Significance
The mode and tempo of extinctions and extirpations after the first contact phase of human settlements is a widely debated topic. As the last major landmass to be settled by humans, New Zealand offers a unique lens through which to study interactions of people and biota. By analyzing ancient DNA from more than 5,000 nondiagnostic and fragmented bones from 38 subfossil assemblages, we describe species and patterns that have been missed by morphological approaches. We report the identification of five species of whale from an archaeological context in New Zealand and describe the prehistoric kākāpō population structure. Taken together, this study demonstrates insights into subsistence practices and extinction processes and demonstrates the value of genetic analyses of fossil assemblages.
Keywords: paleoecology, human impacts, subsistence practices, bulk bone metabarcoding, ancient DNA
Abstract
New Zealand’s geographic isolation, lack of native terrestrial mammals, and Gondwanan origins make it an ideal location to study evolutionary processes. However, since the archipelago was first settled by humans 750 y ago, its unique biodiversity has been under pressure, and today an estimated 49% of the terrestrial avifauna is extinct. Current efforts to conserve the remaining fauna rely on a better understanding of the composition of past ecosystems, as well as the causes and timing of past extinctions. The exact temporal and spatial dynamics of New Zealand’s extinct fauna, however, can be difficult to interpret, as only a small proportion of animals are preserved as morphologically identifiable fossils. Here, we conduct a large-scale genetic survey of subfossil bone assemblages to elucidate the impact of humans on the environment in New Zealand. By genetically identifying more than 5,000 nondiagnostic bone fragments from archaeological and paleontological sites, we reconstruct a rich faunal record of 110 species of birds, fish, reptiles, amphibians, and marine mammals. We report evidence of five whale species rarely reported from New Zealand archaeological middens and characterize extinct lineages of leiopelmatid frog (Leiopelma sp.) and kākāpō (Strigops habroptilus) haplotypes lost from the gene pool. Taken together, this molecular audit of New Zealand’s subfossil record not only contributes to our understanding of past biodiversity and precontact Māori subsistence practices but also provides a more nuanced snapshot of anthropogenic impacts on native fauna after first human arrival.
The early isolation of New Zealand from the Gondwanan mainland 55 million years ago (1), along with extinctions during the cooling events of the Miocene and Pliocene, created an island archipelago free of land mammals (1). Over time, the ecological niches filled by mammals elsewhere were taken over by birds, giving rise to a variety of flightless avifauna found nowhere else in the world, including the iconic moa (Dinornithiformes), kiwi (Apteryx sp.), and kākāpō (Strigops habroptilus). However, since the arrival of Polynesian settlers to the archipelago 750 y ago (2, 3), the terrestrial avifauna has nearly halved, and at least 64 species of New Zealand’s endemic birds and reptiles have become extinct, with new species added to the extinction list each year (4–7).
The impact of human arrival on the local biodiversity across different continents is a hotly debated topic (8–10), with climate, disease, hunting, and human commensals (or a combination thereof) put forward as drivers of extinctions and extirpations. However, the direct impacts of humans are often challenging to tease apart from climatic changes (11) when they coincide. Moreover, in many locations, the archaeological record is blurred by the Holocene sea level rise that consumed many early coastal sites across the globe (12). As the last major landmass to be settled by humans, and with a relatively stable Holocene climate (13), New Zealand continues to offer a unique opportunity to assess the impact of human arrival on island ecosystems.
During the last 150 y, morphological analyses of the Quaternary fossil and subfossil records in New Zealand have provided insights into biodiversity turnover on the islands. For example, the finding of thousands of bones across hundreds of archaeological middens suggests that within a few centuries after the initial Polynesian settlement, a small population (<2,000 individuals) drove the moa to extinction (14–17) and caused the extirpation of sea lion (Phocarctos spp.) and penguins (Megadyptes waitaha) from the New Zealand mainland (18, 19). However, the full extent of human-driven extinctions in New Zealand is still unclear, as morphological analysis of the subfossil record is complicated by a number of documented extirpation/recolonization events (18, 19) and the existence of taxa exhibiting sexual dimorphism (20) or cryptic morphology (21). Furthermore, an important component of New Zealand’s subfossil assemblages remains understudied, as nondiagnostic bones figure prominently in many deposits (22), especially in archaeological middens, where bones are typically fragmented by human processing. As such, morphological approaches might overlook key species when diagnostic features are lost from taphonomic processes.
Here we analyze ∼5,276 bone fragments from archaeological and paleontological sites across New Zealand to reveal past biodiversity, population dynamics, extinction processes, and the impact of subsistence practices on the local fauna. We analyzed undiagnostic, fragmented bones from museum collections in New Zealand, using Bulk Bone Metabarcoding (BBM) (23), which allows for a cost-effective and rapid screening of hundreds of fragments at each site, and thousands of fragments across a landscape, simultaneously. By overlaying genetic data onto the already-existing morphological record, we characterize the unknown fractions of subfossil assemblages across the archipelago, providing an alternative lens through which to view the critical period in which humans and native fauna first interacted.
Results and Discussion
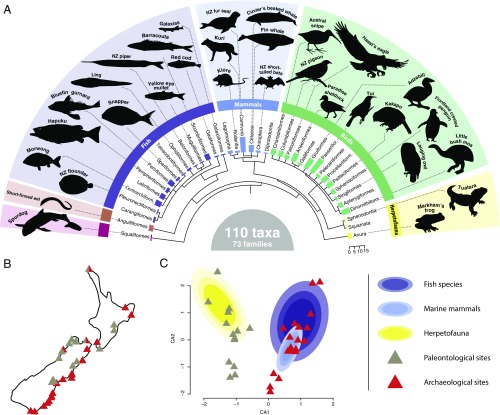
We analyzed ancient DNA extracted from 70 bulk bone powder samples, representing more than 5,276 bone fragments collected from 38 different bone assemblages across New Zealand. Of these, 15 sites are naturally deposited (paleontological) assemblages, 21 assemblages represent archaeological midden deposits, and two sites contain layers of both paleontological and archaeological bone fragments (Fig. 1B and SI Appendix, Table S1). The 38 bone deposits analyzed cover the last 20,000 y of New Zealand’s past, with paleontological sites represented by four Pleistocene deposits and 11 Holocene deposits, whereas the archaeological middens represent prehistoric Māori activities between AD 1300 and 1800 (SI Appendix, Table S1). As the associated level of detail on excavation process and stratigraphy varies between bulk bone samples, we do not distinguish between stratigraphic units within one site, but consider bulk bone samples from one site as a single entity, and the age of those bones as the age range of that bone deposit (Methods).
Fig. 1.
Overall biodiversity of excavated bulk bone. Species composition was analyzed using four metabarcoding assays targeting vertebrate taxa. (A) Dendrogram highlighting the diversity of orders identified in all samples, with examples of taxa identified in silhouettes. Bar sizes represent the number of taxa identified in each order. (B) Sample localities of archaeological midden sites (red triangles) and paleontological deposits (gray triangles). (C) Correspondence analysis based on presence/absence of all taxa identified from archaeological or paleontological sites. The distribution of herpetofauna (Class: Amphibia and Reptilia), fish species (Class: Actinopterygii and Chondrichthyes), and marine mammals (Family: Phocidae and Otariidae and Order: Cetacea) is highlighted by ellipses of incremental confidence intervals of 0.4, 0.6, and 0.8.
Using four different metabarcoding assays (SI Appendix, Table S2), we were able to amplify endogenous DNA in 37 of the 38 assemblages investigated. Next-generation sequencing yielded a total of 1,811,732 reads postfiltering (7,580 reads per sample per assay on average), corresponding to 653 operational taxonomic units. Of these, 436 operational taxonomic units could be assigned to a taxonomic node at family level or below (Methods). We found a highly diverse composition of species across all sites, with 170 taxa representing at least 110 different species (Fig. 1A). Unsurprisingly, birds (class: Aves) make up the largest and most diverse group, with 54 different species, whereas bony fish (class: Actinopterygii) and mammals (class: Mammalia) represent the second and third most diverse groups, with 29 and 21 different species identified, respectively. Other classes represented are Amphibia (three species), Reptilia (two species), and Chondrichthyes (one species).
Correspondence analysis (CA) based on the entire dataset revealed a clear clustering of archaeological and paleontological sites (Fig. 1C), which was confirmed by a canonical correspondence analysis constrained by site type (archaeological/paleontological) showing that 6.7% of the variance in the data (P < 0.001; 999 permutations) is explained by site type. The difference between archaeological and paleontological sites is primarily explained by the high number of bony fish species and marine mammals unique to the archaeological sites. Conversely, amphibians (Leiopelma sp.), skinks (Oligosoma sp.), and owls (Ninox novaeseelandiae and N. albifacies) were found in paleontological sites but were absent in archaeological middens.
In the following sections, we discuss how BBM can inform us about past anthropogenic impacts on the environment by focusing on the four major groups of species identified: marine mammals, bony fish, birds, and reptiles and frogs.
Marine Mammals: Sealing and Whaling in Precontact New Zealand.
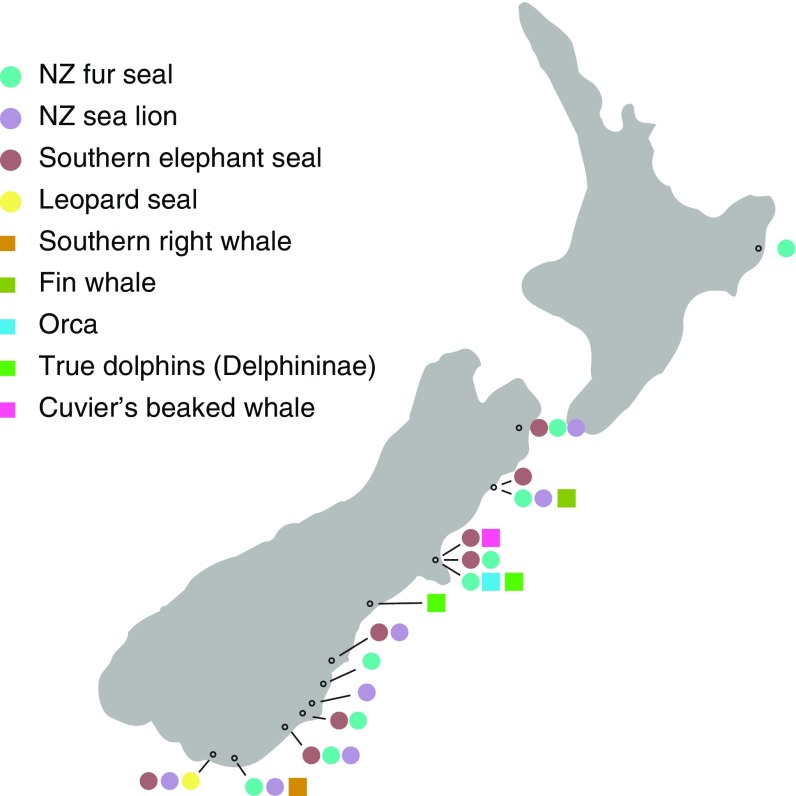
Marine mammals were detected in 71% (15/21) of the archaeological sites, and in total, nine species of marine mammals were definitively identified (Fig. 2 and SI Appendix, Table S3). The most commonly detected taxa are the New Zealand fur seal (Arctocephalus fosteri), the New Zealand sea lion (Phocarctos hookeri), and the southern elephant seal (Mirounga leonina), identified at 9, 7, and 8 different sites, respectively, and recovered consistently from independent bulk bone subsamples. Our BBM data complement previous studies (24, 25) in confirming the importance of sealing to Māori in New Zealand before 1500 AD: of the 14 sites in which pinniped DNA is detected, only a single site is characterized exclusively as late Māori (deposited after 1500 AD). Nonetheless, the frequency of sites where sea lion and elephant seal were detected is higher in the DNA record than in the morphological record. In previous morphological studies, the number of early assemblages where fur seal remains were identified was consistently twice as high as that of elephant seal and sea lion (24), whereas in our study, we detect fur seal at the same frequency as elephant seal and sea lion. This suggests that these species might have played a more prominent role in the subsistence economy of precontact New Zealand than previously believed. This discrepancy could reflect a bias in morphological identifications of highly fragmented seal remains, which might be more likely to follow the current understanding of a dominance of fur seal in Māori middens. Nevertheless, the presence of fur seal and sea lions in early Māori middens supports recent findings demonstrating that both species were hunted extensively immediately after human arrival, causing the extinction of the mainland lineage of seal lions (18) and a near extinction of fur seals (26). For elephant seals, their abundance in middens before 1500 AD strongly suggests that, as opposed to today, elephant seals were breeding in New Zealand at the time of Polynesian arrival.
Fig. 2.
Localities of midden sites in which DNA from marine mammals was identified.
In the morphological record, the most commonly identified cetacean from New Zealand middens is the pilot whale (Globicephala sp.), followed by dolphins (Delphinidae spp.) (27), a pattern that is reflected in the record of modern strandings, where pilot whales are the most abundant by far (28). However, the majority of cetacean bones cannot be identified morphologically because of osteological similarities between whales (24, 29) and the common practice of reworking their bones into tools (30). DNA analysis is not dependent on morphology, and thus, of the five whale species identified here, only Eubalaena australis have been identified before in an archaeological context in New Zealand. Toothed whales are represented in the DNA record by three species: orca (Orcinus orca), true dolphins (subfamily: Delphininae), and Cuvier’s beaked whale (Ziphius cavirostris). The baleen whales are represented by two species: the fin whale (Balaenoptera physalus) and the southern right whale (Eubalaena australis), identified at one site each on the South Island (30) (Fig. 2). Analogous to the identification of seals discussed earlier, the unexpected absence of pilot whale DNA in our data (SI Appendix, Fig. S6) could suggest that the identification of fragmented whale bones is more likely to follow the dominant archaeological paradigm that reflects the modern stranding record, where pilot whale is frequently reported. Our data suggest that the diversity of whales used in precontact New Zealand is far greater than previously believed.
The nature and value of whale products in preindustrial cultures is heavily debated (27, 29, 31): Do they stem from the occasional whale stranding, or is it a result of a more organized strategy to target these species? With an adult weight well above 40 tons, and with the absence of heavy whaling harpoons from the archaeological record (32, 33), it is likely that the two baleen whales identified here represent scavenging of beached whales rather than hunting. The identification of three species of the smaller toothed whales at Redcliffs flat, Canterbury, in contrast, may suggest an organized targeting of these species: from the archaeological record, there is evidence of harpoon heads at the site alongside several morphologically unidentifiable whale bones (34). It is possible that Māori hunted these smaller whales by driving them into shallow waters and harpooning them from the beach (35). Nevertheless, despite the relative abundance of small whales at Redcliffs in our data, it is also possible that these species were scavenged. Single whale strandings are frequent on the shores of the Canterbury region (28), and the Māori harpoon heads from Redcliffs (34) may have been used for a range of potential functions other than whaling.
Fish Species: A Window into Past Fishing Technologies.
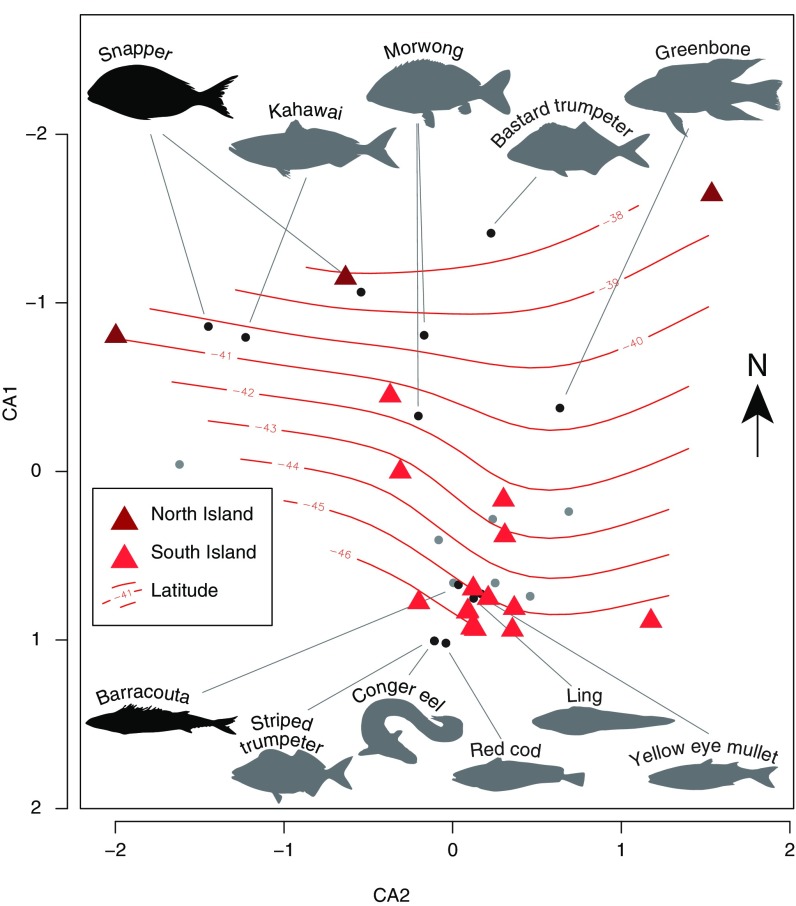
To assess differences in past fishing technologies and geographical distributions of fish communities, we investigated fish species compositions for all archaeological assemblages along a north–south gradient. We found that latitude directly correlates with fish species composition (P < 0.001; 999 permutations; Fig. 3), supporting the hypothesis that Māori were mainly relying on local fishing, rather than trading or traveling long distances to hunt. North Island fish assemblages are characterized by high numbers of snapper (Sparidae, Pagrus auratus), gurnards (Triglidae), and jack mackerels (Trachurus), whereas barracouta (Thyrsites atun), red cod (Pseudophycis), and ling (Genypterus blacodes) are more commonly identified in the South Island assemblages. This structure is consistent with previous studies of NZ subsistence practices (36, 37), demonstrating a clear distinction between fish assemblages between the two islands (SI Appendix, Table S4).
Fig. 3.
Geographic trends in archaeological fish assemblages. Coordination analysis of fish species composition from North Island and South Island assemblages based on presence/absence data, with the latitude of each site overlaid on the plot. The five most extreme taxa on the second axis (CA1) detected at more than one site are highlighted (black dots and gray silhouettes) for high and low latitudes, respectively. Similarly, the most commonly identified taxon on each island is highlighted (black dots and black silhouettes). The distribution of all other taxa detected at more than one site is illustrated by gray dots.
For certain taxa, the taxonomic resolution in the DNA data enables species-level identification that is not typically attainable using morphology. We identify three species of eel not commonly detected in past morphological analyses of Māori middens. From four archaeological sites, one fresh water species (New Zealand short-finned eel, Anguilla australis) and two marine eels (Conger sp. and Gnathophis sp.) were detected (SI Appendix, Table S4). The relative abundance of eel species (4/21 or 19% of sites), and in particular the identification of Gnathophis sp., adds to the current discussion on the importance of eel as a Māori food source before European colonization (37). Because of taphonomy and difficulties in identifying eel remains, it has long been hypothesized that these animals were of greater importance than their bone remains in midden assemblages reflect (38). Our data demonstrate that we can detect previously identified species (Anguilla and Conger) and new species (Gnathophis), using DNA. Still, we do not detect eel frequently. This suggests that in the sites sampled, eel were likely an important seasonal supplement to Māori diet, but not a primary food source (37).
The Avifauna: Extinctions, Endemism, and Endangered Species.
At least 54 different bird species were identified in this study. Of these, 36 species are endemic to New Zealand (67%), and 13 species are extinct (24%; SI Appendix, Table S5 and Fig. S5). Moa species are commonly found in both archaeological (n = 6) and paleontological (n = 9) sites, and are represented by four species in the dataset: South Island giant moa (Dinornis giganteus), eastern moa (Emeus crassus), little bush moa (Anomalopteryx didiformis), and upland moa (Megalapteryx didinus). Furthermore, the different nature of deposition in archaeological middens and subfossil bone assemblages from caves is apparent in the data, with a clear separation of archaeological and paleontological sites based on ordination analysis of bird species composition (P = 0.002; SI Appendix, Fig. S1). Despite geographical differences between sites, the avifauna of archaeological midden assemblages typically include petrels, cormorants, and penguins, whereas paleontological sites are commonly characterized by raptors, kiwi, rails, and the endemic New Zealand parrots (Strigopidae; SI Appendix, Fig. S1).
The New Zealand parrots (Strigopidae) were identified at numerous sites across the North Island and the South Island. Most commonly identified is the kākāpō, detected at seven sites, followed by kea (Nestor notabilis), at three sites, and the kākā (Nestor meridionalis), at two sites. Concordant with morphological studies that frequently identify New Zealand parrots in Māori middens (39), we find DNA evidence of these birds at two midden sites: kea and Nestor sp. were identified at Wairau Bar and Watson’s Beach, respectively. The notion that these birds were hunted by Māori has sparked a debate on the timing, causes, and extent of biodiversity decline for these species, which has been studied in kea (40) and kākāpō (41, 42), based on genetic markers from modern and ancient samples. However, few, if any, ancient samples predating European arrival to New Zealand were included in these studies. Accordingly, as BBM rapidly screens thousands of bones for DNA preservation, the use of bulk bone samples could prove an invaluable tool to increase the amount of ancient material available in such studies, especially for taxa that are comparatively rare (e.g., kea and kākāpō).
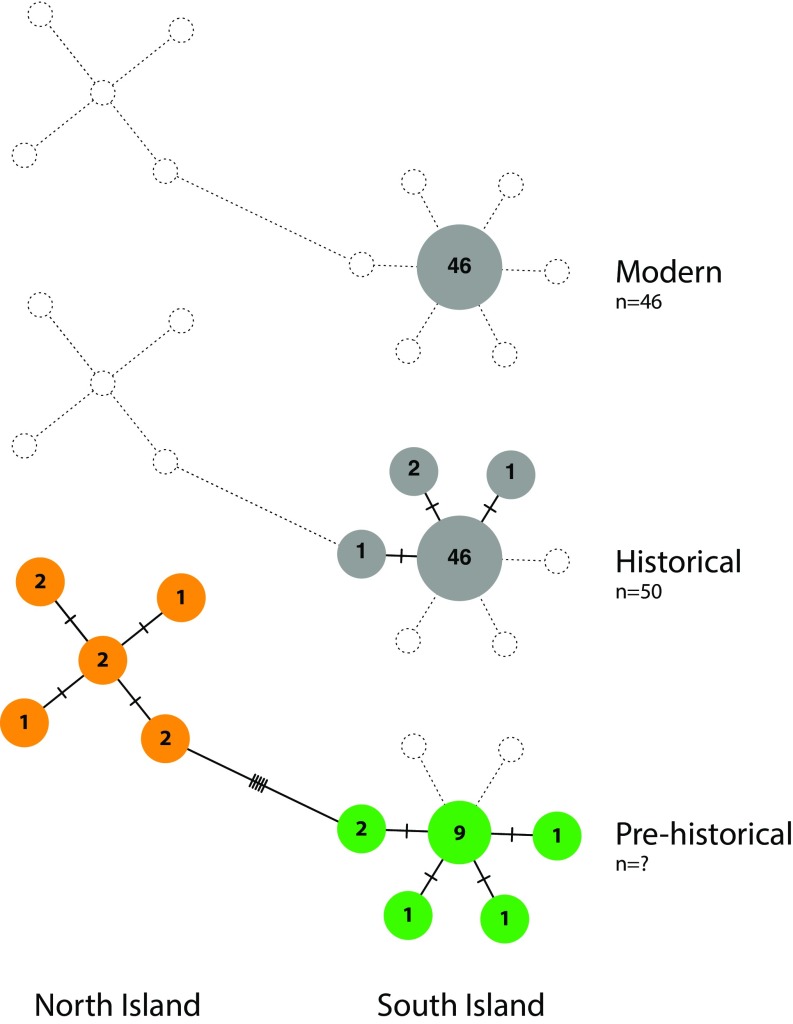
To demonstrate how bulk bone could be used to examine past intraspecific genetic diversity, we reanalyzed the bulk bone extracts in which kākāpō was detected, but this time employed a kākāpō-specific assay targeting a 214-bp stretch of the mitochondrial control region. DNA was successfully amplified in 13 bulk bone extracts, which after strict error filtering (Methods) yielded a total of 10 haplotypes, with a clear separation between the North Island (five haplotypes) and South Island (five haplotypes) clades (Fig. 4). Of these, only two haplotypes have been described previously from modern and historical (AD 1847–1974) samples, using this assay (41). Hence, with the characterization of three previously unknown South Island haplotypes and the description of the North Island clade, these data demonstrate the value of the bulk bone method in providing samples from rarely identified species. Albeit based on a small sample size, these data suggest that a number of kākāpō haplotypes were lost before European arrival, reinforcing the idea that Māori activities also impacted the kākāpō population and that attributing the decline solely to European arrival (41) may be an artifact of ‘missing’ aDNA data.
Fig. 4.
Decline in kākāpō genetic diversity. Haplotype network of a 214-bp mitochondrial control region sequence from modern, historical, and prehistorical kākāpō populations. Gray circles indicate haplotypes identified from single source material in Bergner et al. (41), whereas colored haplotypes were identified from bulk bone samples generated in this study. Hatch marks represents the number of control region mutations between haplotypes. For modern and historical haplotypes, numbers in each circle represent number of individuals in each haplotype, whereas for the prehistorical samples, numbers indicate number of DNA extracts in which each haplotype has been detected.
Reptiles and Amphibians: The Extinct Frog Leiopelma markhami.
We detected five species of reptiles and amphibians: tuatara (Sphenodon punctatus), a skink (Oligosoma sp.), and three leiopelmatid frogs (SI Appendix, Table S6). Despite not targeting herpetofauna specifically, the tuatara is identified by two assays at three paleontological sites and in one midden assemblage. Similarly, the frogs within the genus Leiopelma were detected by two assays at four different paleontological sites. Although cryptic morphology renders species limits within leiopelmatids ambiguous (21), seven species are currently recognized, of which three are extinct. The introduction of the Pacific rat (Rattus exulans) to New Zealand, along with habitat loss, is thought to be the primary cause of population contractions and extinctions within Leiopelmatidae during the past 750 y (6), and today, leiopelmatids survive only on rat-free islands and in isolated areas of the North Island. The fact that these frogs were only found in paleontological sites rather than middens supports the idea that they were not hunted to extinction. The previous widespread distribution of leiopelmatid frogs, as well as the loss in diversity within the genus, is evident from the DNA data, in which we identify several distinct Leiopelma lineages across the South Island. The extinct Leiopelma markhami was identified in six different samples from the sites Graveyard, Eagles Roost, and Cobden Cave. These data suggest that L. markhami was once particularly widespread on the northern half of the South Island. Surprisingly, DNA from the extant L. archeyi and L. hamiltoni was only detected at a single site, Takaka Hill. A phylogeny constructed with these reads including reference sequences from extant taxa and a reference for the extinct L. markhami, generated for this study, places L. markhami as the sister clade to all other species within the genus with high bootstrap support (>0.99). Furthermore, we report a similar phylogenetic spread within the L. markhami clade as in the L. pakeka/L. hamiltoni/L. archeyi complex, which was recently suggested to represent a single species, based on lack of morphological differentiation (21) (SI Appendix, Fig. S3). These results provide additional evidence for the close phylogenetic relationship among L. pakeka, L. hamiltoni, and L. archeyi, but further DNA work is still needed on single-source bones to test whether they should be considered the same species.
Conclusion
Loss of archaeological sites because of rising sea levels coupled with poor DNA preservation have often obscured attempts to gain a more nuanced picture of how humans first interacted with new landscapes. New Zealand remains one of the few places with a recent and well-preserved record spanning a climatically stable time before and after human arrival, and as such can be used as a proxy to understand the mode and tempo of extinctions and extirpations elsewhere in the world. As a large-scale ancient DNA survey of bone assemblages, this study demonstrates that species composition among sites can be reproduced from small samples of unidentifiable bone fragments, using BBM. Furthermore, our study has identified species and patterns typically missed by traditional morphological approaches, including the detection of whales and a high reliance on sea lions and elephant seals by early Māori.
Collectively, these findings highlight the impacts that human arrival had on biodiversity across New Zealand. The identification of moa and pinniped DNA throughout early Māori sites confirm that these species were hunted up until their extinction/extirpation in the 15th century. In contrast, the absence from Māori middens of other extinct species that survived into the 20th century, such as bush wren, kōkako, and laughing owl, indicate that activities by European settlers were likely drivers of their decline. Last, for still-extant species, range contraction and population size decline are reflected in our dataset by the decrease in kākāpō and Leiopelma genetic diversity, along with the detection of tuatara (currently restricted to islands in Cook Strait and off northern New Zealand) in three South Island sites.
The insights into past biodiversity and subsistence practices gained here highlight the value of the nondiagnostic fraction of bone assemblages for the use in ancient DNA studies as a complementary approach to traditional morphological identifications. Although both morphological analysis and BBM are challenged by inherent limitations, the high level of agreement between the morphological record and the genetic data presented here serves as a validation of both. As such, these results demonstrate how future excavations can benefit from genetic approaches to study excavated bones; in particular, in assemblages rich in bones that are challenging to identify morphologically, such as bones from fish, amphibians, micromammals, and small birds.
Materials and Methods
Sampling, Extraction, and Metabarcoding.
Samples were obtained from the collections at Canterbury Museum, University of Otago (Department of Zoology and Department of Anthropology and Archaeology), and the Museum of New Zealand Te Papa Tongarewa (SI Appendix, Table S1). In total, fragmented bones from 14 paleontological, 21 archaeological, and 2 mixed (paleontological and archaeological) sites were subsampled from larger collections of unidentified bones. In addition, previously published BBM data from the paleontological site Finsch’s Folly (43) were included in the analysis to increase the sample size. As the earliest samples were excavated in the 1950s (others were excavated in the last decade), the excavation process and level of detail on stratigraphic information of the deposits vary. Hence, for this study, we do not distinguish between different stratigraphical units within one site, but consider bones from one site as one entity, and the age of those bones as the age range of that bone deposit (SI Appendix, Table S1).
Pre-PCR processing was undertaken at the TRACE (Trace Research Advanced Clean Environment) aDNA facility at Curtin University, Western Australia, following strict aDNA guidelines (44) and including at least two controls for each batch of sample preparations (SI Appendix, Supplementary Information Text and Tables S7 and S8). Bone fragments were subsampled (SI Appendix, Section 4.1) and DNA was extracted using a modified version of the extraction protocol described by Dabney et al. (45). DNA metabarcoding was carried out using four metabarcoding assays (SI Appendix, Table S2), in which barcode regions of several mitochondrial genes (12S and 16S rRNA gene) are amplified with primers targeting vertebrates (12SV5), mammals (Mam16S), fish (Fish16S), and birds (12SAH).
Sequence Analysis.
Sequences were filtered using a custom-made pipeline based on OBItools (46) (https://pythonhosted.org/OBITools/welcome.html#installing-the-obitools; SI Appendix, Section 4.3 and Table S10), and taxonomic assignment of the reads was achieved using a modified version of the getLCA approach described in Seersholm et al. (31) (https://github.com/frederikseersholm/getLCA). Lastly, after raw taxonomic assignments, each taxonomic node was examined and correlated to records of species present in New Zealand, as in Murray et al. (23) (SI Appendix, Section 4.4). Raw data from the taxonomic assignment, along with information on reassigned nodes, are available in Dataset S1.
Statistical Analyses.
Correspondence analyses were performed in R, using the vegan package (https://cran.r-project.org/web/packages/vegan/index.html), based on presence absence data. “Fingerprints” to illustrate distributions of specific subgroups of species were plotted using ordiellipse (vegan) with incremental confidence intervals of 0.4, 0.6, and 0.8 (Fig. 1C) or a single confidence interval of 0.6 (SI Appendix, Fig. S1). Coordination analysis was used to visualize the relationship between latitude and fish assemblage clustering, using ordisurf (vegan) with the maximum likelihood method to estimate the smoothing parameter. To identify significant environmental variables, permutation tests were carried out on ordinations constrained by each variable, using Permutation Test for Constrained Correspondence Analysis (anova.cca) with 999 permutations.
Kākāpō DNA Amplification and Haplotype Analysis.
To assess kākāpō genetic diversity, we amplified a 214-bp region of the kākāpō mitochondrial d-loop (control region), using primers from Bergner et al. (41) (SI Appendix, Table S2). Using this assay, we successfully amplified DNA from 13 of the 17 DNA extracts in which kākāpō was identified previously with metabarcoding. To filter out spurious reads and PCR errors, we only considered reads present in all four replicates at an abundance of more than 1.29% of the most abundant read to be true haplotypes [note that this threshold was calculated from eight separate amplifications of DNA extracted from a single kākāpō bone (47)]. To visualize the relationships between haplotypes, a minimum spanning haplotype network was generated with the R package pegas (ape-package.ird.fr/pegas.html), including modern (n = 50) and historic (n = 46) data published by Bergner et al. (41).
Supplementary Material
Acknowledgments
We thank Shar Briden and Jamie Wood for sample excavation and James Taylor for technical assistance. This study would not have been possible without many important contributions by the late Chris Jacomb on the archaeology of New Zealand. This study was supported by the Australian Research Council Discovery Project DP160104473; Forrest Research Foundation (to F.V.S.), Royal Society of New Zealand Marsden Fund (N.J.R. and L.J.E.), and a Royal Society of New Zealand Rutherford Discovery Fellowship (L.S. and M.K.). Data analysis was carried out with help from the Pawsey Supercomputing Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequencing data reported in this paper are available from Data Dryad (doi.org/10.5061/dryad.ss0k19q) and GenBank (accession nos MH395815–MH395838).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803573115/-/DCSupplemental.
References
- 1.Tennyson AJD. The origin and history of New Zealand’s terrestrial vertebrates. N Z J Ecol. 2010;34:6–27. [Google Scholar]
- 2.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc Natl Acad Sci USA. 2008;105:7676–7680. doi: 10.1073/pnas.0801507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson A. Changing perspectives upon Māori colonisation voyaging. J R Soc N Z. 2017;47:222–231. [Google Scholar]
- 4.Duncan RP, Blackburn TM. Extinction and endemism in New Zealand land birds. Glob Ecol Biogeogr. 2004;13:509–517. [Google Scholar]
- 5.Worthy TH, Holdaway RN. The Lost World of the Moa: Prehistoric Life of New Zealand. Indiana Univ Press; Bloomington, IN: 2002. [Google Scholar]
- 6.Towns DR, Daugherty CH. Patterns of range contractions and extinctions in the New Zealand herpetofauna following human colonisation. N Z J Zool. 1994;21:325–339. [Google Scholar]
- 7.Tennyson A, Martinson P. Extinct Birds of New Zealand. Te Papa Press; Wellington: 2006. [Google Scholar]
- 8.Lorenzen ED, et al. Species-specific responses of late Quaternary megafauna to climate and humans. Nature. 2011;479:359–364. doi: 10.1038/nature10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saltré F, et al. Climate change not to blame for late Quaternary megafauna extinctions in Australia. Nat Commun. 2016;7:10511. doi: 10.1038/ncomms10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart AJ. Late Quaternary megafaunal extinctions on the continents: A short review. Geol J. 2015;50:338–363. [Google Scholar]
- 11.Roberts RG, et al. New ages for the last Australian megafauna: Continent-wide extinction about 46,000 years ago. Science. 2001;292:1888–1892, and erratum (2001) 293:49b. doi: 10.1126/science.1060264. [DOI] [PubMed] [Google Scholar]
- 12.Evans AM, Flatman JC, Flemming NC. Prehistoric Archaeology on the Continental Shelf: A Global Review. Springer; New York: 2014. [Google Scholar]
- 13.Wanner H, et al. Mid- to late Holocene climate change: An overview. Quat Sci Rev. 2008;27:1791–1828. [Google Scholar]
- 14.Holdaway RN, et al. An extremely low-density human population exterminated New Zealand moa. Nat Commun. 2014;5:5436. doi: 10.1038/ncomms6436. [DOI] [PubMed] [Google Scholar]
- 15.Perry GLW, Wheeler AB, Wood JR, Wilmshurst JM. A high-precision chronology for the rapid extinction of New Zealand moa (Aves, Dinornithiformes) Quat Sci Rev. 2014;105:126–135. [Google Scholar]
- 16.Nagaoka L. Declining foraging efficiency and moa carcass exploitation in southern New Zealand. J Archaeol Sci. 2005;32:1328–1338. [Google Scholar]
- 17.Anderson A. Mechanics of overkill in the extinction of New Zealand moas. J Archaeol Sci. 1989;16:137–151. [Google Scholar]
- 18.Collins CJ, et al. Extinction and recolonization of coastal megafauna following human arrival in New Zealand. Proc Biol Sci. 2014;281:20140097. doi: 10.1098/rspb.2014.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boessenkool S, et al. Relict or colonizer? Extinction and range expansion of penguins in southern New Zealand. Proc Biol Sci. 2009;276:815–821. doi: 10.1098/rspb.2008.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunce M, et al. Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature. 2003;425:172–175. doi: 10.1038/nature01871. [DOI] [PubMed] [Google Scholar]
- 21.Easton LJ, et al. Testing species limits of New Zealand’s leiopelmatid frogs through morphometric analyses. Zool J Linn Soc. 2017;183:431–444. [Google Scholar]
- 22.Lyman RL. Vertebrate Taphonomy. Cambridge Univ Press; Cambridge: 1994. [Google Scholar]
- 23.Murray DC, et al. Scrapheap challenge: A novel bulk-bone metabarcoding method to investigate ancient DNA in faunal assemblages. Sci Rep. 2013;3:3371. doi: 10.1038/srep03371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith I. Pre-European Maori exploitation of marine resources in two New Zealand case study areas : Species range and temporal change. J R Soc N Z. 2011;43:1–37. [Google Scholar]
- 25.Waters JM, Fraser CI, Maxwell JJ, Rawlence NJ. Did interaction between human pressure and Little Ice Age drive biological turnover in New Zealand? J Biogeogr. 2017;44:1481–1490. [Google Scholar]
- 26.Salis AT, et al. Myth or relict: Does ancient DNA detect the enigmatic Upland seal? Mol Phylogenet Evol. 2016;97:101–106. doi: 10.1016/j.ympev.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Smith IWG. Maori impact on the marine megafauna: Pre-European distributions of New Zealand sea mammals. In: Sutton DG, editor. Saying So Doesn’t Make It So: Papers in Honour of B Foss Leach. New Zealand Archaeological Association; Dunedin, New Zealand: 1989. pp. 76–108. [Google Scholar]
- 28.Brabyn MW. 1991. An analysis of the New Zealand whale stranding record (Department of Conservation, Wellington, New Zealand)
- 29.Szabo VE. Monstrous Fishes and the Mead-Dark Sea. BRILL; Leiden, The Netherlands: 2008. [Google Scholar]
- 30.Cunliffe EA, Brooks E. Prehistoric whale bone technology in southern New Zealand. Int J Osteoarchaeol. 2016;26:384–396. [Google Scholar]
- 31.Seersholm FV, et al. DNA evidence of bowhead whale exploitation by Greenlandic Paleo-Inuit 4000 years ago. Nat Commun. 2016;7:13389. doi: 10.1038/ncomms13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson J. The Prehistory of New Zealand. Longman Paul Limited; Auckland: 1984. [Google Scholar]
- 33.Trotter MM. Birds of a Feather: Osteological and Archaeological Papers from the South Pacific in Honour of R. J. Scarlett. N Z Archaeol Assoc; Dunedin, New Zealand: 1979. Tai Rua: A Moa-hunter site in North Otago; pp. 203–230. [Google Scholar]
- 34.Trotter MM. Archaeological investigations at Redcliffs, Canterbury, New Zealand. Rec Cantebury Mus. 1975;9:198–220. [Google Scholar]
- 35.Smith IWG. 1985. Sea mammal hunting and prehistoric subsistence in New Zealand. DPhil Dissertation (University of Otago, Dunedin, New Zealand)
- 36.Leach BF, Boocock AS. Prehistoric fish catches in New Zealand. In: Davison DP, editor. British Archaeological Reports. BAR Publishing; Oxford: 1992. [Google Scholar]
- 37.Leach BF. Fishing in pre-european New Zealand. Int J Archaeozoology. 2006;15:9–14. [Google Scholar]
- 38.Marshall Y. Maori mass capture of freshwater Eels : An ethnoarchaeological reconstruction of prehistoric subsistence and social behaviour. N Z J Archaeol. 1987;9:55–79. [Google Scholar]
- 39.Lentini PE, Stirnemann IA, Stojanovic D, Worthy TH, Stein JA. Using fossil records to inform reintroduction of the kakapo as a refugee species. Biol Conserv. 2018;217:157–165. [Google Scholar]
- 40.Dussex N, Rawlence NJ, Robertson BC. Ancient and contemporary DNA reveal a pre-human decline but no population bottleneck associated with recent human persecution in the kea (Nestor notabilis) PLoS One. 2015;10:e0118522. doi: 10.1371/journal.pone.0118522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergner LM, Dussex N, Jamieson IG, Robertson BC. European colonization, not Polynesian arrival, impacted population size and genetic diversity in the critically endangered New Zealand kākāpō. J Hered. 2016;107:593–602. doi: 10.1093/jhered/esw065. [DOI] [PubMed] [Google Scholar]
- 42.Dussex N, VonSeth J, Robertson BC, Dalén L. Full mitogenomes in the critically endangered kākāpō reveal major post-glacial and anthropogenic effects on genetic diversity. Genes (Basel) 2018;9:220. doi: 10.3390/genes9040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grealy AC, et al. A critical evaluation of how ancient DNA bulk bone metabarcoding complements traditional morphological analysis of fossil assemblages. Quat Sci Rev. 2015;128:37–47. [Google Scholar]
- 44.Willerslev E, Cooper A. Ancient DNA. Proc Biol Sci. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dabney J, et al. Complete mitochondrial genome sequence of a middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyer F, et al. obitools: A unix-inspired software package for DNA metabarcoding. Mol Ecol Resour. 2016;16:176–182. doi: 10.1111/1755-0998.12428. [DOI] [PubMed] [Google Scholar]
- 47.Stat M, et al. Ecosystem biomonitoring with eDNA: Metabarcoding across the tree of life in a tropical marine environment. Sci Rep. 2017;7:12240. doi: 10.1038/s41598-017-12501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.