and direction of the effects depend upon complex interactions with gender and environmental conditions and between inversions on independent arms. Our data support the suppressed recombination model for the role of inversions in local adaptation, and—supported by transcriptional and physiological measurements following perturbation with the drug rapamycin—suggest that one mechanism underlying their adaptive role may be the maintenance of energy homeostasis.
Chromosomal inversions are structural mutations causing reversals of gene order arising when a chromosome breaks in two places and rejoins after a 180° rotation. Pioneering population studies of inversions relied on polytene chromosomes found in Drosophila and other dipterans, but the postgenomic era is revealing that inversions have much wider evolutionary impacts across the tree of life (1–4). While direct phenotypic consequences of these mutations can arise from lesions in coding sequences, the creation of fusion genes, or altered regulatory context at inversion breakpoints (5–8), it is their indirect effect on crossing-over that is thought to drive sex chromosome evolution, local adaptation, and speciation (9–12). In inversion heterozygotes, crossing-over between alternative orientations is effectively suppressed (13), due both to physical constraints and to aneuploid recombinant gametes. As a result, beneficial allelic combinations captured by and/or arising on the inverted orientation are protected against homogenization with genetic backgrounds on the opposite (standard) orientation. Under spatially varying selection, suppressed recombination in polymorphic inversions may facilitate local adaptation to environmental heterogeneities even in the face of migration and gene flow (3, 9, 14).
Owing to a rich historical tradition and powerful genetic tools, the most extensive evidence that inversions are adaptive and may be the targets of natural selection comes from studies of the genus Drosophila (1, 4), although evidence from outside this genus is mounting (e.g., refs. 15–20). In Drosophila melanogaster, latitudinal and altitudinal clines in inversion frequencies associated with climatic variation have persisted over decades despite unimpeded population connectivity (21). Clines have both shifted in response to climate change and formed in parallel on multiple continents. Within localities, inversion frequencies cycle predictably with the season. Moreover, inversion polymorphism has been associated with a variety of fitness-related and ecologically relevant traits, including heat and cold tolerance, desiccation resistance, growth rate, body size, fecundity, and longevity (1, 4). Population genomic studies of clinal inversion polymorphisms have revealed patterns of nucleotide variation consistent with an adaptive role for inversions (22, 23). Although the genic targets of selection and the molecular mechanisms underlying inversion-associated phenotypes remain unresolved in most systems (24), there has been some recent progress in the Drosophila model. Using distinctly different species and experimental designs, two studies investigating the effect of chromosomal inversions on patterns of gene expression reached remarkably similar conclusions: (i) inversions significantly affected the transcript abundance of numerous genes, (ii) inversion-affected transcripts were overrepresented inside inverted regions but were not exclusive to these regions, and (iii) there was scant evidence of direct (position) effects near breakpoints (6, 8). These observations support a model of local adaptation premised on the indirect effect of recombination suppression maintaining combinations of differentially expressed genes (DEGs). Functions enriched among inversion-affected genes included chemoreception (8) and sterol transport (6). Investigation of the patterns of karyotype-influenced transcriptional variation outside of Drosophila could provide valuable insights regarding the generality of these findings.
In a recently radiated group of Afrotropical mosquitos known as the Anopheles gambiae complex, most of the morphologically indistinguishable sibling species are separated by at least one fixed chromosomal inversion difference. Several species in this group play minor or no roles in malaria transmission, but three—those that are extensively polymorphic for chromosomal inversions on chromosome 2—are among the most efficient malaria vectors in the world, including the nominal member, An. gambiae sensu stricto (henceforth, An. gambiae). This species is broadly distributed across much of sub-Saharan Africa, spanning contrasting environments from tropical rain forest through Sahel savanna. Six of its common inversion polymorphisms—one on chromosome 2L and the rest on chromosome 2R—vary in frequency along gradients of aridity in Nigeria, Burkina Faso, Cameroon, and elsewhere (20, 25–27). Notably, the frequencies of all six inversions increase as the degree of aridity increases. Assuming independent assortment of left and right arms, this suggests that the 2L inversion and one or more 2R inversions might contribute, additively or epistatically, to phenotypes that resist aridity. A pooled genomic sequencing study of An. gambiae clines of two polymorphic inversion systems in Cameroon, In(2La) and In(2Rb), revealed large and significant levels of differentiation only between rearranged (not collinear) genomic regions at opposite ends of the cline (25; also see ref. 28). Together with evidence of gene flux between inverted and standard orientations of rearrangements in the center of the cline where heterokaryotypes are frequent, this suggests that spatially varying selection targeting 2La and 2Rb maintains the cline. In laboratory studies, the 2La and 2Rb inversions have been associated with body size and resistance to thermal and desiccation stress (29–31), consistent with a role in aridity tolerance. Transcriptional profiling of larvae subjected to heat hardening revealed differences in the intensity and nature of the thermal stress response between 2La and 2L+a larvae, particularly with respect to proteasome, chaperone, and metabolic functions (32). In field studies, inversion 2La also has been associated with epidemiologically relevant traits such as Plasmodium infection rate and indoor resting/biting (20, 33–35), the latter a presumed behavioral response to an arid microclimate (20). The common association between climate and polymorphic inversion frequencies in Anopheles and Drosophila raises the question of whether parallel inversion-associated mechanisms of climatic adaptation exist.
Previously, we performed a genomic analysis based on clinal sequence variation in karyotyped An. gambiae from an aridity gradient in Cameroon to identify candidate functions implicated in climatic adaptation (25). Here, we adopt a complementary systems genetic approach. Beginning with a chromosomally polymorphic laboratory colony recently established from the center of the Cameroon 2La-2Rb inversion cline, four alternative homokaryotypic subcolonies were derived [which we designate (a;b), (a;+b), (+a;b), and (+a;+b)]. Using 8-d-old male and female adults from each subcolony, maintained under controlled climatic regimes (either benign or arid), we measure organismal traits (body mass, lipid and glycogen content, fecundity, longevity, and nighttime foraging behavior) and global transcriptional profiles. We analyze these data to address the following questions: (i) Do inversions significantly influence organismal and transcriptional phenotypes? (ii) Does the phenotypic influence of an inversion depend upon gender, climate, and/or the status of the other inversion? (iii) Are the inversion-affected genes overrepresented in rearranged regions? (iv) What functions are enriched among inversion-affected genes, and can functional hypotheses be supported through pharmacological perturbation? Our results demonstrate that inversions significantly affect phenotypes, but the magnitude and even direction of the effect depends on complex interactions with gender, environment, and the inversions themselves. In agreement with previous studies in this and other systems, inversion-affected genes are overrepresented inside rearranged regions. Our data suggest that lipid metabolism and, more generally, energy homeostasis are associated with inversions, and this hypothesis is broadly supported by perturbation with the drug rapamycin.
Results and Discussion
Organismal Traits Are Strongly Affected by Gender and Environmental Treatment.
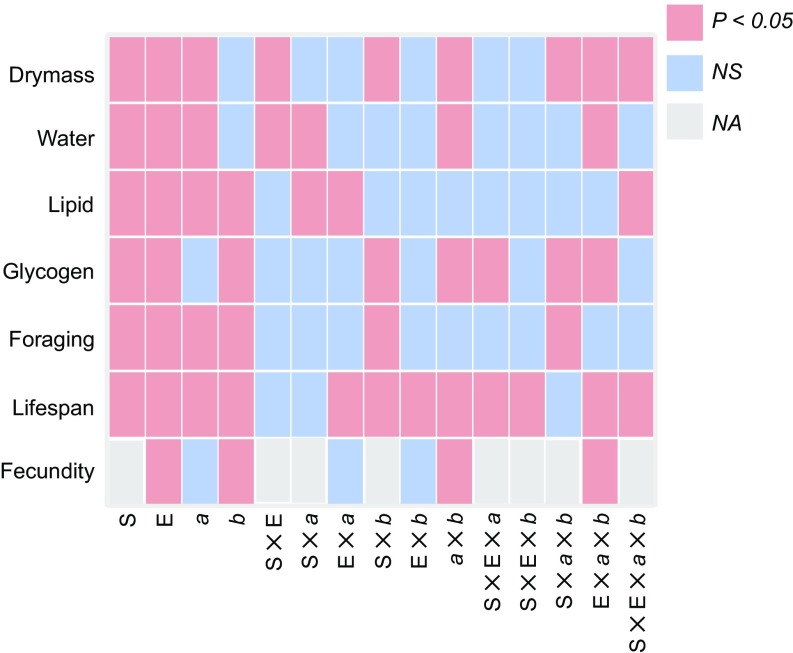
We measured a set of organismal traits in arid-acclimated and benign-treated samples of male and female An. gambiae adults carrying all four homokaryotypic combinations at In(2La) and In(2Rb): (+a;+b), (a;+b), (+a;b), and (a;b). Mean trait values for each group are provided in SI Appendix, Fig. S1 and Table S1. A four-way ANOVA with gender, treatment, and both inversions as main effects indicated that gender and treatment significantly influenced all traits investigated (Fig. 1 and SI Appendix, Table S2). With the exception of life span, the contribution of gender to trait variation (as reflected by effect size, η2) greatly outweighed that of the other factors and interactions (SI Appendix, Table S1). Females were consistently larger (in dry mass) than males, containing more lipid and less glycogen per unit mass than their male counterparts (SI Appendix, Fig. S1 and Table S1). Females also lived significantly longer than males, but prolonged exposure to arid conditions drastically reduced the life span of both sexes and tended to reduce female fecundity, as measured by the number of eggs in the first clutch (SI Appendix, Fig. S1 and Table S1). Arid-acclimation also tended to increase dry mass, reduce standardized water content, and increase nighttime foraging activity in both sexes. Indeed, environmental treatment was second only to gender in terms of its impact on dry mass, water content, and nighttime foraging activity and was the most important source of variation in life span.
Fig. 1.
Summary of ANOVA describing the effects of factors and their interactions on organismal trait means. a, 2La; b, 2Rb; E, environmental condition; NA, not applicable; NS, not significant; S, gender.
Karyotype Is Another Important Influence on Organismal Traits.
One or both inversions also significantly influenced each of the organismal traits we assessed (Fig. 1 and SI Appendix, Fig. S1 and Tables S1 and S2). Inversion 2Rb had a greater effect than environmental treatment on lipid and glycogen content as well as fecundity (SI Appendix, Table S2). Notably, significant interactions between 2Rb and 2La impacted all traits except lipid content and nighttime foraging. Two-way interaction effects were particularly large with respect to glycogen content. In addition, we observed significant higher-order interaction effects.
Our data suggest that arid-acclimated females with (a;+b) and (+a;b) karyotypes are significantly larger in dry mass than arid-acclimated females with (a;b) or (+a;+b) karyotypes (SI Appendix, Fig. S1 and Table S1). In addition, benign-treated (a;+b) females were larger than all other karyotypes under comparable environmental conditions. By contrast, the dry mass of males was statistically indistinguishable by karyotype class irrespective of environmental treatment. Lipid content of benign-treated (+a;+b) females and arid-acclimated (+a;+b) karyotypes of both sexes was significantly higher than that of other karyotypes under equivalent environmental conditions (SI Appendix, Fig. S1 and Table S1). Under either environmental treatment, the (a;b) karyotype tended to have the lowest lipid content in females, the lowest glycogen content in both sexes, and was generally associated with a higher level of nocturnal foraging activity, particularly in males. Under both environmental conditions, (a;b) and (+a;b) females laid significantly fewer eggs in their first clutch than (+a;+b) and (a;+b) females. Benign-treated (a;b) females also had the shortest life spans, but, contrary to expectation if inversions have a simple additive effect, (+a;b) females had the longest life spans, while (+a;+b) and (a;+b) females were intermediate under these conditions.
Karyotype Profoundly Influences the Transcriptional Profile of Females.
Using microarrays, we assessed variation in global transcript abundance across sex, environmental treatment, and the four karyotype classes. Of the 13,123 transcripts on the array, 12,322 (94%) were expressed in our samples and included in the analysis.
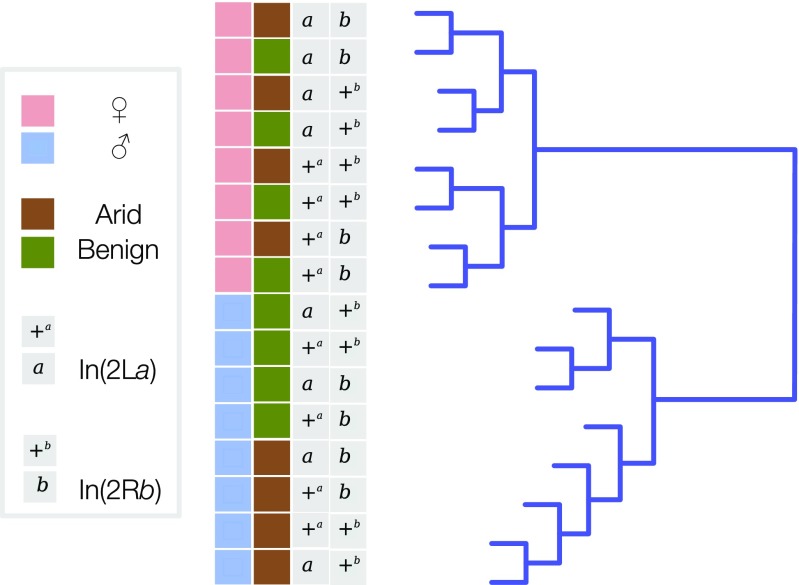
As a first step, we performed unsupervised hierarchical clustering based on overall similarity of transcriptional profiles (Fig. 2). The resulting dendrogram initially bifurcates into two deeply divided main clusters reflecting profound differences in female and male expression profiles. The female cluster further bifurcates according to karyotype rather than environmental treatment. The first female bifurcation corresponds to the In(2La) orientation, +a versus a. Each female In(2La) cluster then bifurcates again according to the In(2Rb) orientation, +b versus b. The male cluster, by contrast, does not bifurcate in a pattern that is easily classified. However, the initial subdivision loosely corresponds to environmental treatment: arid-acclimation versus benign. There is no obvious clustering by karyotype.
Fig. 2.
Hierarchical clustering dendrogram of gene-expression data. Terminal branches (samples) are labeled by gender (pink, female; blue, male), environmental condition (green, benign; brown, arid), and inversion status (+, standard; a or b, inverted).
Next, ANOVA was used to partition the variation in expression among gender, treatment, the two inversions, and their interactions (Table 1 and SI Appendix, Fig. S2). Of the four main effects, gender was the most important. The sex term was significant at a false-discovery rate (FDR) of Q <0.05 for 11,444 (93%) of expressed transcripts, showing pervasive sexual dimorphism for gene expression. A total of 2,862 transcripts had at least a log2 fold change of 1.2 in transcript abundance between sexes. Among the other three main effects, it is noteworthy that 2La affected the expression of a larger fraction of transcripts than arid-acclimation, 4,550 (37%) versus 3,841 (31%). Inversion 2Rb had a smaller impact than the other factors but nevertheless differentially affected 1,576 transcripts (13%).
Table 1.
Genes differentially expressed at Q <0.05
| Source | Genes | Up* | Down* |
| S | 11,444 | 5,704 | 5,740 |
| E | 3,841 | 1,769 | 2,072 |
| a | 4,461 | 2,622 | 1,839 |
| b | 1,576 | 813 | 763 |
| S × E | 4,739 | 2,536 | 2,203 |
| S × a | 339 | 144 | 195 |
| S × b | 328 | 137 | 191 |
| E × a | 0 | — | — |
| E × b | 556 | 184 | 372 |
| a × b | 812 | 375 | 437 |
| S × E × a | 6 | 2 | 4 |
| S × E × b | 311 | 61 | 250 |
| S × a × b | 32 | 11 | 21 |
| E × a × b | 0 | — | — |
| S × E × a × b | 0 | — | — |
a, 2La; b, 2Rb; E, environment; S, sex.
In males vs. females; inverted vs. standard karyotypes; arid vs. benign conditions.
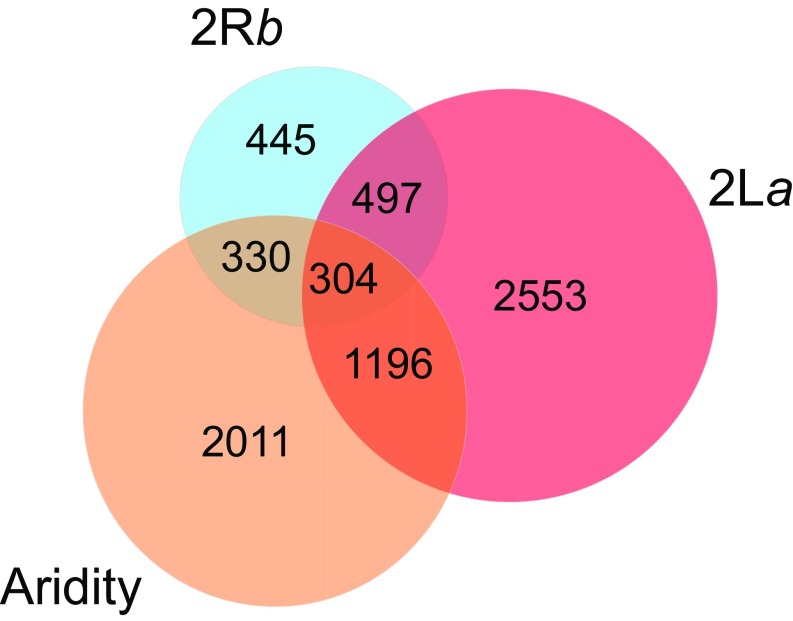
Given the positive correlation between degree of aridity in Africa and the frequencies of the inverted orientations of both inversions, it is perhaps unsurprising that there was overlap in the sets of genes whose expression was influenced by 2La, 2Rb, and aridity (Fig. 3). Inversions 2La and 2Rb jointly affected 802 genes. The overlap between genes influenced by either or both inversions and environmental treatment involved 2,279 genes, of which 308 were significantly affected by all three factors.
Fig. 3.
Venn diagram indicating numbers of genes whose expression was significantly affected by one or more of three factors: 2La, 2Rb, and aridity.
Transcript Abundance Is Affected by Complex Interactions.
The ANOVA revealed numerous pairwise and higher-order interactions among gender, environmental treatment, and inversions that impacted transcript abundance (Table 1 and SI Appendix, Fig. S3), as is consistent with our findings based on organismal phenotypes. For example, the interaction between gender and treatment significantly affected the transcript abundance of 4,739 genes (38%). For a subset of these genes (43%), we detected no significant impact on gene expression of environmental treatment alone, likely because transcriptional abundance was significantly altered by environmental treatment only in males (not females). Additionally, a significant interaction between 2Rb and aridity affected the expression of 556 genes. Unexpectedly, however, there was no indication of a corresponding interaction between 2La and aridity. Consistent with the hierarchical clustering results (Fig. 2), there were also important pairwise interactions between inversions and gender that affected the expression of ∼300 genes in both cases. Finally, we found a significant interaction between 2La and 2Rb that impacted the expression of 812 (6%) genes.
DEGs Are Concentrated in Chromosomal Rearrangements.
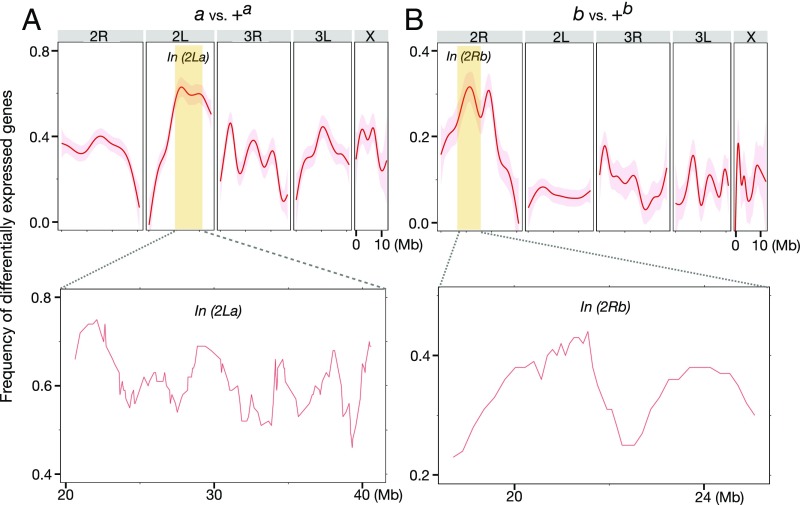
Genes whose expression was significantly altered by 2La were not randomly distributed across the genome. Instead, there was a significant overrepresentation on chromosome 2L (P << 0.001 by Fisher exact test of a 2 × 2 contingency table) (Fig. 4). Similarly, genes whose expression was significantly affected by 2Rb were significantly overrepresented on chromosome 2R (P << 0.001) (Fig. 4). The patterns of significant excess on 2L and 2R were robust both to a more stringent cutoff for differential gene expression (Q <0.01) and to separate consideration of up- and down-regulated genes. The excess of 2La-responsive genes on chromosome 2L and the corresponding excess of 2Rb-responsive genes on chromosome 2R was explained mainly by a disproportionate concentration of such genes between the breakpoints of the ∼22-Mb In(2La) and the ∼7.6-Mb In(2Rb) on chromosomes 2L and 2R, respectively (P << 0.001 by Fisher exact test in both cases). Nevertheless, 2La- and 2Rb-responsive genes were not overrepresented near breakpoints within inversions, as judged by comparing the proportions in the 10% most-distal and most-proximal regions with the 80% in the center (P = 0.2 by Fisher exact test in both cases). This suggests that the pattern is not merely a direct phenotypic consequence of chromosome breakage with resulting position effects. A peak of 2Rb-responsive genes on chromosome 2R also occurs outside of In(2Rb). This peak lies outside the span of two other chromosomal rearrangements polymorphic in West and Central Africa, In(2Rc) and In(2Ru), but it may overlap a third whose breakpoints have not yet been molecularly localized, In(2Rd).
Fig. 4.
Enrichment of DEGs inside In(2La) and In(2Rb). (A and B, Upper) Curves (red lines) and 95% CIs (shaded surrounds) showing the frequency of inversion-affected DEGs along each chromosome were generated by the (stat_smooth) function of R package ggplot2. (Lower) For the focused view inside In(2La) and In(2Rb) (yellow panels), the frequency of DEGs was calculated for windows of 100 genes, slid at a step size of 10 genes. The position of each window was defined as the midpoint of the 100-gene segment.
Coexpression Network Analysis of Genes Suggests Female-Specific Transcriptional Correlations Across Karyotype and Phenotypic Traits.
Complementing differential gene-expression analyses, we also performed an unsupervised weighted network-based analysis (WGCNA) of the complete dataset to identify clusters (modules) of similarly expressed and potentially coregulated genes. We then assessed the correlation of each module “eigengene” with individual phenotypic traits or treatments. The eigengenes of 14 of 22 modules identified were significantly correlated with gender status (after correction for multiple testing), and most of these were also significantly correlated with multiple behavioral and physiological traits (SI Appendix, Fig. S4). The module 8 eigengene was the only one significantly negatively correlated with aridity (i.e., component genes were underexpressed in the arid versus benign treatment); it was also significantly positively correlated with normalized water content, a trait known to affect survival in the face of desiccation stress (36, 37). Likewise, only module 17 was significantly correlated with inversion status, showing the strongest (positive) correlation of any module–trait relationship, but this module was not significantly correlated with any other trait.
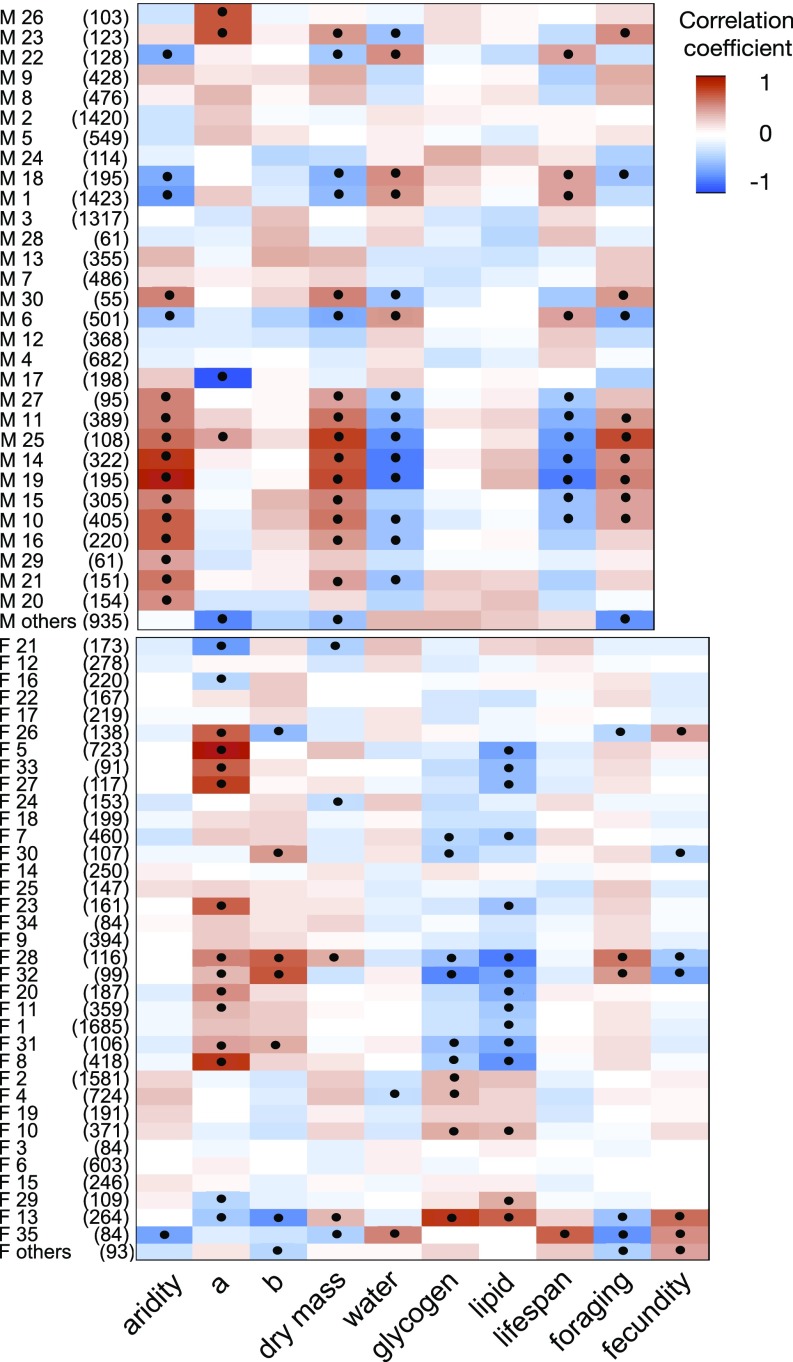
Given the extensive sexual dimorphism seen at the transcriptional level, we constructed separate coexpression networks for females and males and found 35 and 30 modules, respectively (Fig. 5). While the majority (77%) of male modules overlapped with at least one female module (Fisher’s exact test of 2 × 2 contingency tables with P < 0.05 after correction for multiple testing) (SI Appendix, Fig. S5), in most cases the degree of overlap was weak. Of the seven male modules most strongly overlapping female modules (M1, M3, M28, M13, M7, M6, and M17), only two (M1 and M17) were significantly correlated with any traits or treatment (Fig. 5A and SI Appendix, Fig. S5). Overall, the male module–trait relationships revealed significant correlations between six modules and aridity, a subset of which were also strongly correlated with dry mass, normalized water content, lifespan, and nighttime foraging activity but not with inversion status. Indeed, only three male modules were significantly correlated with inversion status (and no other traits), and one of these, M17, is of interest because it is the module most strongly correlated to any trait and strongly overlaps female module F5 (see below).
Fig. 5.
Correlations between WGCNA modules from gene networks and treatment/traits for (A) males only and (B) females only. Modules (rows) are named alphanumerically at the left of each plot, followed by the number of genes in each module (in parentheses). Heatmap colors and intensities reflect the direction and magnitude of correlations between each module and a given organismal trait (columns). Red and blue indicate positive and negative correlations, respectively; significant correlations (after Bonferroni correction) are indicated within the plot.
In striking contrast to male module–trait relationships, only one female module was significantly correlated with aridity, but nine were significantly associated with the status of one or both inversions (Fig. 5B). In turn, subsets of these inversion-correlated modules were also correlated with body traits, but instead of the correlations with dry mass, water content, and lifespan, as was the case for males, the body trait correlations in females mainly involved glycogen and lipid content. Female module F5, like its strongly overlapping male counterpart M17, was the module most strongly correlated with any trait in females, and in both cases the association was with 2La. However, strikingly, the correlations between 2La and female F5 or male M17 are in opposite directions—positive in females, negative in males. Overall, the WGCNA analysis agrees well with the hierarchical clustering analysis (Fig. 2) in revealing the strong gender-based dichotomy in the role of karyotype and environment on male and female gene expression. These results are also in accord with the analysis of organismal traits (Fig. 1) in supporting the relationship between inversions and energy stores.
Functional Enrichment of the Transcriptional Response to Aridity and Karyotype.
To gain insight into biological functions affected by aridity, 2Rb, and 2La, we performed functional enrichment analyses of genes whose expression was significantly impacted (Q <0.05) by these factors according to our ANOVA, dividing each list according to whether gene expression was up- or down-regulated in the focal samples relative to the reference samples [i.e., arid versus benign; inverted orientation versus standard (uninverted) orientation]. Using the Database for Annotation, Visualization and Integrated Discovery (DAVID) to find enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and annotation clusters, we then manually appraised affected genes for functional insights, leveraging the experimental evidence available in FlyBase for those genes with a D. melanogaster ortholog predicted by OrthoDB (38). Detailed results are provided in Dataset S1.
Chronic exposure to aridity up-regulates stress-response genes.
In a previous study, we employed the same aridity-acclimation regimen to explore its impact on cuticular hydrocarbon composition in An. gambiae and its sister species Anopheles coluzzii (39). We showed that longer unbranched alkanes were proportionately more abundant in arid-acclimated mosquitoes than in those maintained under benign conditions, implicating longer alkanes with higher melting points in desiccation resistance. Consistent with those results, here we observed that aridity acclimation was associated with an enrichment of genes involved in lipid biosynthesis (SI Appendix, Table S3); among them were at least six encoding fatty acid elongases. Other enriched annotation terms and pathways (SI Appendix, Table S3) point to a stress response involving chaperones in the Hsp90 and Hsp40 (DnaJ) families and the proteasome, with concomitantly increased cellular energy production and response to oxidative stress (e.g., cytochrome P450s, catalase, peroxidases).
Functions enriched among genes down-regulated by aridity include carbon metabolism and innate immunity.
2Rb up-regulates genes involved in intermediary metabolism and ATP production.
Among genes up-regulated by 2Rb, we detected a number of enriched pathways and annotation clusters related to metabolism and energy production (Table 2). Two other annotation clusters are noteworthy. In the modulation of synaptic transmission cluster are six carboxylesterase genes which have many-to-many orthology relationships to genes in Drosophila that have been implicated in a variety of behaviors, including thermal nociception (40). The DNA-repair cluster contains the master transcriptional regulator p53, which initiates the response to DNA damage, as well as mismatch-repair genes, nucleotide excision-repair genes, DNA polymerase η, and ribosomal protein S3. A robust repair response may be required to repair DNA damage induced by the up-regulated oxidative phosphorylation and/or greater UV exposure encountered at high latitudes in Africa where 2Rb predominates, echoing findings reported for D. melanogaster (41).
Table 2.
Functional enrichment of genes differentially regulated by inversions and their interaction (2La, 2Rb, a × b)
| Up-regulated | Down-regulated | |||||
| KEGG pathway or DAVID cluster | Score | Count | KEGG pathway or DAVID cluster | Score | Count | |
| 2Rb | KEGG: metabolic pathways | 4.9E-8 | 95 | KEGG: glutathione metabolism | 2.4E-3 | 9 |
| KEGG: oxidative phosphorylation | 9.3E-6 | 23 | C-type lectin | 1.8 | 7 | |
| KEGG: citrate cycle (TCA) | 1.9E-3 | 9 | Lipcalin/fatty acid-binding | 1.44 | 4 | |
| KEGG: carbon metabolism | 6.1E-3 | 15 | CHK-kinase | 1.35 | 19 | |
| Oxidoreductase/Cyp450 | 3.12 | 46 | ||||
| Transmembrane helix | 2.3 | 202 | ||||
| 4Fe-4S | 2.07 | 9 | ||||
| TCA | 1.80 | 25 | ||||
| CHK kinase | 1.68 | 22 | ||||
| Proton transporting atpase | 1.66 | 5 | ||||
| Modulation of synaptic transmission | 1.64 | 18 | ||||
| Lipid biosynthesis | 1.57 | 8 | ||||
| Glycolysis/gluconeogenesis | 1.52 | 8 | ||||
| DNA repair | 1.37 | 12 | ||||
| 2La | KEGG: endocytosis | 4.2E-2 | 35 | Alpha-macroglobulin | 2.76 | 8 |
| Lipid recognition (ML) domain | 1.94 | 25 | Serine protease (S1) | 2.53 | 170 | |
| WD40 repeat | 1.62 | 58 | Circadian rhythm | 2.12 | 9 | |
| Amidase | 1.54 | 5 | Innate immunity/ peptidoglycan recognition protein (PGRP) | 2.05 | 9 | |
| Glycoside hydrolase | 1.50 | 37 | Lysozyme | 1.98 | 7 | |
| Sirtuin | 1.80 | 4 | ||||
| Proteolysis | 1.56 | 20 | ||||
| Cell division/cytoskeleton | 1.45 | 8 | ||||
| LDLa | 1.38 | 10 | ||||
| Sensory perception of smell | 1.33 | 15 | ||||
| a × b | KEGG: glutathione metabolism | 2.6E-3 | 7 | Ribosome/translation | 1.54 | 14 |
| KEGG: carbon metabolism | 4.7E-3 | 9 | Lipocalin/fatty acid-binding | 1.34 | 3 | |
| KEGG: TCA | 1.4E-2 | 5 | ||||
| KEGG: retinol metabolism | 2.7E-2 | 3 | ||||
| KEGG: fructose/mannose metabolism | 3.5E-2 | 4 | ||||
| GST | 1.98 | 9 | ||||
| Aminopeptidase | 1.83 | 13 | ||||
| Zymogen | 1.58 | 7 | ||||
| Carbon metabolism | 1.54 | 10 | ||||
| Metabolic process | 1.42 | 11 | ||||
| Trypsin-like serine protease | 1.39 | 37 | ||||
Among the functions enriched in the gene set down-regulated by 2Rb was innate immunity [e.g., C-type lectin genes and TGFβ-activated kinase 1].
2La is implicated in the maintenance of energy balance.
Although the frequencies of 2Rb and 2La covary geographically, their functional effects appear distinctive, with one main exception. Similar to 2Rb (and aridity), 2La down-regulates genes involved in innate immune functions (Table 2). Represented across six enriched annotation clusters are genes encoding thioester-containing proteins, peptidoglycan-recognition proteins, class A scavenger receptors, clip-domain serine proteases, Toll-like receptors, lysozymes, and cecropin antimicrobial peptides, all of which have known or suspected roles in insect immunity (42, 43). Three other annotation clusters based on genes down-regulated by 2La are of special interest in light of the working hypothesis (discussed below) that physiological and behavioral aspects of energy balance are differentially regulated by 2La and 2L+a. The first, circadian rhythm, contains takeout 1 and takeout 3 whose single fly ortholog (takeout) is under circadian control and regulates feeding frequency and locomotory behavior (44, 45). The second contains sirtuin genes, evolutionarily conserved energy sensors that regulate metabolism and energy balance (46, 47). The third (sensory perception of smell) mainly contains genes encoding odorant-binding proteins that have been shown to modulate feeding behavior in Drosophila (48). Given that gustatory receptors (GRs) also modulate feeding behavior (49), it is worth noting that among the 40 genes most strongly down-regulated by 2La (i.e., Q <0.05 and log2 fold change >1.5) was GR31, a gene located on chromosome 2 inside In(2La).
Transcripts significantly up-regulated by 2La were enriched for components of the KEGG pathway endocytosis (Table 2), the process of internalization and intracellular trafficking of cell membrane components and nutrients that modulates cell signaling (50). In addition, DAVID identified four significantly enriched annotation clusters that, taken together, highlight lipid metabolism and integrate it with endocytosis (SI Appendix, Table S4). The lipid recognition (ML) domain cluster contained eight presumptive Niemann–Pick type C2 (npc2) genes whose homologs in humans and fruit flies are implicated in sterol transport (51). Two of these were among the top 31 genes most strongly up-regulated by 2La (Q <0.05 and log2 fold change >1.5). The WD40 repeat domain cluster included genes that encode key components of the TOR complex 1 (TORC1) pathway, which serves in nutrient sensing and promotes both endocytosis and lipid synthesis (52–55). The small amidase cluster was composed of genes annotated as fatty-acid amide hydrolases, enzymes involved in lipid signaling. Two of these are orthologs of Drosophila genes whose products are found in association with lipid droplets (56), organelles that play crucial roles in lipid metabolism (57). Of several genes in the glycoside hydrolase cluster encoding enzymes that breakdown sugars, one α-glucosidase gene stood out as the ortholog of Drosophila target of brain insulin (tobi), a transcriptional target of adipokinetic hormone (AKH) signaling (58, 59).
Drug Perturbation Validates a Role for 2La in Energy Homeostasis.
Overall, the organismal trait data suggest that female mosquitoes carrying inverted arrangements at In(2La) and In(2Rb) contain less lipid and forage more extensively at night than do uninverted karyotypes, regardless of benign or arid environmental conditions. Consistent with these findings, coexpression network analysis in females revealed especially striking transcriptional correlations with 2La status and lipid content and, to a lesser extent, with glycogen content and foraging bouts. These data suggest that 2La maintains a higher energy budget that depends on lipid fuel.
Functional enrichment (DAVID) analysis of DEGs tended to reinforce a connection between 2La status and lipid metabolism, but the limitations of this approach prompted us to look beyond significantly enriched annotation clusters to survey more comprehensively the list of 2La-responsive genes for patterns consistent with our energy-balance hypothesis. We found three lines of support (SI Appendix, Table S4). First, insects maintain energy balance through systemic signaling initiated through peptide hormones binding their G protein-coupled receptors, resulting in the activation of PKA and alteration of physiology (e.g., mobilization of lipid and/or sugar stores) and behavior (e.g., food intake) (60–65). In agreement with expectations, genes encoding AKH, the receptors for AKH, corazonin, and short neuropeptide F, and PKA catalytic and regulatory subunits were significantly up-regulated in the 2La background. Second, we found many additional up-regulated genes that function in TORC1 signaling: components of the TORC1 complex itself, upstream activators, the downstream master lipogenic transcription factor, and the two proteases required to activate this factor. Finally, we noted the up-regulation of several key genes involved in ecdysteroid biosynthesis and transport. These include genes encoding both the most important regulator of prothoracic gland edysteroidogenesis in insects and its receptor (60, 66, 67) as well as sterol transporters, among others. Unexpectedly, because our mosquito samples were neither mated nor blood fed, we also found transcriptional up-regulation of both ovary ecdysteroidogenic hormone and miso genes (SI Appendix, Fig. S3), the latter previously characterized as a female atrial-specific reproductive protein whose expression is triggered by 20-hydroxy-ecdysone transferred by the male during copulation (68). Although both genes have roles in egg maturation (68, 69), our data suggest that they may have additional functions.
Prompted by transcriptional data consistent with a role for 2La in boosting lipid biosynthesis, plausibly through coordination with the TORC1 pathway, we employed pharmacological intervention to reduce TORC1 signaling. Importantly, TORC1 is not required for the basal homeostatic activity of the metabolic pathways it controls; instead, it “acts as a rheostat for cellular metabolism, reinforcing shifts between anabolic and catabolic processes as the nutrient state of the local and systemic environment changes” (55). To inhibit TORC1 signaling, adult female 2La or 2L+a mosquitoes were fed the drug rapamycin for 8 d. We then profiled global gene expression and measured normalized glycogen and lipid content from treated and control samples of both genotypes.
Rapamycin treatment profoundly altered the global transcriptional profile. Considering this treatment as a single factor without regard for alternative orientations of In(2La), we found that 11,091 genes (of the 12,143 expressed) were differentially expressed at the Q <0.05 threshold. Functional enrichment analysis of the 5,863 genes down-regulated by rapamycin revealed the expected overrepresentation of genes involved in translation (SI Appendix, Table S5). Among the slightly smaller number of up-regulated genes (5,228), there was an enrichment of functions related to membrane channels and cell-surface receptors (particularly striking among olfactory receptors), hinting at possible neurosensory and behavioral alterations in response to metabolic perturbation. These same two functions, translation and olfaction, were also the most overrepresented in the 6,347 genes whose transcript levels reflected a significant drug × karyotype interaction (SI Appendix, Table S5). These data suggest that in a 2La background, the consequences of rapamycin treatment may be blunted; translation (and oxidative phosphorylation) may not be as strongly repressed, and, conversely, sensory perception may not be as strongly primed as in the 2L+a background. In this regard, it is notable that the gene reptor was affected by the drug × karyotype interaction, so that its expression was higher in a 2La background. REPTOR is a recently discovered transcription factor acting downstream of TORC1 and is a major effector responsible for ∼90% of the transcriptional induction that occurs in response to TORC1 inhibition (70).
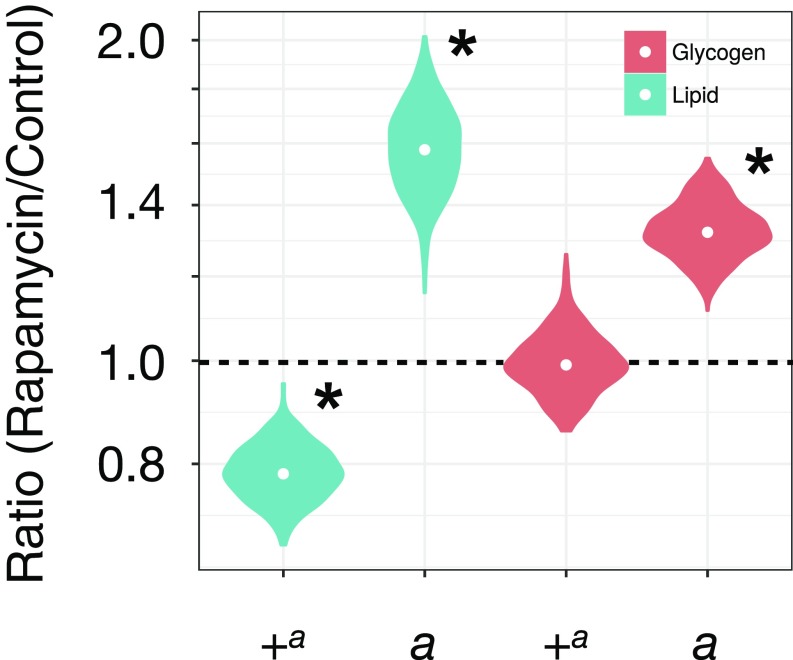
Broadly consistent with our expectation of differences in energy homeostasis between opposite orientations of In(2La), chronic rapamycin treatment had different impacts on lipid and glycogen content in 2La and 2L+a mosquitoes (Fig. 6). Treated 2L+a mosquitoes had significantly less lipid than untreated controls, while glycogen content was not significantly altered. In contrast, treated 2La mosquitoes contained significantly more lipid and glycogen than untreated controls.
Fig. 6.
Effect of rapamycin on lipid and glycogen content. Results are shown as the ratio of rapamycin-treated to untreated control. Ratios were assessed for significance by a Wilcoxon rank sum test (*P < 0.05). Circles inside violin plots represent the median.
Conclusions
The concept of energy-limited tolerance to environmental stress (71) provides a useful framework for integrating the inversion-affected phenotypes observed in this study into a testable working model. Under optimal environmental conditions such as those at the southern endpoints of An. gambiae inversion clines, energy remaining from aerobic metabolism after securing basal metabolic costs would cover other investments, including immune defense, deposition of energy reserves, and reproduction. Moderate (nonlethal) thermal and desiccation stress at the northern endpoints should impose significantly higher basal metabolic costs and limit other energy allocations. These costs accrue not merely due to elevated temperatures per se but also to increased energetic demands of diurnal temperature fluctuations (72), which are substantial at high latitudes (>20 °C) relative to lower latitudes of the forest belt (∼5 °C) (73). In keeping with energy-limited stress tolerance, studies using Drosophila have shown both that elevated temperatures and thermal stress deplete lipid reserves and that the protective but energetically expensive heat shock response is essential to protect flies from long-term body fat decline (74).
Based on the results from our systems genetic analysis and our previous analysis of genomic differentiation along inversion clines (25), we hypothesize that chromosomal inversions may have enabled northern An. gambiae populations to adapt to fluctuating thermal (and associated desiccation) stress through a life-history alteration in energy allocation, increasing basal ATP production, lipid metabolism, and cellular stress responses at the cost of lipid storage (and fecundity, for 2Rb). Consistent with this hypothesis, our previous study of larval An. gambiae and their transcriptional response to heat hardening revealed not only a more aggressive up-regulation of heat shock genes by 2La versus 2L+a but also the exclusive and very strong up-regulation by 2La of an Hsp90 cochaperone (Aha1) that is the most potent stimulator of its activity (75). Under this model, the inversion cline would be maintained by fitness tradeoffs balancing higher survival of inverted karyotypes in the north with higher fecundity of standard karyotypes in the south. The patterns of lipid content, fecundity, and longevity measured in the laboratory are complex (SI Appendix, Fig. S1), and their applicability to the field is not straightforward, given that our experimental design differs in many ways from field conditions. Additional insights could come from incorporating temperature fluctuations, allowing blood meals and mating, and increasing the precision of stored energy measurements (76). Future transcriptional profiling should include heterokaryotypes, assess tissue rather than global transcriptomes, measure miRNA levels, and, importantly, interrogate different developmental stages and multiple time points. Finally, protein-level investigations could complement these data, given that transcript abundance is not a perfect proxy for protein abundance. Despite these limitations, encouraging parallels between this and other systems are accumulating that may suggest common elements of climate adaptation: latitudinal gene expression and body size clines for Drosophila subobscura in Europe implicating the TOR pathway (77), corresponding tradeoffs between metabolism versus reproduction (78), and parallel latitudinal inversion clines in D. melanogaster on two continents implicating the TOR, ecdysone, and EGFR pathways as well as peptide hormone signaling and lipid metabolism (6, 79). Such overlap between hypothesis-generating studies across different systems now justifies more targeted research.
The results presented here have conclusively established that An. gambiae inversions significantly influence organismal traits and transcriptional phenotypes. The disproportionate number of inversion-affected genes in rearranged regions away from breakpoints supports the suppressed recombination model of inversions in local adaptation, in accord with recent studies in other systems (6, 8) and with the transcriptional patterns observed in our earlier study of larval heat hardening (32). Although the candidate heat-responsive genes reported in that study were not significantly enriched inside 2La, 2La-affected genes (defined as differentially expressed by ANOVA at an FDR <0.01) were indeed disproportionately represented 1.2-fold inside 2La (32). These data complement our previous finding that significant nucleotide divergence between populations at opposite endpoints of inversion clines is also concentrated in the rearranged regions (and largely absent elsewhere) (25). Finally, we have also clearly shown that the phenotypic effects of inversions depend on complex interactions with gender, climate, and other inversions. Inversion-affected phenotypes are expressed largely in females, perhaps because male An. gambiae are relatively short-lived, make smaller energetic contributions to reproduction, and store energy mainly as glycogen rather than lipid (SI Appendix, Fig. S1). While climate and 2Rb interact strongly, as anticipated, the absence of an interaction between 2La and climatic conditions is consistent with the hypothesis that 2La controls a life-history shift that preconditions the mosquito to withstand stress rather than merely responding to it.
Historical studies conducted with natural populations of several Drosophila species reported nonrandom associations between independent inversions on different chromosomes or chromosome arms, and a few found that natural selection was the probable cause (80). For example, the nonrandom associations observed by Prakash (81) in a population of Drosophila robusta from Missouri were unlikely to have arisen from nonrandom mating and affected female viability and male fertility. To our knowledge, little attention has been devoted to relationships between the intermediate (transcriptome) phenotypes of independent inversions. Here we have shown interactions between inversions on different arms of An. gambiae chromosome 2 that differentially affect the expression of hundreds of genes, many of which appear to be involved in metabolic processes and translation. It is plausible, if not likely, that such epistatic interactions affect the physiology of An. gambiae in natural populations and may differentially affect fitness in heterogeneous environments. Not only natural An. gambiae populations but also many of the most commonly used laboratory stocks are highly polymorphic for chromosomal inversions. At the very least, these results caution against inversion-blind interpretations of gene expression, physiology, and behavior. Taken together, our data suggest that by providing the means to successfully exploit stressful environments, chromosomal inversions have indirectly but profoundly increased the vectorial capacity of this malignant malaria vector.
Materials and Methods
Mosquito Colonies and Husbandry.
Experiments were performed using four subcolonies derived from the same parental colony of An. gambiae (formerly An. gambiae S molecular form) (82). Established in 2008 from Ndokayo, Cameroon (29) where 2La and 2Rb segregate in An. gambiae populations, the parental NDO colony was polymorphic for both inversions. The four NDO subcolonies carrying all possible homokaryotypic configurations of these two inversions [(+a;+b), (a;+b), (+a;b), and (a;b)] were selected as described previously (39). Experiments were started three generations after their establishment. All subcolonies were maintained in an insectary under controlled benign conditions of 27 °C, 85% relative humidity (RH), and a 12-h:12-h light:dark cycle with 1-h crepuscular transitions, except where otherwise noted. For each generation, eggs were placed in plastic trays (27 × 16 × 6.5 cm) containing 1 L of water purified by reverse osmosis. Within 24 h after hatching, first-instar larvae were transferred into new trays at a density of 200 larvae per tray and were fed daily with 120 mg of 2:1 finely ground tropical fish pellet:bakers’ yeast. Pupae were transferred to 0.2-m3 emergence cages. Upon emergence, adult mosquitoes were supplied with absorbent cotton saturated with 10% corn syrup solution.
Determination of Body Mass, Lipid, and Glycogen in Individual Mosquitoes.
Phenotypic measurements were made exclusively from sets of adults that eclosed on day 8 after hatching, to control for the effect of development time on body characteristics. Upon eclosion, 30 teneral virgin females and 30 males were transferred to each of two single-sex cages, which were randomly assigned to either benign (85% RH, 27 °C) or to sublethal arid (60% RH, 30 °C) insectary conditions and were maintained for 8 d with access to 10% corn syrup ad libitum, in conformity with previous studies of desiccation resistance in the N.J.B. laboratory (30). On day 8, between 14:00 and 16:00, individual mosquitoes were killed at −20 °C and were placed in individual microfuge tubes. Measurements of wet and dry body mass, lipid, and glycogen content were made for individual mosquitoes in 16 treatment groups (2 sexes × 2 environmental conditions × 4 karyotypes), with five replicates each. First, wet and dry mass were determined on a microbalance (accuracy ±0.2 μg; Mettler Toledo) before and after the carcass was dried overnight at 70 °C. The difference in those measures reflected body water content. Next, lipid and glycogen content were determined following method 3.6 from Methods in Anopheles Research, 4th Ed (2014) (www.beiresources.org) and were standardized by body mass.
Nighttime Foraging Activity.
Teneral female or male adult mosquitoes that eclosed 8 d after hatching were placed individually into 25 × 150 mm glass tubes with access to a sugar source (10% corn syrup) at one end. Following Rund et al. (83), individual mosquito activity was sampled with a Locomotor Activity Monitor 25 (LAM 25) system (TriKinetics). The LAM system simultaneously monitored each tube of mosquitoes in a 32-tube matrix (4 vertical × 8 horizontal tubes). Movement was quantified by breaks in an infrared light beam. The data were collected continuously for 8 d from an LAM 25 unit placed in a light-proof box with its own lighting system (12-h:12-h light:dark cycle with 1-h crepuscular transitions) housed in an insectary maintained under constant benign or arid conditions. Data from the first day were excluded, to mitigate the effects of manipulation and handling. Activity measurements were made per individual mosquito in 16 treatment groups (2 sexes × 2 environmental conditions × 4 karyotypes) of 32 mosquitoes per group, replicated five times. We expressed the data in terms of the number of activity bouts per 13-h night (11 h of darkness preceded and followed by 1 h of crepuscular transitions). An activity bout was defined as any number of contiguous minutes in which the infrared beam was broken at least once per minute.
Longevity and Fecundity.
Longevity was defined as the mean number of days to death from the start of the experiment, which commenced with teneral adults that eclosed on day 8 posthatching. Thirty teneral female or male virgins were placed in each of five replicated cages supplied with 10% corn syrup solution. Cages were maintained under constant benign or arid environmental conditions. Dead mosquitoes were removed, the bottoms of the cages were cleaned, and survival was recorded daily. Longevity was estimated for each cohort (30 mosquitoes × 2 sexes × 2 conditions × 4 karyotypes × 5 replicates).
For the purposes of this study, fecundity was defined as the number of eggs in the first clutch; we did not attempt to record lifetime fecundity. Teneral adults that eclosed 8 d after hatching were placed in gender-specific cages supplied with 10% corn syrup solution and were held for 8 d under constant benign or arid environmental conditions. On the eighth day, 30 females and 30 males were cointroduced into each of five replicated cages. After a 24-h period to allow for mating, females were offered a blood meal. Two days following the blood meal, females were placed in individual oviposition tubes and were allowed to lay eggs over a 48-h period. All females tested laid eggs, and all clutches were observed to hatch. Mean clutch size was calculated for each cohort (30 gravid females × 2 conditions × 4 karyotypes × 5 replicates).
Rapamycin Treatment.
As our focus was the role of 2La in energy homeostasis, experiments with rapamycin were conducted on only two NDO subcolonies, (+a;+b) and (a;+b), each maintained under benign (27 °C, 85% RH) or arid (30 °C, 60% RH) conditions. Following Bjedov et al. (84), rapamycin (LC Laboratories) was dissolved in ethanol and added to adult food (10% corn syrup solution) at a concentration of 200 μM. (Immatures were reared on normal food.) Adults were fed rapamycin chronically (via saturated cotton balls) from eclosion until mosquitoes were killed at day 8.
Gene-Expression Profiling.
Adult mosquitoes that were the source of RNA were maintained as described for collecting measurements of body characteristics: teneral adults that eclosed on day 8 posthatching were cultured for 8 d as virgins (without blood meals but with constant access to 10% corn syrup) under benign or arid conditions in single-sex cages. On day 8, they were flash-frozen in liquid nitrogen and stored at −80 °C until RNA isolation. All mosquito samples were killed between 14:00 and 16:00 to minimize between-sample shifts in transcriptional profiles related to circadian rhythm. Following Cassone et al. (32), total RNA was extracted from four replicate pools of 24 females (males). Each pool was converted to cDNA and was hybridized to 12 × 135 custom NimbleGen arrays for An. gambiae designed from genebuild AgamP3.5 (32). Probe sets on this array were mapped to the AgamP3.8 gene set using the BioMart tool provided by VectorBase (85). Experiments were run so that labeled cDNA samples from the different treatments were evenly distributed across the 12-plex chips; in no case were batch effects detected. Arrays were washed and scanned as described previously (32).
A total of 64 arrays were hybridized for the main gene-expression profiling experiment that considered gender, environmental condition, and all four karyotypes (2 sexes × 2 conditions × 4 karyotypes × 4 replicates); these data have been deposited with ArrayExpress under accession no. E-MTAB-2463. In addition, we hybridized 12 arrays to assess the effect of rapamycin treatment on the expression profile of females homokaryotypic for alternative orientations of In(2La) on a uniformly 2R+b background [i.e., (+a;+b) vs. (a;+b), thus 2 treatments × 2 karyotypes × 3 replicates]. This dataset has been deposited with ArrayExpress under accession no. E-MTAB-6034.
XYS files containing the raw intensity values were imported into Bioconductor (86). The quality of each array was assessed using the Bioconductor package arrayQualityMetrics (87). The data were filtered to omit oligos that were misannotated or affected by physical imperfections, as previously described (32). Filtered probes were subjected to background subtraction, normalization, and summarization using the Robust Multi-Array Average (RMA) algorithm. A transcript was tagged as not expressed if its expression values were within the bottom quantile (25%) of expression values across all genes and arrays. Only expressed transcripts were used for subsequent analysis.
The R package nlme was used to partition variation in gene expression using a linear mixed model with four main fixed factors and their interactions (gender, environmental condition, 2La, 2Rb) and replicate as a random factor. The significance of each factor was defined at an FDR <0.05 for this and all other analyses unless otherwise specified.
Tests of Functional Enrichment.
Functional enrichment of Gene Ontology and other annotation terms in candidate gene lists was explored using the DAVID functional annotation tool (https://david.ncifcrf.gov/) (88). DAVID 6.8 with updated Knowledgebase was used. The DAVID clustering module identifies functionally related gene groups based on their annotations and combines them into annotation clusters. The clusters are assigned an enrichment score which represents the minus log-transformed geometric mean of the modified Fisher Exact (EASE) scores within the cluster (89). Significantly enriched annotation clusters were defined as those assigned an enrichment score ≥1.3 (P < 0.05). DAVID also was used to identify enriched KEGG pathways (https://www.genome.jp/kegg/pathway.html).
WGCNA.
As a complementary approach to standard differential expression analysis, gene-expression data were explored using the R package WGCNA (90, 91). Similarity of expression profiles between pairs of genes across all experimental conditions was assessed by Pearson’s correlation coefficient. These pairwise coexpression-similarity measures (the value of the Pearson product moment correlation) were transformed into weighted adjacencies (values between 0 and 1 that reflect the network connection strength between genes), using a soft power adjacency function whose threshold parameters resulted in approximately scale-free network topology (92). The gene-coexpression network constructed from a matrix of weighted adjacencies was analyzed to detect groups of densely interconnected positively correlated genes (modules), which potentially are coregulated. Modules were identified by calculating the topological overlap for all pairs of genes in the network and then selecting those genes strongly connected to the same group of genes in the network. The module eigengene, representative of a weighted average expression profile of a given module, was defined as the first principal component of the correlation matrix of the gene-expression data from that module, after mean centering and unit variance scaling. Module eigengenes were tested for association with gender, environment, karyotype, and phenotypic traits using a P value obtained from a univariate regression model (FDR < 0.05).
Supplementary Material
Acknowledgments
We thank E. Kern-Lovick and L. Xiao for assistance with phenotypic assays, M. Kern and J. Niedbalski for assistance with mosquito rearing, and G. Duffield for access to the LAM 25 system. This work was supported by NIH Grants R01AI076584 and R01AI125360.
Footnotes
Conflict of interest statement: J.C.T. is currently affiliated with Roche-Madison, which produced the microarrays used in this study.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the ArrayExpress gene expression database (accession nos. E-MTAB-2463 and E-MTAB-6034).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806760115/-/DCSupplemental.
References
- 1.Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biol. 2010;8:e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobzhansky T. Genetics of the Evolutionary Process. Columbia Univ Press; New York: 1970. [Google Scholar]
- 4.Krimbas CB, Powell JR. Drosophila Inversion Polymorphism. CRC; London: 1992. [Google Scholar]
- 5.Puig M, Cáceres M, Ruiz A. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc Natl Acad Sci USA. 2004;101:9013–9018. doi: 10.1073/pnas.0403090101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavington E, Kern AD. The effect of common inversion polymorphisms In(2L)t and In(3R)Mo on patterns of transcriptional variation in Drosophila melanogaster. G3 (Bethesda) 2017;7:3659–3668. doi: 10.1534/g3.117.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puig M, et al. Functional impact and evolution of a novel human polymorphic inversion that disrupts a gene and creates a fusion transcript. PLoS Genet. 2015;11:e1005495. doi: 10.1371/journal.pgen.1005495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller ZL, Haynes GD, Richards S, Schaeffer SW. Genomics of natural populations: How differentially expressed genes shape the evolution of chromosomal inversions in Drosophila pseudoobscura. Genetics. 2016;204:287–301. doi: 10.1534/genetics.116.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 11.Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- 13.Dobzhansky T, Epling C. The suppression of crossing over in inversion heterozygotes of Drosophila pseudoobscura. Proc Natl Acad Sci USA. 1948;34:137–141. doi: 10.1073/pnas.34.4.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer SW. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution. 2008;62:3082–3099. doi: 10.1111/j.1558-5646.2008.00504.x. [DOI] [PubMed] [Google Scholar]
- 15.Jones FC, et al. Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joron M, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477:203–206. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 19.Ayala D, et al. Chromosome inversions and ecological plasticity in the main African malaria mosquitoes. Evolution. 2017;71:686–701. doi: 10.1111/evo.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 21.Adrion JR, Hahn MW, Cooper BS. Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. 2015;31:434–444. doi: 10.1016/j.tig.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapun M, Fabian DK, Goudet J, Flatt T. Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Mol Biol Evol. 2016;33:1317–1336. doi: 10.1093/molbev/msw016. [DOI] [PubMed] [Google Scholar]
- 23.Rane RV, Rako L, Kapun M, Lee SF, Hoffmann AA. Genomic evidence for role of inversion 3RP of Drosophila melanogaster in facilitating climate change adaptation. Mol Ecol. 2015;24:2423–2432. doi: 10.1111/mec.13161. [DOI] [PubMed] [Google Scholar]
- 24.Kirkpatrick M, Kern A. Where’s the money? Inversions, genes, and the hunt for genomic targets of selection. Genetics. 2012;190:1153–1155. doi: 10.1534/genetics.112.139899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng C, et al. Ecological genomics of Anopheles gambiae along a latitudinal cline: A population-resequencing approach. Genetics. 2012;190:1417–1432. doi: 10.1534/genetics.111.137794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costantini C, et al. Living at the edge: Biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simard F, et al. Ecological niche partitioning between the M and S molecular forms of Anopheles gambiae in Cameroon: The ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White BJ, et al. Localization of candidate regions maintaining a common polymorphic inversion (2La) in Anopheles gambiae. PLoS Genet. 2007;3:e217. doi: 10.1371/journal.pgen.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fouet C, Gray E, Besansky NJ, Costantini C. Adaptation to aridity in the malaria mosquito Anopheles gambiae: Chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One. 2012;7:e34841. doi: 10.1371/journal.pone.0034841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray EM, Rocca KA, Costantini C, Besansky NJ. Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malar J. 2009;8:215. doi: 10.1186/1475-2875-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rocca KA, Gray EM, Costantini C, Besansky NJ. 2La chromosomal inversion enhances thermal tolerance of Anopheles gambiae larvae. Malar J. 2009;8:147. doi: 10.1186/1475-2875-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassone BJ, et al. Divergent transcriptional response to thermal stress by Anopheles gambiae larvae carrying alternative arrangements of inversion 2La. Mol Ecol. 2011;20:2567–2580. doi: 10.1111/j.1365-294X.2011.05114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrarca V, Beier JC. Intraspecific chromosomal polymorphism in the Anopheles gambiae complex as a factor affecting malaria transmission in the Kisumu area of Kenya. Am J Trop Med Hyg. 1992;46:229–237. doi: 10.4269/ajtmh.1992.46.229. [DOI] [PubMed] [Google Scholar]
- 34.Riehle MM, et al. The Anopheles gambiae 2La chromosome inversion is associated with susceptibility to Plasmodium falciparum in Africa. eLife. 2017;6:e25813. doi: 10.7554/eLife.25813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Behavioural divergences between mosquitoes with different inversion karyotypes in polymorphic populations of the Anopheles gambiae complex. Nature. 1977;266:832–833. doi: 10.1038/266832a0. [DOI] [PubMed] [Google Scholar]
- 36.Benoit JB, Denlinger DL. Meeting the challenges of on-host and off-host water balance in blood-feeding arthropods. J Insect Physiol. 2010;56:1366–1376. doi: 10.1016/j.jinsphys.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hadley NF. Water Relations of Terrestrial Arthropods. Academic; San Diego: 1994. [Google Scholar]
- 38.Zdobnov EM, et al. OrthoDB v9.1: Cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 2017;45:D744–D749. doi: 10.1093/nar/gkw1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reidenbach KR, et al. Cuticular differences associated with aridity acclimation in African malaria vectors carrying alternative arrangements of inversion 2La. Parasit Vectors. 2014;7:176. doi: 10.1186/1756-3305-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honjo K, Mauthner SE, Wang Y, Skene JHP, Tracey WD., Jr Nociceptor-enriched genes required for normal thermal nociception. Cell Rep. 2016;16:295–303. doi: 10.1016/j.celrep.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svetec N, Cridland JM, Zhao L, Begun DJ. The adaptive significance of natural genetic variation in the DNA damage response of Drosophila melanogaster. PLoS Genet. 2016;12:e1005869. doi: 10.1371/journal.pgen.1005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillyer JF. Insect immunology and hematopoiesis. Dev Comp Immunol. 2016;58:102–118. doi: 10.1016/j.dci.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santiago PB, et al. Proteases of haematophagous arthropod vectors are involved in blood-feeding, yolk formation and immunity–A review. Parasit Vectors. 2017;10:79. doi: 10.1186/s13071-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meunier N, Belgacem YH, Martin JR. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J Exp Biol. 2007;210:1424–1434. doi: 10.1242/jeb.02755. [DOI] [PubMed] [Google Scholar]
- 45.Wong R, Piper MD, Wertheim B, Partridge L. Quantification of food intake in Drosophila. PLoS One. 2009;4:e6063. doi: 10.1371/journal.pone.0006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye X, et al. Sirtuins in glucose and lipid metabolism. Oncotarget. 2017;8:1845–1859. doi: 10.18632/oncotarget.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swarup S, Morozova TV, Sridhar S, Nokes M, Anholt RR. Modulation of feeding behavior by odorant-binding proteins in Drosophila melanogaster. Chem Senses. 2014;39:125–132. doi: 10.1093/chemse/bjt061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyamoto T, Slone J, Song X, Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell. 2012;151:1113–1125. doi: 10.1016/j.cell.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- 51.Huang X, Warren JT, Buchanan J, Gilbert LI, Scott MP. Drosophila Niemann-Pick type C-2 genes control sterol homeostasis and steroid biosynthesis: A model of human neurodegenerative disease. Development. 2007;134:3733–3742. doi: 10.1242/dev.004572. [DOI] [PubMed] [Google Scholar]
- 52.Teleman AA. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem J. 2009;425:13–26. doi: 10.1042/BJ20091181. [DOI] [PubMed] [Google Scholar]
- 53.Grewal SS. Insulin/TOR signaling in growth and homeostasis: A view from the fly world. Int J Biochem Cell Biol. 2009;41:1006–1010. doi: 10.1016/j.biocel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 54.Kennedy BK, Lamming DW. The mechanistic target of rapamycin: The grand conducTOR of metabolism and aging. Cell Metab. 2016;23:990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 57.Hashemi HF, Goodman JM. The life cycle of lipid droplets. Curr Opin Cell Biol. 2015;33:119–124. doi: 10.1016/j.ceb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buch S, Melcher C, Bauer M, Katzenberger J, Pankratz MJ. Opposing effects of dietary protein and sugar regulate a transcriptional target of Drosophila insulin-like peptide signaling. Cell Metab. 2008;7:321–332. doi: 10.1016/j.cmet.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 59.Kim J, Neufeld TP. Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat Commun. 2015;6:6846. doi: 10.1038/ncomms7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strand MR, Brown MR, Vogel KJ. Mosquito peptide hormones: Diversity, production, and function. Prog Mosq Res. 2016;51:145–188. doi: 10.1016/bs.aiip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gáliková M, et al. Energy homeostasis control in Drosophila adipokinetic hormone mutants. Genetics. 2015;201:665–683. doi: 10.1534/genetics.115.178897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grönke S, et al. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5:e137. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kubrak OI, Lushchak OV, Zandawala M, Nässel DR. Systemic corazonin signalling modulates stress responses and metabolism in Drosophila. Open Biol. 2016;6:160152. doi: 10.1098/rsob.160152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu Y, et al. Regulation of starvation-induced hyperactivity by insulin and glucagon signaling in adult Drosophila. eLife. 2016;5:e15693. doi: 10.7554/eLife.15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pool AH, Scott K. Feeding regulation in Drosophila. Curr Opin Neurobiol. 2014;29:57–63. doi: 10.1016/j.conb.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marchal E, et al. Control of ecdysteroidogenesis in prothoracic glands of insects: A review. Peptides. 2010;31:506–519. doi: 10.1016/j.peptides.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 67.Rewitz KF, Yamanaka N, Gilbert LI, O’Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326:1403–1405. doi: 10.1126/science.1176450. [DOI] [PubMed] [Google Scholar]
- 68.Baldini F, et al. The interaction between a sexually transferred steroid hormone and a female protein regulates oogenesis in the malaria mosquito Anopheles gambiae. PLoS Biol. 2013;11:e1001695. doi: 10.1371/journal.pbio.1001695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhara A, et al. Ovary ecdysteroidogenic hormone functions independently of the insulin receptor in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2013;43:1100–1108. doi: 10.1016/j.ibmb.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tiebe M, et al. REPTOR and REPTOR-BP regulate organismal metabolism and transcription downstream of TORC1. Dev Cell. 2015;33:272–284. doi: 10.1016/j.devcel.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sokolova IM. Energy-limited tolerance to stress as a conceptual framework to integrate the effects of multiple stressors. Integr Comp Biol. 2013;53:597–608. doi: 10.1093/icb/ict028. [DOI] [PubMed] [Google Scholar]
- 72.Denny M. The fallacy of the average: On the ubiquity, utility and continuing novelty of Jensen’s inequality. J Exp Biol. 2017;220:139–146. doi: 10.1242/jeb.140368. [DOI] [PubMed] [Google Scholar]
- 73.Paaijmans KP, et al. Influence of climate on malaria transmission depends on daily temperature variation. Proc Natl Acad Sci USA. 2010;107:15135–15139. doi: 10.1073/pnas.1006422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klepsatel P, Gáliková M, Xu Y, Kühnlein RP. Thermal stress depletes energy reserves in Drosophila. Sci Rep. 2016;6:33667. doi: 10.1038/srep33667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolmarans A, Lee B, Spyracopoulos L, LaPointe P. The mechanism of Hsp90 ATPase stimulation by Aha1. Sci Rep. 2016;6:33179. doi: 10.1038/srep33179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Foray V, et al. A handbook for uncovering the complete energetic budget in insects: The van Handel’s method (1985) revisited. Physiol Entomol. 2012;37:295–302. [Google Scholar]
- 77.De Jong G, Bochdanovits Z. Latitudinal clines in Drosophila melanogaster: Body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J Genet. 2003;82:207–223. doi: 10.1007/BF02715819. [DOI] [PubMed] [Google Scholar]
- 78.Porcelli D, et al. Gene expression clines reveal local adaptation and associated trade-offs at a continental scale. Sci Rep. 2016;6:32975. doi: 10.1038/srep32975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fabian DK, et al. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 2012;21:4748–4769. doi: 10.1111/j.1365-294X.2012.05731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh BN. Chromosome inversions and linkage disequilibrium in Drosophila. Curr Sci. 2008;94:459–464. [Google Scholar]
- 81.Prakash S. Chromosome interactions in Drosophila robusta. Genetics. 1967;57:385–400. doi: 10.1093/genetics/57.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coetzee M, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–274. [PubMed] [Google Scholar]
- 83.Rund SS, Lee SJ, Bush BR, Duffield GE. Strain- and sex-specific differences in daily flight activity and the circadian clock of Anopheles gambiae mosquitoes. J Insect Physiol. 2012;58:1609–1619. doi: 10.1016/j.jinsphys.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 84.Bjedov I, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giraldo-Calderón GI, et al. VectorBase Consortium VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43:D707–D713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kauffmann A, et al. Importing ArrayExpress datasets into R/Bioconductor. Bioinformatics. 2009;25:2092–2094. doi: 10.1093/bioinformatics/btp354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 89.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012;46:i11. [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.