Significance
Viral populations exhibit an extraordinary ability to survive abrupt changes in host environment by rapidly generating adaptive mutations. However, our understanding of how viral populations respond to selection pressure and the underlying molecular mechanisms supporting viral adaptation in vivo is limited. Here, we report a set of clinical data sampled from subjects chronically infected by hepatitis C virus (HCV). The data show rapid expansion and turnover of drug-resistant viruses following treatment with an HCV protease inhibitor. By fitting mathematical models to the data, we propose that superinfection and cure of infected cells play critical roles in facilitating the rapid expansion and turnover of viral populations. Our results highlight the importance of considering intracellular viral competition in understanding rapid viral adaptation.
Keywords: virus evolution, virus persistence, hepatitis C virus, phylodynamic modeling, mathematical modeling
Abstract
RNA viruses exist as a genetically diverse quasispecies with extraordinary ability to adapt to abrupt changes in the host environment. However, the molecular mechanisms that contribute to their rapid adaptation and persistence in vivo are not well studied. Here, we probe hepatitis C virus (HCV) persistence by analyzing clinical samples taken from subjects who were treated with a second-generation HCV protease inhibitor. Frequent longitudinal viral load determinations and large-scale single-genome sequence analyses revealed rapid antiviral resistance development, and surprisingly, dynamic turnover of dominant drug-resistant mutant populations long after treatment cessation. We fitted mathematical models to both the viral load and the viral sequencing data, and the results provided strong support for the critical roles that superinfection and cure of infected cells play in facilitating the rapid turnover and persistence of viral populations. More broadly, our results highlight the importance of considering viral dynamics and competition at the intracellular level in understanding rapid viral adaptation. Thus, we propose a theoretical framework integrating viral and molecular mechanisms to explain rapid viral evolution, resistance, and persistence despite antiviral treatment and host immune responses.
Many RNA viruses, including global pathogens of major medical importance such as hepatitis C virus (HCV), HIV, and ebolavirus, exist within infected hosts as large populations of genetically related viral variants commonly referred to as a quasispecies (1). The diverse nature of the viral quasispecies allows viral populations to evolve rapidly to adapt to abrupt changes in the host environment (2); examples include escape from adaptive immune pressures (3, 4) and development of resistance to antivirals (5, 6). Despite the importance of viral adaptation, our understanding of how the viral quasispecies responds to selection pressure and the underlying molecular mechanisms supporting viral adaptation in vivo is limited by a lack of frequent longitudinal viral sequence data generated by methods that retain linkage across genes and genomes (7).
Previously, much attention has been devoted to estimating the effectiveness of antivirals against viral variants in the quasispecies (8–10); however, the mechanisms that drive an adaptive/fitter variant (e.g., a drug-resistant mutant) at an initially low frequency to a high frequency are less well studied. Theoretical work suggests that fitter variants at a low frequency cannot expand unless there are sufficient numbers of target cells, that is, “replication space” (5, 11–14). This is analogous to the concept of competitive release in ecological “niche” theory, that is, a species is not able to expand unless there is a niche vacated by other species (15). Previous models have in general assumed that, once a variant infects a cell, the cell is occupied by the variant for its remaining lifetime, and thus the replication space is provided by generation of new target cells (5, 8, 13, 16–18). Under this framework, viral variants compete for infecting newly generated target cells, and thus, the expansion of a low-frequency fitter mutant is determined by how quickly infected cells die and are replaced by new target cells. This framework has been successfully applied to understand HIV drug resistance (6, 13, 16, 18). However, the source of replication space for other rapidly evolving viruses, and in particular HCV, is unclear.
HCV is a positive-strand RNA virus belonging to the flaviviridae, a family of medically important single-strand RNA viruses. HCV infects ∼80–170 million people worldwide (19). Chronic infection can lead to cirrhosis and hepatocellular carcinoma. Current treatments for HCV infection involve combinations of direct acting antivirals (DAAs), and they have achieved remarkably high cure rates (20–22). Ironically, because of this clinical effectiveness, combination treatments cannot generally be used to probe the molecular mechanisms that contribute to virus persistence. However, this information may be critical for designing effective vaccination strategies for HCV and therapeutic regimens and vaccines for other members of the flaviviridae family. Here, we analyzed clinical samples collected from a phase I/IIa monotherapy trial (23) of a second-generation HCV protease inhibitor, MK-5172 (also called grazoprevir) (22, 24–26), that is extremely potent but susceptible to drug resistance development through multiple viral genetic pathways of escape. Measuring viral loads and analyzing viral populations longitudinally by single-genome sequencing (7) allowed us to follow the population dynamics and evolutionary responses of HCV to drug pressure and its removal in vivo at high resolution. We analyzed this unique dataset by combining mathematical modeling and phylogenetic analysis [termed “phylodynamic” analysis (27)]. We provide strong evidence that cure and superinfection of infected cells contribute essentially to the replication space, allowing resistance mutant expansion and evolution.
Results
Study Subjects and Single-Genome Sequence Analysis.
We studied eight participants from a dose-ranging monotherapy trial of MK-5172 (23). Five were treated with MK-5172 (50 or 800 mg) once daily for 7 d (subjects 1–5, genotypes 1a, n = 4; 1b, n = 1) and three were treated with placebo (subjects 6–8, genotypes 1a, n = 2; 1b, n = 1) (SI Appendix, Table S1). All of the participants were followed for 7–8 wk after treatment cessation. We measured viral loads longitudinally and analyzed viral populations using single-genome sequencing (7).
Samples taken immediately before treatment initiation, designated as day 0, were analyzed in all eight subjects for genotypic complexity and baseline drug resistance mutations (DRMs). At each sampling timepoint, ∼100 viral genomes were sequenced in each subject (Fig. 1 and SI Appendix, Figs. S1–S7). Day 0 sequences of each subject showed patterns of virus diversity typical of chronic infection with broad genotypic heterogeneity (SI Appendix, Table S1). Maximum pairwise diversity and mean pairwise diversity ranged from 1.17 to 5.64% and 0.53 to 3.45%, respectively. No drug resistance-associated mutations were identified in the pretreatment sequences, although a Q80K substitution was found in all sequences in subjects 2 and 3. Q80K is a common polymorphism that confers no resistance to MK-5172.
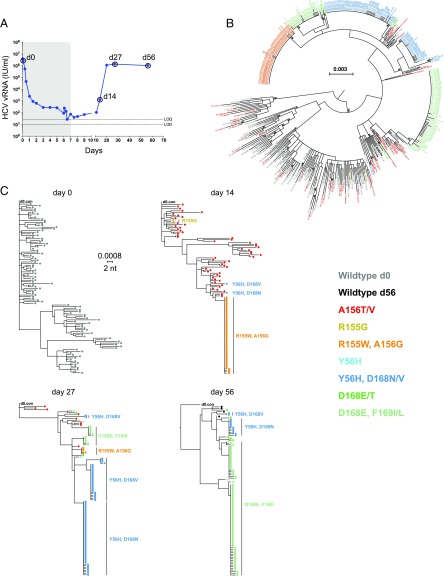
Fig. 1.
Sequential plasma virus load and sequences from subject 1. (A) Time course of treatment with MK-5172 (shaded area, days 1–7), viral load determinations (blue solid dots), and viral sequence analyses (open circles at days 0, 14, 27, and 56). (B) A maximum-likelihood (ML) phylogenetic tree of all viral sequences sampled from subject 1 from all time points. Tree tips are color coded according to the known resistant mutations they bear. (C) ML phylogenetic trees of viral sequences sampled from subject 1 at each time point.
In the five subjects treated with MK-5172, viral load decreased over 5 logs and went below (or close to) the limit of quantification (=25 IU/mL) during the 7-d treatment period (Fig. 1A and SI Appendix, Figs. S1–S4). At the end of this period, viral load was too low to permit sequencing analyses. By day 27, 20 d after treatment was stopped, the viral load rebounded to levels (106 to 107 IU/mL) similar to the baseline level before treatment in all treated subjects and could be quantified as early as day 14 (7 d after stopping treatment) in subjects 1 and 4. Follow-up single-genome sequencing analyses were conducted on samples from days 14 to 62.
After therapy was stopped, the viral populations that emerged carried drug resistance mutations. Surprisingly, the dynamics of mutant virus populations exhibited stringent population bottlenecks and rapid turnover of the dominant resistant mutants during the follow-up period, even in the presence of relatively stable viral load (e.g., subject 1 between days 27 and 56; Fig. 1). Extremely rapid and continuous turnover of the resistant mutants, after therapy cessation, was observed in subjects 1 and 4 (Fig. 1 and SI Appendix, Fig. S3). In contrast to these dramatic changes in plasma viral RNA (vRNA) load in subjects treated with MK-5172, the three subjects who received placebo had no changes in plasma vRNA levels and no sequences containing DRMs. Viral sequences from these individuals were obtained on days 0 and 27 or 34 (SI Appendix, Figs. S5–S7). Below, we describe the evolutionary patterns in subject 1 in detail. The patterns in other treated subjects are described in SI Appendix, SI Methods and Results.
In subject 1, none of 103 single genome sequences on day 0 contained drug resistance-associated mutations (Fig. 1C). Conversely, on day 14 (i.e., 7 d after therapy was stopped), all 111 single-genome sequences analyzed contained one or more drug resistance-associated mutations. Fifty-six of the 111 sequences contained single A156T or V mutations listed in color on the right-hand side of Fig. 1C and two sequences contained a single R155G mutation. These 58 sequences containing single drug resistance mutations were widely dispersed throughout the phylogenetic tree of pretreatment sequences (Fig. 1B). This indicates that nearly all of these drug-resistant sequences represented distinct viral lineages that traced back 14 d earlier to infected hepatocytes that carried A156T or A156V mutations in widely divergent genetic backgrounds. In marked contrast, 51 of 111 sequences (46%) contained a R155W-plus-A156G double mutation, in a nearly homogeneous genetic background where sequence diversity ranged from 0 to 1 nucleotide substitution over a span of 2,210 bp (<0.1% diversity). This result suggests that ∼46% of hepatocytes, releasing HCV virions into the circulation at 14 d after the initiation of MK-5172 treatment (and 7 d after its discontinuation), were infected by HCV genomes that had evolved from a single R155W-plus-A156G drug-resistant viral sequence lineage. In turn, this lineage most likely emanated from a single or a few productively infected hepatocytes that existed before treatment initiation (Fig. 1C). This inference is drawn based of the identity or near identity (1 mutation in 2,210 nt) of the genetic background in which the R155W-plus-A156G drug resistance mutation expanded. These findings are consistent with model predictions of the expected frequency of a single mutation or any set of two mutations in an HCV vRNA quasispecies in the absence of selection (5, 28). In addition, one sequence with Y56H-plus-D168V and one with Y56H-plus-D168N double mutations were also detected. At day 27 when the plasma viral load had returned to baseline (>106 vRNA/mL), there was a striking contraction in both the R155W-plus-A156G and A156T or V populations and a remarkable expansion of the discrete homogeneous Y56H-plus-D168V and Y56H-plus-D168N double-mutant populations (already detected at day 14). A single dominant Y56H-plus-D168N double-mutant population and three dominant Y56H-plus-D168V double-mutant populations together represented 70% of the sequences. This finding is all of the more remarkable given that, by this time, steady-state plasma vRNA load had returned to its pretreatment setpoint of >106 vRNA copies per mL and drug therapy had been stopped 20 d previously. Two additional monophyletic lineages, each containing the double-mutant D168E-plus-F169I, were present on day 27. By day 56, the D168E-plus-F169I mutant lineages (first detected at day 27) expanded to comprise 79% of the sequences, whereas the previously dominant Y56H-plus-D168N and Y56H-plus-D168V lineages contracted and now comprised only 12% of the sequences. Four wild-type sequences, that is, sequences with no known resistance mutations, were detected for the first time posttreatment at day 56.
Fitting a Baseline Viral Dynamic Model to the Viral Load and Sequence Data.
The rapid viral load decline during treatment and continuous turnover of resistant mutants after treatment cessation observed in the treated subjects raise intriguing questions, such as how do resistance clones expand so rapidly and how can resistant mutants dominate the viral population for such a long period after treatment cessation without being replaced by the nonresistant virus? To address these questions, we first constructed a “baseline” multistrain HCV model, similar to a previous standard viral dynamic model that incorporates competition for target cells (5) (Methods). The strains in the model are grouped according to the shared drug resistance mutations in the clinical samples as described in the section above and as color-coded in Fig. 1 and SI Appendix, Figs. S1–S7 (see SI Appendix, Table S2, for the mutants modeled). In this model, we assume that infected cells are lost at a per-capita rate δ and that target cells proliferate following a logistic growth term to replace them. Cure and superinfection of infected cells are not included in this model.
We fitted this model to the data from all five treated subjects. This model describes both the viral load and the sequence data well (SI Appendix, Fig. S8). However, the mean loss rate of infected cells, δ, estimated across all five treated subjects was 0.68 d−1 with a SD of 0.09 d−1 (see SI Appendix, Tables S3–S7, for the best-fit values of δ). In our model, the loss of infected cells corresponds to death of these cells. A large death rate is needed to explain the rapid second-phase viral load decline during treatment and rapid turnover of dominant mutants after treatment. However, such a large death rate of infected cells (0.68 d−1) does not seem physiological, as it implies a half-life for infected hepatocytes of 1 d. In stark contrast, the death rate of infected cells has been estimated as 0.14 d−1 from IFN therapy (29, 30) and previously used by Guedj et al. (31) and Lau et al. (21) to explain viral declines seen with other DAAs. To confirm the necessity of a high loss rate in this model, we refitted the model fixing δ at 0.14 d−1 and found this alternative parameterization explains the data poorly (SI Appendix, Fig. S9). These results strongly suggest that other mechanisms (in addition to death of infected cells) must play essential roles in driving the patterns seen in the data.
Potential Roles for Infected Cell Cure and Intracellular Viral Competition Through Superinfection.
We hypothesize that cure of infected cells is the primary cause for the loss of infected cells during the rapid second-phase decline observed in our data and other studies (5, 8, 32). In addition, we hypothesize that the continuous turnover of dominant mutants in the absence of treatment is mostly driven by intracellular competition among viral strains, as a result of superinfection (rather than competition for newly generated target cells). Previously, cure of infected cells has been demonstrated in in vitro studies (33–37). In vivo, the rapid second-phase viral declines under treatment were suggested to be attributable to cure of infected cells (32, 38). For superinfection, in vitro experiments showed that multiple viruses can enter the same cell, although limiting host resources necessary for viral replication and/or translation may restrict active production of multiple viruses (39, 40). Interestingly, a recent study demonstrated that a fitter HCV strain can enter already-infected cells and outcompete the resident strain (41), suggesting that multiple HCVs can enter a cell and compete for intracellular resources.
To test these hypotheses, we extended the baseline model by incorporating infected cell cure and superinfection, and fixing the rate of infected cell death at 0.14 d−1 as estimated under IFN therapy (29, 30) (Methods). Here, we assumed that infected cells are cured and become target cells again under MK-5172 treatment at per-capita rate , where is a rate constant and is the drug efficacy against the ith strain (see Methods for detail). To model the intracellular viral competition due to superinfection, we assume, for simplicity, that once an infected cell is superinfected, the fitter strain can outcompete the less fit strain intracellularly, and the cell becomes a cell infected by the fitter strain at rate ksuper.
We used three variations of the extended model to test the importance of cure and superinfection in explaining the kinetic patterns in the data: (i) we allowed cure but no superinfection (denoted as the “cure” model), or (ii) superinfection but no cure (denoted as the “superinfection” model), or (iii) both processes (denoted as the “full” model) (see SI Appendix, Figs. S10 and S11 and Fig. 2, respectively, for model fits and SI Appendix, Tables S3–S7 for the best-fit parameter values). To statistically test the importance of cure or/and superinfection in explaining the clinical data, we performed model selection using the corrected Akaike information criterion (AICc) (Table 1). The results suggest that cure of infected cells is needed to explain the rapid second-phase viral load decline during treatment seen in all five subjects (SI Appendix, Fig. S12). It also helps to explain the rapid selection of the resistant mutants seen at day 14 in subjects 1 and 4 (compare the fits between solid lines and dashed lines to data in SI Appendix, Fig. S13). Based on the best-fit parameters in the best model for each subject (SI Appendix, Table S8), we estimated the mean rate of cure of cells infected by baseline viruses under MK-5172 treatment [], where is the drug efficacy against the baseline viruses) to be 0.56 d−1. On average, it takes about 1.9 d to cure a cell under MK-5172 treatment (SI Appendix, Table S8) and the loss of infected cells is primarily through cure rather than death of infected cells (assumed to occur at rate of 0.14 d−1).
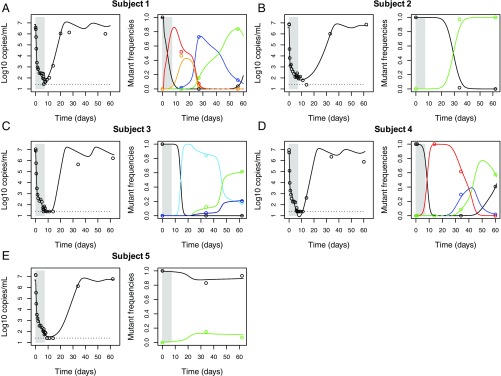
Fig. 2.
Best fit of the “full” model (lines) to the clinical data (circles) from five subjects treated with MK-5172. (A–E) Schematic diagrams of the evolutionary dynamics in subjects 1–5, respectively. A–E, the data and simulation results for viral loads are shown in open circles and black lines, respectively, on the Left; the data and simulation results for mutant frequencies are shown in colored open circles and lines, respectively, on the Right. The color coding for each mutant considered is shown in SI Appendix, Table S2.
Table 1.
Summary of the model characteristics and the fitting results (i.e., AICc scores) of each model for each subject
| Model | Model characteristics | Fitting results (AICc) | ||||||
| kcure | ksuper | Subject 1 | Subject 2 | Subject 3 | Subject 4 | Subject 5 | Total | |
| Baseline model with = 0.14 d−1 | 0.0 | 0.0 | −16.2 | −22.1 | −8.1 | 5.0 | −24.9 | −66.3 |
| Cure model | Fitted | 0.0 | −32.8 | −53.2 | −47.6 | −42.6 | −86.5 | −262.7 |
| Superinfection model | 0.0 | Fitted | −12.0 | −17.8 | −9.5 | −5.1 | −21.2 | −65.6 |
| Full model | Fitted | Fitted | −63.8 | −51.8 | −44.5 | −90.6 | −83.6 | −334.3 |
Bolded AICc scores denote the best model fit among all models for the five subjects.
The AICc results suggest that intracellular competition through superinfection is needed to explain the rapid and continuous turnover of dominant resistant strains after treatment stops in subjects 1 and 4 (SI Appendix, Fig. S14). This is because after treatment cessation, drug is eliminated and drug-induced cure is no longer possible. As a result, new target cells become available at a much slower rate than the rate during treatment. However, superinfection allows fitter strains to enter already-infected cells and compete intracellularly, greatly increasing the rate at which a fitter strain rises to a high frequency. In contrast, without superinfection and intracellular competition, a fitter strain is predicted to increase in frequency at a much lower rate (see the changes in the frequencies of the mutant in green during the period of days 30–60 in SI Appendix, Fig. S14A).
We performed uncertainty analysis in the best model for each subject using likelihood profiling (42) (SI Appendix, Tables S3–S7). These analyses indicated that the data allow us to estimate accurately the effectiveness of MK-5172 against the baseline virus, ε1, the rate of viral clearance, c, and the cure rate kcure in all five subjects, and the value of superinfection constant ksuper in subjects 1 and 4. In addition, we explored whether models with alternative assumptions can explain the data. This included a model assuming a constant rate of target cell generation and models assuming a DAA-independent cure of infected cells (SI Appendix, SI Methods and Results). Fitting results show that the full model presented in the main text is the best model, and the conclusions about the role of superinfection and cure of infected cells are robust against these model variations.
Compensatory Mutations, Clonal Interference of HCV-Resistant Mutants.
To understand better the evolutionary dynamics of the viral strains considered in our model, we analyzed how the strains at different time points are related to each other. Here, we define strains phenotypically, that is, each “strain” consists of a group of related viral sequences in the clinical samples sharing drug resistance mutations. We calculated the genetic distances between sequences belonging to different groups at the different sampling time points (see Methods and SI Appendix, Figs. S15–S17). We then combined the mutational pattern derived from the sequence data with the results from fitting the dynamical models to generate a diagram that summarizes the evolutionary dynamics of the drug-resistant strains over the period of the study (Fig. 3).
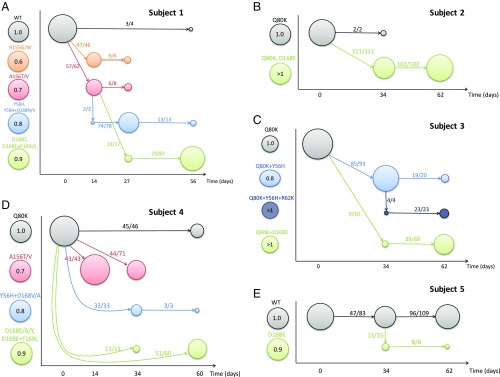
Fig. 3.
Evolutionary dynamics of HCV before and after treatment with MK-5172. (A–E) Schematic diagrams of the evolutionary dynamics in subjects 1–5, respectively. Each mutant is denoted as a colored circle, and the color coding is shown at the left-hand side of the y axis in each panel. The numbers within the circles denote the fitness of the mutant relative to the baseline strain in the absence of treatment according to the best-fit parameter values in the best model for each subject. The x axis shows the time when sequence data are taken. The size of the circle is scaled according to the frequency of the mutant in a given sample. Solid arrows show the strain (where the arrow starts; say, strain a) to which the majority of the sequences in a mutant strain of interest (where the arrow ends; say, strain b) are most closely related, and the numbers on each arrow show the number of sequences in strain b that are mostly closely related to strain a and the total number of sequences in strain b.
The diagram reveals how the HCV population responds to MK-5172 treatment. First, viral load rebounded rapidly in subjects 1 and 4 to high levels that permitted sequencing by day 14, presumably because of selection of the resistant mutants bearing A156T/V or R155G/W in subject 1 and A156T/V in subject 4. We estimated that these mutants are highly resistant, yet have high fitness costs (Fig. 3), which is consistent with results shown in previous in vitro studies (24–26). Because of their short genetic distance to the wild-type viruses (SI Appendix, Figs. S15 and S17), they are likely present at low frequencies before treatment. During and shortly after treatment, they rise to high frequencies transiently before being replaced by other resistant mutants with higher fitness while drug is cleared. Second, mutants with the Y56H mutation are observed transiently in subjects 1, 3, and 4 between days 27 and 34. Our analysis suggests that the transient appearance of these mutants is due to their intermediate resistance and fitness values. These mutants gave way to fitter strains with resistant mutations at position 168 (e.g., D168E, D168A, or D168Y; see refs. 24 and 25) that dominated the viral population in subjects 1–4 at the end of follow-up period, that is, 7–8 wk after treatment cessation. These dynamics resemble the classical pattern, termed “clonal interference,” as suggested by population genetic models for the evolution of asexual organisms (43, 44).
The fitnesses of the resistant mutants observed at late time points are predicted to be close to or higher than those of the baseline viruses, which remained at low frequencies in subjects 1–4 (Fig. 3). A likely explanation for the late appearance of these high-fitness variants is that mutation at position 168 on the baseline virus background resulted in low fitness (and thus these mutants are at low frequency before treatment). However, growth of these low-fitness resistant mutants generated compensatory mutations that increased their fitness, and allowed their frequency to increase and stabilize at a high level in the population.
Last, we observe that drug-sensitive viruses, that is, viruses with no known drug-resistant mutations, reappeared in the sequences at the last time point of follow-up in subjects 1, 4, and 5. Statistical analysis shows that these viruses are likely to be descendants of drug-sensitive viruses present before treatment rather than reversions from resistant variants (SI Appendix, SI Methods and Results). This suggests that the drug-sensitive viruses were suppressed to low frequencies under treatment and only grow to high frequencies long after treatment cessation.
Discussion
Here, we have analyzed the evolutionary dynamics of the HCV population in response to 7-d treatment with the protease inhibitor grazoprevir (MK-5172) in five subjects using single-genome sequencing and phylodynamic modeling. HCV-resistant variants underwent rapid expansion after drug treatment with surprisingly fast sequential turnover of dominant resistant mutants in the 7- to 8-wk follow-up period, a distinct feature of viral persistence. Using mathematical models, we showed that superinfection and cure of infected cells are likely to be crucial mechanisms driving the extremely rapid expansion and turnover of resistant variants observed in the clinical data. Although superinfection and cure of infected cells have been demonstrated in vitro (33–37, 41), our work suggests that these processes occur and play important roles in driving viral adaptation and persistence in vivo.
We estimated the rate of cure of cells infected by wild-type viruses under grazoprevir to be 0.56 d−1 on average. Further, it takes about 1.9 d on average to cure a cell under MK-5172 treatment. This high rate of infected cell cure under treatment is consistent with the clinical data in this study and from previous studies (8, 30, 32, 45), where rapid second-phase viral load declines have been observed. Previous mathematical models suggested that the second-phase decline reflects both the death rate of infected cells and the rate of intracellular vRNA degradation (31, 32, 38). Thus, this rapid cure of infected cells is likely to be a result of the rapid loss of intracellular HCV RNAs under treatment. Our model shows that, due to the rapid cure, drug-sensitive viruses were at a much lower frequency than resistant mutants when treatment stopped. That coupled with the rapid expansion of resistant mutants and generation of compensatory mutations led to the disappearance or a delay in the reappearance of drug-sensitive virus.
Superinfection has been demonstrated in vivo indirectly by the identification of recombinant forms of HCV in patients (46–49). Although recombination between HCV genomes is apparently rare (50, 51), this does not mean that superinfection of hepatocytes is infrequent, because the formation of distinct intracellular HCV replication complexes and a processive RNA polymerase may prevent recombination. In line with a recent in vitro study showing that a fitter HCV strain can enter already-infected cells and outcompete the resident strain (41), our results suggest that superinfection and intracellular competition occur frequently in vivo, especially when different strains exhibit a difference in intracellular replication fitness.
We argue that superinfection and intracellular competition are critical for the establishment and expansion of adaptive mutants and thus for the persistence of HCV under selection pressures. For viruses that largely transmit by cell-to-cell infection, such as HCV (52–55), transmission is often limited from an infected cell to a few neighboring cells. When most of the cells are infected by resident viral variants, without superinfection, the probability that an adaptive mutant finds an uninfected cell is low. Thus, adaptive mutants would likely be lost, and for those mutants that do establish infection, the rate at which they expand would be low. In contrast, with superinfection and intracellular competition, an adaptive mutant can enter all neighboring cells to compete with any resident virus, which substantially increases the probability of establishment and the rate at which the adaptive mutant expands. While previous theoretical work discussed the role of superinfection in viral evolution at the host level (56, 57), our work highlights the important role that superinfection plays in facilitating viral adaptation at the cellular level.
By combining phylogenetic analysis and mathematical modeling, we provided an integrated understanding of the HCV evolutionary and population dynamics. First, the HCV within-host evolution of HCV resembles the classical pattern for the evolution of asexual organisms as suggested by population genetic models, termed clonal interference (43, 44). This emphasizes the usefulness of population genetic models in understanding viral evolutionary dynamics. Second, our results suggest that compensatory mutations are likely to be generated after treatment, allowing for stabilization and persistence of resistant mutants in the viral population long after treatment cessation. This highlights the need to maintain a high level of adherence to combination therapies of DAAs. Treatment interruption would allow rapid expansion of viral mutants resistant to one or more DAAs and possible generation of compensatory mutations that stabilize the resistant mutations at a high frequency, and thus creating opportunity for transmission of resistant mutants. In situations where treatment is interrupted frequently, multiple mutations may accumulate such that the evolved virus may become resistant to all DAAs in the combination therapy (12).
Although beyond the scope of this work, a couple of predictions of our study can be further tested experimentally. First, our models suggest that superinfection and intracellular competition are important mechanisms facilitating the rapid expansion and turnover of resistant mutants in vivo after treatment cessation. Although testing this prediction in vivo is not possible, intracellular viral competition experiments as in ref. 41 can be performed to compete pairs of laboratory viruses with the resistant mutations identified in this study. The results would reconfirm the occurrence of superinfection and determine the relative fitness of those mutants at the intracellular scale. Second, our analysis suggests that compensatory mutations are involved in stabilizing the resistant mutations at amino acid 168 seen in subjects 1–4. This can be tested by first performing whole-genome sequencing on the viral isolates and then identifying the shared mutations of the viruses with a mutation at position 168 in each subject. These shared mutations would be candidate compensatory mutations for the mutation at amino acid 168. Introducing one or a combination of these mutations into viral replicons, and then testing and comparing the replication fitness of the replicons (as shown in refs. 24–26), would be a means to identify compensatory mutations.
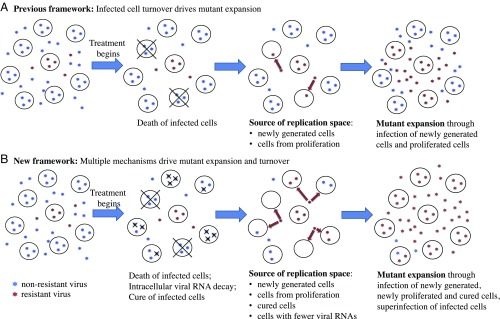
To summarize, the results in our study elucidate important, yet previously unrecognized, mechanisms that we suggest operate in vivo, drive within-host viral evolution, and allow the adaptation and persistence of HCV in the face of drug pressure and immune response. A previous framework frequently used in viral dynamic models (5, 8, 13, 16–18) assumed that viruses compete for the replication space needed for adaptation to changing host environments only through infection of newly generated target cells (Fig. 4A). Consequently, the rate of adaptation was limited by the rate of target cell generation. In contrast, our framework emphasizes that both superinfection and cure of infected cells (in addition to infection of new target cells) can be important mechanisms contributing to the replication space needed for mutant virus expansion (Fig. 4B). This mechanism can greatly accelerate the rate of within-host viral adaptation beyond the rate set by the generation of target cells. This framework may be important in elucidating the contributions and mechanisms of HCV containment and/or elimination mediated by the host immune system that occur naturally or following vaccination (58). More broadly, our work points toward the important role molecular mechanisms play in facilitating viral evolution and persistence. Frequent cell-to-cell infection and high multiplicity of infection, that is, mechanisms that provide the means for intracellular competition, have been reported recently for HIV, HCV, influenza, and other viruses (54, 59–61). We argue that our framework and the consideration of viral competition at both the intracellular and host level in particular, is critical for the understanding of the evolutionary and adaptive dynamics of viral population at the within-host level (62, 63). Thus, it opens the door to the next generation of viral dynamic models and quantitative frameworks to understand viral evolution, interpret clinical datasets, and predict treatment outcomes.
Fig. 4.
Conceptual frameworks for virus evolution under antiviral pressure. Resistant mutants expand through occupying/competing for available replication space. (A) Previous modeling mostly assumed that, once a cell is infected by a virus, the cell remains infected until death. The replication space arises through generation of new target cells. Under this framework, resistant mutants expand through infection of newly generated target cells, and the rate of the increase of mutant frequency is mostly set by the rate at which infected cells die and are replaced by newly generated cells. (B) Our results suggest a conceptual framework where the replication space arises from multiple sources. In the presence of potent antivirals, the level of intracellular HCV RNAs decreases, leading to cure of infection in some cells. Replication space thus arises from both newly generated cells and cured cells. In addition, superinfection makes replication space available by allowing resistant viruses to enter an already-infected cell and compete for intracellular resources. Thus, cure and superinfection of cells allow resistant mutant expansion and turnover to occur at a much faster rate than the rate set by the death and replacement of infected cells only.
Methods
Experimental Model and Subject Details.
For this study, Merck Sharp and Dohme Corporation provided preexisting, deidentified human blood plasma specimens collected from a multiinstitutional experimental drug treatment protocol (MK-5172-004) designed to test the antiviral activity of the HCV protease inhibitor grazoprevir/MK-5172 (ClinicalTrials.gov identifier NCT00998985). The specimens were collected, stored, and inventoried with study subjects’ informed consent and approval for future use in the analysis of the effects of grazoprevir/MK-5172 on HCV replication, although the specimens were not collected specifically for the research project described here. Based on these stipulations, the research described in the current report was deemed by the Institutional Review Board of the University of Pennsylvania not to constitute human subjects in research.
Single-Genome Sequencing.
The partial NS2 and complete NS3 and NS4A gene sequences from all eight subjects were generated using the single-genome sequencing method previously described (50). The primers used for the nested PCR included the following: (i) genotype 1a: first-round sense primer 1aNS2.F2, 5′-ACCCGRCTTTGGTATTTGACATCACC-3′ (nucleotides 2983–3008, H77); first-round antisense primer 1aNS4B.R3, 5′-TATTGTATCCCACTGATGAAGTTCCACAT-3′ (nucleotides 5634–5662, H77); second-round sense primer 1aNS2.F3, 5′-AAAGTGCCCTACTTYGTGCGCGT-3′ (nucleotides 3063–3085, H77); and second-round antisense primer 1aNS4B.R4, 5′-AGGGCCTTCTGCTTGAACTGCTC-3′ (nucleotides 5517–5539, H77); (ii) genotype 1b: first-round sense primer 1bNS2.F2, 5′-GCCCGTCGTCTTYTCTGACATGGA-3′ (nucleotides 3257–3280, H77); first-round antisense primer 1bNS4B.R3, 5′-TTCCACATGTGCTTCGCCCA-3′ (nucleotides 5622–5641, H77); second-round sense primer 1bNS2.F3, 5′-TCATCACCTGGGGGGCAGACA-3′ (nucleotides 3289–3309, H77); and second-round antisense primer 1bNS4B.R4, 5′-CGAGCGCCTTCTGCTTGAATTG-3′ (nucleotides 5520–5541, H77).
Viral Load and Sequence Diversity Analysis.
The vRNA measurement was done by Merck and Co., Inc., using the Roche Cobas TaqMAN 2.0 assay with a lower limit of quantification of 25 IU/mL and a limit of detection of 10 IU/mL. A total of 2,755 NS2–NS3–NS4 genomes were generated and analyzed from the eight subjects. The median number of sequences analyzed per time point was 107 (mean, 106; range, 43–144) (SI Appendix, Table S1). Sequences alignments were initially made with clustalW and then hand-checked using Geneious for correct codon alignments. The maximum, minimum, median, and mean diversity for each sequence set was calculated based on Hamming distance (SI Appendix, Table S1). For each subject, phylogenetic trees were generated by maximum-likelihood methods using PhyML. The combined phylogenetic tree and the tree from each time point were rooted with consensus sequences from day 0.
Construction of a Baseline Multistrain HCV Model.
We construct a viral dynamic model for within-host infection by multiple strains of HCV based on previous published models (5, 29, 64). In this model, we keep track of n different HCV strains. The ordinary differential equations (ODEs) for the model are as follows:
Target cells (T), presumably a subset of hepatocytes, die at a per-capita rate, d. Existing target cells can proliferate, and this proliferation is modeled using a logistic term, , as in Rong et al. (5), where is a proliferation rate constant, Ii are cells infected with virus strain i, N is the concentration of hepatocytes that are not target cells, and Tmax is the liver carrying capacity. Target cells are infected by the ith strain of HCV () at rate . Infected cells die at per-capita rate, . Drug-sensitive viruses, represented by strain i = 1, are produced from infected cells at rate p per cell in the absence of treatment. We assume that the ith strain has a fitness of ri relative to the drug-sensitive viruses (r1 = 1). This difference in fitness is reflected only in the differences in viral production in our model. Thus, the production rate of the ith strain is in the absence of treatment. Under treatment, we assume that viral production of the ith strain is reduced by a factor , where corresponds to a 100% effective drug. The drug effectiveness εi is modeled as a function of the drug concentration and the EC50 value for each strain (EC50,i) according to an Emax model (65). We assume that the drug concentration, D, stays constant during the first 7 d of treatment, since drug concentration reaches its maximal concentration within 2–4 h (23). After treatment stops on day 7, we assume the drug concentration declines exponentially at rate, w, obtained previously (23). All viruses are cleared at per-capita rate c. The values of the parameters are shown in SI Appendix, Table S9.
Construction of Models with Cure and Superinfection of Infected Cells.
We constructed a model incorporating cure and superinfection of infected cells by extending the basic model. The ODEs for this model are as follows:
To model cure of infected cells, we assumed that the rate of cure of infected cells is linearly dependent on the log10 of the efficacy of the drug, , and the rate constant is kcure. This assumption is based on a previous finding with the protease inhibitor telaprevir that the rate of second phase of viral load decline increased linearly with − (32).
The superinfection we track here includes the infection of a cell already infected with strain i with a virus of strain j, and the subsequent conversion of the cell into a cell that produces virus of strain j. We assume, for simplicity, that, if a fitter virus, strain i, enters a cell infected with a less fit strain, strain j (modeled using the term ), it can outcompete strain j, and then the cell is converted to a cell productively infected by strain i. We further assume the rate of this conversion is linearly dependent on the fitness difference () of the two strains, , and ksuper is a constant for the efficiency of this conversion.
Supplementary Material
Acknowledgments
We thank Gerald Learn for assistance with DNA sequence compilations. We gratefully acknowledge Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., for providing the samples analyzed in this work. Portions of this work were performed under the auspices of the US Department of Energy under Contract DE-AC52-06NA25396. This work was funded by National Institutes of Health Grants R01-OD011095, R01-AI028433, R01-AI078881 (to A.S.P.), R01-AI116868 (to R.M.R.), and U19-AI088791 and P30-AI45008 (to G.M.S.). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc.
Footnotes
Conflict of interest statement: R.J.O.B. is an employee and shareholder of Merck & Co., Inc. This work was funded by a grant from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc. (to G.M.S.). A.S.P. consulted for Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MF282014–MF284666).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805267115/-/DCSupplemental.
References
- 1.Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev. 2012;76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lauring AS, Frydman J, Andino R. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol. 2013;11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 4.Erickson AL, et al. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity. 2001;15:883–895. doi: 10.1016/s1074-7613(01)00245-x. [DOI] [PubMed] [Google Scholar]
- 5.Rong L, Dahari H, Ribeiro RM, Perelson AS. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010;2:30ra32. doi: 10.1126/scitranslmed.3000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenbloom DI, Hill AL, Rabi SA, Siliciano RF, Nowak MA. Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nat Med. 2012;18:1378–1385. doi: 10.1038/nm.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adiwijaya BS, et al. A multi-variant, viral dynamic model of genotype 1 HCV to assess the in vivo evolution of protease-inhibitor resistant variants. PLoS Comput Biol. 2010;6:e1000745. doi: 10.1371/journal.pcbi.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridell RA, et al. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: In vitro and in vivo correlations. Hepatology. 2011;54:1924–1935. doi: 10.1002/hep.24594. [DOI] [PubMed] [Google Scholar]
- 10.Ke R, et al. Modelling clinical data shows active tissue concentration of daclatasvir is 10-fold lower than its plasma concentration. J Antimicrob Chemother. 2014;69:724–727. doi: 10.1093/jac/dkt423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YY, Summers J. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J Virol. 2000;74:5257–5265. doi: 10.1128/jvi.74.11.5257-5265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke R, Loverdo C, Qi H, Sun R, Lloyd-Smith JO. Rational design and adaptive management of combination therapies for hepatitis C virus infection. PLoS Comput Biol. 2015;11:e1004040. doi: 10.1371/journal.pcbi.1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander HK, Bonhoeffer S. Pre-existence and emergence of drug resistance in a generalized model of intra-host viral dynamics. Epidemics. 2012;4:187–202. doi: 10.1016/j.epidem.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Guedj J, Rong L, Dahari H, Perelson AS. A perspective on modelling hepatitis C virus infection. J Viral Hepat. 2010;17:825–833. doi: 10.1111/j.1365-2893.2010.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd-Smith JO. Vacated niches, competitive release and the community ecology of pathogen eradication. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120150. doi: 10.1098/rstb.2012.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonhoeffer S, Nowak MA. Pre-existence and emergence of drug resistance in HIV-1 infection. Proc Biol Sci. 1997;264:631–637. doi: 10.1098/rspb.1997.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciupe SM, Ribeiro RM, Nelson PW, Dusheiko G, Perelson AS. The role of cells refractory to productive infection in acute hepatitis B viral dynamics. Proc Natl Acad Sci USA. 2007;104:5050–5055. doi: 10.1073/pnas.0603626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perelson AS, Nelson PW. Mathematical analysis of HIV-1 dynamics in vivo. SIAM Rev. 1999;41:3–44. [Google Scholar]
- 19.Thomas DL. Global control of hepatitis C: Where challenge meets opportunity. Nat Med. 2013;19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feld JJ, et al. ASTRAL-1 Investigators Sofosbuvir and Velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med. 2015;373:2599–2607. doi: 10.1056/NEJMoa1512610. [DOI] [PubMed] [Google Scholar]
- 21.Lau G, et al. Efficacy and safety of 3-week response-guided triple direct-acting antiviral therapy for chronic hepatitis C infection: A phase 2, open-label, proof-of-concept study. Lancet Gastroenterol Hepatol. 2016;1:97–104. doi: 10.1016/S2468-1253(16)30015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulkowski M, et al. Efficacy and safety of 8 weeks versus 12 weeks of treatment with grazoprevir (MK-5172) and elbasvir (MK-8742) with or without ribavirin in patients with hepatitis C virus genotype 1 mono-infection and HIV/hepatitis C virus co-infection (C-WORTHY): A randomised, open-label phase 2 trial. Lancet. 2015;385:1087–1097. doi: 10.1016/S0140-6736(14)61793-1. [DOI] [PubMed] [Google Scholar]
- 23.Brainard DM, et al. Safety and antiviral activity of MK-5172, a novel HCV NS3/4a protease inhibitor with potent activity against known resistance mutants, in genotype 1 and 3 HCV-infected patients. Hepatology. 2010;52:706A–707A. [Google Scholar]
- 24.Howe AY, et al. Virologic resistance analysis from a phase 2 study of MK-5172 combined with pegylated interferon/ribavirin in treatment-naive patients with hepatitis C virus genotype 1 infection. Clin Infect Dis. 2014;59:1657–1665. doi: 10.1093/cid/ciu696. [DOI] [PubMed] [Google Scholar]
- 25.Shimakami T, et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology. 2011;140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Summa V, et al. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob Agents Chemother. 2012;56:4161–4167, and erratum (2014) 58:4995. doi: 10.1128/AAC.00324-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grenfell BT, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 28.Ribeiro RM, et al. Quantifying the diversification of hepatitis C virus (HCV) during primary infection: Estimates of the in vivo mutation rate. PLoS Pathog. 2012;8:e1002881. doi: 10.1371/journal.ppat.1002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann AU, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 30.Snoeck E, et al. A comprehensive hepatitis C viral kinetic model explaining cure. Clin Pharmacol Ther. 2010;87:706–713. doi: 10.1038/clpt.2010.35. [DOI] [PubMed] [Google Scholar]
- 31.Guedj J, et al. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci USA. 2013;110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guedj J, Perelson AS. Second-phase hepatitis C virus RNA decline during telaprevir-based therapy increases with drug effectiveness: Implications for treatment duration. Hepatology. 2011;53:1801–1808. doi: 10.1002/hep.24272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin K, Kwong AD, Lin C. Combination of a hepatitis C virus NS3-NS4A protease inhibitor and alpha interferon synergistically inhibits viral RNA replication and facilitates viral RNA clearance in replicon cells. Antimicrob Agents Chemother. 2004;48:4784–4792. doi: 10.1128/AAC.48.12.4784-4792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vliegen I, Paeshuyse J, Zhong W, Neyts J. In vitro combinations containing Tegobuvir are highly efficient in curing cells from HCV replicon and in delaying/preventing the development of drug resistance. Antiviral Res. 2015;120:112–121. doi: 10.1016/j.antiviral.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 38.Guedj J, Neumann AU. Understanding hepatitis C viral dynamics with direct-acting antiviral agents due to the interplay between intracellular replication and cellular infection dynamics. J Theor Biol. 2010;267:330–340. doi: 10.1016/j.jtbi.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 39.Schaller T, et al. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J Virol. 2007;81:4591–4603. doi: 10.1128/JVI.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tscherne DM, et al. Superinfection exclusion in cells infected with hepatitis C virus. J Virol. 2007;81:3693–3703. doi: 10.1128/JVI.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster B, Ott M, Greene WC. Evasion of superinfection exclusion and elimination of primary viral RNA by an adapted strain of hepatitis C virus. J Virol. 2013;87:13354–13369. doi: 10.1128/JVI.02465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolker BM. Ecological Models and Data in R. Princeton Univ Press; Princeton: 2008. [Google Scholar]
- 43.Desai MM, Fisher DS. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics. 2007;176:1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102-103:127–144. [PubMed] [Google Scholar]
- 45.Rong L, et al. Analysis of hepatitis C virus decline during treatment with the protease inhibitor danoprevir using a multiscale model. PLoS Comput Biol. 2013;9:e1002959. doi: 10.1371/journal.pcbi.1002959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedskog C, et al. Characterization of hepatitis C virus intergenotypic recombinant strains and associated virological response to sofosbuvir/ribavirin. Hepatology. 2015;61:471–480. doi: 10.1002/hep.27361. [DOI] [PubMed] [Google Scholar]
- 47.Iles JC, et al. Characterization of hepatitis C virus recombination in Cameroon by use of nonspecific next-generation sequencing. J Clin Microbiol. 2015;53:3155–3164. doi: 10.1128/JCM.00483-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morel V, et al. Genetic recombination of the hepatitis C virus: Clinical implications. J Viral Hepat. 2011;18:77–83. doi: 10.1111/j.1365-2893.2010.01367.x. [DOI] [PubMed] [Google Scholar]
- 49.Raghwani J, et al. Origin and evolution of the unique hepatitis C virus circulating recombinant form 2k/1b. J Virol. 2012;86:2212–2220. doi: 10.1128/JVI.06184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, et al. Elucidation of hepatitis C virus transmission and early diversification by single genome sequencing. PLoS Pathog. 2012;8:e1002880. doi: 10.1371/journal.ppat.1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoddard MB, et al. Identification, molecular cloning, and analysis of full-length hepatitis C virus transmitted/founder genotypes 1, 3, and 4. MBio. 2015;6:e02518. doi: 10.1128/mBio.02518-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graw F, et al. Inferring viral dynamics in chronically HCV infected patients from the spatial distribution of infected hepatocytes. PLoS Comput Biol. 2014;10:e1003934. doi: 10.1371/journal.pcbi.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graw F, Martin DN, Perelson AS, Uprichard SL, Dahari H. Quantification of hepatitis C virus cell-to-cell spread using a stochastic modeling approach. J Virol. 2015;89:6551–6561. doi: 10.1128/JVI.00016-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kandathil AJ, et al. Use of laser capture microdissection to map hepatitis C virus-positive hepatocytes in human liver. Gastroenterology. 2013;145:1404–1413.e10. doi: 10.1053/j.gastro.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wieland S, et al. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology. 2014;59:2121–2130. doi: 10.1002/hep.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.May RM, Nowak MA. Coinfection and the evolution of parasite virulence. Proc Biol Sci. 1995;261:209–215. doi: 10.1098/rspb.1995.0138. [DOI] [PubMed] [Google Scholar]
- 57.Nowak MA, May RM. Superinfection and the evolution of parasite virulence. Proc Biol Sci. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- 58.Bailey JR, et al. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight. 2017;2:92872. doi: 10.1172/jci.insight.92872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graw F, Perelson AS. Modeling viral spread. Annu Rev Virol. 2016;3:555–572. doi: 10.1146/annurev-virology-110615-042249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts KL, Manicassamy B, Lamb RA. Influenza A virus uses intercellular connections to spread to neighboring cells. J Virol. 2015;89:1537–1549. doi: 10.1128/JVI.03306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sattentau Q. Avoiding the void: Cell-to-cell spread of human viruses. Nat Rev Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 62.Loverdo C, Park M, Schreiber SJ, Lloyd-Smith JO. Influence of viral replication mechanisms on within-host evolutionary dynamics. Evolution. 2012;66:3462–3471. doi: 10.1111/j.1558-5646.2012.01687.x. [DOI] [PubMed] [Google Scholar]
- 63.Ke R, Lloyd-Smith JO. Evolutionary analysis of human immunodeficiency virus type 1 therapies based on conditionally replicating vectors. PLoS Comput Biol. 2012;8:e1002744. doi: 10.1371/journal.pcbi.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nowak MA, May RM. Virus Dynamics. 1st Ed Oxford Univ Press; Oxford: 2001. [Google Scholar]
- 65.Felmlee MA, Morris ME, Mager DE. Mechanism-based pharmacodynamic modeling. Methods Mol Biol. 2012;929:583–600. doi: 10.1007/978-1-62703-050-2_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.