Significance
Duchenne muscular dystrophy (DMD) is characterized by a catastrophic progression of muscle degeneration that leads to early death. Currently, there is neither a cure for DMD nor any treatments that effectively halt muscle degeneration. Novel methods that assess disease activity non-invasively would greatly accelerate the development of effective therapies. Here we present a novel preclinical animal model that uses bioluminescence imaging as a read-out of muscle degeneration, therefore constituting a non-invasive method to assess disease progression and response to experimental gene therapy in the mdx model of DMD. This model should be widely applicable for monitoring disease activity and responses to therapy in mouse models of muscular dystrophy.
Keywords: monitoring disease activity, DMD, BLI, muscle degeneration, muscle regeneration
Abstract
Duchenne muscular dystrophy (DMD) is a rare, muscle degenerative disease resulting from the absence of the dystrophin protein. DMD is characterized by progressive loss of muscle fibers, muscle weakness, and eventually loss of ambulation and premature death. Currently, there is no cure for DMD and improved methods of disease monitoring are crucial for the development of novel treatments. In this study, we describe a new method of assessing disease progression noninvasively in the mdx model of DMD. The reporter mice, which we term the dystrophic Degeneration Reporter strains, contain an inducible CRE-responsive luciferase reporter active in mature myofibers. In these mice, muscle degeneration is reflected in changes in the level of luciferase expression, which can be monitored using noninvasive, bioluminescence imaging. We monitored the natural history and disease progression in these dystrophic report mice and found that decreases in luciferase signals directly correlated with muscle degeneration. We further demonstrated that this reporter strain, as well as a previously reported Regeneration Reporter strain, successfully reveals the effectiveness of a gene therapy treatment following systemic administration of a recombinant adeno-associated virus-6 (rAAV-6) encoding a microdystrophin construct. Our data demonstrate the value of these noninvasive imaging modalities for monitoring disease progression and response to therapy in mouse models of muscular dystrophy.
Duchenne muscular dystrophy (DMD) is the most common muscle genetic disorder, with an incidence of ∼1 in 5,000 live male births (1, 2). The disease is caused by mutations in the gene coding for dystrophin (3). The lack of dystrophin protein leads to membrane damage associated with fiber necrosis and inflammation, ultimately resulting in progressive muscle degeneration and weakness (4–7). Symptom onset begins in early childhood, usually between the ages of 3 and 5 (3, 8). Boys with DMD present with inability to run and difficulty in rising from the floor, in the first 5 y of life. The combination of muscle weakness, degeneration, and contractures leads to loss of independent walking (9, 10).
There is currently no cure for DMD, nor is there any effective treatment to reverse or halt the muscle degeneration. Since 1987, when the genetic defect was identified (3, 11), many promising therapeutic strategies, including pharmacological, genetic, and cell-based approaches, have been tested in several animal models of DMD (12–17). Of the hundreds of experimental therapies that have been tested, only a few have been approved by the Food and Drug Administration and are currently in phase III clinical trials (18).
One of the hurdles in developing cures or treatments for DMD is the lack of efficient, time-saving, and simple assays that assess the effectiveness of a therapeutic intervention. Most commonly, the outcomes of therapeutic interventions are determined by histologic analysis of acute or cumulative muscle damage, such as areas of necrosis, regeneration, and fibrosis; serum markers of muscle breakdown, such as creatine kinase levels; or muscle function in either isolated muscles or living animals, such as grip strength. Although all of these methods have been used extensively, they present disadvantages in that they are labor intensive, poorly sensitive to changes in disease activity and therefore of limited quantitative value, terminal for the experimental animals, or some combination of these. Thus, the development of new methods to monitor disease progression is crucial for the assessment of novel treatments for DMD.
Noninvasive imaging modalities have a number of distinct advantages, including offering the potential to monitor disease activity in living animals. This allows for repetitive serial measurements without having to sacrifice animals at each time point, and it permits longitudinal monitoring of response to a therapeutic intervention. To date, there are a very limited number of approaches for monitoring disease activity in dystrophic muscles in living animals. Among the noninvasive imaging modalities that have been used to follow disease progression in the muscular dystrophies are magnetic resonance imaging (MRI) and fluorescence optical imaging (19–23). Bioluminescence imaging (BLI) has been applied extensively to the monitoring of many different diseases, particularly cancer, in living animals because of its remarkable sensitivity combined with relative special resolution. Although requiring transgenesis, this technology has been widely applied to studies of temporal progression of disease and it offers great advantages over the other approaches. It is accessible, accurate, relatively inexpensive, and easy to use (24, 25). Furthermore, BLI offers fast, sensitive imaging, even detecting microscopic activity (26, 27). This thereby permits disease activity to be tracked temporally and noninvasively.
Recently, we developed a transgenic reporter mouse strain (which we termed the “regeneration reporter” strain) to test whether we could assess muscle disease activity and progression noninvasively in living animals over time (26). By conditionally expressing luciferase in muscle stem cells (MuSCs), or satellite cells, of the SJL strain [a dysferlin-deficient mouse strain that is an animal model for the human disease, limb girdle muscular dystrophy 2B (LGMD2B)], we could monitor muscle regeneration as a surrogate marker of disease activity using BLI of MuSC activity. Indeed, the BLI readout correlated with the temporal progression and spatial distribution of muscle degeneration. This strain is thus a reliable and quantitative reporter, even if indirect, of disease activity.
In this report, we describe a model, which we term the “degeneration reporter” strain, in which muscle degeneration can be directly monitored using BLI by conditionally expressing luciferase in the myofiber compartment of muscle. Crossing this strain with a dystrophic mouse strain (the mdx mouse), we were able to monitor disease activity directly using BLI. Furthermore, we validated the value of this strain (as well as the previously described regeneration reporter) to faithfully report changes in disease progression in response to experimental therapy. By delivering microdystrophin to the muscle using an rAAV vector (28), we found that the decline (for the degeneration reporter) or increase (for the regeneration reporter) of the BLI signal ceased following effective gene therapy. These data demonstrate that these reporter strains are reliable and are quantitative tools to monitor the efficacy of experimental therapeutics for mouse models of muscular dystrophy noninvasively in living mice.
Results
Monitoring Muscle Degeneration by Noninvasive, Bioluminescence Imaging.
To monitor muscle degeneration of adult skeletal muscle by BLI, we wanted to conditionally and selectively express luciferase in the myofiber compartment of skeletal muscle. Toward that end, we used a mouse strain (29), designated HSACreER, in which the conditional Cre recombinase is expressed under the control of the muscle-specific human skeletal actin (HSA) promoter (Fig. 1A). We bred that strain with the Rosa26LuSEAP mouse reporter in which luciferase is expressed in a Cre-dependent manner (30) (Fig. 1A). Upon administration of tamoxifen to the offspring (HSACreER/Rosa26LuSEAP), which we term degeneration reporter mice, luciferase is permanently expressed in the existing myofibers.
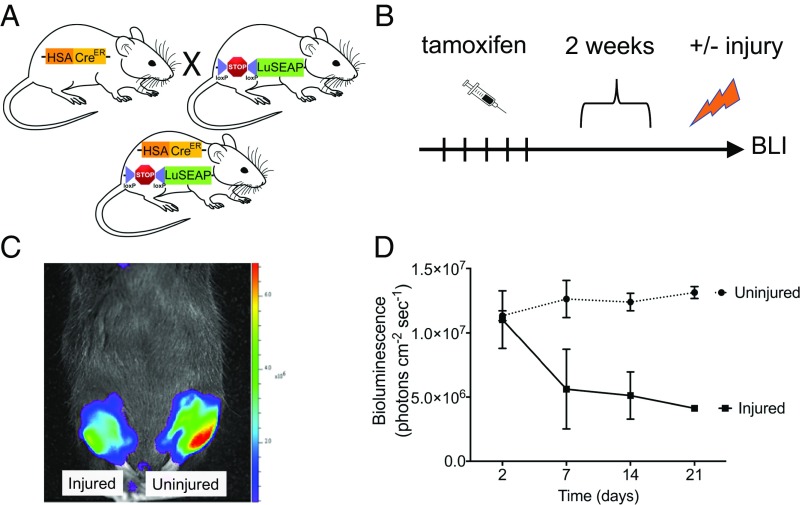
Fig. 1.
Characterization of the degeneration reporter strain. (A) Schematic of the breeding strategy used to obtain mice with the tg-HSACreER transgene with the luciferase gene flanked by loxP sites. In the tg-HSACreER/Rosa26LuSEAP/wt progeny, the STOP sequence is deleted following tamoxifen injection and luciferase expression is observed in the reporter mice. (B) Scheme of tamoxifen injection and sample analysis. (C) BLI images of representative wild-type mice 7 d after an acute injury and control uninjured hindlimb. The scale bar to the Right of the image represents the photon emissions from the tissue surface. (D) Luciferase expression of degeneration reporter mice monitored over 3 wk. Bioluminescence values are indicated as radiance (p·cm−2·s−1). Data represent the mean ± SEM; n = 3.
To test whether this reporter strain could be used to monitor the degeneration of skeletal muscle, 2-mo-old degeneration reporter mice were injected for 5 d with tamoxifen. We injured the right tibialis anterior (TA) muscles, and the BLI signals from the muscles (injured and uninjured) were monitored over the subsequent 3 wk (Fig. 1B). As expected, the luciferase signals remained constant in uninjured TA muscles (Fig. 1 C and D). On the contrary, injured TA muscles showed a dramatic reduction in luciferase signals over the course of the first week (Fig. 1D). Thus, the degeneration reporter strain is a reliable assay system to follow muscle degeneration in vivo.
As noted previously, we had constructed the regeneration reporter strain (Pax7CreER/Rosa26LuSEAP) as an indirect readout of muscle fiber degeneration, since the regenerative response follows muscle degeneration (26). Indeed, BLI of the regeneration reporter (SI Appendix, Fig. S1A) after the same kind of muscle injury revealed a mirror-image signal of that seen with the degeneration reporter; the signal increased over the first week and then plateaued (SI Appendix, Fig. S1 B–D). Together, these data support the value of both strains, one direct and the other indirect, in monitoring the degeneration of muscle fibers in vivo in living animals.
Monitoring the Progression of Muscular Dystrophy by Bioluminescence Imaging.
To assess whether the degeneration reporter strain would be a valuable noninvasive tool for monitoring the progressive degeneration of muscle fibers in a dystrophic model, we crossed this reporter with the most widely studied animal model of DMD, the mdx strain. We utilized the mdx5cv mouse model that has ∼10-fold fewer revertant dystrophin+ve myofibers at both 2 and 6 mo of age (31). In the absence of tamoxifen treatment, neither the wild-type nor the dystrophic strain exhibited any significant BLI signal. However, both strains exhibited robust BLI signals following tamoxifen treatment (SI Appendix, Fig. S2).
We imaged mdx degeneration reporter mice monthly from the age of 2 mo up to the age of 12 mo (Fig. 2). The BLI signal declined progressively, most notably over the first 6 mo and then more gradually over the subsequent 6 mo (Fig. 2 A and B), paralleling what has been described histologically during the first year of life of the mdx strain (32–35). The nondystrophic strain exhibited no change in the BLI signal over the same time course (Fig. 2B). The pattern of declining BLI signal over 10 mo in the dystrophic strain was also reflected by the levels of luciferase activity in the muscle as measured by biochemical analysis (Fig. 2C).
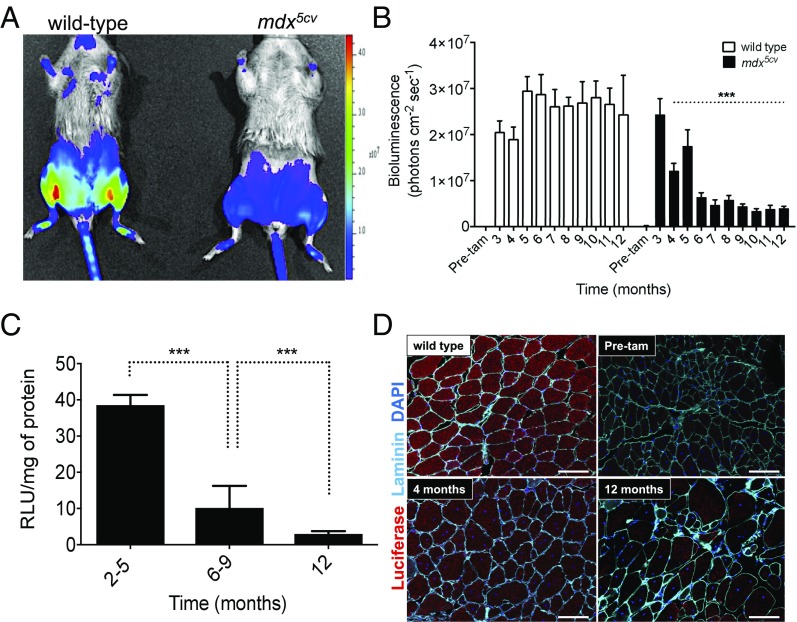
Fig. 2.
Disease activity monitored in the mdx5cv degeneration reporter mice. (A) Representative examples of wild-type and mdx5cv degeneration reporter strains imaged at 5 mo of age following tamoxifen injection at 3 mo of age. (B) BLI data from hindlimb muscles of both wild-type and mdx5cv degeneration reporter mice before tamoxifen injection (3 mo of age; “Pre-tam”) and each month thereafter, up to 12 mo of age. Data represent the mean ± SEM; n = 12; ***P < 0.0001, relative to the respective control group. (C) Luciferase enzymatic activity from hindlimb muscles of degeneration reporter mice of different age ranges following tamoxifen injection at 2 mo of age. (D) Immunostaining for luciferase (red) and laminin (cyan) in gastrocnemius muscles of mdx5cv degeneration reporter mice at different ages following tamoxifen injection at 2 mo of age. 4,6-diamidino-2-phenylindole appears as blue. (Scale bar: 50 µm.)
The results of the luciferase measurements (by both BLI and biochemical analysis) were further confirmed by histological examination of the muscle tissues. Cryosections of gastrocnemius muscles were stained for luciferase protein at 4 and 12 mo of age (Fig. 2D). As expected, we observed a reduction of luciferase expression over time in the dystrophic strain. Together, these results indicate that the degeneration report is a valuable research tool for monitoring the natural history and progression of disease activity over time in mdx5cv mice.
We previously reported that the regeneration reporter strain reliably reflected disease activity in the dysferlin-deficient SJL strain (26). To confirm that this reporter also reliably reflected disease activity in the mdx5cv strain, we repeated the natural history experiments using the regeneration reporter crossed onto the mdx5cv background. Indeed, just as we observed with the degeneration reporter, this reporter strain reliably reflected disease activity in the mdx strain, although of course with patterns inverse to those seen with the degeneration reporter, i.e., increases in BLI signals, increases in luciferase activity, and increases in luciferase+ve fibers (SI Appendix, Fig. S3 A–D). These data demonstrate that the regeneration reporter reliable reflects disease activity in the mdx5cv strain as it does in the SJL strain.
Monitoring Disease Progression Following Treatment with an AAV-Microdystrophin Construct.
In light of these findings, we asked whether these two reporter strains would be useful in monitoring changes in disease progression in response to a therapeutic intervention. We chose, as a gold standard, the restoration of dystrophin expression as the therapeutic intervention since this should, in principle, halt disease activity. We used the delivery of microdystrophin by an rAAV vector since this has been used extensively in preclinical studies in mice to halt disease activity (14).
The left hindlimbs of tamoxifen-treated mdx5cv degeneration reporter and mdx5cv regeneration reporter mice received the rAAV6-microdystrophin vector using hydrodynamic limb vein injection (36). The right hindlimbs were injected only with saline. Wild-type degeneration reporter and regeneration reporter mice were also used as controls. The mice were analyzed at 3, 6, and 9 mo after injection (rAAV-microdystrophin or saline) by BLI imaging (Fig. 3). The saline-treated hindlimbs showed the same patterns as described for uninjected mice (Fig. 2 and SI Appendix, Fig. S3). By contrast, hindlimbs of dystrophic reporter mice injected with the rAAV-microdystrophin construct revealed a cessation of progressive disease activity as evidenced by a plateauing of BLI imaging (Fig. 3 A–D).
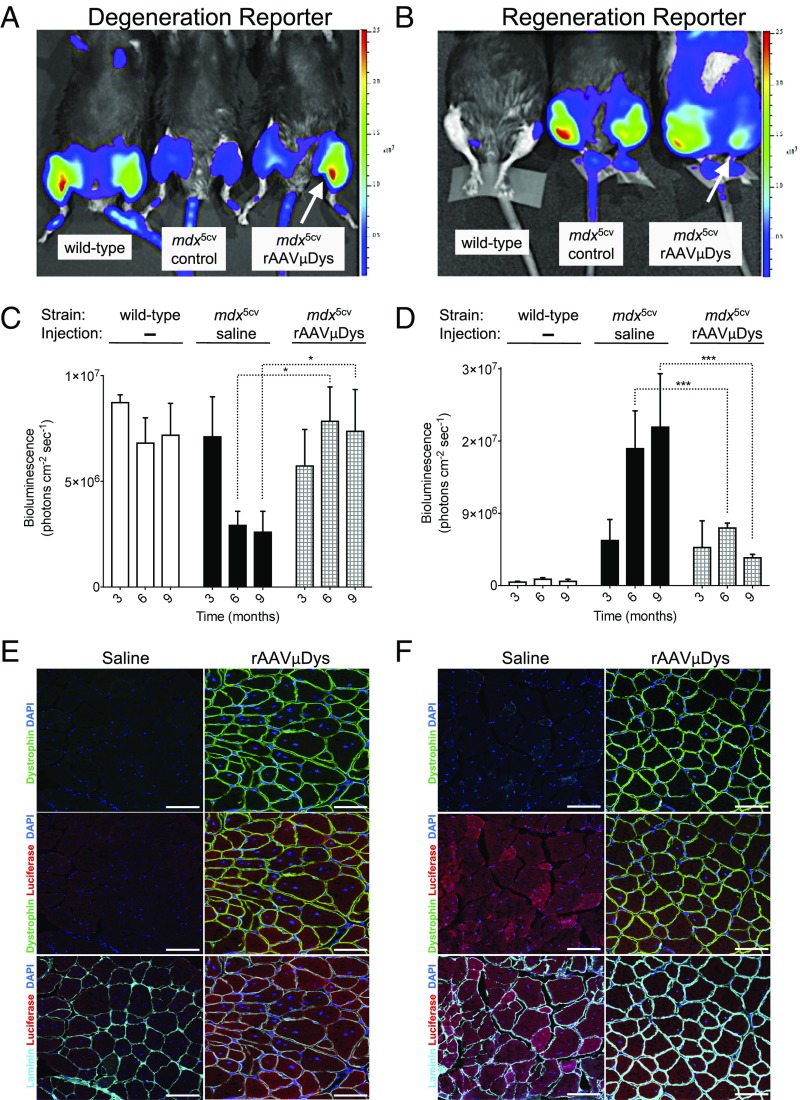
Fig. 3.
Evaluation of the therapeutic effectiveness of the rAAV-microdystrophin treatment of mdx5cv degeneration and regeneration reporter strains. (A and B) Representative BLI images of (A) degeneration reporter and (B) regeneration reporter strains imaged at 9 mo after the gene therapy treatment. Mice are wild type (Left), control (saline injected) mdx5cv (Middle), and mdx5cv where the left hindlimb (white arrow) had been injected with rAAV6-microdystrophin virus (Right). Tamoxifen treatments were done at 2 mo of age; control and rAAV injections were performed 2 wk after last dose of tamoxifen. (C and D) Quantitation of BLI signals at 3, 6, and 9 mo after rAAV injection from groups of mice illustrated in A and B. Data represent the mean ± SEM; n = 3–5; *P < 0.05; ***P < 0.0001. (E and F) Immunostaining for luciferase (red), dystrophin (green), and laminin (cyan) of cryosections from tibialis anterior muscles of tamoxifen-treated degeneration (E) and regeneration (F) reporter mice, 9 mo after hindlimb injections with saline or rAAV6-microdystrophin. 4,6-diamidino-2-phenylindole appears as blue. (Scale bar: 50 µm.)
At the conclusion of the BLI imaging time course (9 mo after injections), mice were killed to assess the status of the muscles histologically. For both reporter strains, immunofluorescence analysis of the muscles revealed the presence of dystrophin+ve fibers throughout the muscles (Fig. 3 E and F), confirming the efficacy of the rAAV delivery, as previously reported (28). Furthermore, 9 mo after treatment with the rAAV-microdystrophin construct, the degeneration reporter hindlimbs revealed a preservation of luciferasehigh fibers and the regeneration reporter hindlimbs revealed a preservation of luciferaselow fibers (Fig. 3 E and F), both indicative of a cessation of disease activity. These findings confirm the reliability of both strains to monitor disease activity, albeit using different readouts, and support the contention that these reporter strains can serve as effective noninvasive, longitudinal readouts of disease activity in living mice.
Discussion
Here we present a mouse model in which muscle degeneration, a primary pathogenic consequence of the lack of dystrophin (15), can be followed noninvasively using BLI. We demonstrated the value of the dystrophic degeneration and regeneration reporter strains by delivering microdystrophin to the muscle using an rAAV6 vector (28). Both models demonstrated that changes in disease activity are reflected in the BLI readout.
To accelerate the development of new therapies for DMD, it is necessary to utilize methods to detect disease activity as well as to evaluate the outcomes of therapeutic interventions, thereby reducing cost and development time (37, 38). Noninvasive techniques emerged as an indispensable tool in biological research, and only a few imaging techniques have been developed to noninvasively monitor muscular dystrophy in vivo (21–23). Among the live imaging modalities tested in mdx mice are MRI technologies (21, 22) and near infrared fluorescence optical imaging (23). While each of these reliably reflects disease progression, each also has practical limitations. MRI has several limitations, including cost, sensitivity for changes in muscle tissue, and speed of operation (38, 39). The near infrared imaging approach uses a protease-activatable fluorescence polymer, which is activated by the cathepsins, a family of proteases. Although the protease cathepsin B (CTSB) has been reported to be up-regulated in DMD muscles, it is primarily localized to areas within which infiltrating macrophages are present (40, 41), it is highly up-regulated in fusing myoblasts (42), and it can be detected in many other nonmuscle cell types that dramatically increase during muscle repair (43–46). Therefore, the fluorescence readout of this method is primarily correlated with muscle regeneration and inflammatory responses. The reporter strains described here of course are limited to the transgenic strains developed, but the mdx mouse strain plays a crucial role in DMD research and in particular for the testing and development of all therapeutic modalities. The degeneration and regeneration reporters are valuable for monitoring disease activity, progression, and response to therapy because the technology is inexpensive, highly sensitive to degenerative changes in the tissue, and specific to the pathogenetic processes that occur in DMD.
Interestingly, when the disease activity was monitored over 12 mo in the mdx degeneration reporter, we observed persistent luciferase signals indicating an attenuation of the disease progression with aging. This is in agreement with previous studies (based on histological sections) which showed a plateau of attenuation of the disease activity accompanied by minimal myofiber necrosis in aging mdx mice (<5% of muscle area) (32, 35, 47, 48).
Traditionally, the efficacy of drug treatments for the muscular dystrophy has been assayed using strength measurements, serum-biomarker concentrations, and changes in histopathology, either alone or in combination. As an alternative, the degeneration reporter system (as well as the regeneration reporter strain) will be an invaluable alternative mouse model for testing pharmacological and genetic treatments for DMD. Reliable quantitation, the ability to use each animal as its own control, and the elimination of the need to perform labor-intensive and time-consuming histopathology all will allow for rapid testing and development of therapeutic modalities for DMD.
Methods
Animal Studies.
The Pax7CreER and Rosa26LuSEAP mice were gifts from Charles Keller, Oregon Health and Science University, Portland, OR (30). The tg-HSACreER mice were a gift from Pierre Chambon, Universite Louis Pasteur, College de France, Illkirch-Cedex, France (29). Mdx5cv mice were obtained from The Jackson Laboratories (stock no. 002379).
Bioluminescence Imaging.
Bioluminescence imaging was performed using the IVIS Spectrum Imaging System from Xenogen (Caliper Life Sciences). The mice were anesthetized using 2% isoflurane and 100% oxygen with a flow rate of 2.5 L/min. Sterile d-luciferin at a concentration of 50 mg/mL was administered by i.p. injection at 300 μL per mouse and mice were imaged 23 min later. All hindlimb hair was removed using Nair before imaging. The images were collected for 30 s at the maximum light collection (f-stop 1) and the highest resolution (small binning). The images were saved for analysis. The image analysis was performed using Living Image 4.5.2 (Caliper Life Sciences). A manually generated circle [region-of-interest (ROI) function] was placed upon the region of interest in the hindlimb of the mouse. The circle was then duplicated and moved to the dystrophic mouse. Bioluminescence values are indicated as radiance (p·cm−2·s−1).
Luciferase Enzymatic Activity.
At the specified time points, mice were killed using CO2 asphyxiation followed by cervical dislocation. The muscles were weighed immediately following isolation and then placed into 5 mL of a 2 mg/mL solution of collagenase II for 1 h at 37 °C with agitation. The muscles were then triturated, and the samples were centrifuged. Supernatants were aspirated, and muscle tissue was lysed as per the manufacturer’s instructions for the luciferase assay system (Promega, Inc.).
Skeletal Muscle Injury and Tamoxifen Treatment.
Mice were anesthetized using a combination of 2% isoflurane and 100% oxygen at a flow rate of 2.5 L/min. The hindlimbs were shaved, and 25 µL of 1.2% BaCl2 was injected into the TA muscle from just above the ankle using 0.3 mL, 31-gauge insulin syringes (BD Biosciences).
For tamoxifen induction, mice were injected with tamoxifen (20 mg/mL) for 5 consecutive days. Control mice received corn oil only.
Generation of rAAV6-Microdystrophin.
The CMV-ΔR4-R23/ΔCT expression vector (28) was cotransfected with the pDGM6 packaging plasmid into HEK293 cells to generate recombinant AAV vectors comprising serotype six capsids that were harvested, purified, and quantitated as described previously (49). Enriched (up to ∼1 × 1014 vg/mL) preparations of rAAV6 vector were stored in PBS at −80 °C until thawed for use.
Hydrodynamic Limb Vein Injection.
Hydrodynamic limb vein injection (HLV) was performed as described with some modifications (36). Briefly, mice were anesthetized using 2% isoflurane and 100% oxygen at a flow rate of 2.5 L/min and hair was removed from both hindlimbs. A tourniquet was secured around the upper hindlimb to restrict the blood flow for 2 min before the injection and 2 min after the injection. The hindlimb was sterilized using 70% ethanol, and an incision was made using surgical scissors on the medial surface of the leg to expose the saphenous vein. A 1/2 inch 30-gauge needle connected by a catheter to a syringe was then inserted in an anterograde direction into the vein. The rAAV6 vector, at a dose of 1.75 × 1011 viral genomes, was injected in a solution of 1 mL HBSS at a rate of 7 mL/min by a programmable syringe pump (KD Scientific). Two minutes after the injection, the tourniquet was released and the incision was closed with absorbable 6-0 silk sutures (Ethicon). Mice recovered on a 37 °C heating pad.
Immunofluorescence of Tissue Sections.
The mice were killed at specific time points by CO2 asphyxiation followed by cervical dislocation. Hindlimb muscles were submerged in 0.5% PFA for 5 h and then dehydrated in 20% sucrose overnight at 4 °C. The muscles were patted dry, submerged in OCT (Sakura Finetek USA, Inc.), and immediately frozen for 30 s in liquid nitrogen chilled in isopentane. Muscle cryosections (10-µm thick) were blocked with 10% donkey serum and M.O.M basic kit blocking reagent (Vector Labs, Burlingame, CA, www.vectorlabs.com) for 1 h, and then incubated with primary antibodies for 1 h including: rabbit anti-luciferase (1:3,000; Sigma-Aldrich L0159), mouse MANEX1011B 1C7 (1:100; Developmental Studies Hybridoma Bank), and rat anti-laminin (1:1,000, L0663). After the primary antibody incubation step, the slides were rinsed twice in PBS with 0.3% Tween-20 and then incubated in the M.O.M. biotinylated anti-mouse IgG reagent for 15 min. Alexa Fluor 488 streptavidin, 555 donkey anti-rabbit, and 647 goat anti-rat (all from Molecular Probes) were used as secondary antibodies. A Zenon antibody labeling kit (Invitrogen) was used to directly label mouse dystrophin with Alexa 488. All secondary antibodies were diluted 1:1,000 and incubated for 45 min at room temperature. The 4,6-diamidino-2-phenylindole (Molecular Probes) was used to counterstain nuclei. Control tissues were processed simultaneously in the same manner. Staining was visualized by laser confocal scanning (Zeiss LSM880).
Statistical Analysis.
All quantitative data are represented as means, and error bars indicate the SEM. Data were subjected to either a two-way ANOVA test (for disease progression and rAAV treatment studies) or an ordinary one-way ANOVA test (for enzyme assays) to determine the significance of calculated differences. These differences were considered statistically significant at P < 0.05.
Study Approval.
All animals were housed and handled in accordance with the guidelines set forth by the Veterinary Medical Unit at the Veterans Affairs Palo Alto Health Care System, and all procedures were preapproved by the Institutional Animal Care and Use Committee before being performed.
Supplementary Material
Acknowledgments
We thank Dr. Martin Guess for technical assistance with the hydrodynamic tail vein injections. This work was supported by grants from the Muscular Dystrophy Association and the Duchenne Parent Project Netherlands (to T.A.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802425115/-/DCSupplemental.
References
- 1.Romitti PA, et al. MD STARnet Prevalence of Duchenne and Becker muscular dystrophies in the United States. Pediatrics. 2015;135:513–521. doi: 10.1542/peds.2014-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendell JR, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol. 2012;71:304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 4.Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 5.Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 6.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn RD, Campbell KP. Molecular basis of muscular dystrophies. Muscle Nerve. 2000;23:1456–1471. doi: 10.1002/1097-4598(200010)23:10<1456::aid-mus2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 8.Wallace GQ, McNally EM. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu Rev Physiol. 2009;71:37–57. doi: 10.1146/annurev.physiol.010908.163216. [DOI] [PubMed] [Google Scholar]
- 9.Muntoni F. Cardiomyopathy in muscular dystrophies. Curr Opin Neurol. 2003;16:577–583. doi: 10.1097/01.wco.0000093100.34793.81. [DOI] [PubMed] [Google Scholar]
- 10.Frankel KA, Rosser RJ. The pathology of the heart in progressive muscular dystrophy: Epimyocardial fibrosis. Hum Pathol. 1976;7:375–386. doi: 10.1016/s0046-8177(76)80053-6. [DOI] [PubMed] [Google Scholar]
- 11.Monaco AP, et al. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 12.Abdul-Razak H, Malerba A, Dickson G. Advances in gene therapy for muscular dystrophies. F1000 Res. 2016;5:2030. doi: 10.12688/f1000research.8735.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson-Hamm JN, Gersbach CA. Gene therapies that restore dystrophin expression for the treatment of Duchenne muscular dystrophy. Hum Genet. 2016;135:1029–1040. doi: 10.1007/s00439-016-1725-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamberlain JR, Chamberlain JS. Progress toward gene therapy for Duchenne muscular dystrophy. Mol Ther. 2017;25:1125–1131. doi: 10.1016/j.ymthe.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge TA. Impending therapies for Duchenne muscular dystrophy. Curr Opin Neurol. 2011;24:415–422. doi: 10.1097/WCO.0b013e32834aa3f1. [DOI] [PubMed] [Google Scholar]
- 16.Ostrovidov S, et al. Stem cell differentiation toward the myogenic lineage for muscle tissue regeneration: A focus on muscular dystrophy. Stem Cell Rev. 2015;11:866–884. doi: 10.1007/s12015-015-9618-4. [DOI] [PubMed] [Google Scholar]
- 17.Skuk D, Tremblay JP. Cell therapy in muscular dystrophies: Many promises in mice and dogs, few facts in patients. Expert Opin Biol Ther. 2015;15:1307–1319. doi: 10.1517/14712598.2015.1057564. [DOI] [PubMed] [Google Scholar]
- 18.Guiraud S, Davies KE. Pharmacological advances for treatment in Duchenne muscular dystrophy. Curr Opin Pharmacol. 2017;34:36–48. doi: 10.1016/j.coph.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Straub V, et al. Contrast agent-enhanced magnetic resonance imaging of skeletal muscle damage in animal models of muscular dystrophy. Magn Reson Med. 2000;44:655–659. doi: 10.1002/1522-2594(200010)44:4<655::aid-mrm22>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 20.Dunn JF, Zaim-Wadghiri Y. Quantitative magnetic resonance imaging of the mdx mouse model of Duchenne muscular dystrophy. Muscle Nerve. 1999;22:1367–1371. doi: 10.1002/(sici)1097-4598(199910)22:10<1367::aid-mus5>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 21.Heier CR, et al. Non-invasive MRI and spectroscopy of mdx mice reveal temporal changes in dystrophic muscle imaging and in energy deficits. PLoS One. 2014;9:e112477-12. doi: 10.1371/journal.pone.0112477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J, Wicki J, Knoblaugh SE, Chamberlain JS, Lee D. Multi-parametric MRI at 14T for muscular dystrophy mice treated with AAV vector-mediated gene therapy. PLoS One. 2015;10:e0124914. doi: 10.1371/journal.pone.0124914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baudy AR, et al. Non-invasive optical imaging of muscle pathology in mdx mice using cathepsin caged near-infrared imaging. Mol Imaging Biol. 2011;13:462–470. doi: 10.1007/s11307-010-0376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Deiry WS, Sigman CC, Kelloff GJ. Imaging and oncologic drug development. J Clin Oncol. 2006;24:3261–3273. doi: 10.1200/JCO.2006.06.5623. [DOI] [PubMed] [Google Scholar]
- 25.Rehemtulla A, et al. Rapid and quantitative assessment of cancer treatment response using in vivo bioluminescence imaging. Neoplasia. 2000;2:491–495. doi: 10.1038/sj.neo.7900121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maguire KK, Lim L, Speedy S, Rando TA. Assessment of disease activity in muscular dystrophies by noninvasive imaging. J Clin Invest. 2013;123:2298–2305. doi: 10.1172/JCI68458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negrin RS, Contag CH. In vivo imaging using bioluminescence: A tool for probing graft-versus-host disease. Nat Rev Immunol. 2006;6:484–490. doi: 10.1038/nri1879. [DOI] [PubMed] [Google Scholar]
- 28.Banks GB, Judge LM, Allen JM, Chamberlain JS. The polyproline site in hinge 2 influences the functional capacity of truncated dystrophins. PLoS Genet. 2010;6:e1000958-11. doi: 10.1371/journal.pgen.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuler M, Ali F, Metzger E, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in skeletal muscles of the mouse. Genesis. 2005;41:165–170. doi: 10.1002/gene.20107. [DOI] [PubMed] [Google Scholar]
- 30.Nishijo K, et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23:2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danko I, Chapman V, Wolff JA. The frequency of revertants in mdx mouse genetic models for Duchenne muscular dystrophy. Pediatr Res. 1992;32:128–131. doi: 10.1203/00006450-199207000-00025. [DOI] [PubMed] [Google Scholar]
- 32.Coulton GR, Morgan JE, Partridge TA, Sloper JC. The mdx mouse skeletal muscle myopathy: I. A histological, morphometric and biochemical investigation. Neuropathol Appl Neurobiol. 1988;14:53–70. doi: 10.1111/j.1365-2990.1988.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 33.Anderson JE, Ovalle WK, Bressler BH. Electron microscopic and autoradiographic characterization of hindlimb muscle regeneration in the mdx mouse. Anat Rec. 1987;219:243–257. doi: 10.1002/ar.1092190305. [DOI] [PubMed] [Google Scholar]
- 34.McGeachie JK, Grounds MD, Partridge TA, Morgan JE. Age-related changes in replication of myogenic cells in mdx mice: Quantitative autoradiographic studies. J Neurol Sci. 1993;119:169–179. doi: 10.1016/0022-510x(93)90130-q. [DOI] [PubMed] [Google Scholar]
- 35.Duddy W, et al. Muscular dystrophy in the mdx mouse is a severe myopathy compounded by hypotrophy, hypertrophy and hyperplasia. Skelet Muscle. 2015;5:16. doi: 10.1186/s13395-015-0041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guess MG, Barthel KK, Pugach EK, Leinwand LA. Measuring microRNA reporter activity in skeletal muscle using hydrodynamic limb vein injection of plasmid DNA combined with in vivo imaging. Skelet Muscle. 2013;3:19. doi: 10.1186/2044-5040-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pien HH, Fischman AJ, Thrall JH, Sorensen AG. Using imaging biomarkers to accelerate drug development and clinical trials. Drug Discov Today. 2005;10:259–266. doi: 10.1016/S1359-6446(04)03334-3. [DOI] [PubMed] [Google Scholar]
- 38.Grounds MD, Radley HG, Lynch GS, Nagaraju K, De Luca A. Towards developing standard operating procedures for pre-clinical testing in the mdx mouse model of Duchenne muscular dystrophy. Neurobiol Dis. 2008;31:1–19. doi: 10.1016/j.nbd.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brockmann MA, Kemmling A, Groden C. Current issues and perspectives in small rodent magnetic resonance imaging using clinical MRI scanners. Methods. 2007;43:79–87. doi: 10.1016/j.ymeth.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Kar NC, Pearson CM. Muscular dystrophy and activation of proteinases. Muscle Nerve. 1978;1:308–313. doi: 10.1002/mus.880010407. [DOI] [PubMed] [Google Scholar]
- 41.Takeda A, et al. Demonstration of cathepsins B, H and L in xenografts of normal and Duchenne-muscular-dystrophy muscles transplanted into nude mice. Biochem J. 1992;288:643–648. doi: 10.1042/bj2880643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jane DT, et al. Cathepsin B localizes to plasma membrane caveolae of differentiating myoblasts and is secreted in an active form at physiological pH. Biol Chem. 2006;387:223–234. doi: 10.1515/BC.2006.030. [DOI] [PubMed] [Google Scholar]
- 43.Gounaris E, et al. Live imaging of cysteine-cathepsin activity reveals dynamics of focal inflammation, angiogenesis, and polyp growth. PLoS One. 2008;3:e2916. doi: 10.1371/journal.pone.0002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bratlie KM, et al. Rapid biocompatibility analysis of materials via in vivo fluorescence imaging of mouse models. PLoS One. 2010;5:e10032. doi: 10.1371/journal.pone.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sciorati C, Rigamonti E, Manfredi AA, Rovere-Querini P. Cell death, clearance and immunity in the skeletal muscle. Cell Death Differ. 2016;23:927–937. doi: 10.1038/cdd.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kharraz Y, Guerra J, Mann CJ, Serrano AL, Muñoz-Cánoves P. Macrophage plasticity and the role of inflammation in skeletal muscle repair. Mediators Inflamm. 2013;2013:491497. doi: 10.1155/2013/491497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson JE, Bressler BH, Ovalle WK. Functional regeneration in the hindlimb skeletal muscle of the mdx mouse. J Muscle Res Cell Motil. 1988;9:499–515. doi: 10.1007/BF01738755. [DOI] [PubMed] [Google Scholar]
- 48.Spurney CF, et al. Preclinical drug trials in the mdx mouse: Assessment of reliable and sensitive outcome measures. Muscle Nerve. 2009;39:591–602. doi: 10.1002/mus.21211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregorevic P, et al. rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat Med. 2006;12:787–789. doi: 10.1038/nm1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.