Significance
Chromosomes of most organisms have regions of high GC content interspersed with regions of low GC content. We constructed three variants of the yeast URA3 gene with GC contents of 31%, 43%, and 63%. We found that the high-GC URA3 gene had a substantially elevated rate of mutations, both single-base substitutions and deletions. The elevated base substitutions require an error-prone DNA polymerase, and the high rate of deletions occurs as a consequence of DNA polymerase slippage. The high-GC gene also had substantially elevated rates of mitotic and meiotic recombination. These observations indicate that GC content is an important parameter influencing genome evolution.
Keywords: mutations, error-prone DNA polymerase, mitotic recombination, meiotic recombination, high GC content
Abstract
The chromosomes of many eukaryotes have regions of high GC content interspersed with regions of low GC content. In the yeast Saccharomyces cerevisiae, high-GC regions are often associated with high levels of meiotic recombination. In this study, we constructed URA3 genes that differ substantially in their base composition [URA3-AT (31% GC), URA3-WT (43% GC), and URA3-GC (63% GC)] but encode proteins with the same amino acid sequence. The strain with URA3-GC had an approximately sevenfold elevated rate of ura3 mutations compared with the strains with URA3-WT or URA3-AT. About half of these mutations were single-base substitutions and were dependent on the error-prone DNA polymerase ζ. About 30% were deletions or duplications between short (5–10 base) direct repeats resulting from DNA polymerase slippage. The URA3-GC gene also had elevated rates of meiotic and mitotic recombination relative to the URA3-AT or URA3-WT genes. Thus, base composition has a substantial effect on the basic parameters of genome stability and evolution.
The genomes of many eukaryotes, including Saccharomyces cerevisiae, are mosaics of regions with high- and low-GC base composition (1). In our research, we examine the effects of altering the base composition of a yeast gene on two important genetic properties: mutation and recombination rates.
Mutation rates are often determined using reversion assays. This method generally selects for only one type of mutation. Broader mutational spectra can be determined using genes in which forward mutations can be selected, such as URA3, CAN1, and SUP4-o, or in nonselective mutation-accumulation experiments (e.g., ref. 2). The rate of mutations per gene is a complex function of its length and the fraction of base substitutions that result in loss of gene function (3). Since microsatellites tend to be hotspots for small insertions and deletions (indels), the number and length of such microsatellites within the coding sequence also affects the rate of mutations (4). There are multiple sources of spontaneous mutations, including misincorporation errors during DNA synthesis, error-prone repair of oxidative damage of DNA bases, and DNA polymerase slippage within microsatellites. Many (although not all) of these events are mutagenic because the initiating DNA damage/mismatch is acted on by error-prone DNA polymerases. About two-thirds of spontaneous mutations are dependent on the error-prone DNA polymerase ζ (5–7).
Mutations that affect any of the replicative DNA polymerases or other proteins involved in DNA replication frequently elevate the rate of spontaneous mutations, and this elevation is dependent on polymerase ζ (8–10). Mutations in POL3 (encoding the catalytic subunit of DNA polymerase δ) often elevate both base substitutions and large deletions occurring between short direct repeats (8, 11, 12). As described below, the replication of a high-GC template (URA3-GC) by WT DNA polymerases results in a similar spectrum of mutations.
The mutation rate is also substantially affected by the rate of transcription (13). For example, the rate of mutations at the CAN1 locus is elevated 10-fold by transcription, and the mutation spectrum is skewed to small deletions under conditions of high transcription (14). The elevated rate of base substitutions under conditions of high transcription is largely dependent on DNA polymerase ζ, whereas the elevated rate of small (2- to 5-bp) deletions is largely independent of DNA polymerase ζ but requires topoisomerase 1 (14).
Genomic sites associated with high levels of dsDNA breaks (DSBs), (for example, those resulting from certain trinucleotide repeats or palindromic sequences) are often associated with elevated rates of mutations in adjacent sequences. These DSB-induced mutations are often dependent on DNA polymerase ζ (15). In addition, certain types of recombination (such as break-induced replication) lead to high levels of mutations (16).
The effect of genomic context on mutation rates in yeast has been examined in several studies. Ito-Harashima et al. (17) showed that eight identical tRNA-Tyr genes had mutation rates that varied by more than a factor of five. At least part of the variation in mutation rates in different regions of the genome is a consequence of regional differences in mismatch repair efficiency (2, 18). Lang and Murray (19) demonstrated that the rate of mutations of URA3 varied about sixfold in different locations on chromosome VI. The insertion sites that were replicated early in the S phase had lower mutation rates than those that replicated late, and they suggested that late-replicating regions were more susceptible to mutations caused by error-prone DNA polymerases. In addition, Lujan et al. (2) found that mutation rates are affected by proximity to replication origins, the direction of the replication fork, the positions of nucleosomes, and other factors. Furthermore, differences in mutation rates dependent on genome context have been observed in mammalian cells, particularly in studies of cell lines derived from tumors (20). In the experiments described below, we examine mutation rates and spectra in URA3 genes with different base compositions located in the same chromosomal context.
In organisms from yeast to humans, GC richness correlates with elevated rates of meiotic recombination (21–23). One explanation of this observation is that high-GC regions are more susceptible to the enzymatic machinery that creates the recombinogenic DSBs (24). Alternatively, regions with high levels of recombination result in elevated levels of heteroduplexes with base–base mismatches. In some organisms, A/G, T/C, A/C, or T/G mismatches are repaired with a bias toward the formation of a G-C pair rather than an A-T pair (25). These properties of repair could result in the evolution of recombination hotspots toward elevated levels of GC. Although we will not review all the arguments relevant to these two explanations, in previous studies we showed that the GC-rich β-lactamase gene of the Tn3 transposon acted as a strong meiotic recombination hotspot in yeast (26), an observation consistent with the hypothesis that GC-rich sequences act as meiotic recombination hotspots. In our current study, we show that a GC-rich gene stimulates not only meiotic recombination but also mitotic exchange.
Results
Elevated Mutation Rates in a GC-Rich Version of URA3.
We first examined the effect of base composition on mutation rates. The WT URA3 gene in S. cerevisiae (URA3-WT) has a GC content of 43.4%. Because of the redundancy of the genetic code, we were able to construct two genes with very different base compositions from the canonical URA3 gene (URA3-GC, 63% GC; URA3-AT, 31% GC) but with amino acid sequences that were identical to the protein encoded by URA3-WT (SI Appendix, Figs. S1 and S2). DNA fragments containing the three different URA3 alleles were introduced by transformation into the endogenous URA3 locus of the W303-1A–derived strain W1588-4C, replacing the ura3-1 allele. The resulting three isogenic strains (DKy18 with URA3-WT, DKy39 with URA3-GC, and DKy40 with URA3-AT; strain genotypes/constructions are shown in Datasets S1 and S2) grew at similar rates in medium lacking uracil (SI Appendix, Fig. S3). Thus, the altered DNA sequences do not adversely affect the function of the Ura3p. In addition, all three strains were sensitive to 5-fluoroorotate (5-FOA), a drug that prevents the growth of cells with the WT Ura3p (27).
We determined the mutation rate at the URA3 locus in the three strains by measuring the frequency of 5-FOA–resistant (5-FOAR) derivatives in at least 30 independent cultures and then converting these frequency measurements into a rate (see Materials and Methods for details). In our experiments, as well as in a previous study, a small fraction (about 10%) of the 5-FOAR strains were not Ura− and did not contain mutations in the ura3 coding sequence (3). Therefore we corrected the rate estimates by multiplying the rate of 5-FOAR derivatives by the fraction of those derivatives that were Ura− (Dataset S3). This correction had only a small effect. The rate of ura3 mutations in the URA3-GC strain (4.9 × 10−8 per division, 95% confidence limits of 3.6–6.3 × 10−8) was elevated more than sixfold relative to the URA3-WT strain (7.3 × 10−9 per division, 95% confidence limit of 4–11 × 10−9), an increase that was statistically significant by the Mann– Whitney U test (P = 0.0001). Although the strain with the URA3-AT gene had a slightly reduced rate of mutations (5.2 × 10−9 per division, 95% confidence limits of 2.7–8.1 × 10−9 per division) compared with URA3-WT, this reduction was not statistically significant (P = 0.84 by Mann–Whitney U test). These results are summarized in Fig. 1A and Table 1.
Fig. 1.
Rates and types of ura3 mutations in strains with URA3-AT, URA3-WT, and URA3-GC alleles. (A) Rates of ura3 mutations. From measurements of the frequency of 5-FOAR derivatives in multiple cultures, we calculated the rates of mutation in strains with URA3-AT, URA3-WT, and URA3-GC (see Materials and Methods for details). The 95% confidence limits are shown for each measurement. (B) By DNA sequencing, we determined whether the ura3 mutations were single-base substitutions, long (≥5 bp) indels, single-base indels, or complex mutations (more than one mutant substitution). The proportion of each of these classes was multiplied by the rate of ura3 mutations to generate the rates for these classes.
Table 1.
Rates of various types of ura3 mutations in strains with URA3-WT, URA3-GC or URA3-AT
| URA3 allele* | Strain genotype* | Location of URA3 gene† | Rate of base substitutions (μ × 10−9)‡ | Rates of long deletions/insertions (μ × 10−9)‡,§ | Rates of short indels (μ × 10−9)‡,§ | Rates of other mutations (μ × 10−9)¶ | Total rates of ura3 mutations (μ × 10−9)# |
| URA3-WT | WT | V | 5.9 (3.2–9.2) | 0 | 0.6 (0.2–1.3) | 0.7 (0.2–1.5) | 7.3 (4–11) |
| 0 | 0.1 (0–0.6) | ||||||
| URA3-GC | WT | V | 25.5 (16.9–34.7) | 12.5 (7.2–18.8) | 5.4 (2.4–10.1) | 2.7 (0.9–6.5) | 49.4 (36–64) |
| 3.3 (1.2–7.2) | 0 | ||||||
| URA3-AT | WT | V | 3.8 (1.9–6) | 0.3 (0.1–0.8) | 0.3 (0.1–0.8) | 0.6 (0.2–1.1) | 5.2 (2.7–8.1) |
| 0.1 (0–0.3) | 0.1 (0–0.4) | ||||||
| URA3-WT | rev3 | V | 2.8 (1.3–4.6) | 0.2 (0–0.5) | 0.3 (0.1–0.7) | 0.1 (0–0.4) | 3.5 (1.7–5.7) |
| 0.04 (0–0.2) | 0 | ||||||
| URA3-GC | rev3 | V | 2.9 (1.5–4.6) | 8.5 (5.2–12.2) | 0.3 (0–1.0) | 0.8 (0.2–1.7) | 12.9 (8.2–18) |
| 0.5 (0.1–1.3) | 0 | ||||||
| URA3-WT | yku70 | V | 5.7 (2.9–9) | 0.1 (0.0.5) | 0.7 (0.2–1.4) | 0.1 (0–0.6) | 6.7 (3.5–11) |
| 0 | 0.1 (0–0.6) | ||||||
| URA3-GC | yku70 | V | 6.2 (3–10.1) | 22.9 (13.9–33.1) | 0 | 1.5 (0.4–3.6) | 31.4 (20–45) |
| 0.9 (0.2–2.7) | 0 | ||||||
| URA3-WT | dnl4 | V | 5.7 (3–8.9) | 0 | 0.7 (0.2–1.3) | 0.4 (0.1–1) | 7.0 (3.8–11) |
| 0.3 (0.2–0.7) | 0.1 (0.1–0.5) | ||||||
| URA3-GC | dnl4 | V | 19.5 (12–27.9) | 27 (17.6–37.3) | 5.9 (2.7–10.8) | 3.2 (1.1–7.3) | 57.8 (41–76) |
| 2.2 (0.6–5.8) | 0 | ||||||
| URA3-WT | rad1 | V | 35.1 (24.4–46.5) | 0.5 (0–3) | 10.4 (5.9–16) | 3.8 (1.5–7.6) | 50.7 (37–66) |
| 0.5 (0–3) | 0.5 (0–3) | ||||||
| URA3-GC | rad1 | V | 45.9 (32.6–60.0) | 13.1 (7.2–21.3) | 31.1 (20.8–42.8) | 4.9 (1.8–11) | 100 (78–123) |
| 2.5 (0.6–7.6) | 2.5 (0.6–7.6) | ||||||
| URA3-WT-OR1 | WT | III | 8.1 (4.6–12.3) | 0.5 (0.1–1.3) | 1.6 (0–2.4) | 0.2 (0–0.9) | 11.1 (7–17) |
| 0.2 (0–0.9) | 0.5 (0–1) | ||||||
| URA3-GC-OR1 | WT | III | 17.3 (11.1–24) | 10 (5.8–14.9) | 4.8 (2.4–8.3) | 0.7 (0.1–2.7) | 34.2 (24–46) |
| 1 (0.2–3.2) | 0.3 (0–2.2) | ||||||
| URA3-WT-OR2 | WT | III | 10.3 (6.2–14.9) | 0.5 (0.1–1.4) | 2.3 (1–3.9) | 0.5 (0.1–1.4) | 14.1 (8.8–20) |
| 0.2 (0–0.9) | 0.5 (0.1–1.4) | ||||||
| URA3-GC-OR2 | WT | III | 17.5 (11.1–24.7) | 15.7 (9.7–22.4) | 4.6 (2–8.6) | 1.8 (0.5–4.9) | 42.5 (30–56) |
| 2.8 (1–6.2) | 0 |
The different URA3 alleles and the strain genotypes are described in the text.
In most of the strains, the URA3 gene was located at its normal position on the left arm of chromosome V. In the four strains with the URA3 gene on III, the gene was located in two orientations near the efficiently utilized ARS306 origin (Fig. 4).
Rates were calculated as described in Materials and Methods. Numbers in parentheses are 95% confidence limits.
The upper line in each entry shows the rate of deletions, and the lower line shows the rate of insertions.
Other events include those with more than one mutation per isolate.
Sum of rates for all types of ura3 mutations.
Base Composition-Dependent Alterations in the Mutational Spectra of URA3-WT, URA3-GC, and URA3-AT.
We sequenced between 75 and 94 independent ura3 mutant isolates per strain from the three strains. The resulting data are summarized in Dataset S3. The locations of individual base substitutions and single-base indels are given in Dataset S4, and the sequences of deletions greater than four bases are given in Dataset S5. The mutation spectrum for URA3-WT is very similar to that previously reported by Lang and Murray (3). In our study, 81% of the mutations were single-base substitutions, 9% were single-base indels, and 9% were complex events (multiple base changes, indels plus base changes) (Table 1). No deletions greater than 4 bp were observed in our study or that of Lang and Murray.
Large indels (≥5 bp) were a prominent component of the mutation spectra for URA3-GC, representing more than 30% of the mutational alterations. Because no deletions were observed for URA3-WT, it is difficult to calculate the relative increase in deletions in URA3-GC accurately. However, if we assume the existence of a single deletion among the 75 Ura− strains, we calculate that there was a 129-fold elevated rate of long deletions in the URA3-GC strain. The positions of the deletions for the mutant derivatives of URA3-GC and URA3-AT are given in Dataset S5. The elevated number of large indels for URA3-GC relative to URA3-WT is highly significant (P < 0.0001 by Fisher exact test). The relative reduction of single-base substitutions relative to other types of mutations for URA3-GC compared with URA3-WT was also highly significant (P < 0.0001 by Fisher exact test). It should be emphasized, however, that because of the sixfold elevation in the rate of ura3 mutations for URA3-GC compared with URA3-WT, the rate of single-base substitutions for URA3-GC was still about fourfold higher than for URA3-WT (Fig. 1B and Table 1).
Although the rate of ura3 mutations for URA3-AT was about 70% of that observed for URA3-WT, the mutation spectrum of URA3-AT shared some similarities with that of URA3-GC. In particular, there was a significant elevation in the frequency of large deletions in the URA3-AT strain relative to the URA3-WT strain (P = 0.02 by Fisher exact test). However, the reduction in the rate of base substitutions in the URA3-AT strain was not statistically significant (P = 0.27).
All the long deletions derived as mutant isolates from URA3-GC (23 of 23) and URA3-AT (6 of 6) occur between short direct repeats 4–9 bp in length (Dataset S5). One example is shown in Fig. 2A. In this event, one repeat of GCGGCCA and the intervening sequences are removed. The lengths of the deletions varied between 24 and 222 bp for the URA3-GC allele and between 24 and 423 bp for the URA3-AT allele (Dataset S5). For most of the deletion events (21 of 29), the deletion occurred between two identical sequences (Fig. 2A). However, in 8 of 29 deletions, the deletion could be explained by an interaction between longer imperfect repeats. For example, one deletion was consistent with an interaction between 5-bp perfect repeats of CAAGT or between a 9-bp imperfect repeat with sequences CCGcCAAGT and CCGtCAAGT (Fig. 2B).
Fig. 2.
Examples of deletions and duplications detected in URA3-GC. The red letters indicate the sequences deleted (A and B) or duplicated (C), and boxes outline the repeats located at the deletion/duplication breakpoints. The numbers are the coordinates of URA3-GC with 1 representing the first base. (A) Deletion of 24 bp involving 7-bp perfect repeats. (B) Deletion of 57 bp involving 9-bp imperfect or 5-bp perfect repeats. (C) Duplication of 24 bp involving 7-bp repeats. Note that the repeats are identical to those associated with the deletion in A.
As discussed below, it is likely that the deletions reflect DNA polymerase slippage and therefore hybridization between the repeats. Thus, the stability of the duplex formed will influence the frequency of deletions. Using a program that compares melting temperatures of various lengths of oligonucleotides with and without a mismatch (ITD OligoAnalyzer 3.1; https://www.idtdna.com/calc/analyzer), we found that the melting temperature of a longer duplex with a single mismatch is usually greater than the melting temperature of a shorter duplex without mismatches. In our analysis in Dataset S5, we allowed a single mismatch within the repeats if the mismatch was followed by at least two matched bases. In previous studies of deletions formed in yeast strains with perturbed DNA replication, deletion formation often involved imperfect repeats (11, 28–30).
In addition to the deletions, we found six duplications in the URA3-GC strain and one duplication in the URA3-AT strain. In six of the seven isolates, the duplication was bounded by a short direct repeat (Dataset S5). An example of one of these duplications is shown in Fig. 2C; the duplication involved the same repeats utilized in formation of the deletion shown in Fig. 2A. Although other mechanisms are not precluded, both deletions and duplications can be explained by DNA polymerase slippage (Fig. 3). Deletions reflect a slippage event in which a replicated repeat dissociates from the template and reassociates with a different unreplicated repeat on the template (31). In contrast, a duplication is generated by a dissociation of a replicated repeat from the template followed by a reassociation with a repeat that has already been replicated (Fig. 3B).
Fig. 3.
DNA polymerase slippage as a mechanism for the generation of deletions and duplications. The depicted sequences are those shown in Fig. 2 A and C. Repeats involved in the deletion and duplication are boxed. (A) Deletion produced by polymerase slippage. The primer strand (Top) dissociates from Repeat 1 of the template strand (Bottom) and reassociates with Repeat 2. The resulting intermediate has a 24-base single-stranded loop. Replication of this intermediate would produce a chromosome with a deletion (left side of figure) and a chromosome that retains the WT sequence (right side of figure). (B) Duplication produced by polymerase slippage. Following replication of the template up to Repeat 2, the primer strand slips back to Repeat 1 and then continues synthesis (blue sequences). Replication of the resulting looped intermediate results in one chromosome with a duplication and one with the original sequence.
The Elevated Rate of Base Substitutions, but Not the Elevated Rate of Large Deletions, in URA3-GC Is Dependent on the Error-Prone DNA Polymerase ζ.
In S. cerevisiae the error-prone DNA polymerase ζ is responsible for about half of the spontaneous mutations and more than 90% of UV-induced mutations (7). We found that deletion of REV3, encoding the catalytic subunit of DNA polymerase ζ (32), decreased the ura3 mutation rate of URA3-WT about twofold (Table 1). The rate of base substitutions for URA3-WT was also reduced by a factor of two. In contrast, the rate of long deletions was not decreased in the URA3-WT gene in the rev3 strain compared with the WT strain (Table 1). We observed five long deletions out of 88 total ura3 mutations in the rev3 strain versus 0 of 75 in the WT strain. By the Fisher exact test, this difference is not statistically significant (P = 0.06).
The rev3 deletion resulted in a statistically significant fourfold decrease in the rate of ura3 mutations in URA3-GC compared with URA3-GC in the WT strain (Table 1). Single-base substitutions were reduced ninefold with only a small statistically insignificant reduction (35%) in the rate of large deletions. The positions of mutations in the rev3 strains are shown in Dataset S4, and the sizes and locations of long indels are shown in Dataset S5. In summary, compared with the URA3-WT allele, the URA3-GC allele had an elevated rate of base substitutions that was substantially dependent on DNA polymerase ζ and a greatly elevated rate of large deletions that was largely independent of DNA polymerase ζ.
Influence of the Mutations Affecting Nonhomologous End Joining and the Single-Strand Annealing Pathway on the Mutation Rates and Types of Mutations in the URA3-WT and URA3-GC Genes.
Although the long indels detected in the URA3-GC strain have the properties expected for DNA polymerase slippage events, such alterations could also be a consequence of the repair of a DSB by nonhomologous end-joining (NHEJ). In NHEJ events, the broken ends are rejoined by a mechanism that involves little or no sequence homology (33–35). There are two varieties of NHEJ that are distinguishable by the nature of their products and their genetic requirements. The classic type of NHEJ requires the end-binding proteins Yku70p and Yku80p, the specialized DNA ligase Dnl4p, and the Mre11p/Rad50p/Xrs2p (MRX) complex; microhomology-mediated end-joining (MMEJ) occurs independently of the Ku proteins (34, 35). The absence of the MRX complex has little effect on MMEJ in some studies (36) and a substantial effect in others (37). The Rad1/Rad10 endonuclease is required for efficient MMEJ (37, 38). In the classic NHEJ pathway, the breakpoints are often associated with short (1- to 4-bp) additions or deletions, whereas the MMEJ pathway often results in larger deletions occurring between 5- to 25-bp repeats with frequent insertions of nucleotides (33).
Mutations in the YKU70 or DNL4 genes had no statistically significant effect on the rate of long deletions in the WT or URA3-GC strains (Table 1), indicating that these deletions are not the result of classic NHEJ. Surprisingly, the rate of single-base substitutions in URA3-GC was reduced in the yku70 strain. One interpretation of this result is that Yku70p could be involved in recruiting DNA polymerase ζ, although other explanations are also possible. The mutational spectra for the yku70 and dnl4 strains are shown in Datasets S3–S5.
To determine whether the observed deletions could reflect MMEJ, we examined URA3-WT and URA3-GC mutations in strains with the rad1 mutation. An intermediate in the deletion pathway occurring by MMEJ would be expected to have single-stranded branches that would require processing by the Rad1/Rad10 endonuclease (33). In the rad1 strains with the URA3-WT and URA3-GC alleles, the rates of 5-FOAR were elevated about sevenfold and twofold, respectively (Dataset S3). A fivefold elevated mutation rate in rad1 strains for a plasmid-borne SUP4-o gene was previously reported (39). Importantly, the rates of long deletions in the URA3-GC allele in the WT and rad1 strains were almost identical, 12.5 × 10−9 per division and 13.1 × 10−9 per division, respectively (Table 1). These results strongly argue that the long deletions are not a consequence of MMEJ. In addition, in yeast strains with mutations in RFA1, elevated rates of large deletions were detected, and, partly based on their Rad10p dependency, it was suggested that single-strand annealing (SSA) was involved (28). Since Rad1p and Rad10p would be expected to act in the same pathway, our observations indicate that the URA3-GC–associated long deletions do not involve the SSA pathway. In summary, our results strongly support the hypothesis that the long deletions observed in the URA3-GC gene reflect DNA polymerase slippage rather than MMEJ or SSA.
Analysis of the Influence of Chromosomal Context on the Mutation Rates and Mutational Spectra for URA3-WT and URA3-GC.
In some yeast studies, the spectra of mutations are affected by the transcriptional orientation of the reporter gene with respect to the replication fork (11, 40). In our first experiments, the strains had the URA3 genes located at their normal positions on the left arm of chromosome V. Since the URA3 gene in this location is located approximately equidistant from AR508 and ARS510, the direction of the replication fork through the gene was unclear. Therefore we constructed strains in which URA3-WT or URA-GC were inserted in two different orientations 1 kb centromere-proximal to ARS306 (Fig. 4A); previous experiments showed that insertions at this position were replicated by forks moving rightward from ARS306 (41). In orientation 1 (OR1) the replication forks proceed in the same direction as URA3 transcription, and in orientation 2 (OR2) the forks move in direction opposed to transcription.
Fig. 4.
Rate of ura3 mutations in strains in which the reporter genes (URA3-WT or URA3-GC) are in different orientations relative to the ARS306 replication origin. (A) Four strains used to determine whether the transcription orientation of the reporter gene relative to the replication origin affected mutation rates. (B) The rates of mutations of URA3-WT and URA3-GC in the four strains described above.
The rate of 5-FOAR Ura− was about threefold greater for URA3-GC than for URA3-WT in both orientations, and the rates were unaffected by orientation (Fig. 4B). The rates of single-base substitutions for both the URA3-GC-OR1 and URA3-GC-OR2 were about twofold higher than those observed for the URA3-WT strains. Long deletions/duplications were elevated between 20- and 30-fold for the URA3-GC strains compared with the URA3-WT strains. As observed in our previous experiments, large indels represented a substantial fraction of the total mutations for the URA3-GC strains (32/99 for URA3-GC-OR1 and 40/92 for URA3-GC-OR2) but a smaller fraction for the URA3-WT strains (6/93 for URA3-WT-OR1 and 4/93 for URA3-WT-OR2) (Dataset S3). The types of deletions (size of the relevant repeats and deletion size) observed in the URA3-GC strains on chromosome III are similar to those observed on chromosome V (Dataset S5). There are more large indels relative to other mutational changes in the URA3-WT gene on chromosome III (10 indels and 176 other events) than in the URA3-WT gene on chromosome V (0 indels and 75 other events), although this difference is not statistically significant (P = 0.06 by Fisher exact test).
In summary, the orientation of the URA3 reporter gene relative to the direction of fork movement does not affect the frequency of mutations or the mutation spectra. However, the difference in mutation rates between URA3-GC and URA-WT were reduced from about sixfold on chromosome V to about threefold on chromosome III.
URA3-GC Is a Hotspot for Mitotic Recombination.
In our previous studies, we found that G4-quadruplex sequences were overrepresented at the breakpoints of mitotic recombination events (42). In addition, some GC-rich trinucleotide repeats are associated with elevated levels of mitotic recombination in yeast (43). To determine more directly the relationship between mitotic recombination and GC content, we measured the rate of mitotic recombination between heteroalleles in diploid strains with URA3-AT, URA3-WT, and URA3-GC sequences. In each diploid, one chromosome had a mutation located at coordinate 89 of URA3, and at the allelic position on the other homolog the ura3 gene contained a mutation at coordinate 777. Two of the four strains (DKy147 and DKy149) had mutant alleles derived from URA3-AT, one strain had mutant alleles derived from URA3-WT (DKy143), and one strain had mutant alleles derived from URA3-GC (DKy145). The specific genotypes (given in parentheses) for each strain were DKy147 (ura3-at-C89T/ura3-at-T777G), DKy149 (ura3-at-C89G/ura3-at-T777G), DKy143 (ura3-wt-C89T/ura3-wt-T777A), and DKy145 (ura3-gc-C89T/ura3-gc-T777A).
Since the mutations in the four strains do not show intragenic complementation, the diploids were Ura−, and we measured the rate of alterations to Ura+ by fluctuation analysis as described in Materials and Methods. There are two likely pathways of homologous recombination that could generate Ura+ derivatives (Fig. 5A). A mitotic gene conversion event could transfer WT information from one allele to the other, resulting in the loss of either mutant allele. Alternatively, there could be a crossover between the heteroalleles, resulting in one WT gene and one gene with two mutations. In previous studies of heteroallelic recombination (44) it was shown that most prototrophs result from gene conversion of one of the alleles rather than by crossovers between the alleles. Although a prototrophic diploid could also result from reversion of one of the mutant alleles, we observed that the Ura+ reversion rates in all seven haploids used in constructing the diploids for the mitotic recombination experiments were less than 10−7 per division, at least 50-fold below the rate of prototroph formation in the diploids.
Fig. 5.
Mitotic recombination rates in diploids with ura3 heteroalleles. In all strains examined in this analysis, the diploid had noncomplementing mutations in both copies of URA3-GC, URA3-WT, or URA3-AT. Recombination rates were determined by measuring the rate of Ura+ derivatives from the starting strain. (A) Pathways for generating a WT URA3 gene by mitotic recombination. In previous studies, it is known that the generation of a prototroph from a heteroallelic diploid usually involves gene conversion rather than a reciprocal crossover (44). (B) Rates of mitotic Ura+ derivatives from diploids heteroallelic for mutations in ura3-wt (blue), ura3-at (gray), or ura3-gc (orange). Error bars indicate 95% confidence limits. (C) Percentages of Ura+ spores following sporulation of heteroallelic diploids. Most tetrads segregated zero Ura+ to four Ura− spores; the percentage of tetrads with one Ura+ spore, resulting from meiotic gene conversion, is shown. (D) Percentages of tetrads with meiotic gene conversion events (three Ura+ to one Ura− or one Ura+ to three Ura− tetrads) in diploids heterozygous for ura3-at, ura3-wt, or ura3-gc mutations.
The rate of Ura+ derivatives (Fig. 5B) in the strain with the URA3-GC–derived heteroalleles (95% confidence limits are given in parentheses) was 3.9 × 10−5 per division (3.4–4.4 × 10−5), about 19-fold greater than that in the strain with the URA3-WT–derived alleles (2 × 10−6 per division; 1.6–2.4 × 10−6) and about 14-fold greater than those in the strains with the URA3-AT–derived alleles [2.6 × 10−6 per division (2-3.1 × 10−6) for DKy147; 2.8 × 10−5 per division (2.2–3.4 × 10−6) for DKy149]. Since most spontaneous mitotic recombination events are initiated by dsDNA breaks or ssDNA gaps (45), these observations suggest that GC-rich genes are more prone to the formation of recombinogenic DNA lesions than yeast genes of average GC content.
URA3-GC Does Not Cause a Strong Replication Fork Block or a High Level of DSBs.
Elevated rates of mitotic recombination are sometimes associated with sequences that stall replication forks (46). Sites in the genome that result in substantial stalling of the replication fork (a stall in >5% of the cells) can be detected by 2D gel electrophoresis followed by Southern blotting. Sequences that result in fork stalling produce a pronounced spot on the Y-arc that represents DNA replication forks at various stages of elongation (47). For this analysis, we isolated DNA from bar1 derivatives of isogenic haploid strains that have URA3-AT, URA3-WT, or URA3-GC alleles. Cells were synchronized with alpha pheromone, and DNA was isolated 40 min after release from arrest. DNA isolation protocols and the conditions of gel electrophoresis are described in Kim et al. (48). No strong stall was observed in any of the strains (Fig. 6A). This result was not unexpected because the rates of mitotic recombination induced by URA3-GC and the level of URA-GC–specific mutations were low, 3.9 × 10−5 and 4.9 × 10−8 per division, respectively. We should point out, however, that fork stalling is quite an indirect measurement of the recombinogenic or mutagenic properties of a sequence. For example, poly GAA/CTT repeats cause a polar fork stall (49) but stimulate recombination in both orientations (50). In addition, telomeric repeats result in a very strong fork stall (51) but result in relatively low rates of recombination and mutation (about 10−6 per division) (52).
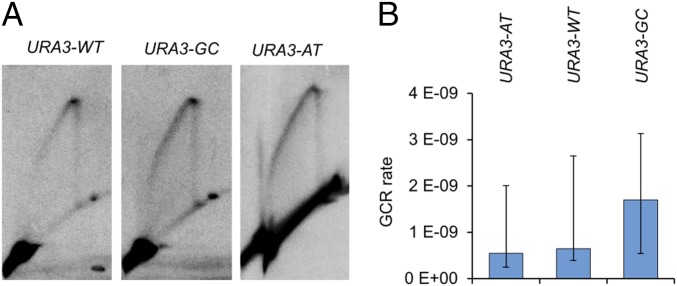
Fig. 6.
URA3-GC does not result in a strong replication fork stall or a high rate of DSBs. (A) Analysis of replication fork stalling at URA3-GC, URA3-AT, and URA3-WT by 2D gel electrophoresis (see Materials and Methods for details). Genomic DNA was isolated from S-synchronized cells, treated with BamHI, and examined by 2D electrophoresis. No spot of hybridization was observed at a position where a stalled replication fork was expected in any of the strains. (B) Rates of GCR in strains in which the URA3 genes were relocated adjacent to CAN1. GCR rates were measured by determining the rates of 5-FOAR CanR derivatives. Error bars indicate 95% confidence limits.
We also used a gross chromosome rearrangements (GCR) assay to determine whether the URA3-GC gene was associated with elevated DSBs. In this assay (53), a URA3 gene is inserted about 1 kb centromere-proximal to the CAN1 gene near the left telomere of chromosome V in a haploid strain; there are no essential genes located between CAN1 and the left telomere. Derivatives that are 5-FOAR and canavanine resistant (CanR) have a variety of GCRs including terminal deletions, large interstitial deletions, and translocations (53). Since these alterations are likely initiated by DSBs, we constructed three haploid strains in which URA3-AT, URA3-WT, and URA3-GC, respectively were relocated near CAN1. The rates of 5-FOAR CanR derivatives were low for all three strains (95% confidence limits are given in parentheses): 5.4 × 10−10 (4–8.3 × 10−10) for the URA3-AT strain; 6.5 × 10−10 (6.3–6.8 × 10−10) for the URA3-WT strain; and 1.7 × 10−9 (0.3–2.8 × 10−9) for the URA3-GC strain. Although the rate for the URA3-GC strain is about two- to threefold higher than for the other two strains, the difference is not significant (P > 0.1 by the Mann–Whitney U test).
Although the URA3-GC gene does not significantly elevate the GCR rate, the 15- to 20-fold stimulation in mitotic recombination observed in heteroallelic recombination argues that the URA3-GC gene has a higher level of recombinogenic lesions than the WT or URA3-AT genes. Two factors are likely relevant for the different results in the two assays. First, to stimulate heteroallelic recombination, the initiating DSB must be located within or very close to the ura3 heteroalleles, whereas a DSB anywhere between CAN1 and the first essential gene to CAN1 (located 11 kb away) can produce a 5-FOAR CanR derivative. Second, the heteroallelic recombination assay requires homologous recombination events following DSB formation. To produce a GCR event, the broken chromosome must be capped by a telomeric sequence or joined to another broken chromosome by NHEJ (53). Thus, different results in the two assays are not surprising.
URA3-GC Is a Hotspot for Meiotic Recombination.
Although the intermediates formed during mitotic and meiotic recombination are likely to be similar (54), the rate of meiotic recombination is four to five orders higher than the rate of mitotic recombination (55). In addition, meiotic DSBs are initiated by programmed DSBs produced by Spo11p and associated proteins (54). In previous studies of meiotic recombination in yeast, GC-rich sequences were found to have higher levels of recombination than regions with normal or GC-poor base composition (22, 23). As discussed in the Introduction, this relationship can be interpreted as indicating that high-GC regions are preferred substrates for Spo11p-induced DSBs or as indicating that regions with high levels of recombination tend to evolve to high-GC content because of biased gene conversion. As described below, our results strongly support the first of these alternatives.
Two types of meiotic analysis were done. First, we sporulated the heteroallelic diploids DKy147 (ura3-at-C89T/ura3-at-T777G), DKy143 (ura3-wt-C89T/ura3-wt-T777A), and DKy145 (ura3-gc-C89T/ura3-gc-CT777A). As observed for mitotic recombination events, Ura+ alleles generated by meiotic recombination usually reflect gene-conversion events rather than crossovers (56). As expected, most tetrads segregated 0 Ura+ to 4 Ura− spores. The number of tetrads with a Ura+ spore divided by the total number of four-spored tetrads for each strain are 2/263 (DKy147), 2/198 (DKy143), and 18/255 (DKy145). The strain with the GC-rich heteroalleles had significantly more Ura+ spores than the other two strains (P < 0.01 by Fisher exact test) (Fig. 5C).
To verify our results, we generated and sporulated diploids that had one WT URA3 gene and one mutant ura3 gene. The strain names and genotypes were DKy141 (ura3-at-C89G/URA3-AT), DKy139 (ura3-at-C89T/URA3-AT), DKy137 (ura3-at-T777G/URA3-AT), DKy129 (ura3-wt-T777A/URA3-WT), DKy131 (ura3-wt-C89G/URA3-WT), DKy133 (ura3-gc-C777A/URA3-GC), and DKy135 (ura3-gc-C89T/URA3-GC). Gene-conversion events at the URA3 locus are detected as tetrads that segregate three Ura+ to one Ura− or one Ura+ to three Ura− spores instead of the two Ura+ to two Ura− spores expected from Mendelian segregation. The number of tetrads showing the non-Mendelian segregation patterns divided by the total number of four-spored tetrads examined for each strain were DKy141 (1/159), DKy139 (1/165), DKy137 (2/240), DKy129 (2/225), DKy131 (2/197), DKy133 (20/229), and DKy135 (22/264). The high-GC alleles have approximately sevenfold more recombinational events than the alleles with normal or low GC content (Fig. 5D). By the Fisher exact test, the numbers of three Ura+ to one Ura− or one Ura+ to three Ura− tetrads in the strains with the high-GC alleles was significantly (P < 0.002) greater than the numbers for all the other strains. In summary, our results show that at least one GC-rich sequence results in higher rates of both mitotic and meiotic recombination.
Discussion
The main conclusion of this research is that the base composition of a gene can substantially affect its mutation rate and its recombinational properties. We found that a high-GC (63%) URA3 gene had elevated rates of mutations relative to a WT URA3 gene (43% GC) or one with low-GC content (31%). Two different mechanisms were responsible for the mutator phenotype: base substitutions introduced by the error-prone DNA polymerase ζ and deletion/duplication mutations that likely reflect DNA polymerase slippage. In addition, the high-GC URA3 gene had elevated levels of both mitotic and meiotic recombination relative to a WT URA3 gene or a low-GC URA3 gene.
Elevated Rate of Point Mutations in URA3-GC.
As discussed in Results, the elevated rate of base substitutions in URA3-GC was dependent on DNA polymerase ζ. This observation may be a consequence of increased recruitment of DNA polymerase ζ to a DNA lesion in URA3-GC or to a stalled replication fork in the absence of a DNA lesion (57). By 2D gel analysis, there was no evidence for a stalled replication fork, but this assay requires that the stall occur in a substantial fraction (>5%) of the cells. Although the GCR assay indicated that the rate of DNA lesions was threefold greater in the URA3-GC–containing strain than in strains with URA3-WT or URA3-AT, this difference was not statistically significant; the detection of a GCR event requires both a DSB and a subsequent acquisition of a telomere onto the broken end. In contrast, our mitotic recombination assay indicated that the URA3-GC gene is associated with a 10- to 20-fold elevated level of recombinogenic DNA lesions. Thus, DNA polymerase ζ could be recruited to the damaged template, resulting in the point mutations. Alternatively, since the increased DNA polymerase slippage indicates that the replicative DNA polymerases may be less processive on a high-GC template (discussed further below), it is possible that loss of processivity of the replicative polymerases allows increased recruitment of DNA polymerase ζ.
In several studies (e.g., ref. 12), rev3 strains have reduced rates of GC-to-CG changes and complex mutations. Although the rate of GC-to-CG alterations was reduced about 40-fold in our study, we also found that the URA3-GC allele in the rev3 strain had significantly reduced rates of most other types of single-base substitutions (AT-to-GC, AT-to-CG, GC-to-AT, and GC-to-TA) relative to the WT strain (Dataset S6). The rate of complex events was also about fourfold lower in the rev3 strain than in the WT strain, but this reduction was not statistically significant. These data indicate that DNA polymerase ζ can generate a variety of single-base substitutions even in the presence of functional replicative DNA polymerases.
It is important to stress that the mutations in URA3-GC were distributed throughout the gene rather than being concentrated in a few hotspots. In addition, the base substitutions that we made to produce URA3-GC altered 153 codons (57%) and left 114 (43%) unaltered compared with the WT gene. If the altered codons were more readily mutated to produce a nonfunctional URA3 gene, they should be overrepresented among the observed mutant substitutions. However, only 38% of the observed mutations were in the altered codons, and 62% were in the unaltered codons, indicating that the elevated mutation rate of URA3-GC was not a consequence of more mutable codons generated by our construction but was a consequence of the GC richness of URA3-GC. Last, Lang and Murray (3) developed a method of calculating target size for individual genes (an estimate of the number of bases that, when altered, produce a mutant phenotype). Using this method, we calculated target sizes for URA3-WT, URA3-AT, and URA3-GC of 115, 132, and 96 bp, respectively, from our data. Thus, the base substitutions that were used in the construction of URA3-GC did not create a gene that was a larger target for mutations.
Elevated Level of Deletions and Duplications in URA3-GC.
In addition to the elevated level of point mutations, the strain with the URA3-GC allele had a greatly elevated level of deletions with short direct repeats at their endpoints. Although similar deletions could be produced by SSA or MMEJ, the Rad1p independence of the deletions is most consistent with their generation by DNA polymerase slippage. We suggest that these deletions reflect slippage by one of the replicative DNA polymerases since they occur in strains lacking Rev3p. Similar deletions are observed in yeast strains with mutations affecting the catalytic subunit of DNA polymerase δ (8, 11, 12, 58), in strains with low levels of DNA polymerase δ (29, 30), and in strains that lack Pol32p (59), a subunit of DNA polymerase δ and ζ.
In addition to the in vivo studies that show elevated rates of indels in strains with mutant forms of DNA polymerase, DNA polymerases α (60), δ (11, 61), and ε (62) have substantial rates of slippage in vitro. The rates of indels produced by gap-filling DNA synthesis are about 3 × 10−5, 1 × 10−5, and 5 × 10−7 for DNA polymerases α, δ, and ε, respectively (63). Although many of these indels involve loss or gain of a single base in a homopolymeric run, the deletions generated by DNA polymerase δ are often larger and occur between direct repeats (61). Based on these observations, we suggest that the deletions observed in URA3-GC may be generated by DNA polymerase δ.
We suggest that the high GC content of URA3-GC results in reduced DNA polymerase processivity and, consequently, elevated DNA polymerase slippage. One type of evidence consistent with this possibility is that poly (G) tracts are less stable than poly(A) tracts in mismatch-repair–defective isolates of both yeast (64) and mammalian cells (65). In addition, the bacterial polymerase pol II polymerizes more efficiently on an AT-rich template than on a GC-rich template (66). Although these studies are consistent with the possibility that DNA polymerases are less processive on GC-rich templates, a definitive analysis of the processivity of DNA polymerase as a function of base composition has not yet been undertaken.
An alternative possibility is that other features of the URA3-GC sequence rather than GC content per se are responsible for the deletions. It is difficult to exclude this possibility completely; however, the elevated deletion rate is not a consequence of a much larger number of short perfect repeats in the URA3-GC gene compared with URA3-AT and URA3-WT. The numbers of perfect repeats between 4 and 11 bases in the URA3-WT, URA3-GC, and URA3-AT were 2,192, 3,021, and 3,957, respectively (calculated using the wordcount program; www.bioinformatics.nl/cgi-bin/emboss/wordcount), and the rates of large deletions in the URA3-WT, URA3-GC, and URA3-AT WT strains were 0, 12.5 × 10−9 per division, and 0.3 × 10−9 per division, respectively (Table 1). It should also be pointed out that among the natural S. cerevisiae genes, SRX1 has the highest GC content (59%), and SPG3 has the lowest (26%) (https://www.yeastgenome.org). Since these values are close to the GC content of URA3-GC and URA3-AT, respectively, the base compositions of our constructed URA3 genes are within realistic natural limits.
Another appealing feature of the DNA polymerase slippage model is that slippage events can produce either a deletion or a duplication. If the nascent (primer) strand slips forward, a single-stranded loop is formed on the template strand, resulting in a deletion (Fig. 3A). If the nascent strand slips backward, a loop is formed on the template strand, resulting in a duplication (Fig. 3B). Considering only those deletions or duplications that occur between perfect direct repeats in the URA3-GC strains, we found 193 deletions and 19 duplications. The 10-fold preference in favor of deletions suggests that the primer strand tends to slip forward to unreplicated sequences rather than backward to previously replicated sequences.
If deletions reflect DNA polymerase slippage, we expect that deletions between closely spaced repeats would likely be more frequent than deletions between more widely spaced repeats. To determine the expected sizes of the deletions based on the distribution of repeats within URA3-GC, we used the wordcount software described previously. For this analysis, the length of the repeat was defined by perfectly matched contiguous bases. In addition, repeats that were within larger repeats were not included. Finally, we considered only repeats of 4 bp or larger; 90% of the observed deletions were included in our analysis. The numbers of repeats that fulfilled these criteria (repeat size is in parentheses) were 1,024 (4 bp), 271 (5 bp), 83 (6 bp), 33 (7 bp), 9 (8 bp), 4 (9 bp), 3 (10 bp), and 1 (11 bp). We then determined the expected sizes of deletions from events that occurred between pairs of repeats for all repeat sizes (Dataset S7) and the observed deletion sizes (Dataset S8). These data are summarized in SI Appendix, Table S1. In this table we also calculated the expected average and median deletion size for each repeat class for all the strains containing URA3-GC.
The observed sizes of deletions are substantially smaller than the expected sizes (assuming a random interaction between repeats of the same size) for all repeat sizes between 4 and 9 bp; these differences are very significant (P < 0.0001) by χ2 analysis (SI Appendix, Table S1). Within each repeat class, there are also striking differences. For example, there are four classes of deletions predicted for 9-bp repeats from the sequence: 63, 153, 222, and 760 bp (Dataset S7). Of the observed 44 deletions involving 9-bp repeats, 27 were 63-bp deletions, and only one was a 760-bp deletion. In summary, these observations suggest that the deletions usually occur between closely spaced repeats in URA3-GC, as expected if the deletions are formed by DNA polymerase slippage.
In a previous study of a strain with the pol3-Y708A mutation, Northam et al. (12) found most of the deletions in the CAN1 gene were less than 40 bases, considerably shorter than those we observed in the WT strain with URA3-GC (median size of 87 bp). In the double-mutant pol3-Y708A rev3 strain, can1 deletions were larger with about 40% exceeding 40 bases. In our URA3-GC rev3 strain, the sizes of the deletions were not significantly larger than in the WT URA3-GC strain (median size, 93 bp; P = 0.53 by Mann–Whitney U test comparison). Thus, the slippage events induced by a mutant polymerase δ in CAN1 (GC content of 41%) are qualitatively different from those induced by WT DNA polymerases on a GC-rich template.
Since a slippage event involved base pairing, we would expect that longer repeats would be a better substrate for a slippage event. As described above, we determined the expected number of repeats of various sizes (4–11 bp) in URA3-GC. If all these repeats are equally likely to be involved in deletion events, we can calculate the expected proportions and numbers of deletions of various sizes. In SI Appendix, Table S2, we compare the expected numbers of deletions with the observed numbers of deletions for these classes. The deletion events involving repeats of 8–11 bp are overrepresented by 17- to 56-fold, and deletions involving repeats of 7 bp are overrepresented by about sixfold; the P values for all these comparisons (χ2 analysis) are highly significant (<0.0001). Deletion events involving 5- or 6-bp repeats are not significantly over- or underrepresented, and deletion events involving 4-bp repeats are very significantly underrepresented. This analysis suggests that repeats >6 bp are preferred substrates for slippage; however, deletions can occur between smaller repeats. In summary, the frequency of different types of deletions is regulated by the length of the repeats and the distance between repeats, as well as (presumably) the number of repeats within the target sequence.
As described in Results, many of the observed deletions could be explained by slippage events involving imperfect repeats rather than perfect repeats; similar deletions between imperfect repeats have been noted previously (11). In our study, of 297 deletions between repeats of all sizes, 193 involved perfect repeats, and 104 involved imperfect repeats (Dataset S5). Since the stability of duplexes of various sizes with various mismatches has been determined only in vitro studies using short oligonucleotides, in our analysis described above we considered only the perfectly paired regions of imperfect repeats. For example, in the imperfect repeat shown in Fig. 2B (CCGcCAAGT/CCGtCAAGT), for our analysis in SI Appendix, Tables S1 and S2, we considered this repeat as a 5-bp perfect repeat of CAAGT. Nonetheless, it is possible that the additional bases of imperfect repeats may stabilize the slippage intermediates.
GC Content and Recombination Hotspots.
Our analysis shows that high-GC regions are associated with elevated levels of both meiotic and mitotic recombination. In yeast and several other eukaryotes, the rate of meiotic recombination is correlated with the local and regional GC content (67). We argued that this relationship might be a consequence of a histone modification at GC-rich regions that attracts Spo11p and associated proteins or that GC-rich motifs directly interact with the Spo11p complex (24). In contrast, Birdsell (25) argued that GC-biased gene-conversion events would produce high-GC regions at recombination hotspots. The observations described above strongly suggest that, at least in S. cerevisiae, high-GC regions create a meiotic recombination hotspot rather than vice versa.
Although the information about sequences that elevate mitotic recombination is limited, recombination rates are elevated by high levels of transcription and by sequences that stall replication forks (45). Both DSBs and single-stranded nicks/gaps stimulate mitotic recombination, although which of these DNA lesions accounts for most spontaneous recombination is unclear. Based on observations of Rad52p foci, DSBs occur more frequently in S/G2 than in G1 (68). However, at least half of spontaneous crossovers between homologs are initiated by DSBs formed in G1 (42). It is unclear whether the recombinogenic DNA lesion associated with URA3-GC is a DSB or a single-stranded nick/gap and whether the lesion is formed in G1 or S/G2. Although we did not observe stalling of the replication fork by URA3-GC, subtle pausing of the fork would not be detectable by gel electrophoresis. Regardless of the details of the mechanism, our results indicate that high GC content in yeast can substantially elevate the frequency of local mitotic recombination. This association has not been established previously.
Summary
Our studies demonstrate that regions of high-GC content in yeast elevate mutation rates (both single-base substitutions and deletions) and recombination (both meiotic and mitotic exchange). These observations are likely relevant to understanding the trajectory of genome evolution. Because of the elevated mutation rate in high-GC genomic regions, we expect that these regions will evolve at a higher rate than low-GC regions. This effect may be partly counterbalanced by the hyperrecombination phenotype associated with high-GC regions. If a diploid is heterozygous for a high-GC gene and a low-GC gene, the recombinogenic DSB will likely occur in the high-GC gene, resulting in its replacement by gene conversion with the low-GC gene. It should also be emphasized that the GC content is only one of many parameters likely to influence genome evolution. In a mutation-accumulation study, Lujan et al. (2) found that mutation rates vary depending on replication timing, proximity to replication origins, direction of the replication fork, location with respect to nucleosomes, and other factors. A logical extension of our study would be a genome-wide examination of the density of mutational alterations as a function of base composition in sequence-diverged S. cerevisiae strains isolated from the wild.
Materials and Methods
Strains and Growth Conditions.
All yeast strains were derivatives of W1588-4C (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1; ref. 69) or other RAD5 derivatives of W303-1A. The genotypes and strain constructions are described in Dataset S1. Primers used in strain constructions and diagnosis of mutational alterations are in Dataset S2. Ura− derivatives were selected with medium containing 0.1% 5-FOA (27). Strains with deletions of both URA3 and CAN1 were selected using synthetic medium lacking arginine and containing 0.1% 5-FOA and 120 mg/L canavanine. Cells were grown at 30 °C in all experiments.
Estimation of Mutation Rates.
As in previous studies (70), we determined ura3 mutation rates by calculating the frequency of 5-FOAR mutations in multiple (>20) independent cultures. There was no difference in the sensitivity of strains containing URA3-WT, URA3-GC, or URA3-AT on medium containing 5-FOA. The frequency data were converted to rate data as described below. We confirmed that the 5-FOAR derivatives were Ura− by replica-plating these derivatives to uracil omission medium. GCR rates were determined by a similar protocol except that the selective medium contained both 5-FOA and canavanine.
Statistical Analysis.
Mutations rates and their CIs were calculated using the Ma-Sandri-Sarkar Maximum Likelihood Estimator (MSS-MLE) method and the FALCOR tool (www.keshavsingh.org/protocols/FALCOR.html) (71).
The CIs for proportions and the χ2, Fisher exact, and the Mann–Whitney U tests were calculated using VassarStats tools (www.vassarstats.net/). CIs for individual mutation events were calculated using formula CLx = the square root of [(CLMx/M)2 + (CLPx/P)2], where CL is a 95% confidence limit (either upper or lower, indicated by subscript x) of a given event, M is a median of total mutation rate, CLM is the 95% confidence limit for the median rate, P is a proportion of a given event, and CLP is the 95% confidence limit for the proportion (ipl.physics.harvard.edu/wp-uploads/2013/03/PS3_Error_Propagation_sp13.pdf).
Molecular Techniques.
The URA3-GC and URA3-AT genes were designed by the authors and synthesized by GeneArt AG (Life Sciences Solutions/Thermo Fisher Scientific). The synthesized genes were inserted into the ampR-containing vector pMA (URA3-AT) or the kanR-containing vector pMK0RQ-Bb (URA3-GC) (Life Sciences Solutions/Thermo Fisher Scientific). Mutant ura3 genes were sequenced by Sanger sequencing (Eton Bioscience Inc.) or by PacBio sequencing (Duke Center for Genomic and Computational Biology). The oligonucleotides used for PacBio sequencing are listed in Dataset S2.
We looked for replication fork stalling in three bar1 derivatives of DKY18, DKY39, and DKY40 (strains KT632, KT634, and KT636, respectively). Strains were synchronized in G1 with alpha pheromone. Following this treatment, the cells were incubated in rich medium, and DNA was harvested 40 min later. Genomic DNA containing replication intermediates was prepared as described by Friedman and Brewer (47). Samples were treated with BamHI before analysis with 2D gels. Southern analysis was performed using a URA3-specific probe generated by PCR using the primers uranae-31 and uranae-52 (Dataset S2).
Supplementary Material
Acknowledgments
We thank Sue Jinks-Robertson, Peter Burgers, Tom Kunkel, Youri Pavlov, Myron Goodman, Kristin Eckert, and Polina Shcherbakova for useful discussions; Fred Dietrich for his analysis of high-GC yeast genes; and Sue Jinks-Robertson, Natasha Degtyareva, and Dmitry Gordenin for comments on the manuscript. The research was supported by NIH Grants R01GM24110, R01GM52319, and R35GM118020 (to T.D.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807334115/-/DCSupplemental.
References
- 1.Costantini M, Musto H. The isochores as a fundamental level of genome structure and organization: A general overview. J Mol Evol. 2017;84:93–103. doi: 10.1007/s00239-017-9785-9. [DOI] [PubMed] [Google Scholar]
- 2.Lujan SA, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maki H. Origins of spontaneous mutations: Specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu Rev Genet. 2002;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- 5.Cassier C, Chanet R, Henriques JA, Moustacchi E. The effects of three PSO genes on induced mutagenesis: A novel class of mutationally defective yeast. Genetics. 1980;96:841–857. doi: 10.1093/genetics/96.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quah SK, von Borstel RC, Hastings PJ. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980;96:819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova AV, Burgers PM. Eukaryotic DNA polymerase ζ. DNA Repair (Amst) 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Borstel RC, et al. The mutator mut7-1 of Saccharomyces cerevisiae. Mutat Res. 1993;289:97–106. doi: 10.1016/0027-5107(93)90135-3. [DOI] [PubMed] [Google Scholar]
- 9.Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 2006;25:4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraszewska J, Garbacz M, Jonczyk P, Fijalkowska IJ, Jaszczur M. Defect of Dpb2p, a noncatalytic subunit of DNA polymerase ε, promotes error prone replication of undamaged chromosomal DNA in Saccharomyces cerevisiae. Mutat Res. 2012;737:34–42. doi: 10.1016/j.mrfmmm.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Tran HT, et al. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Northam MR, Robinson HA, Kochenova OV, Shcherbakova PV. Participation of DNA polymerase ζ in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 2010;184:27–42. doi: 10.1534/genetics.109.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinks-Robertson S, Bhagwat AS. Transcription-associated mutagenesis. Annu Rev Genet. 2014;48:341–359. doi: 10.1146/annurev-genet-120213-092015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lippert MJ, et al. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci USA. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah KA, Mirkin SM. The hidden side of unstable DNA repeats: Mutagenesis at a distance. DNA Repair (Amst) 2015;32:106–112. doi: 10.1016/j.dnarep.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 17.Ito-Harashima S, Hartzog PE, Sinha H, McCusker JH. The tRNA-Tyr gene family of Saccharomyces cerevisiae: Agents of phenotypic variation and position effects on mutation frequency. Genetics. 2002;161:1395–1410. doi: 10.1093/genetics/161.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawk JD, Stefanovic L, Boyer JC, Petes TD, Farber RA. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc Natl Acad Sci USA. 2005;102:8639–8643. doi: 10.1073/pnas.0503415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang GI, Murray AW. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol Evol. 2011;3:799–811. doi: 10.1093/gbe/evr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: Footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyre-Walker A. Recombination and mammalian genome evolution. Proc Biol Sci. 1993;252:237–243. doi: 10.1098/rspb.1993.0071. [DOI] [PubMed] [Google Scholar]
- 22.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 23.Gerton JL, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 25.Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- 26.Stapleton A, Petes TD. The Tn3 beta-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics. 1991;127:39–51. doi: 10.1093/genetics/127.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-Fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Umezu K, Kolodner RD. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 29.Kokoska RJ, Stefanovic L, DeMai J, Petes TD. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol Cell Biol. 2000;20:7490–7504. doi: 10.1128/mcb.20.20.7490-7504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng DQ, Zhang K, Wu XC, Mieczkowski PA, Petes TD. Global analysis of genomic instability caused by DNA replication stress in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2016;113:E8114–E8121. doi: 10.1073/pnas.1618129113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streisinger G, et al. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Morrison A, et al. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sfeir A, Symington LS. Microhomology-mediated end joining: A back-up survival mechanism or dedicated pathway? Trends Biochem Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha S, Villarreal D, Shim EY, Lee SE. Risky business: Microhomology-mediated end joining. Mutat Res. 2016;788:17–24. doi: 10.1016/j.mrfmmm.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roche H, Gietz RD, Kunz BA. Specificity of the yeast rev3 delta antimutator and REV3 dependency of the mutator resulting from a defect (rad1 delta) in nucleotide excision repair. Genetics. 1994;137:637–646. doi: 10.1093/genetics/137.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst) 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shishkin AA, et al. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St Charles J, Petes TD. High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 Mb arm of yeast chromosome IV. PLoS Genet. 2013;9:e1003434. doi: 10.1371/journal.pgen.1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 44.Esposito MS, Wagstaff JE. Mechanisms of mitotic recombination. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1981. pp. 341–370. [Google Scholar]
- 45.Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 47.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 48.Kim H-M, et al. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang W, et al. Friedreich’s ataxia (GAA)n•(TTC)n repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisae. PLoS Genet. 2011;7:e1001270. doi: 10.1371/journal.pgen.1001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anand RP, et al. Overcoming natural replication barriers: Differential helicase requirements. Nucleic Acids Res. 2012;40:1091–1105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aksenova AY, et al. Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc Natl Acad Sci USA. 2013;110:19866–19871. doi: 10.1073/pnas.1319313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 54.Andersen SL, Sekelsky J. Meiotic versus mitotic recombination: Two different routes for double-strand break repair: The different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. BioEssays. 2010;32:1058–1066. doi: 10.1002/bies.201000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbera MA, Petes TD. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:12819–12824. doi: 10.1073/pnas.0605778103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fogel S, Mortimer RK. Informational transfer in meiotic gene conversion. Proc Natl Acad Sci USA. 1969;62:96–103. doi: 10.1073/pnas.62.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Northam MR, et al. DNA polymerases ζ and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014;42:290–306. doi: 10.1093/nar/gkt830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saini N, et al. Fragile DNA motifs trigger mutagenesis at distant chromosomal loci in saccharomyces cerevisiae. PLoS Genet. 2013;9:e1003551. doi: 10.1371/journal.pgen.1003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang ME, Rio AG, Galibert MD, Galibert F. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase δ, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002;160:1409–1422. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kunkel TA, Hamatake RK, Motto-Fox J, Fitzgerald MP, Sugino A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:4447–4458. doi: 10.1128/mcb.9.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fortune JM, et al. Saccharomyces cerevisiae DNA polymerase delta: High fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 62.Shcherbakova PV, et al. Unique error signature of the four-subunit yeast DNA polymerase ε. J Biol Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 63.Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burkholder AB, et al. Muver, a computational framework for accurately calling accumulated mutations. BMC Genomics. 2018;19:345–363. doi: 10.1186/s12864-018-4753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boyer JC, et al. Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum Mol Genet. 2002;11:707–713. doi: 10.1093/hmg/11.6.707. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z, Lazarov E, O’Donnell M, Goodman MF. Resolving a fidelity paradox: Why Escherichia coli DNA polymerase II makes more base substitution errors in AT- compared with GC-rich DNA. J Biol Chem. 2002;277:4446–4454. doi: 10.1074/jbc.M110006200. [DOI] [PubMed] [Google Scholar]
- 67.Marsolier-Kergoat MC, Yeramian E. GC content and recombination: Reassessing the causal effects for the Saccharomyces cerevisiae genome. Genetics. 2009;183:31–38. doi: 10.1534/genetics.109.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 70.Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: Dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: A web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.