Significance
This work dissects how cells monitor failure of proteasomes and trigger signaling responses defining whether cells survive proteotoxic stress or undergo apoptosis. The monitoring mechanism involves detection of a buildup of abnormal polypeptides released from ribosomes. Accordingly, the system simultaneously monitors effectiveness of several major processes, including protein synthesis, folding, and degradation. A special scaffold complex composed of heat shock protein 70 (Hsp70) and its cofactor Bcl-2–associated athanogene 3 (Bag3) links accumulation of abnormal polypeptide species with a number of protein kinases involved in various signal-transduction pathways. A startling finding is that an Hsp70–Bag3–regulated kinase, LATS1, regulates very early events of formation of protein aggregates; thus protein aggregation appears to be a tightly regulated process rather than the simple collapse of abnormal proteins.
Keywords: proteasome inhibition, DRiPs, aggresome, Hippo pathway, stress kinases
Abstract
Protein abnormalities in cells are the cause of major pathologies, and a number of adaptive responses have evolved to relieve the toxicity of misfolded polypeptides. To trigger these responses, cells must detect the buildup of aberrant proteins which often associate with proteasome failure, but the sensing mechanism is poorly understood. Here we demonstrate that this mechanism involves the heat shock protein 70–Bcl-2–associated athanogene 3 (Hsp70–Bag3) complex, which upon proteasome suppression responds to the accumulation of defective ribosomal products, preferentially recognizing the stalled polypeptides. Components of the ribosome quality control system LTN1 and VCP and the ribosome-associated chaperone NAC are necessary for the interaction of these species with the Hsp70–Bag3 complex. This complex regulates important signaling pathways, including the Hippo pathway effectors LATS1/2 and the p38 and JNK stress kinases. Furthermore, under proteotoxic stress Hsp70–Bag3–LATS1/2 signaling regulates protein aggregation. We established that the regulated step was the emergence and growth of abnormal protein oligomers containing only a few molecules, indicating that aggregation is regulated at very early stages. The Hsp70–Bag3 complex therefore functions as an important signaling node that senses proteotoxicity and triggers multiple pathways that control cell physiology, including activation of protein aggregation.
In aging and in many protein conformation disorders organisms experience suppression or failure of the ubiquitin–proteasome system (UPS), which causes a major buildup of abnormal proteins. Such abnormal species increase the load on the protein degradation and folding machineries. They can form toxic aggregates and further exacerbate aging and facilitate age-related protein conformation disorders (1). In response to proteasome suppression, cells activate pathways that can relieve the proteotoxic stress, including aggresome formation and autophagy. Other pathways activated under these conditions can trigger programmed cell death or regulate various aspects of cell physiology, e.g., the stress kinases JNK and p38 (2), the heat shock response (3), the unfolded protein response (4), or transcription factor NRF2 (5). How mammalian cells sense the proteasome’s failure to modulate many of these pathways is not well understood. Such knowledge would significantly enhance our understanding of the biology of stress response and the molecular basis of many protein conformation disorders. Here we hypothesized that the proteotoxic stress-sensing mechanism involves a complex of the heat shock protein 70 (Hsp70) and a cochaperone BCL-2–associated athanogene 3 (Bag3) (the HB complex).
Our prior work has shown that the HB complex controls several cancer-related signaling pathways (6, 7). However, involvement of this complex in the regulation of other pathways has not been explored. Bag3 is a multidomain protein that interacts with Hsp70 family members through a specialized Bag domain and with small heat shock protein (sHSP) family members through a site in its so-called “M-domain” (8). Bag3 has a PXXP motif and a WW domain responsible for its interaction with a number of signaling factors. The domain structure of Bag3 suggests that it can serve as a scaffold that links Hsp70 with multiple signaling pathways.
Considering that the HB complex has the capacity to interact with abnormal protein molecules (9, 10), we hypothesized that this module could act as a sensor of protein abnormalities in the cell and transduce signals to downstream pathways. Here we describe three pathways (i.e., JNK, p38, and Lats1/2) which are regulated under proteotoxic conditions via interactions with the HB complex. We found that upon proteasome failure this complex primarily senses the buildup of defective ribosomal products (DRiPs) and that components of the ribosome-associated quality control (RQC) system and a ribosome-associated chaperone, nascent polypeptide-associated complex (NAC), play an important role in the sensing process. Finally, via modulation of Lats1/2 activity under these conditions, the HB complex regulates the early stages of the aggregation of abnormal proteins before their transport to the aggresome.
Results
The Hsp70–Bag3 Complex Mediates the Activation of Signaling Pathways in Response to Proteasome Inhibition.
Multiple regulatory systems respond to proteasome failure and other proteotoxic stresses, but how these stimuli are sensed remains poorly understood. We hypothesized that the Hsp70–Bag3 module responds to the accumulation of abnormal polypeptides by triggering various signaling events. To test the idea that the HB complex can regulate numerous signaling responses to proteotoxic stress, we assessed the effects of proteasome inhibition on the spectrum of Bag3 partners. HeLa cells were transfected with plasmids expressing His-tagged Bag3 or its ΔC deletion mutant that lacks the Bag-domain essential for interaction with Hsp70. After treatment with the proteasome inhibitor MG132, Bag3 was isolated together with associated proteins using cobalt affinity resin (SI Appendix, Fig. S1A), and the complexes were analyzed by mass spectrometry. An isolate from cells transfected with an empty vector was used as a negative control. In agreement with a previous report (11), levels of both major Hsp70 family members were dramatically reduced in ΔC pull-downs compared with full-length Bag3 (Dataset S1), indicating the specificity and quantification reliability of the analysis. Overall, Dataset S1 demonstrates that Bag3 associates with a large number of cellular proteins, including those involved in signaling. Understandably, some proteins in Dataset S1 could be associated with Bag3 indirectly as a part of multiprotein complexes. Notably, deletion of the Hsp70-binding domain prevented many of these interactions, suggesting that these Bag3 partners associate with the HB complex as a whole. Importantly, inhibition of proteasome activity significantly affected the interactions of Bag3 with many partners, including several regulatory proteins (Dataset S1), suggesting the involvement of HB complex in various regulatory processes triggered by proteotoxic challenges.
Among the signaling proteins that associate with Bag3 in an Hsp70-dependent way, we detected mitogen-activated protein kinase p38 (Dataset S1). Since proteasome inhibitors can stimulate both p38 and homologous JNK kinases (2), we reasoned that JNK may also associate with Bag3 but escape detection by mass spectrometry, which could happen for multiple reasons, e.g., poor ionization of peptides. Indeed, we detected JNK in the Bag3 pull-downs by immunoblotting (SI Appendix, Fig. S1B). As with p38, the association of Bag3 and JNK was reduced by the deletion of the Bag domain, suggesting that Hsp70 affects the interaction between Bag3 and JNK. To test whether Bag3 mediates p38 and JNK responses upon proteasome inhibition, we depleted Bag3 in MCF10A cells and found that activation of these kinases by MG132 treatment indeed was reduced significantly in the depleted cells (Fig. 1A). To examine the role of Hsp70 in this signaling, we depleted HspA8, the major constitutively expressed Hsp70 family member, and simultaneously depleted the heat shock transcription factor Hsf1 to avoid a compensatory up-regulation of the inducible Hsp70 isoform HspA1 (SI Appendix, Fig. S1C) (12). Indeed, similar to the depletion of Bag3, the reduction of HspA8 levels suppressed the activation of p38 and JNK by proteasome inhibitors (SI Appendix, Fig. S1D). These data therefore indicate that the HB complex plays a critical role in the regulation of stress kinases in response to proteasome inhibition.
Fig. 1.
The HB complex mediates LATS1/2 suppression in response to proteasome inhibition. The antibodies for immunoblots are indicated on the right. (A) Bag3 depletion reverses the effects of proteasome inhibition on JNK and p38 activity. Control MCF10A cells and MCF10A cells depleted of Bag3 were treated with MG132 under the indicated conditions. Here and throughout the figures, the results of immunoblotting of total cellular lysates were independently reproduced at least three times. (B) The effects of the deletion of various functional domains in Bag3 on its association with LATS1. HeLa cells were transfected with plasmids encoding His-tagged full-length Bag3, with Bag3 deletion mutants, or with empty plasmid (vector, V). Here and throughout this work HeLa cells were utilized because, unlike MCF10A cells, they could be efficiently transfected with a plasmid DNA. On the next day the cells were treated for 75 min with 5 μM MG132 or were left untreated. Then the cells were incubated for 10 min with 1.2% formaldehyde, and His-tagged constructs were isolated using cobalt affinity resin. To calculate relative amounts of LATS1 associated with each His-tagged construct, the quantified LATS1 signal in each lane was normalized by a FLAG signal representing the amount of pulled-down Bag3 in the same sample. No detectable signal was seen in the isolate from the cells transfected with empty vector (V). The results of this pull-down are typical of three independent experiments. ΔC, mutant with a deleted C terminus containing the Bag domain critical for interaction with Hsp70; ΔM, mutant with a deleted M-domain; ΔP, mutant with a deleted PxxP motif; ΔWW, mutant with a deleted WW domain; WT, full-length Bag3. (C) Proteasome inhibition leads to the suppression of LATS1/2 activity as monitored by phosphorylation of YAP. MCF10A cells were incubated for 3.5 h with the indicated concentrations of MG132. (D) Bag3 depletion reduces the effects of proteasome inhibition on LATS1/2 activity. Control MCF10A cells and cells depleted of Bag3 were treated with MG132 under the indicated conditions. Quantification (expressed in relative units) of the phospho-YAP signal normalized by total YAP in the same samples is shown for each of the series. (E) Hsp70 depletion blocks the effects of proteasome inhibition on LATS1/2 activity. MCF10A cells were infected with HSF1 shRNA, briefly selected, and 3 d after the infection were transfected with either HspA8 or control siRNA. Two days later the cells were treated with MG132. Quantification (expressed in relative units) of the phospho-YAP signal normalized by total YAP in the same samples is shown for each of the series. (F) The Bag domain of Bag3 is critical for the regulation of LATS1/2 by proteasome inhibition. MCF10A cells were infected with retroviruses encoding full-length Bag3 (WT) or its ΔC-truncated form (both with mutations making them resistant to the Bag3 siRNA used in this experiment) or with an empty vector. After selection, the cells were transfected with either Bag3 (Lower), or control (Upper) siRNA and 2 d later were treated for 3.5 h with 0.6 μM MG132. Quantification (expressed in relative units) of the phospho-YAP signal normalized by total YAP signal in the same samples is shown for each of the series. Results of an analogous experiment are shown in SI Appendix, Fig. S1G.
Several signaling factors, including LATS1/2, a component of the Hippo pathway, had been reported to interact with Bag3 (13). First, we confirmed the association of endogenous LATS1 with Bag3 and analyzed how deletions of various functional Bag3 domains affect its association with LATS1. As described above, we pulled down His-tagged Bag3 constructs, including WT and deletion mutants, and analyzed the isolated complexes by immunoblotting. While LATS1 was found in association with WT Bag3, none of this protein was pulled down with the ΔWW deletion construct, indicating, in line with the previous report (13), that this domain is critical for the interaction. Importantly, the ΔC deletion also reduced the association of Bag3 with LATS1, suggesting that LATS1 is preferentially recruited to the HB complex rather than to Bag3 alone. In contrast, deletion of the sHSP-binding domain (ΔM) of Bag3 did not significantly affect its association with LATS1 (Fig. 1B). These data confirm that association between Hsp70 and Bag3 plays a role in the recruitment of LATS1.
We hypothesized that, as with JNK and p38, LATS1/2 responses could be triggered by a buildup of abnormal proteins and that the triggering of these responses is mediated by the HB complex. First, we tested whether proteasome inhibition affects LATS1/2 activity. We incubated MCF10A cells with MG132 and assessed LATS1/2 activity by measuring the phosphorylation levels of one of its substrates, the transcriptional regulator YAP. Indeed, YAP phosphorylation by LATS1/2 was significantly suppressed in response to the proteasome inhibitor (Fig. 1C), indicating activation of the Hippo pathway under these conditions. Consistent with this activation, we observed that expression of the YAP target gene CTGF was up-regulated upon exposure to MG132 (SI Appendix, Fig. S1E).
To test if the LATS1/2 response is mediated by the HB complex, we measured the effects of MG132 treatment in cells depleted of either of the two elements of the complex. Depletion of Bag3 reversed the suppression of LATS1/2 activity upon proteasome inhibition (Fig. 1D). Similarly, depletion of HspA8 (against the background of Hsf1 depletion) blocked the regulation of LATS1/2 by proteasome inhibitors (Fig. 1E), suggesting that the HB complex is critical for this signaling process.
To investigate further the role of the Hsp70–Bag3 complex in the regulation of LATS1/2, we depleted endogenous Bag3 with siRNA and replaced it with siRNA-resistant forms of either WT Bag3 or a construct missing the Hsp70-interacting Bag domain (ΔC) (SI Appendix, Fig. S1F). Expression of WT Bag3 completely reversed the effects of Bag3 siRNA on LATS1/2 suppression (Fig. 1F and SI Appendix, Fig. S1G). In contrast, the ΔC mutant could not reverse the effects of MG132 on LATS1/2 activity (Fig. 1F). Furthermore, acting as a dominant-negative mutant, it partially inhibited the effects of MG132 on LATS1/2 activity even in the control cells with unaltered levels of endogenous Bag3. In line with this finding, YM1, a small molecule that targets Hsp70 family members and disrupts their interaction with Bag3 (6), completely reversed the effect of MG132 on LATS1/2 activity (Fig. 2A, last lane). These observations indicate that the interaction between Bag3 and Hsp70 plays a critical role in the regulation of LATS1/2 upon proteasome inhibition. Because of the WW domain-dependent interaction of Bag3 with LATS1/2, the effects of the HB module on LATS1/2 activity are likely to be direct, although we cannot exclude the possibility of the indirect effects. Together, these data indicate that the HB complex monitors the effectiveness of protein degradation in a cell to modulate a number of signaling pathways.
Fig. 2.
DRiPs interact with the HB complex to activate the response to proteasome failure. (A) Incubation with the proline analog AZC enhances the effects of proteasome inhibition. MCF10A cells were left untreated or were incubated for 3 h with the indicated concentrations of MG132 alone or combined with 2.5 mM AZC. YM1, an inhibitor of the HB complex, reverses the effects of proteasome inhibition on LATS1/2 activity. Cells were preincubated with YM1 for 3 h and were incubated with 5 μM MG132 combined with 5 μM YM1. (B) Slowing down translation lessens the effects of proteasome inhibition on the suppression of LATS1/2. MCF10A cells were left untreated or were incubated for 3 h with 1 μM MG132 alone or combined with 0.15 μM emetine. Results of an analogous experiment with the addition of cycloheximide are shown in SI Appendix, Fig. S2A. (C) The effects of proteasome inhibition, inhibition of translation, and disruption of the HB complex on the association of Hsp70 and LATS1 with ubiquitinated species. MCF10A cells were incubated for 2 h with the indicated combinations of 2.5 μM MG132, 0.5 μM emetine, or 10 μM YM1. All cells incubated with YM1 were preincubated for 1.5 h. After the treatments, cells were incubated for 10 min with 1.2% formaldehyde to cross-link protein complexes. Ubiquitinated species were isolated from the cell lysates and analyzed by immunoblotting. To measure Hsp70, we used a mixture of antibodies against HspA1 and HspA8. Typical results of two independent experiments are shown.
Sensing of DRiPs by the HB Complex.
We examined the signaling events upstream of the HB complex. Upon proteasome inhibition, the HB complex could sense either the accumulation of a specific, short-lived regulatory protein(s) or a general buildup of abnormal polypeptides. To assess these possibilities, we took advantage of a recently described effect of mild inhibition of translation elongation on the quality of ribosome output. We and others have shown that at concentrations that only mildly suppress protein synthesis, these inhibitors significantly improve cotranslational folding, thus decreasing the fraction of aberrant polypeptides in the pool of newly generated proteins (14–16). We added 150 nM of the translation inhibitor emetine, which decreases the ribosomal output by about 50% (17), and found that under these conditions the effects of MG132 on the down-regulation of LATS1/2 activity were almost completely suppressed (Fig. 2B). We confirmed this finding with another inhibitor of translation elongation, cycloheximide, at concentrations that inhibited translation by less than 50% (SI Appendix, Fig. S2A). These experiments indicate that LATS1/2 suppression by proteasome inhibition depends on the de novo generation of polypeptides. The fact that the response is blocked by improved folding of newly synthesized polypeptides suggests that this suppression is triggered by the buildup of aberrant proteins released from ribosomes, although the possibility remains that the mild inhibition of translation was sufficient to suppress the buildup of a putative short-lived regulator of the LATS1/2 response. To further test the role of abnormal newly synthesized polypeptides in the regulation of LATS1/2, we employed the proline analog l-Azetidine-2-carboxylic acid (AZC), which incorporates into translated polypeptides and prevents their folding. Accordingly, incorporation of AZC would have opposite effects on the levels of the two possible triggers of regulation: The generation of any new functional protein, including a putative short-lived regulator, would be reduced, while the levels of abnormal polypeptides would increase. In fact, AZC significantly enhanced the effects of MG132 on LATS1/2 activity (Fig. 2A), indicating that a buildup of aberrant proteins triggers the response.
To address the mechanism of detecting DRiPs, we first tested whether the entire HB-LATS complex can associate with them.
Notably, it was reported that Bag3 can bind newly synthesized unstable proteins in an Hsp70-dependent manner (18). We confirmed this observation in our model by showing that deletion of the Hsp70-binding Bag domain reduces the association of Bag3 with ubiquitinated proteins, most of which represent newly synthesized polypeptides (SI Appendix, Fig. S2B) (17). To further address the association of the HB complex with DRiPs, we radiolabeled newly synthesized proteins with S35-methionine and S35-cysteine for 10 min. Under these conditions, short-lived polypeptides, including DRiPs, form a large fraction of all labeled proteins. We analyzed their association with His-tagged Bag3 pulled down from HeLa cells as described above. Beside the major band of Bag3, the pull-down samples contained a number of bands that appear to represent DRiPs (SI Appendix, Fig. S2C). Indeed, quantification of the gel showed that the fraction of the labeled species associated with Bag3 decreased during a 1 h chase, while in the cells labeled in the presence of MG132 the association increased (Fig. 2C and SI Appendix).
To test whether LATS1 can associate with DRiPs as a part of the HB complex, we pulled down ubiquitinated proteins from lysates of MCF10A cells and assessed the amounts of associated Hsp70 and LATS1 (we could not conclusively assess Bag3 in these pull-downs because of significant nonspecific interactions). Amounts of copurified LATS1, as well as Hsp70, increased upon proteasome inhibition, and this effect was reversed upon mild inhibition of translation by emetine (Fig. 2C). Importantly, the addition of YM1 (Fig. 2C), which blocks the Hsp70–Bag3 interaction, strongly reduced the amounts of LATS1 associated with DRiPs, suggesting that the association depends on the HB complex.
Detection of Stalled Ribosomal Polypeptides by the HB Complex.
A subset of DRiPs formed by stalled polypeptides remains bound to the large ribosome subunit and requires special RQC machinery, including the ubiquitin ligase LTN1, for dissociation and subsequent degradation by a proteasome (19). We sought to investigate whether this subset of DRiPs plays a role in the suppression of LATS1/2 activity upon proteasome inhibition in the absence of additional protein-damaging conditions, e.g., AZC.
If stalled polypeptides represent a sizable fraction of the DRiPs involved in this signaling, the sensing of proteasome inhibition may be affected by the depletion of LTN1. In fact, the inhibitory effects of MG132 on LATS1/2 were almost completely blocked in cells depleted of LTN1 (Fig. 3A). A similar outcome was observed upon depletion of VCP (SI Appendix, Fig. S3A), another factor involved in the release of stalled polypeptides from the large ribosomal subunit (19). These data suggest that in the absence of additional protein-damaging challenges, e.g., exposure to AZC, the stalled polypeptides play a major role in sensing the proteasome inhibition. Moreover, since LTN1 and VCP are necessary for the release of these species from the large ribosome subunit, these data further suggest that the DRiPs are detected by the HB complex after their release from the ribosomes. However, it should be cautiously noted that VCP affects multiple processes in protein homeostasis, and therefore its effects on LATS1/2 regulation may be indirect.
Fig. 3.
Stalled proteins interact with and regulate the HB complex. (A) Effects of depletion of LTN1, NACA, or DNAJC2 (ZUO1) on the suppression of LATS1/2 upon proteasome inhibition. MCF10A cells with the indicated depletions were incubated for the indicated times with 3 μM MG132, and the lysates were analyzed by immunoblot. (B) Effects of proteasome inhibition and depletion of LTN1 or NACA on levels of GFP-NS. One day after siRNA transfection, HeLa cells were transfected with plasmids encoding either GFP-NS or WT EGFP (this plasmid was 10-fold diluted with an empty vector). Indicated samples were treated for 4 h with 5 μM MG132. (C) The effects of LTN1 or NACA depletion on the association of GFP-NS with the HB complex were assessed by the Duolink assay. HeLa cells transfected for 1 d with one of the indicated siRNAs were transfected with a mixture of plasmids encoding GFP-NS and FLAG-Bag3 (3:1) and 1 d later were treated with MG132 or were left untreated. The cells transfected with only the GFP-NS–encoding plasmid or only the FLAG-Bag3–encoding plasmid were used as negative controls and did not produce any measurable signal. The cells grown on a glass-bottomed plate were treated according to the Duolink protocol with anti-GFP and anti-FLAG antibodies. Images of the Duolink signal and of DAPI nuclear staining were taken from at least 10 randomly chosen images with a fluorescence microscope with a 20× objective. Duolink images were quantified using NIH ImageJ software, and the data were normalized by the total number of cells in all fields chosen for each sample. The error bars represent the SD. For images of cells, see SI Appendix, Fig. S3. (D) Effects of LTN1 or NACA depletion on the association of GFP-NS with the HB complex assessed by the modified ActivSignal protocol. The cells were grown and treated as in Fig. 3B, but here the cells were plated on a plastic 96-well plate for incubation with MG132 and for a following protein–protein interaction assay (Materials and Methods). The qPCR results are presented. The results for a signal produced with the pair of anti-GFP and anti-FLAG antibodies (reflecting the amount of GFP-NS associated with FLAG-Bag3) were normalized by a signal produced with the pair of anti-Bag3 and anti-FLAG antibodies (reflecting the relative amount of FLAG-Bag3) for the same cells. The signals generated from the cells transfected only with a vector were used as a baseline. The error bars represent the SD. For the effect of LTN1 depletion (compared with control), P < 10−6; for NACA, P < 10−5.
To assess direct interactions of these species with the HB complex, we generated a model of nonstop polypeptide, GFP-NS, similar to one used with yeast (20). As with other stalled polypeptides, expression of GFP-NS was low, and depletion of LTN1 or inhibition of the proteasome resulted in increased levels of this polypeptide (Fig. 3B). Nonetheless, even under these conditions, levels of GFP-NS were significantly lower than levels of normal GFP (Fig. 3B), most likely due to the rapid degradation of mRNA. At these levels we were unable to detect GFP-NS in Bag3 pull-downs.
To test if Bag3 associates with the model stalled polypeptide released from ribosome, we employed a Duolink proximity ligation assay (PLA). We cotransfected HeLa cells with plasmids encoding GFP-NS and FLAG-Bag3, treated them with MG132, and fixed them with formaldehyde. The cells were incubated with anti-GFP and anti-FLAG antibodies raised in rabbit and mouse, respectively. The Duolink PLA allows microscopic detection of these two antibodies when they bind to the antigens in close proximity. No signal was detected in the cells transfected with only one of the plasmids (SI Appendix, Fig. S3B). Similarly, association between GFP-NS and Bag3 was almost undetectable in naive cells (Fig. 3C). However, upon incubation with MG132 the association became evident (Fig. 3C). Notably, depletion of LTN1 significantly reduced the association between Bag3 and GFP-NS (Fig. 3C), even though the cellular levels of the latter were increased by the depletion (Fig. 3B).
To better quantify association between GFP-NS and Bag3, we developed a modified proximity ligation protocol based on the ActivSignal, Inc. approach, which allowed ultrasensitive detection of a complex by qPCR. We cotransfected HeLa cells with plasmids encoding GFP-NS and FLAG-Bag3, treated the cells on a 96-well plate with MG132, fixed them, and incubated them with antibodies against GFP and FLAG. Then cells were treated with secondary antibodies against FLAG (murine) and GFP (leporine) tagged with distinctive oligonucleotides. These oligonucleotides could be ligated via a special linker only when they were brought into proximity in a protein complex. We analyzed the ligation products by qPCR. The experiment confirmed that incubation with MG132 results in an association between GFP-NS and Bag3 (Fig. 3D). With this approach, we also confirmed that the association between GFP-NS and Bag3 depends on LTN1 (Fig. 3D). The finding that LTN1 is necessary both for the association of the model stalled polypeptide with the HB complex and for downstream signaling (Fig. 3A) suggests that ubiquitination of stalled polypeptides and most probably their subsequent dissociation from the large ribosomal subunit are important for binding to the HB complex.
NAC Is Involved in Sensing of DRiPs.
Since the HB complex appears to detect defective polypeptides soon after their release from ribosomes, ribosome-associated chaperones that interact with nascent polypeptides may also play a role in the sensing mechanism. The chaperone complex NAC, composed of NACA and NACB, interacts with emerging polypeptides and facilitates their cotranslational folding. We tested if NAC is involved in the sorting of GFP-NS onto the HB complex. Indeed, we found that depletion of NACA significantly reduced the association of GFP-NS with Bag3 when monitored using the PLAs (Fig. 3 C and D). Furthermore, depletion of either NACA or NACB strongly reduced the effect of MG132 on LATS1/2 activity (Fig. 3A and SI Appendix, Fig. S3A). In addition to NAC, another ribosome-associated chaperone complex, RAC, also binds to a subset of nascent polypeptides and promotes their folding. Studies in yeast indicated that RAC facilitates the folding of about 80% of the ribosomal products, and only 20% involve NAC (21). Therefore, we tested whether RAC also has a role in the function of the HB complex. Unlike depletion of NAC components, depletion of DNAJC2 (ZUO1), the integral component of the RAC complex, did not affect the suppression of LATS1/2 upon proteasome inhibition (Fig. 3D). These data suggest that NAC is involved in generating the signal transmitted via the HB complex upon proteasome inhibition.
The Hsp70–Bag3 Module Activates Aggresome Formation in Response to Proteasome Inhibition.
Proteasome inhibition can trigger aggresome formation (22), and here we sought to understand the role of the HB complex in this protective response. The Hsp70–Bag3 module has been implicated in aggresome formation (23, 24). The main paradigm proposed in prior publications was that Hsp70 physically binds to aggregated proteins, while Bag3 links this complex with the dynein motor complex, either directly or via 14-3-3 proteins (25, 26), for further transport to the aggresome. On the other hand, we previously reported that the aggresome response is sensitive to a partial inhibition of translation and can be stimulated by amino acid analogs (for the proline analog AZC, see SI Appendix, Fig. S4A), suggesting that sensing of DRiPs can play a role in aggresome formation (17). Accordingly, here we hypothesized that the HB complex may modulate signaling that triggers aggresome formation.
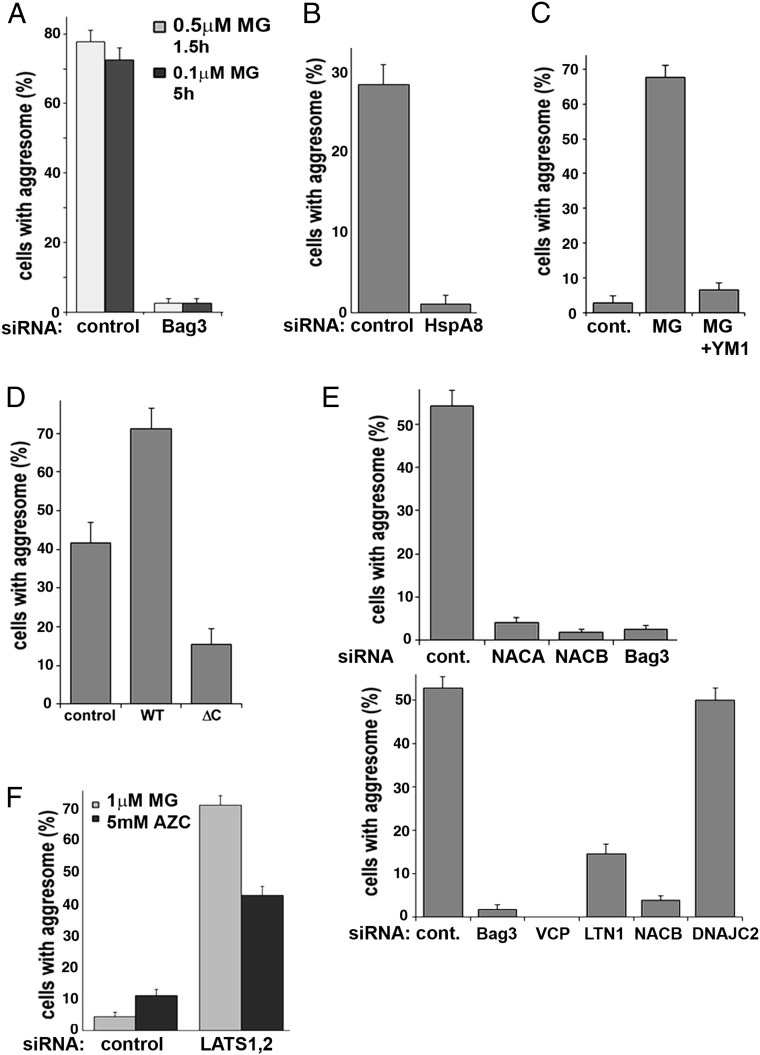
To monitor aggresome formation, we utilized the GFP-tagged synphilin 1 reporter (Syn-GFP) (27) expressed in MCF10A cells. In this cell line the aggresome response is very sensitive to proteasome inhibition, and we used submicromolar concentrations of MG132 to trigger the response. In line with previous reports, aggresome formation was both Bag3- and Hsp70-dependent (Fig. 4 A and B and SI Appendix, Fig. S4B). Moreover, the addition of YM1, which disrupts the HB complex, strongly suppressed aggresome formation (Fig. 4C), indicating the HB complex has a role in aggresome triggering. Furthermore, while Bag3 overexpression did not trigger aggresome formation on its own, it strongly accelerated this process upon proteasome inhibition (Fig. 4D). In contrast, the ΔC mutant missing the Bag domain reproducibly demonstrated an inhibitory effect (Fig. 4D).
Fig. 4.
The HB complex and its partners control aggresome formation. In all experiments cells were fixed with formaldehyde, and the fraction of cells with an aggresome was counted under a fluorescence microscope; in all experiments less than 5% of naive cells formed an aggresome. Error bars on the graphs depicting aggresome formation represent SEs. (A) Bag3 depletion suppresses aggresome formation. MCF10A cells stably expressing Syn-GFP with the indicated depletions were treated with MG132 under the indicated conditions. (B) Hsp70 depletion suppresses aggresome formation. MCF10A cells stably expressing Syn-GFP were infected with HSF1 shRNA and were briefly selected and transfected with either HspA8 or control siRNA. Two days later the cells were treated for 3.5 h with 45 nM MG132. (C) Blocking the Hsp70–Bag3 interaction by the small molecule YM1 suppresses aggresome formation. MCF10A cells stably expressing Syn-GFP were treated for 4.5 h with 100 nM MG132 with or without the addition of 5 μM YM1 or were left untreated. YM1 was added 1.5 h before the addition of MG132. (D) The effects of Bag3 on the aggresome require an intact Bag domain. HeLa cells stably expressing Syn-GFP and transfected with plasmids encoding full-length Bag3 (WT), with its ΔC deletion mutant (ΔC), or with empty vector were treated for 5 h with 2 μM MG132. The fraction of cells with an aggresome was counted only among the transfected cells. (E) The effects of ribosome-associated factors involved in handling DRiPs on aggresome formation. MCF10A cells stably expressing Syn-GFP with the indicated depletions were treated with 90 nM MG132 for 3 h (Upper) or with 50 nM MG132 for 5 h (Lower). (F) Depletion of LATS1 stimulates aggresome formation in response to proteasome inhibition. MCF10A cells stably expressing Syn-GFP depleted of LATS1 were treated with 30 nM MG132 for 2.5 h.
We tested whether the factors that promote the release of DRiPs from ribosome are required for aggresome formation. Indeed, depletion of either LTN1 or VCP significantly reduced this response (Fig. 4E). Furthermore, in line with the role suggested above for NAC in sensing DRiPs, knocking down either NACA or NACB dramatically reduced aggresome formation. In contrast, depletion of DNAJC2, an integral component of RAC, had no effect on the aggresome (Fig. 4E), further demonstrating the specificity of NAC in cellular responses to the buildup of DRiPs. These data strongly suggest that the release of DRiPs and their detection by the HB complex are required for the induction of aggresome formation.
To test whether the discovered HB-dependent regulation of LATS1/2 is relevant to the aggresome response, we depleted both LATS1 and LATS2 from MCF10A cells using a mixture of corresponding siRNAs. Even though LATS1/2 depletion in naive cells did not result in significant aggresome formation, it dramatically enhanced the process upon exposure to low concentrations of MG132. In fact, after 2.5 h of incubation with 30 nM MG132, we could detect only a few aggresomes in control MCF10A cells, while more than a third of the LATS1/2-depleted cells developed aggresomes (Fig. 4F). Similarly, in HeLa cells the aggresome response to 1 μM MG132 was negligible, but depletion of LATS1/2 from these cells resulted in a remarkable up-regulation of aggresome formation (SI Appendix, Fig. S4C). Importantly, expression of the siRNA-resistant form of LATS1 reversed the aggresome-stimulating effect of LATS1/2 depletion (SI Appendix, Fig. S4D). The observed effect of LATS1/2 depletion on aggresome formation was temporary, and ultimately most of the MG132-treated control cells formed aggresomes. Of note, we observed that an acceleration of aggresome formation in MCF10A cells was seen with siRNAs against LATS1, while siRNA against LATS2 had little effect on the aggresome (SI Appendix, Fig. S4E). We verified that aggresome formation triggered by other DRiP-generating conditions is also regulated by LATS1/2. Indeed, we observed significant acceleration of AZC-induced aggresome formation in cells depleted of LATS1 (SI Appendix, Fig. S4C). Therefore, suppression of LATS1 activity via the HB complex upon the buildup of abnormal polypeptides accelerates the aggresome response.
Hsp70–Bag3–LATS1 Signaling Regulates Early Events in Protein Aggregation.
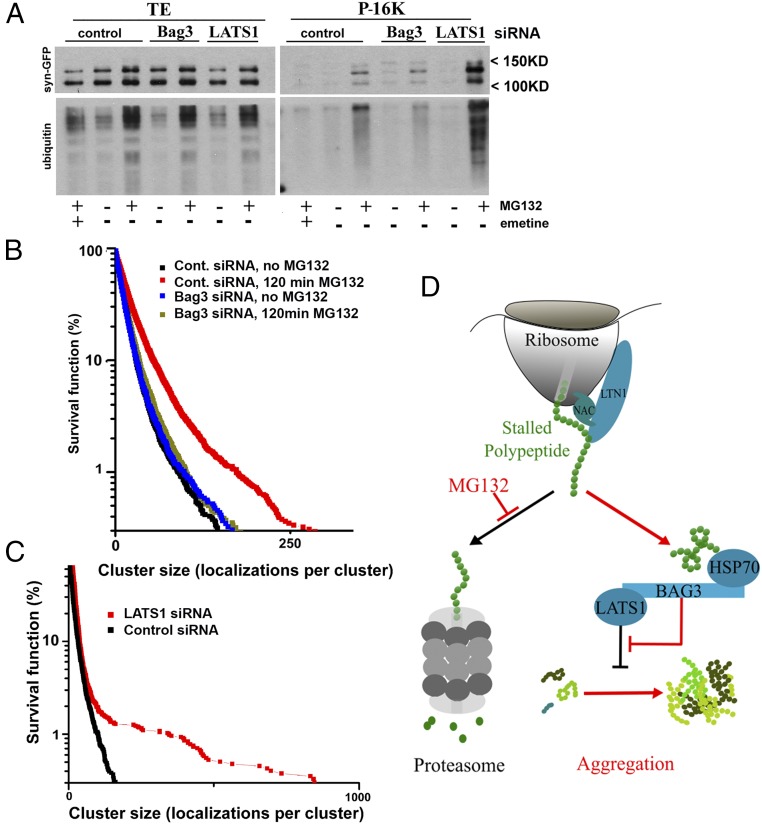
If the sole function of Bag3 is promoting the transport of aggregates to the aggresome, depletion of this protein should cause the accumulation in the cytoplasm of multiple aggregates that cannot be transported to the aggresome. However, Bag3 depletion, while significantly decreasing the number and size of aggresomes, did not increase the number of visible aggregates in the cells (SI Appendix, Fig. S4B), suggesting that Bag3 controls the formation of aggregates in addition to, or rather than, their transport to aggresome. Therefore, we further assessed the effects of Bag3 as well as of LATS1 on aggregate formation. MCF10A control cells and cells depleted of these proteins were incubated with MG132 or were left untreated, and the aggregate fractions in preclarified lysates were separated by centrifugation at 16,000 × g for 15 min. The samples were analyzed by immunoblot with anti-GFP antibody to detect Syn-GFP (Fig. 5A) and with anti-ubiquitin antibody to detect ubiquitinated polypeptides representing endogenous defective and short-lived proteins. Upon proteasome inhibition, more Syn-GFP as well as the ubiquitinated species could be seen in the insoluble fraction (for Ponceau S-stained membranes see SI Appendix, Fig. S5A). However, the addition of the translation inhibitor emetine obliterated this redistribution (Fig. 5A), suggesting that the aggregation of Syn-GFP and ubiquitinated polypeptides is prompted by the accumulation of DRiPs.
Fig. 5.
Bag3 and LATS1 regulate protein aggregation in response to proteasome inhibition. (A) Depletion of Bag3 reduces and depletion of LATS1 increases the association of Syn-GFP and ubiquitinated species with the particulate fraction. MCF10A cells stably expressing Syn-GFP with the indicated depletions were treated for 2 h with 2.5 μM MG132 with or without 0.5 μM emetine. The preclarified lysates were subjected to 15-min centrifugation at 16,000 × g. The pellets were washed once with a lysis buffer and then were dissolved in Laemmli sample buffer for immunoblotting with the antibodies indicated on the left. Note that both bands developed with anti-GFP antibody are specific. (B) The effects of incubation with 1 μM MG132 on the distribution of cluster sizes in naive and Bag3-depleted cells. Survival plots delineate a fraction of clusters containing at least N (indicated on the x axis) localization counts. Each plot was generated from 10 cells and represents the statistical distribution of 9,000–10,000 identified aggregates. (C) Survival plot showing the effects of LATS1 depletion on the distribution of cluster sizes in naive cells. Each plot was generated from 10 cells and represents the statistical distribution of 4,000 (LATS1) or 7,000 (control) identified aggregates. (D) A model for DRiPs sensing by the HB complex. Blue indicates elements of the pathway regulating the aggregation response. Red indicates changes upon proteasome inhibition; under these conditions the HB complex suppresses LATS1/2, thus removing its inhibition of protein aggregation.
In agreement with the proposed role of Bag3 in protein aggregation, its depletion significantly reduced amounts of both Syn-GFP and ubiquitinated species associated with the insoluble fraction (Fig. 5A). Furthermore, in line with the inhibitory role of LATS1 in aggresome formation, depletion of this kinase promoted the redistribution of these proteins into the particulate fraction (Fig. 5A). Of note, we observed similar effects of Bag3 and LATS1 on the distribution of Syn-GFP and of ubiquitinated species to the low-speed centrifugation pellets (200 × g) used to clarify the lysates and representing larger aggregates (SI Appendix, Fig. S5B). Overall, these data indicate that the HB complex regulates LATS1 kinase to control the early stages of aggregate formation before their transport to the aggresome.
To verify these conclusions, we monitored the effects of Bag3 and LATS1 depletions on synphilin 1 aggregation microscopically. First, we examined these effects in nocodazole-treated cells, in which Syn-GFP aggregates emerged upon MG132 treatment but could not be transported to the aggresome. Depletion of Bag3 significantly decreased the number of fluorescent aggregates (SI Appendix, Fig. S5C, Upper), while depletion of LATS1 boosted the aggregation (SI Appendix, Fig. S5C, Lower).
To better understand HB complex regulation of protein aggregation, we utilized superresolution imaging by photoactivation localization microscopy (PALM) to resolve early stages of aggregate formation that are undetectable using conventional fluorescence microscopy. In these experiments, we replaced GFP on the C terminus of synphilin 1 with photo-convertible Dendra-2 (Syn-DEN). MCF10A cells stably expressing Syn-DEN were fixed, and the localization of Syn-DEN molecules was imaged. PALM images were constructed by stacking up to 10,000 single frames, each collected with a low-intensity excitation. To quantify the population of aggregates and to study their evolution under the experimental conditions, we developed a computational image-analysis code that would scan the PALM imaging-generated localization maps (localization of the Syn-DEN fluorophore emissions) using an image-analysis algorithm DBSCAN (density-based spatial clustering of applications with noise) (https://www.biorxiv.org/content/early/2017/06/09/148395). On the localization maps DBSCAN grouped the points with high local density of emission events into clusters (localizations) while eliminating outlier points that were surrounded by fewer neighbors and thus represented monomers. This method allows the identification of clusters containing as few as four or five Syn-DEN molecules, allowing monitoring of early events in protein aggregation (28) (https://www.biorxiv.org/content/early/2017/06/09/148395). An example of the clusters found by DBSCAN in a typical Syn-DEN localization map is shown in SI Appendix, Fig. S5D. In addition to the small clusters below subdiffraction limits (invisible by conventional microscopy), the superresolution localization map allowed us to observe conventionally visible aggregates and aggresomes (SI Appendix, Fig. S5E). We found that treatment with MG132 led to a significant shift in the distribution curve of Syn-DEN cluster sizes at this scale (black vs. red curves in Fig. 5B), indicating intensification of cluster growth. Depletion of Bag3 did not affect cluster distribution in untreated cells (black and blue curves in Fig. 5B). On the other hand, in cells depleted of Bag3 we did not observe any cluster growth in the course of 2-h incubation with MG132 (blue vs. green curves in Fig. 5B). Therefore, cluster growth triggered by proteasome inhibition depends on Bag3 even at the smallest size tier.
We further tested the effects of LATS1, another element of the signaling machinery. In line with the biochemical and conventional microscopy data, depletion of this kinase led to significantly increased cluster sizes compared with the control cells (Fig. 5C), indicating that this protein represses the early stages of protein aggregation.
In a related study, cluster distribution analysis allowed us to establish that the kinetics of protein aggregation is similar to vapor condensation, where the system is in the supersaturated state even in naive cells (29). This finding, in turn, allowed establishing a critical cluster size below which clusters spontaneously dissolve but above which clusters spontaneously grow. Changes in critical cluster size reflect changes in the concentration of aggregating monomers and changes in the energy of interactions between monomers. Mathematical analysis of experiments in Fig. 5 B and C indicated that MG132 treatment reduces the critical size and that Bag3 depletion prevents this effect (SI Appendix, Fig. S5F). On the other hand, depletion of LATS1 reduced the critical size even in naive cells. Given that neither Bag3 nor LATS1 depletion significantly affected total levels of synphilin or ubiquitinated species (Fig. 5A), the effects on cluster critical size strongly suggest that these mutations regulate the energy of the interaction between the aggregating species. In other words, Bag3 is necessary for MG132-dependent stimulation of the interaction between the monomers, and LATS1 activity reduces this interaction.
Collectively, these data and the regulation of LATS1/2 by the HB complex described above imply that in response to proteotoxic stresses this system allows the initial stages of protein aggregation, which eventually leads to aggresome formation (Fig. 5D).
Discussion
Previously, we reported that disruption of the HB complex either genetically or pharmacologically dramatically affected multiple signaling pathways involved in cancer development, which suggested that the HB complex has an important function in cell signaling. In this work, we hypothesized that the HB module serves a general role in detecting protein abnormalities following proteasome inhibition and transmits signals to signaling kinases. Indeed, we found that LATS kinases are regulated by proteasome inhibition, and we present multiple lines of evidence indicating that the HB complex plays an important role in this signal-transduction process. Similarly, this complex mediated the response of stress kinases to proteasome inhibition. Therefore, the HB complex may play a general role in the cellular response to an upsurge of abnormal polypeptides.
The buildup of abnormal proteins upon proteasome inhibition depends mostly on defective polypeptides constantly generated by the ribosome. Accordingly, AZC, which increases the generation of such proteins, stimulated the LATS1/2 response. On the other hand, translation elongation inhibitors, which at low concentrations improve the quality of ribosomes’ output (17), blocked the LATS1/2 response. Here we observed the direct interaction of the HB complex with DRiPs. Indeed, in a Bag3 pull-down we saw both newly translated ubiquitinated species and radioactive species labeled during a short pulse. Furthermore, proteasome inhibition, which blocks the degradation of DRiPs, increased their association with Bag3, while a “chase” with unlabeled amino acids decreased this association. The association of DRiPs with the HB complex was dependent on Hsp70, which most likely recruits them to Bag3, since disruption of the Bag3 interaction with Hsp70 significantly reduced the association. Notably, only a small fraction of DRiPs was found in the complex with Bag3, which is an expected result based on our mechanistic knowledge of the interaction of Bag3 and Hsp70. Indeed, Bag3 interacts with Hsp70 only in its ATP-bound form, i.e., in the open conformation, when polypeptides rapidly associate with Hsp70 and rapidly dissociate from it. Therefore, the association of the HB complex with DRiPs must be very dynamic, and at any moment only a fraction of DRiPs would be in the complex.
A surprising finding here was that in the absence of additional challenges, for example AZC, stalled polypeptides represent a pool of abnormal ribosomal products which are preferentially involved in sensing the proteasome failure by the HB complex. Indeed, depletion of the ribosome-associated ubiquitin ligase LTN1 or the chaperone VCP, which are responsible for the ubiquitination and release of nonstop and stalled polypeptides from the ribosome, reduced the response to proteasome inhibition. These data are in line with previous reports that Paget disease-related mutations in VCP suppress aggresome formation (30) and that a temperature-sensitive mutation in the VCP ortholog Cdc48 blocks the formation of aggresome-like structures in yeast (31). Together these findings suggest that the HB-mediated sensing system is tuned to monitor not only proteasome activity but also the fidelity of translation. Therefore, the HB complex appears to serve a very general role in monitoring major physiological processes in the cell. Interestingly, the finding of the role of HB complex in sensing DRiPs is complemented by a recent report that this module plays a role in targeting puromycil polypeptides for degradation (32).
Here we also report that NAC plays a role in sensing proteotoxicity. Depletion of NAC elements significantly reduced the association of a model stalled polypeptide with the HB complex and suppressed both the regulation of LATS1/2 upon inhibition of the proteasome and aggresome formation, suggesting that NAC facilitates the association of stalled polypeptides with the HB complex. Interestingly, structural studies identified the site of NAC interaction with the ribosome in close proximity to a location of the LTN1 domain responsible for the interaction with polypeptides emerging from the large subunit pore (33, 34). It is possible that NAC may cooperate with LTN1 in the ubiquitination of nonstop and stalled polypeptides and their release.
Overall, our model is that normally stalled polypeptides are ubiquitinated by LTN1 and released and delivered to proteasome for degradation. However, upon proteasome inhibition they interact with the HB complex in an NAC-dependent manner. The interaction of these polypeptides with the HB complex modulates the activities of various associated kinases and possibly other signaling factors.
It should be noted that the HB complex may be able to detect a variety of protein abnormalities that are not necessarily DRiPs. In fact, this complex was reported to be involved in mechanotransduction by sensing denatured filamin A (13).
The ability to sense a decline in proteasome activity has an important physiological meaning. In aging or certain protein conformation diseases the proteasome is down-regulated, and cells activate protective responses or trigger apoptosis. Some of the protective responses, e.g., induction of the genes encoding proteasome proteins, involve stabilization and the accumulation of a specific regulator, NRF2. However, the regulation of other responses, including apoptosis, involves the activation of JNK (2) and therefore is mediated by the HB complex. Similarly, activation of the protective aggresome response involves LATS1 and is also mediated by the HB complex. Therefore, the HB complex is involved in multiple processes that allow cells to adjust to proteotoxic conditions.
The accepted paradigm for the role of Hsp70 and Bag3 in the processes of aggresome formation is that Bag3 recruits Hsp70-bound protein aggregates and links them to the dynein motor complex that in turn drives the microtubule-dependent transport. Here we show that signaling of the HB complex via LATS1 plays a significant role in aggresome formation. Notably, regulation of YAP or any other transcription factor by LATS1/2 is not critical for the response, since the transcription inhibitor actinomycin D does not inhibit the process (17). Therefore, in regulating aggresome formation, LATS1 must control a different target that is not a transcription factor.
An unexpected discovery here was that aggresome formation could be regulated at the level of initial protein aggregation. The concept that protein aggregation is controlled by signaling pathways is counterintuitive, but here we identified signaling elements that regulate this process.
The data obtained with PALM microscopy provided important mechanistic insights into the early events leading to the appearance of aggregates undetectable with conventional fluorescence microscopy. Using this methodology, we demonstrate that proteasome inhibition facilitates the formation and growth of clusters comprising only several Syn-DEN molecules. Importantly, depletion of Bag3 blocked the effects of MG132 on the cluster size, while depletion of LATS1 mimicked effects of proteasome inhibition in untreated cells. The data obtained below subdiffraction limits were in line with the effects of Bag3 and LATS1 depletion on the distribution of synphilin 1 and ubiquitinated polypeptides into the particulate fractions measured biochemically. Given that neither Bag3 nor LATS1 depletion affected the overall levels of these species in the cell (Fig. 5A, Left), these data suggest that the HB–LATS1 complex signals to enhance the strength of the interaction between monomers and the forming aggregates. The simplest possibility is that LATS1 phosphorylates either aggregating species or a factor(s) that mediates their interactions. Overall, these data indicate that the Hsp70–Bag3–LATS1 system transmits the signal of proteasome inhibition to trigger protein aggregation.
Materials and Methods
Reagents and Antibodies.
MG132 and nocodazole were purchased from Biomol, emetine and AZC were purchased from Sigma-Aldrich, and 16% formaldehyde was purchased from Thermo Scientific. YM-1 was developed and supplied by Jason Gestwicki, University of California, San Francisco. Antibodies against YAP, phospho-YAP (S127), phospho-JNK, phospho-p38, anti-LATS1, and β-actin were from Cell Signaling Technology; anti-HspA1, anti-HspA8, and anti-multiubiquitin (FK2) were from Enzo Life Sciences; anti-BAG3 and anti-puromycin were from Millipore; anti-GFP (peptide) used for immunoblotting was from Clontech; anti-GFP (full protein) used in the Duolink assay (Sigma-Aldrich) and the modified ActivSignal assay was from GeneTex; anti-FLAG (M2) was from Sigma-Aldrich; anti–V5-tag was from Thermo Fisher Scientific (Pierce); anti-NACB was from Santa Cruz Biotechnology; and anti-DNAJC2 was from Abgent.
Constructs and Oligonucleotides.
The retroviral expression construct with C-terminally tagged synphilin 1 (Syn-GFP) subcloned into the pCXbsr vector was described previously (27). For PALM imaging EGFP at the C terminus of the construct was replaced by Dendra-2. pcDNA3.1-based plasmids used for overexpression of N-terminally FLAG-tagged Bag3 or its ΔC, ΔP, and ΔW deletion mutants were described previously (6). Additionally, we constructed a ΔM mutant missing a stretch between amino acids 86–214. For expression in MCF10A cells, we cloned some of these constructs in retroviral vector pBabe (puro). For pull-down experiments we replaced FLAG on the N terminus of these constructs with the four following sequences connected by short linkers: FLAG-tag, S-tag, SBP-tag, and (His)9-tag. To generate the GFP-NS construct, we deleted the stop codon of the EGFP gene in pEGFP-N1 plasmid and with small deletions and frame shifts eliminated any in-frame stop codons in the flanking downstream sequence of about 200 bp while preserving the downstream polyadenylation signals. For a reporter of NACA association with Bag3, we synthesized and cloned in pcDNA3.1(+) a gene encoding V5–FLAG–NACa1-b.
The retroviral Hsf1 shRNA construct and control vector were described previously (35). We used the following siGENOME siRNAs purchased from Dharmacon: nontargeting siRNA no. 5, BAG3 (D-011957-01), NACB (D-016634-02), HSPA8 (M-017609-01), LTN1 (M-006968-00), VCP (M-008727-01), LATS2 (M-003865-02), and DNAJC2 (M-025435-02). LATS1 (SASI_Hs01_00046130) and NACA (SASI_Hs02_00325613) were purchased from Sigma. For the rescue of Bag3 depletion by its overexpression, we synthesized siRNA homologous to the Bag3 shRNA, which was described previously (6) and to which the exogenous Bag3 constructs are resistant.
Cell Culture.
HeLa (cervix carcinoma) cells were grown in DMEM supplemented with 10% FBS, and MCF10A (human breast epithelial) cells were grown in DMEM/F-12 50/50 medium supplemented with 5% horse serum, 20 ng/mL epidermal growth factor, 0.5 μg/mL hydrocortisone, 10 μg/mL human insulin, and 100 ng/mL cholera toxin. All cultures were supplemented with l-glutamine as well as with MycoZap Plus-CL (Lonza) (for HeLa cells) or Plasmocin (InvivoGen), penicillin, and streptomycin (for MCF10A cells), and were grown at 37 °C with 5% CO2. For analysis with a fluorescence microscope, cells were grown on Nunc Lab-Tek chambered cover glasses (Thermo Fisher Scientific) pretreated with poly-l-lysine (Sigma).
For transient plasmid transfection of HeLa cells with Lipofectamine 2000 reagent (Invitrogen, Thermo Fisher Scientific), we followed the manufacturer’s protocol. In a 35-mm dish, 10 μL of the reagent were mixed with 3 μg of plasmid(s). Cells were transfected for 2 h. One hour after the end of transfection the cells were plated for an experiment conducted the next day. For siRNA transfection, we used Lipofectamine RNAiMAX (Invitrogen, Thermo Fisher Scientific) and followed the manufacturer’s reverse-transfection protocol. In a well on a 24-well plate, we mixed 0.4 μL of the reagent with 2 μL of 10 μM siRNA in 100 μL of OptiMEM and added the mixture to 400 μL of a cell suspension in the well. Twenty-four to twenty-eight hours later the transfection was stopped, and the cells were plated for an experiment conducted the next day.
Duolink Assay.
The transfected HeLa cells were grown, treated with MG132, fixed with 4% formaldehyde, and permeabilized with 0.2% Triton X-100 on a Corning 96-Well Half-Area High-Content Imaging Glass Bottom Microplate. We followed the manufacturer’s protocol using Duolink In Situ Detection Reagent Red, Duolink In Situ PLA Probe Anti-Mouse PLUS, and Duolink In Situ PLA Probe Anti-Rabbit MINUS purchased from Sigma-Aldrich. The primary antibodies used were anti-GFP and anti-FLAG antibodies raised in rabbit and mouse, respectively. The resulting images were obtained with a fluorescence microscope with a 20× objective from at least 10 blindly chosen fields with a total at least 250 cells. We measured the combined fluorescence of all cells with the NIH ImageJ program and normalized it by the number of all counted cells stained with DAPI. In the images presented in SI Appendix, Fig. S3, we chose an exposure time that provided visible red fluorescence in less than 5% of the cells in control samples.
The PLA Based on a Modified ActivSignal Protocol.
Transfected HeLa cells were grown, treated with MG132, fixed with 4% formaldehyde, and permeabilized with 0.2% Triton X-100 on a 96-well plastic plate. The samples were left in buffer B (ActivSignal) at 4 °C overnight. All the following steps were conducted at room temperature. The samples were incubated for 2 h with a pair of primary antibodies diluted in buffer B. After three washes with PBS and Tween-20 (PBST), consecutive 1-h incubations in buffer B with each of the secondary antibodies were done. After two washes with PBST followed by two washes with TE buffer (10 mM Tris-HCl, 1 mM EDTA), a ligation mixture including a linker oligo (50 nM) and T4 DNA ligase (5 U/mL) was added to the wells, and the reaction was conducted for 30 min. The wells were washed once with PBST and twice with TE, 50 μL of TE were added to each well, and the ligated conjugates were extracted by 30-min incubation of the plate at 95 °C. The eluates were assessed by qPCR using All-in-One qPCR Mix (GeneCopoeia) according to the manufacturer’s protocol.
Donkey anti-rabbit (711-005-152) and donkey anti-mouse (715-005-150) secondary antibodies were purchased from Jackson ImmunoResearch, Inc. and were conjugated with a pair of oligonucleotides by ActivSignal, Inc. The linker for the ligation reaction was provided by ActivSignal, Inc.
Total RNA Preparation and qRT-PCR.
Total RNAs from cells were isolated using the RNeasy Mini Kit (Qiagen) and were reverse transcribed with SuperScript IV VILO Master Mix with ezDNase Enzyme (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s instructions. qRT-PCR was performed using All-in-One qPCR Mix (GeneCopoeia).
Conditions for cell lysis, immunoprecipitation, pull-down analysis, radiolabeling, mass spectroscopy, and microscopy are described in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01 CA176326 and NIH Clinical and Translational Science Institute Parental Award 1UL1TR001430 (to M.Y.S.). X.V. is supported by Department of Defense Congressionally Directed Medical Research Program Award W81XWH-14-1-0336 and NIH Grant R01 HL124392.
Footnotes
Conflict of interest statement: I.A. is cofounder of ActivSignal. The authors declare no other conflicts of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803130115/-/DCSupplemental.
References
- 1.Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: A cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 2.Meriin AB, Gabai VL, Yaglom J, Shifrin VI, Sherman MY. Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J Biol Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- 3.Lee DH, Goldberg AL. Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:30–38. doi: 10.1128/mcb.18.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 5.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 6.Colvin TA, et al. Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 2014;74:4731–4740. doi: 10.1158/0008-5472.CAN-14-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, et al. Validation of the Hsp70-Bag3 protein-protein interaction as a potential therapeutic target in cancer. Mol Cancer Ther. 2015;14:642–648. doi: 10.1158/1535-7163.MCT-14-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carra S. The stress-inducible HspB8-Bag3 complex induces the eIF2alpha kinase pathway: Implications for protein quality control and viral factory degradation? Autophagy. 2009;5:428–429. doi: 10.4161/auto.5.3.7894. [DOI] [PubMed] [Google Scholar]
- 9.Behl C. Breaking BAG: The co-chaperone BAG3 in health and disease. Trends Pharmacol Sci. 2016;37:672–688. doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Carra S, Seguin SJ, Landry J. HspB8 and Bag3: A new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy. 2008;4:237–239. doi: 10.4161/auto.5407. [DOI] [PubMed] [Google Scholar]
- 11.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 12.Morimoto RI. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 13.Ulbricht A, et al. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol. 2013;23:430–435. doi: 10.1016/j.cub.2013.01.064. [DOI] [PubMed] [Google Scholar]
- 14.Meriin AB, et al. A novel approach to recovery of function of mutant proteins by slowing down translation. J Biol Chem. 2012;287:34264–34272. doi: 10.1074/jbc.M112.397307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman MY, Qian S-B. Less is more: Improving proteostasis by translation slow down. Trends Biochem Sci. 2013;38:585–591. doi: 10.1016/j.tibs.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Conn CS, Qian SB. Nutrient signaling in protein homeostasis: An increase in quantity at the expense of quality. Sci Signal. 2013;6:ra24. doi: 10.1126/scisignal.2003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meriin AB, Zaarur N, Sherman MY. Association of translation factor eEF1A with defective ribosomal products generates a signal for aggresome formation. J Cell Sci. 2012;125:2665–2674. doi: 10.1242/jcs.098954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minoia M, et al. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy. 2014;10:1603–1621. doi: 10.4161/auto.29409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Defenouillère Q, et al. Rqc1 and Ltn1 prevent C-terminal alanine-threonine tail (CAT-tail)-induced protein aggregation by efficient recruitment of Cdc48 on stalled 60S subunits. J Biol Chem. 2016;291:12245–12253. doi: 10.1074/jbc.M116.722264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.del Alamo M, et al. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston JA, Ward CL, Kopito RR. Aggresomes: A cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Qian SB. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol Biol Cell. 2011;22:3277–3288. doi: 10.1091/mbc.E11-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z, et al. 14-3-3 protein targets misfolded chaperone-associated proteins to aggresomes. J Cell Sci. 2013;126:4173–4186. doi: 10.1242/jcs.126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia B, Wu Y, Zhou Y. 14-3-3 and aggresome formation: Implications in neurodegenerative diseases. Prion. 2014;8:28123. doi: 10.4161/pri.28123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaarur N, Meriin AB, Gabai VL, Sherman MY. Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin 1. J Biol Chem. 2008;283:27575–27584. doi: 10.1074/jbc.M802216200. [DOI] [PubMed] [Google Scholar]
- 28.Izeddin I, et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife. 2014;3:02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayanan A, Meriin AB, Sherman MY, Cisse II. 2017. A first order phase transition underlies the formation of sub-diffractive protein aggregates in mammalian cells. bioRxiv:148395. Preprint, posted June 19, 2017.
- 30.Ju J-S, Miller SE, Hanson PI, Weihl CC. Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J Biol Chem. 2008;283:30289–30299. doi: 10.1074/jbc.M805517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, et al. Abnormal proteins can form aggresome in yeast: Aggresome-targeting signals and components of the machinery. FASEB J. 2009;23:451–463. doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganassi M, et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell. 2016;63:796–810. doi: 10.1016/j.molcel.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Lyumkis D, et al. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc Natl Acad Sci USA. 2014;111:15981–15986. doi: 10.1073/pnas.1413882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegrzyn RD, et al. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J Biol Chem. 2006;281:2847–2857. doi: 10.1074/jbc.M511420200. [DOI] [PubMed] [Google Scholar]
- 35.Kim G, et al. The heat shock transcription factor Hsf1 is downregulated in DNA damage-associated senescence, contributing to the maintenance of senescence phenotype. Aging Cell. 2012;11:617–627. doi: 10.1111/j.1474-9726.2012.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.