Significance
Inflammation is a protective response of the body’s immune system against harmful stimuli such as pathogenic microorganisms, toxins, or damaged cells. However, if excessive or prolonged, inflammation may be harmful and therefore has to be regulated. Soluble CD52 is a natural sialoglycopeptide and immune regulator that suppresses inflammatory responses. We elucidated the mechanism of this effect by showing that soluble CD52 first sequesters a mediator of inflammation called HMGB1; in turn, this promotes binding of the sialylated CD52 glycan to an inhibitory receptor, sialic acid-binding immunoglobulin-like lectin (Siglec)-10, present on activated T cells and other immune cells. This concerted antiinflammatory mechanism driven by soluble CD52 may contribute to immune-inflammatory homeostasis and underscores the therapeutic potential of soluble CD52.
Keywords: CD52, HMGB1, Siglec-10, sialoglycan, T cell
Abstract
CD52, a glycophosphatidylinositol (GPI)-anchored glycoprotein, is released in a soluble form following T cell activation and binds to the Siglec (sialic acid-binding Ig-like lectin)-10 receptor on T cells to suppress their function. We show that binding of CD52-Fc to Siglec-10 and T cell suppression requires the damage-associated molecular pattern (DAMP) protein, high-mobility group box 1 (HMGB1). CD52-Fc bound specifically to the proinflammatory Box B domain of HMGB1, and this in turn promoted binding of the CD52 N-linked glycan, in α-2,3 sialic acid linkage with galactose, to Siglec-10. Suppression of T cell function was blocked by anti-HMGB1 antibody or the antiinflammatory Box A domain of HMGB1. CD52-Fc induced tyrosine phosphorylation of Siglec-10 and was recovered from T cells complexed with HMGB1 and Siglec-10 in association with SHP1 phosphatase and the T cell receptor (TCR). Thus, soluble CD52 exerts a concerted immunosuppressive effect by first sequestering HMGB1 to nullify its proinflammatory Box B, followed by binding to the inhibitory Siglec-10 receptor, triggering recruitment of SHP1 to the intracellular immunoreceptor tyrosine-based inhibitory motif of Siglec-10 and its interaction with the TCR. This mechanism may contribute to immune-inflammatory homeostasis in pathophysiologic states and underscores the potential of soluble CD52 as a therapeutic agent.
CD52 is a GPI-anchored glycoprotein, expressed in the hematopoietic system on B and T cells, monocytes, macrophages, eosinophils, natural killer cells, and dendritic cells (1, 2) and in the male reproductive tract by epithelial cells of the epididymis from which it is released into the seminal fluid to be taken up by sperm (3, 4). The mature human CD52 protein, comprising just 12 amino acids, acts as a scaffold for the posttranslational addition of a GPI anchor and an N-glycan (1, 2). CD52 was identified originally as the target of the lymphocyte-depleting rat monoclonal antibody, CAMPATH (1, 2), and subsequently humanized as alemtuzumab. We reported that CD4+ T cells with high expression of CD52 have suppressor activity upon activation, which is mediated by the release of soluble CD52, and that sialic acid-binding immunoglobulin-like lectin (Siglec-10) on T cells is a receptor for soluble CD52 (5). Ligation of Siglec-10 by soluble CD52 led to a decrease in phosphorylation of the T cell receptor (TCR)-associated tyrosine kinases Lck and ZAP-70 and suppression of T cell function (5). The Siglecs function as inhibitory receptors, in some cases via cytoplasmic immunoreceptor tyrosine-based inhibitory motifs (ITIMs) that suppress phosphorylation-based signaling. Following their phosphorylation by Src family kinases, ITIMs recruit Src homology 2 (SH2) domain-containing protein tyrosine phosphatases (SHP1 or SHP2), and in some cases the inositol phosphatase SHIP (6, 7). For Siglec-7 and Siglec-9, the association of their ITIMs with SHP1 leads to coupled dephosphorylation of the TCR (8). More recently, we reported that soluble CD52 (CD52-Fc) suppresses NF-κB–mediated signaling in innate immune monocytes, macrophages, and dendritic cells and at high concentrations depletes the short-lived prosurvival protein Mcl-1, activating intrinsic apoptotic cell death (9).
Several lines of evidence support a key role for CD52 in immune homeostasis. In a T cell transfer model, depletion of CD52high cells markedly accelerated the onset of autoimmune diabetes (5). Although a phenotype had not been identified in CD52 knockout mice (10), we found that cytokine and hypothermic responses to injection of low-dose lipopolysaccharide (LPS) were significantly increased in our CD52 gene knockout mice (9). Moreover, in wild-type mice, injection of CD52-Fc suppressed responses to LPS (9). The therapeutic application of alemtuzumab in chronic lymphocytic leukemia, T cell lymphoma, and autoimmune diseases such as multiple sclerosis has been based on its ability to trigger complement-mediated lysis and deplete lymphocytes (2). However, because it would preferentially target CD52high CD4+ T cells and bind soluble CD52, alemtuzumab should also enhance T cell immunity. This may be beneficial in cancer and may explain why autoimmune diseases appear de novo in at least a third of patients with multiple sclerosis treated with alemtuzumab (11). Finally, the immune regulatory function of CD52 is likely to extend to the reproductive tract, where the presence of soluble CD52 in semen (3, 4) may account for the immune suppression that prevents rejection of the sperm allograft by the female host.
Another GPI-anchored protein, CD24, with a gene organization structure resembling that of CD52 (12), was reported to bind Siglec-10 by first associating with damage-associated molecular pattern (DAMP) proteins, including high mobility group box 1 (HMGB1) (13). HMGB1 is a nonhistone nuclear DNA binding protein with transcriptional regulatory properties now recognized as a DAMP that drives a range of immune-inflammatory states, including sepsis (14, 15). The key role of HMGB1 in sepsis was confirmed in HMGB1 knockout mice (16) and by treatment with blocking antibodies to HMGB1 (14, 15). HMGB1 comprises two domains, the antiinflammatory Box A and proinflammatory Box B domains, and a C-terminal acidic tail (14, 15). It is passively released from cells undergoing necrosis and pyroptosis, and actively secreted by monocytes and macrophages. HMGB1 is normally present in the circulation (17) and increases with inflammation (14, 15), but how its complex functions are regulated to maintain immune-inflammatory homeostasis is not fully understood. We observed that CD52-Fc failed to suppress T cells in serum-free medium and queried whether this could be explained by a requirement for DAMPs present in serum. This led us to discover that soluble CD52 sequesters HMGB1 to abrogate its proinflammatory function, thereby promoting binding of CD52 to the inhibitory Siglec-10 receptor.
Results
HMGB1 Box B Is Required for T Cell Suppression by Soluble CD52.
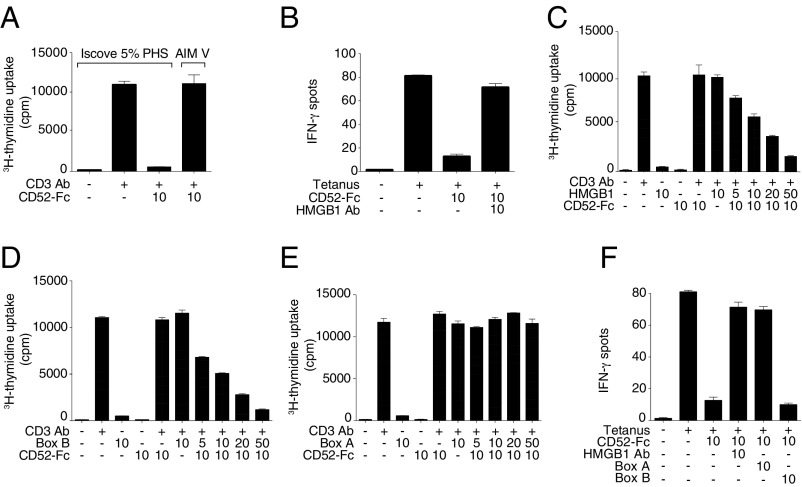
Initial studies showing suppression by CD52-Fc in vitro (5) were performed in serum-containing medium. Subsequently, we observed that CD52-Fc had no suppressive effect in serum-free medium (Fig. 1A). Furthermore, addition of antibody to HMGB1 reversed the suppressive effect of CD52-Fc in serum-containing medium (Fig. 1B), and addition of HMGB1 to serum-free medium restored the suppressive effect of CD52-Fc in a concentration-dependent manner (Fig. 1C). The disulfide form of HMGB1 used in these studies was biologically active in stimulating cytokine secretion from either PBMCs or the human THP-1 monocyte cell line, but inactive after disulfide reduction or as the 3S form in which cysteines are replaced by serines (SI Appendix, Fig. S1 A and B). This effect of HMGB1 was recapitulated by the Box B domain of HMGB1 (Fig. 1D) but not by the Box A domain (Fig. 1E). In fact, like antibody to HMGB1, Box A blocked suppression by CD52-Fc in serum-containing medium (Fig. 1F). Thus, we concluded that the proinflammatory Box B of HMGB1 is necessary for suppression by CD52-Fc, whereas the antiinflammatory Box A blocks this effect.
Fig. 1.
T cell suppression by soluble CD52 requires HMGB1 Box B and is antagonized by HMGB1 Box A. (A) Proliferation of human PBMCs incubated in serum-containing IP5 medium (Iscove’s modified Dulbecco’s medium with 5% PHS) or serum-free medium (AIM V) for 48 h alone or with plate-bound anti-CD3 antibody (1 μg/mL) and CD52-Fc. (B) IFN-γ production measured by ELISpot from human PBMCs (1 × 105) incubated in serum-containing medium with no antigen or tetanus toxoid (10 Lfu/mL) in the presence of CD52-Fc and neutralizing antibody to HMGB1. (C) Proliferation of human PBMCs incubated in serum-free medium alone or with plate-bound anti-CD3 antibody (1 μg/mL), CD52-Fc and with increasing concentrations of disulfide-HMGB1, or HMGB1 Box B (D), or HMGB1 Box A (E). Proliferation was measured as [3H] thymidine uptake. (F) IFN-γ production measured by ELISpot from human PBMCs (1 × 105) incubated in serum-containing medium in the presence of HMGB1 Box A or HMGB1 Box B recombinant proteins and neutralizing antibody to HMGB1. Concentrations of recombinant proteins in every figure are in μg/mL. Serum-free medium was used in experiments in A, C, D, and E.
HMGB1 Box B Promotes Binding of Soluble CD52 to Siglec-10.
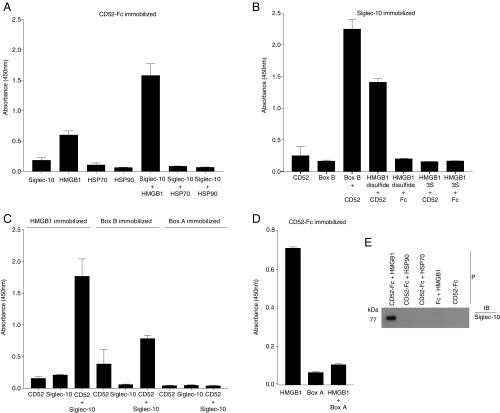
Solid phase assays in microtiter plates showed that soluble Siglec-10-Fc bound to plate-coated CD52-Fc only in the presence of HMGB1 (Fig. 2A). This was corroborated with other coating combinations, in which HMGB1 (disulfide, but not 3S form) or HMGB1 Box B, but not Box A, was shown to bind CD52-Fc and promote its binding to Siglec-10-Fc (Fig. 2 B–E). Moreover, Box A blocked the binding of HMGB1 to CD52-Fc (Fig. 2D). The effect of HMGB1 to promote binding of CD52-Fc to Siglec-10-Fc was not observed with the other DAMPs, HSP70 and HSP90 (Fig. 2A). This was also demonstrated by solution phase binding and precipitation (Fig. 2E).
Fig. 2.
HMGB1 promotes binding of soluble CD52-Fc to Siglec-10. Binding of CD52-Fc to recombinant HMGB1 and Siglec-10-Fc is shown. Proteins immobilized in flat bottom 96-well microtiter plate wells were as follows: CD52-Fc (at 10 μg/mL) (A and D), Siglec-10-Fc (at 10 μg/mL) (B), HMGB1, HMGB1 Box B, and HMGB1 Box A (each at 20 μg/mL) (C). After blocking the plate with binding buffer, different proteins as indicated were added (50 μL per well, 10 μg/mL) to triplicate wells, and binding was detected as described in SI Appendix, SI Materials and Methods. Results are mean ± SD of three separate experiments. (E) CD52-Fc or Fc (each 20 μg/mL) and Siglec-10-Fc (10 μg/mL) were incubated separately or together with HMGB1 or HSPs 70 or 90 (each 20 μg/mL) overnight at 4 °C. CD52-Fc (or Fc) was then precipitated (P) with Strep-Tactin beads and the pellet fractionated by SDS/PAGE and immunoblotted (IB) with anti-Siglec-10 antibody.
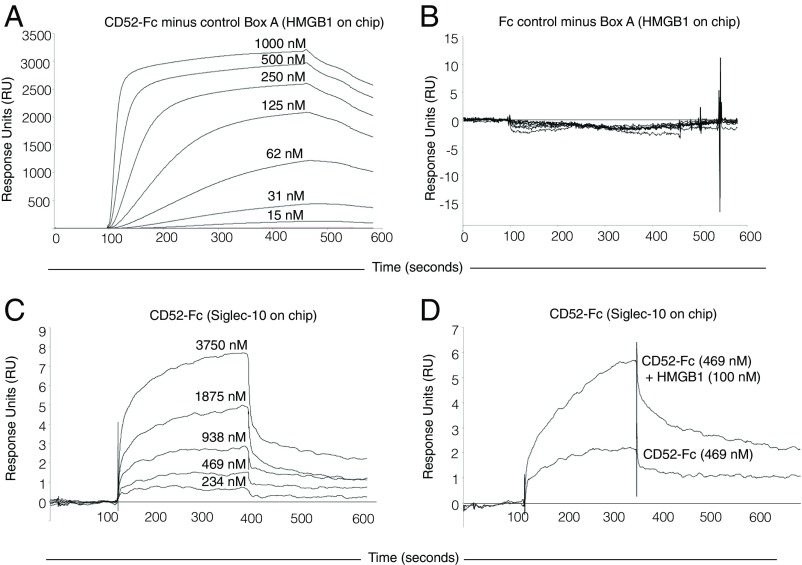
Binding of CD52-Fc to HMGB1 and Siglec-10 was then quantified by surface plasmon resonance (SPR) (Fig. 3). In steady-state binding experiments, CD52-Fc bound to immobilized HMGB1 in a concentration-dependent manner, with an estimated Kd of 130 nM (Fig. 3A). The control Fc did not bind HMGB1 (Fig. 3B). CD52-Fc bound to Siglec-10-Fc with an estimated Kd of 565 nM (Fig. 3C). This direct binding of CD52-Fc to immobilized Siglec-10-Fc, not seen with plate binding, may be due to greater sensitivity of the SPR sensor chip, but in addition the coating concentration of Siglec-10-Fc for the SPR chip (25 µg/mL) was higher than for the microtiter plate (10 µg/mL). Notably, after addition of HMGB1 the binding of CD52-Fc to Siglec-10 increased (Fig. 3D), confirming the plate assay results.
Fig. 3.
Surface plasmon resonance quantifies binding of CD52-Fc to HMGB1. Binding of CD52-Fc (A) or Fc control protein to HMGB1 (B), CD52-Fc to Siglec-10-Fc (C), and CD52-Fc with or without HMGB1 to Siglec-10-Fc (D). HMGB1 (100 µg/mL) or Siglec-10-Fc (25 µg/mL) were immobilized on CM5 dextran sensor chips. As a control for nonspecific binding, recombinant Box A was used in A and B and recombinant Fc in C and D. Data are representative of three experiments. Further details are provided in SI Appendix, SI Materials and Methods.
Sialylated CD52-Fc Glycan, in α-2,3 Linkage with Galactose, Is Required for Binding to HMGB1 and Suppressor Function.
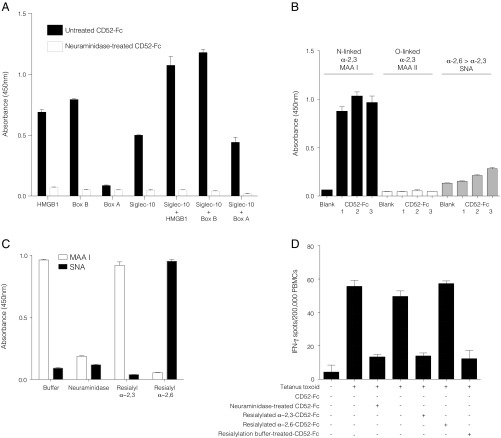
The terminal sialic acids on the N-linked glycan of CD52-Fc are required for T cell suppression, consistent with binding to Siglec receptors (5). Having now shown that CD52-Fc first binds to HMGB1, we investigated the requirement of sialic acid for this interaction. Treatment of CD52-Fc with neuraminidase to cleave its sialic acids abolished its binding to HMGB1 or Box B (Fig. 4A).
Fig. 4.
CD52-Fc glycan sialic acid is required for binding to HMGB1 and Siglec-10. (A) Binding of CD52-Fc to HMGB1, Box A, Box B, and Siglec-10 pretreated with Clostridium perfringens type V neuraminidase or with vehicle alone to plate-bound proteins as indicated. (B) Binding of untreated CD52-Fc to lectins (MAA I, MAA II, and SNA) immobilized on a microtiter plate, showing that CD52-Fc binds MAA I, a lectin that recognizes sialic acid in α-2,3 linkage with galactose. (C) Resialylation of neuraminidase-treated CD52-Fc with either α-2,3 (CstII) or α-2,6 (PdST6Ga1I) sialyltransferases, confirmed by binding to MAA I lectin (α-2,3 linkage) or SNA lectin (α-2,6 linkage). (D) ELISpot assay demonstrating functional activity of CD52-Fc reconstituted with sialic acid in α-2,3 linkage with galactose. Data are mean ± SEM of three independent experiments.
Siglecs exhibit specificity for the different linkages between terminal sialic acid and the underlying galactose of interacting glycans, usually either in α-2,3 or α-2,6 positions (18). To determine the nature of this linkage in CD52, we first measured the binding of CD52-Fc to lectins that recognize specific sialyl-galactose linkages (19). CD52-Fc bound strongly to Maackia amurensis lectin I (MAAI) (Fig. 4B), which recognizes N-linked glycans containing terminal sialic acid in α-2,3 linkage (Siaα2–3Galβ1). Binding was not observed to MAAII, which recognizes O-linked glycans with terminal sialic acids in α-2,3 linkage or to Sambucus nigra agglutinin (SNA), which recognizes sialic acids in α-2,6 linkage. The requirement of the α-2,3 sialic acid conformation for the function of CD52-Fc was confirmed by desialylation of CD52-Fc with neuraminidase followed by resialylation with specific sialyltransferases (Fig. 4C); the sialyltransferase that adds the α-2,3 linkage restored full suppressive activity (Fig. 4D). Similarly, HMGB1 at increasing concentrations bound to CD52-Fc after α-2,3 resialylation, but at higher concentrations also bound to a lesser extent to α-2,6 resialylated CD52-Fc; Siglec-10, however, exhibited absolute specificity for α-2,3 resialylated CD52-Fc (SI Appendix, Fig. S2 A and B).
CD52-Fc Induces Tyrosine Phosphorylation of Siglec-10.
Most Siglecs have ITIMs, which in the case of Siglec-7 and Siglec-9 negatively regulate TCR signaling (8). The cytoplasmic tail of Siglec-10 contains four tyrosines, two of which (Y597 and Y667) are embedded in ITIM-signaling motifs and likely to be involved in intracellular signaling (20). As there are no reports of Siglec-10 phosphorylation, we determined whether CD52-Fc induced tyrosine phosphorylation of Siglec-10. In human PBMCs, in which T cells were activated through the TCR with anti-CD3/CD28 antibody beads, we observed that CD52-Fc rapidly induced phosphorylation of Siglec-10 (SI Appendix, Fig. S3 A and B).
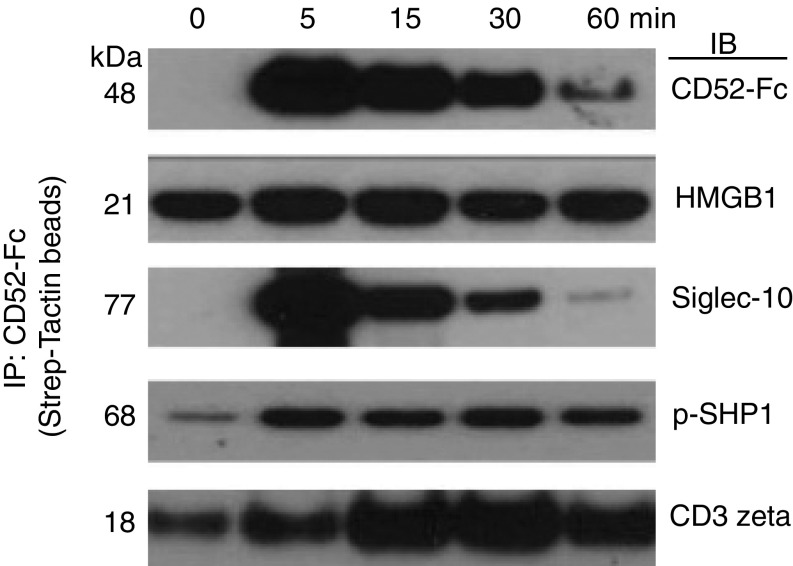
The Trimolecular Complex of CD52-Fc–HMGB1–Siglec-10 Interacts with the T Cell Receptor.
To identify CD52-Fc interacting elements at the cellular level, flow-sorted CD3+CD4+ T cells were activated for different times by anti-CD3/CD28 antibodies in the presence of CD52-Fc. Cells were lysed in digitonin buffer, and CD52-Fc was precipitated with Strep-Tactin beads, fractionated by SDS/PAGE under reducing conditions, and immunoblotted with antibodies to CD52, HMGB1, Siglec-10, p-SHP1, or TCR CD3 zeta. This demonstrated that CD52-Fc could be recovered from cells in association with HMGB1, Siglec-10, p-SHP1, and the TCR (Fig. 5). The association with HMGB1, Siglec-10, and p-SHP1 was maximal within 5 min, followed by association with TCR CD3 zeta. HMGB1 was associated with CD52-Fc from baseline, most likely because HMGB1 is present in serum in the medium.
Fig. 5.
CD52–HMGB1–Siglec-10 trimolecular complex interacts with the T cell receptor. Flow-sorted CD3+CD4+ T cells were incubated with anti-CD3/CD28 antibody beads for the indicated times in the presence of CD52-Fc (40 µg/mL) at 37 °C. The reaction was stopped with ice-cold PBS containing protease and phosphatase inhibitors. Cells were washed in cold PBS with inhibitors and solubilized in 1% digitonin buffer. CD52-Fc was precipitated (IP) with Strep-Tactin beads, fractionated in SDS/PAGE under reducing conditions, and immunoblotted (IB) with antibodies to CD52, HMGB1, Siglec-10, p-SHP1, or TCR CD3 zeta. Coomassie Brilliant Blue protein staining was similar in all lanes.
Discussion
We show that binding of the proinflammatory DAMP protein HMGB1 by soluble CD52 (CD52-Fc) promotes ligation of soluble CD52 with the Siglec-10 receptor and suppression of T cell function. CD52-Fc bound specifically to the proinflammatory Box B of HMGB1, not the Box A; however, Box A inhibited binding of HMGB1 to CD52-Fc and suppression by CD52-Fc. We found no evidence that HMGB1 interacts directly with Siglec-10, and in the absence of HMGB1 there was little or no T cell suppression by CD52-Fc. Binding of HMGB1 to CD52-Fc occurred at a high affinity (Kd 130 nM), which is in the concentration range of free HMGB1 detected in the circulation in the absence of masking proteins (17). CD52-Fc bound only HMGB1 and not other DAMPs, HSPs 70 and 90, which along with HMGB1 were reported to bind the GPI-linked CD24 glycoprotein that also interacts with Siglec-10 (13). Binding to HMGB1 and Siglec-10 required terminal sialylation of the CD52 N-glycan in an α-2,3 linkage to galactose. In activated T cells, Siglec-10 was phosphorylated by CD52-Fc in the presence of HMGB1, followed by its association with SHP1 phosphatase and the TCR. These findings identify a critical role for HMGB1 in the suppressor function of soluble CD52 mediated by Siglec-10. Similar to Siglec-7 and Siglec-9 (8), the interaction of Siglec-10 with SHP1 and the TCR following binding of CD52-Fc–HMGB1 can account for the effect of CD52-Fc to decrease phosphorylation of TCR-associated tyrosine kinases (5).
HMGB1 exists as fully reduced, disulfide and sulfonyl isoforms (14, 15, 21). The cysteine in position 106 in the Box B is indispensable for its activity and is abolished by oxidation or mutation of this amino acid. The “disulfide” form of HMGB1 used in the present study, the active, proinflammatory form that was bound by CD52-Fc, has a disulfide bridge between cysteine residues 23 and 45, but the cysteine residue at position 106 remains reduced. Extracellular HMGB1 has been shown to bind several cell membrane signaling molecules including Toll-like receptors (TLRs) 2, 4, and 9 (22) and the receptor for advanced glycation end products (RAGE) and Mac-1 (23), but the structural basis and physiological significance of these interactions are unknown. However, it is clear that the proinflammatory activity of HMGB1 and its Box B is redox state-dependent and is antagonized by its Box A (14, 15). Here, we observed antagonism between the Boxes B and A in regard to binding to CD52-Fc and T cell suppression by CD52-Fc. As well as suppressing T cell signaling and function, we recently reported that soluble CD52 also suppresses innate immune responses in vitro and in vivo, including to HMGB1 itself (9), also shown here. In rodent models, sepsis is mediated in part by the cellular release of HMGB1 and is ameliorated by injection of blocking antibodies to HMGB1 (14, 15). In light of our present findings, we suggest that the protective effect of CD52-Fc that we observed in the LPS sepsis model is explained first by sequestration of HMGB1 by CD52. The role of soluble CD52 as a regulator of immune inflammation therefore encompasses both innate and adaptive immunity.
Apart from CD52, other soluble regulators are known to bind HMGB1 and subvert its inflammatory actions. Haptoglobin, an acute phase protein that captures extracellular hemoglobin, can form complexes with HMGB1 that disrupt the proinflammatory macrophage scavenger receptor CD163 (24). C1q, the first component of the complement pathway, was shown to bind HMGB1 to then form higher order complexes with cell surface RAGE and LAIR-1 and shift the differentiation of monocytes toward antiinflammatory M2-like macrophages (25) In fact, analogous to the mechanism of action of CD52–HMGB1 complexes to bind and signal through Siglec-10, C1q–HMGB1 complexes bind to LAIR-1 to augment phosphorylation of cytoplasmic ITIMs and recruitment of SHP-1 (25). In addition, the detection of circulating HMGB1 was shown to be masked by Ig (17), consistent with a mechanism whereby Ig binds extracellular HMGB1 to buffer its proinflammatory effects.
Lectin binding and sialoside reconstitution studies revealed that α-2,3 sialic acid is the major sialic acid-galactose linkage in recombinant CD52-Fc, responsible for its binding to Siglec-10 and bioactivity. The α-2,6 form was a minor component of CD52-Fc and did not bind to Siglec-10. However, while CD52-Fc preferentially bound HMGB1 via α-2,3-linked sialic acid, binding was also observed with reconstituted α-2,6–linked sialic acid, and it is possible that this form is present in native CD52 (26). A caveat of our study is that recombinant CD52-Fc produced in HEK 293 cells may not necessarily mimic native CD52 produced by a variety of cell types in vivo. Furthermore, how a complex sialylated glycan on the short CD52 peptide recognizes both HMGB1 and Siglec-10 remains to be determined. While the N-linked sialylated glycan is required for Siglec-10 binding and bioactivity (5), the CD52 peptide contains several serine and threonine sites for potential O-linked glycosylation, the function of which has not been investigated.
Previously, we found that CD52-Fc binds Siglec-10 and suppresses activation of CD4+ T cells (5). Here, we demonstrate that this occurs by a concerted mechanism whereby CD52-Fc first sequesters and inhibits bioactive disulfide HMGB1, which promotes binding of CD52-Fc to Siglec-10. It should be noted however that Siglec-10, like Siglec-7 and Siglec-9, is also expressed by a range of myeloid cells (6, 7). Therefore, in a mixed cell population such as PBMCs, CD52-Fc could also suppress T cells indirectly via an effect on antigen-presenting cells, even when T cells are directly activated with anti-CD3 antibody. Our findings that Siglec-10 in primary human cells is phosphorylated by ligation with CD52-Fc, recruits SHP1, and interacts with the TCR to suppress T cell signaling mirror those reported for Siglec-7 and Siglec-9 transfected into the Jurkat T cell line (8) and provide a structural basis for understanding how Siglec-10 suppresses T cell signaling.
Materials and Methods
Complete details of reagents and blood donors and methods for cell purification and culture, cell proliferation and ELISpot assays, analysis of CD52-Fc and HMGB1 binding to Siglec-10, lectin binding, desialylation and resialylation of recombinant CD52-Fc, immunoprecipitation, and immunoblotting are provided in SI Appendix, SI Materials and Methods. Deidentified venous blood was obtained from healthy young adult donors, with informed consent, through the Volunteer Blood Donor Registry of The Walter and Eliza Hall Institute of Medical Research (WEHI), following approval by WEHI and Melbourne Health Human Ethics Committees.
Supplementary Material
Acknowledgments
We thank Dr. Ajit Varki for helpful discussions. This work was supported by Australian National Health and Medical Research Council (NHMRC) Program Grant 1037321 and the Walter and Eliza Hall Institute Catalyst Fund. L.C.H. is the recipient of a NHMRC Senior Principal Research Fellowship 1080887. We acknowledge TrendBio for access to the automated, quantitative Western blot analysis instrument (WES) system. The work was made possible through Victorian State Government Operational Infrastructure Support and NHMRC Research Institute Infrastructure Support Scheme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.J.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722056115/-/DCSupplemental.
References
- 1.Xia MQ, Tone M, Packman L, Hale G, Waldmann H. Characterization of the CAMPATH-1 (CDw52) antigen: Biochemical analysis and cDNA cloning reveal an unusually small peptide backbone. Eur J Immunol. 1991;21:1677–1684. doi: 10.1002/eji.1830210714. [DOI] [PubMed] [Google Scholar]
- 2.Hale G. CD52 (CAMPATH1) J Biol Regul Homeost Agents. 2001;15:386–391. [PubMed] [Google Scholar]
- 3.Kirchhoff C, Schröter S. New insights into the origin, structure and role of CD52: A major component of the mammalian sperm glycocalyx. Cells Tissues Organs. 2001;168:93–104. doi: 10.1159/000016810. [DOI] [PubMed] [Google Scholar]
- 4.Ito K, Hasegawa A, Komori S, Koyama K. Biochemical property and immunogenicity of mouse male reproductive tract CD52 (mrt-CD52) J Reprod Immunol. 2007;75:32–39. doi: 10.1016/j.jri.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Bandala-Sanchez E, et al. T cell regulation mediated by interaction of soluble CD52 with the inhibitory receptor Siglec-10. Nat Immunol. 2013;14:741–748. doi: 10.1038/ni.2610. [DOI] [PubMed] [Google Scholar]
- 6.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 7.Pillai S, Netravali IA, Cariappa A, Mattoo H. Siglecs and immune regulation. Annu Rev Immunol. 2012;30:357–392. doi: 10.1146/annurev-immunol-020711-075018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikehara Y, Ikehara SK, Paulson JC. Negative regulation of T cell receptor signaling by Siglec-7 (p70/AIRM) and Siglec-9. J Biol Chem. 2004;279:43117–43125. doi: 10.1074/jbc.M403538200. [DOI] [PubMed] [Google Scholar]
- 9.Rashidi M, et al. CD52 inhibits Toll-like receptor activation of NF-κB and triggers apoptosis to suppress inflammation. Cell Death Differ. 2018;25:392–405. doi: 10.1038/cdd.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi R, et al. Cd52, known as a major maturation-associated sperm membrane antigen secreted from the epididymis, is not required for fertilization in the mouse. Genes Cells. 2008;13:851–861. doi: 10.1111/j.1365-2443.2008.01210.x. [DOI] [PubMed] [Google Scholar]
- 11.Willis MD, Robertson NP. Alemtuzumab for multiple sclerosis. Curr Neurol Neurosci Rep. 2016;16:84. doi: 10.1007/s11910-016-0685-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tone M, et al. Structure and chromosomal location of mouse and human CD52 genes. Biochim Biophys Acta. 1999;1446:334–340. doi: 10.1016/s0167-4781(99)00103-7. [DOI] [PubMed] [Google Scholar]
- 13.Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 2009;323:1722–1725. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 15.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanai H, et al. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc Natl Acad Sci USA. 2013;110:20699–20704. doi: 10.1073/pnas.1320808110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urbonaviciute V, et al. Factors masking HMGB1 in human serum and plasma. J Leukoc Biol. 2007;81:67–74. doi: 10.1189/jlb.0306196. [DOI] [PubMed] [Google Scholar]
- 18.Rillahan CD, Paulson JC. Glycan microarrays for decoding the glycome. Annu Rev Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisler C, Jarvis DL. Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology. 2011;21:988–993. doi: 10.1093/glycob/cwr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munday J, et al. Identification, characterization and leucocyte expression of Siglec-10, a novel human sialic acid-binding receptor. Biochem J. 2001;355:489–497. doi: 10.1042/0264-6021:3550489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoine DJ, Harris HE, Andersson U, Tracey KJ, Bianchi ME. A systematic nomenclature for the redox states of high mobility group box (HMGB) proteins. Mol Med. 2014;20:135–137. doi: 10.2119/molmed.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JS, et al. High mobility group box 1 protein interacts with multiple Toll-like receptors. Am J Physiol Cell Physiol. 2006;290:C917–C924. doi: 10.1152/ajpcell.00401.2005. [DOI] [PubMed] [Google Scholar]
- 23.Orlova VV, et al. A novel pathway of HMGB1-mediated inflammatory cell recruitment that requires Mac-1-integrin. EMBO J. 2007;26:1129–1139. doi: 10.1038/sj.emboj.7601552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, et al. Identification of CD163 as an antiinflammatory receptor for HMGB1-haptoglobin complexes. JCI Insight. 2016;1:e85375. doi: 10.1172/jci.insight.85375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Son M, et al. C1q and HMGB1 reciprocally regulate human macrophage polarization. Blood. 2016;128:2218–2228. doi: 10.1182/blood-2016-05-719757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treumann A, Lifely MR, Schneider P, Ferguson MA. Primary structure of CD52. J Biol Chem. 1995;270:6088–6099. doi: 10.1074/jbc.270.11.6088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.