Significance
The transcription coactivator LEDGF/p75 contributes to regulation of gene expression by tethering other factors to actively transcribed genes on chromatin. Its chromatin-tethering activity is hijacked in two important disease settings, HIV and mixed-lineage leukemia; however, the basis for the biological regulation of LEDGF/p75’s interaction to binding partners has remained unknown. This has represented a gap in our understanding of LEDGF/p75’s fundamental biological function and a major limitation for development of therapeutic targeting of LEDGF/p75 in human disease. Our work provides a mechanistic understanding of how the lens epithelium-derived growth factor interaction network is regulated at the molecular level.
Keywords: LEDGF/p75, disordered proteins, protein–protein interactions, phosphorylation, leukemia
Abstract
Lens epithelium-derived growth factor/p75 (LEDGF/p75, or PSIP1) is a transcriptional coactivator that tethers other proteins to gene bodies. The chromatin tethering function of LEDGF/p75 is hijacked by HIV integrase to ensure viral integration at sites of active transcription. LEDGF/p75 is also important for the development of mixed-lineage leukemia (MLL), where it tethers the MLL1 fusion complex at aberrant MLL targets, inducing malignant transformation. However, little is known about how the LEDGF/p75 protein interaction network is regulated. Here, we obtained solution structures of the complete interfaces between the LEDGF/p75 integrase binding domain (IBD) and its cellular binding partners and validated another binding partner, Mediator subunit 1 (MED1). We reveal that structurally conserved IBD-binding motifs (IBMs) on known LEDGF/p75 binding partners can be regulated by phosphorylation, permitting switching between low- and high-affinity states. Finally, we show that elimination of IBM phosphorylation sites on MLL1 disrupts the oncogenic potential of primary MLL1-rearranged leukemic cells. Our results demonstrate that kinase-dependent phosphorylation of MLL1 represents a previously unknown oncogenic dependency that may be harnessed in the treatment of MLL-rearranged leukemia.
Lens epithelium-derived growth factor/p75 (LEDGF/p75 or PSIP1) is a transcriptional coactivator that works in conjunction with other factors. LEDGF/p75 contributes to transcription regulation by tethering other proteins to sites of H3K36me2/3-marked chromatin using its N-terminal PWWP domain (1). While most studies revealed that the PWWP domain of LEDGF/p75 preferentially binds H3K36me2 and me3 marks (2–4), the LEDGF/p75 chromatin binding profile does not completely overlap with the genome-wide distribution of these marks, alluding to the existence of additional interaction determinants. Interactions with other factors mainly occur at the C-terminal portion of LEDGF/p75, which harbors a protein-binding scaffold known to recruit different cargo proteins. This domain is commonly referred to as the integrase binding domain (IBD) because HIV integrase was the first identified binding partner of this domain (5). We and others demonstrated that HIV hijacks the LEDGF/p75 chromatin-tethering function to direct viral integration into active chromatin (6–9). This research resulted in the discovery of potent LEDGF/p75-integrase inhibitors (10) that are currently in clinical development.
In addition to HIV integrase, a diverse network of physiological IBD interaction partners has been discovered, including the PI3K regulator JPO2 (CDCA7L) (11–13), the zinc-finger protein POGZ (14), the cell-cycle regulator ASK (DBF4) (15), the MLL1-MENIN binary complex (16), and the transcription elongation regulator IWS1 (17). We previously revealed that all known cellular partners of the LEDGF/p75 IBD contain an intrinsically disordered IBD-binding motif (IBM) (17, 18).
Several LEDGF/p75 binding partners play important roles in human disease. Mutations in POGZ are associated with intellectual disability and autism spectrum disorder (19). POGZ was also found to be disrupted in individuals with congenital heart disease and neurodevelopmental disorders (20). JPO2 is up-regulated in medulloblastoma and contributes to metastatic medulloblastoma in humans by potentiating Myc-dependent transformation (21). Alongside HIV integrase, perhaps the best-studied disease-related binding partner of the LEDGF/p75 IBD is MLL1 (KMT2A) (16). Interaction of LEDGF/p75 and MLL1 is normally not associated with pathology, but LEDGF/p75 plays a crucial role in acute leukemia development. In mixed-lineage leukemia (MLL), the MLL1 gene frequently undergoes chromosomal translocations or partial tandem duplications resulting in oncogenic fusions (22, 23). A key feature of these MLL1 fusions is their ability to efficiently transform hematopoietic progenitor cells into leukemic stem cells by activating HOX gene expression (24, 25). The oncogenic function of MLL1 fusion proteins therefore is critically dependent on chromatin tethering by LEDGF/p75 (16, 26).
Because the IBD of LEDGF/p75 represents a promising drug target for HIV infection and MLL, it has been the subject of extensive structural studies. The solution structure of the IBD has revealed a compact right-handed bundle composed of five α-helices (27), comparable to other members of the TFIIS N-terminal domain family (TND; InterProID IPR017923) (5, 28). Unstructured IBMs, which recognize the IBD, are composed of two distinct parts: a disordered stretch containing acidic residues followed by a highly conserved hydrophobic FxGF motif. Despite several attempts (17, 18, 29), characterization of the interface between the flexible acidic stretch and the IBD has remained elusive due to its disordered character (17, 18).
Incomplete mapping of the interface also impedes our understanding of how the associations between LEDGF/p75 and its interaction partners may be regulated. Each IBM represents a typical example of a short linear motif (SLiM), an evolutionary conserved unstructured protein module that adopts a characteristic structure upon binding its target site. Several examples of SLiMs have been reported (e.g., SH2, 14-3-3, and PDZ domains), which are modulated by switch-like posttranslational modifications to tune their interaction affinities (30–33). Based on the conservation of the charged residues within the acidic linker in IBMs, we hypothesized that the addition of extra negative charges through phosphorylation could modulate the affinity of binding partners for LEDGF/p75.
To test this hypothesis, we performed here an extensive structural analysis of the LEDGF/p75 IBD interactome at the molecular level. Our approach revealed structural features that were previously obscured and confirmed that all binding partners recognize the IBD in a structurally conserved manner shared with other TNDs. In addition, we validated Mediator subunit 1 as an LEDGF/p75 interaction partner and discovered a second binding motif in ASK. Importantly, we demonstrated that (i) the affinities of the LEDGF/p75 IBD and its interaction partners are modulated by phosphorylation, (ii) IBM components of this network are subject to phosphorylation both in vitro and in cellulo, (iii) IBM can be phosphorylated by casein kinase 2 (CK2), and (iv) elimination of IBM phosphorylation reduces the oncogenic potential of MLL1-ENL leukemic cells.
Results
Structural Conservation of LEDGF/p75–IBM Interfaces.
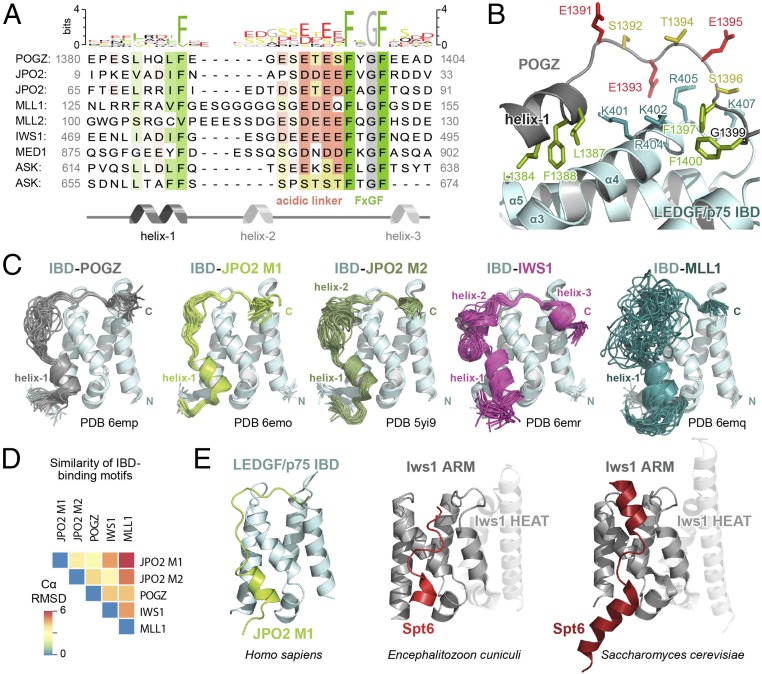
To identify the comprehensive set of sequence-encoded features important for LEDGF/p75 IBD binding, we extended our previous bioinformatics search for IBMs (17) by permitting gaps in the multiple sequence alignments used for motif discovery. Relaxing this requirement led to the identification of a previously undetected conserved region, which we refer to as helix-1 (Fig. 1A). Helix-1 is characterized by high alpha-helical propensity and a conserved phenylalanine upstream of the previously described charged linker region and FxGF motif. This analysis revealed the existence of a second motif in ASK, in addition to the previous motifs discovered in a variety of transcription regulators such as MLL1, MLL2, IWS1, and others. We validated this approach by NMR spectroscopy of JPO2, a factor with a high-affinity IBM. NMR resonance assignment of 3D HNCO spectra for JPO21–130 in the presence and absence of the LEDGF/p75 IBD revealed a significant shift in the helix-1 region at positions 65–73 where helix-1 is found in motif 2 (SI Appendix, Fig. S1 A and B), consistent with the binding of helix-1 to the LEDGF/p75 IBD. We conclude that, in addition to the downstream FxGF motif, helix-1 is an important conserved element of the IBM.
Fig. 1.
LEDGF/p75 IBD binding partners interact in a structurally conserved manner. (A) Multiple sequence alignment illustrating the conserved LEDGF/p75 IBD binding motif (IBM). Helix-1, the acidic linker, and the FxGF region are indicated below the alignment. (B) Detail of the POGZ IBM–IBD interface from the obtained solution structure. Conserved side chains within POGZ supporting the interaction with IBD are presented as sticks and labeled. The linker region stabilizes the interaction by nonspecific electrostatic contacts with the positively charged patch on the IBD surface. (C) Solution structures of the IBD in complex with the binding motifs from POGZ, JPO2 motif 1 and 2 (M1, M2), IWS1, and MLL1 determined by NMR spectroscopy. (D) Rmsd values comparing structural similarity of each pairwise combination of IBD-bound IBM. (E) The related ARM subdomain of Iws1 from different species interacting with Spt6 (PDB ID codes 3oak and 2xpo, refs. 34 and 35) highlights binding mode similarity to LEDGF/p75 IBD–IBM interactions as exemplified by JPO2 M1.
We set out to characterize the structure of the interface, including helix-1. To overcome the modest affinity (Kd > 10 μM) (17, 18) and stabilize the IBD–IBM interface, we designed covalently linked chimeric proteins combining the IBD with the IBMs of POGZ (amino acids 1370–1404), MLL1 (amino acids 111–160), IWS1 (amino acids 446–548), JPO2 motif 1 (M1, amino acids 1–32), and JPO2 motif 2 (M2, amino acids 56–91), respectively. The flexible linker between the IBD and IBM is formed by intrinsically disordered natural sequences of the original proteins already present in the sequences identified above (except for the IBD-MLL1 chimera, where we used a 12-residue GS-repeat to prevent proteolysis). We prepared 13C/15N-labeled recombinant chimeric proteins and obtained complete 15N, 13C, and 1H resonance assignments that allowed automated assignment of the NOEs identified in the 3D 15N- and 13C-edited NOESY spectra, which we used for structural determination. Statistics for the final water-refined sets of structures are provided in SI Appendix, Table S1.
Detailed structural analysis of the protein complexes revealed the presence of a conserved α-helix (helix-1), confirming our motif analysis described above (Fig. 1 A and B). This helix was not detected in the absence of the IBD (SI Appendix, Fig. S1C), indicating that it is only formed upon binding to the IBD. Obtained solution structures revealed that the overall binding interface is highly conserved across each member of the LEDGF/p75 IBD interaction network (Fig. 1C). Helix-1 harbors three conserved bulky amino acid residues, anchoring it in the hydrophobic pocket created by α3, α4, and α5 of the IBD (Fig. 1B). Helix-1 is followed by the flexible acidic linker with variable length spanning 8–16 aa residues. The acidic linker recognizes the positively charged surface formed by residues K401, K402, R404, R405, and K407 (Fig. 1B and SI Appendix, Fig. S2). The third conserved IBM region, the FxGF motif, is anchored between the two interhelical loops of the LEDGF/p75 IBD (amino acids I359 to D369 and K402 to M413) via the two phenylalanines. The aromatic ring of first FxGF phenylalanine resides in a relatively shallow hydrophobic pocket formed by L363, L368, I403, F406, K407, and V408 of the LEDGF/p75 IBD. The second phenylalanine is buried in a deeper pocket defined by I359, K360, L363, T399, K402, and I403 (SI Appendix, Fig. S2).
To quantitatively compare the newly resolved structures, we measured rmsd values of mean Cα residue coordinates for each pairwise combination of IBMs (Fig. 1D). The analysis shows that the motifs of POGZ and JPO2 M1 have the most similar binding mode (rmsd = 2.9 Å), and the MLL1 motif is generally the most divergent [mean rmsd = 5.6 (± 0.8 SD) Å], likely arising from its longer unstructured region. We observed some variation between individual structures, for example additional helices in JPO2 and IWS1 and a flexible glycine repeat in MLL1 (SI Appendix, Fig. S3 A and B). Despite these differences, the positions of conserved residues in helix-1 and the FxGF motif, as well as the charged linker bridging these regions, remain consistent throughout the structures we obtained. Therefore, the IBD–IBM interface is structurally highly conserved across the LEDGF/p75 interactome.
To assess the similarity between LEDGF/p75 IBD–IBM interactions and other members of the TFIIS TND family, we compared our structures to the previously obtained structures of the Iws1 ARM domain in complex with its binding partner in two different species (34, 35). Comparison of Iws1 ARM–Spt6 complexes from Saccharomyces cerevisiae and Encephalitozoon cuniculi with our LEDGF/p75 IBD–IBM complexes revealed a striking similarity between the molecular interfaces (Fig. 1E). Like all IBMs, the binding motif on Spt6 consists of an alpha-helix and an acidic flexible linker followed by a hydrophobic anchoring region. Therefore, in addition to the structural conservation of LEDGF/p75 IBD–IBM complexes identified above, we find that interactions involving distantly related TNDs employ similar binding modes. These results show that interactions between TNDs and their binding partners have been largely preserved throughout evolution.
Helix-1 Is Essential for Interaction with LEDGF/p75.
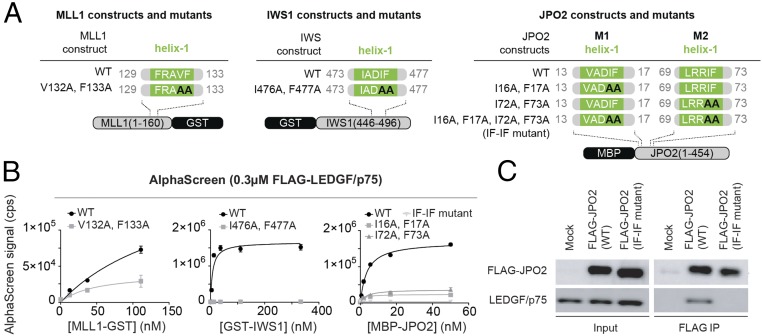
To assess the importance of helix-1 for interaction with LEDGF/p75, we introduced alanine mutations at positions (i) I476 and F477 in recombinant GST-tagged IWS1446–496, (ii) V132 and F133 in GST-tagged MLL11–160, and (iii) I16, F17 (M1) and/or I72, F73 (M2) in maltose binding protein (MBP)-tagged JPO2 (Fig. 2A). We then analyzed the interaction of WT and mutant proteins with LEDGF/p75 using an AlphaScreen assay. In the case of IWS1, mutations completely abolished the interaction with LEDGF/p75, while the effect was less pronounced for MLL1. Because JPO2 has two IBMs, mutation of helix-1 in both IBMs was required to fully abrogate the interaction (Fig. 2B). Together these data confirm the importance of helix-1 residues for maintaining the interaction.
Fig. 2.
Helix-1 in the IBM is essential to maintain interaction with LEDGF/p75 IBD. (A) Diagrams of MLL1, IWS1, and JPO2 constructs and mutants used in B and C. (B) The WT and/or mutant recombinant MLL1, IWS1, and JPO2 protein constructs were titrated against FLAG-tagged LEDGF/p75 in an AlphaScreen assay (counts per second). Error bars represent SDs calculated from independent measurements (n = 4). (C) HEK 293T cells were transfected with FLAG-tagged WT JPO2 and/or the IF-IF mutant. After immunoprecipitation with anti-FLAG antibody the presence of FLAG-tagged proteins and endogenous LEDGF/p75 was confirmed by Western blot.
To evaluate the significance of helix-1 for LEDGF/p75 binding in a cellular context, we expressed FLAG-tagged JPO2 or JPO2 I16A, F17A, I72A, F73A (“IF-IF mutant”) in HEK293T cells and probed coimmunoprecipitation of endogenous LEDGF/p75 (Fig. 2C). Whereas WT JPO2 was able to precipitate LEDGF/p75, we did not detect interaction with the IF-IF mutant JPO2.
Phosphorylated Side Chains of IBM Sample the Positively Charged Region on the LEDGF/p75 IBD Surface.
Since all known IBD interactions are mutually exclusive (17), we investigated their potential regulatory mechanisms. Because of its observed plasticity in our structural studies, we hypothesized that the linker bridging helix-1 and the FxGF motif may serve as a regulatory switch, increasing or decreasing the affinity of an IBM by varying its charge density. Specifically, the presence of multiple Ser/Thr residues amenable to phosphorylation and negatively charged amino acids in the linker region may contribute ionic interactions (Fig. 3A).
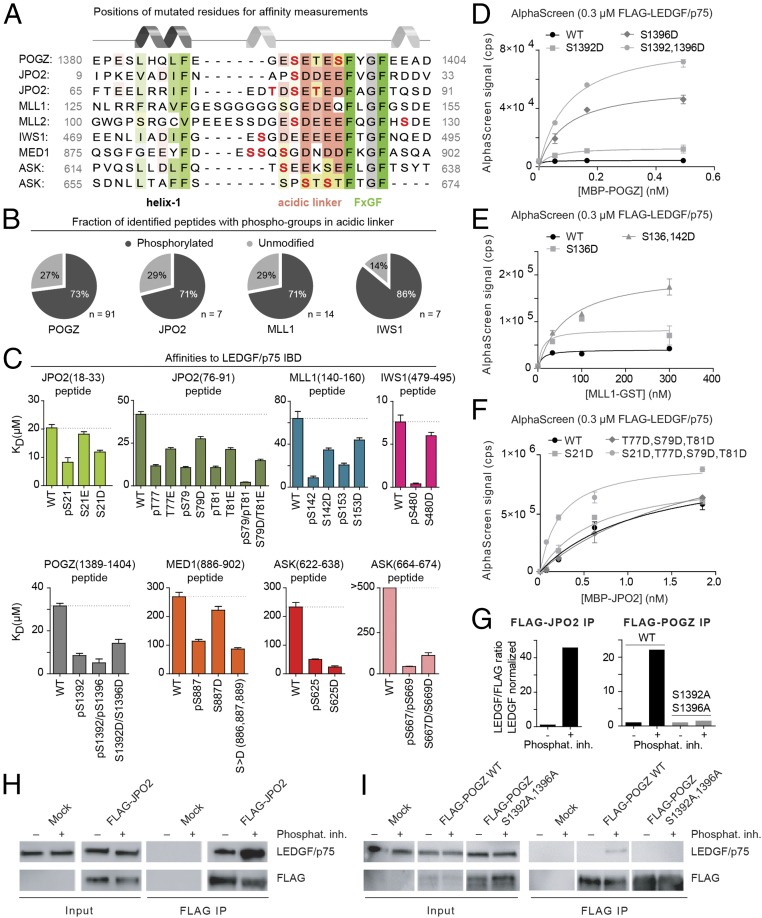
Fig. 3.
Phosphorylation increases the affinity of IBMs for the LEDGF/p75 IBD. (A) Multiple sequence alignment of the LEDGF/p75 IBD binding motif (IBM) highlighting residues modified in B–D and F in red. (B) The abundance of phosphorylation in the JPO2, POGZ, MLL1, and IWS1 IBMs as measured by mass spectroscopy. Peptides carrying at least one phospho-group in the acidic linker region and unmodified peptides were quantified. (C) The dissociation constants of WT IBMs compared with phosphorylated or phosphomimetic versions of the peptides. Kd values were obtained by NMR titration. Error bars represent the error of the fit for the 10 most perturbed residues. WT or phosphomimetic mutants of POGZ (D), MLL1 (E), and JPO2 (F) were titrated against FLAG-tagged LEDGF/p75 in AlphaScreen. Error bars represent SDs calculated from independent measurements (n = 4). (G) Densitometry of Western blot bands in H and I. Data are plotted as the ratio of LEDGF/p75 over FLAG-tagged protein, normalized by WT LEDGF/p75. (H and I) HEK 293T cells were transfected with FLAG-tagged WT JPO2 or POGZ and the proteins were immunoprecipitated in the absence (−) or presence (+) of phosphatase inhibitors with anti-FLAG antibody. For FLAG-POGZ, a mutant with modified Ser residues in the IBM region (PogZ S1392A, S1396A) was used as an additional control. The presence of FLAG-tagged proteins and endogenous LEDGF/p75 was confirmed using specific antibodies by Western blot.
In support of this hypothesis, the PhosphoSitePlus database (36–39) includes several entries for phosphorylated serines and threonines in the IBM region of different LEDGF/p75 interaction partners (SI Appendix, Table S2). To systematically evaluate phosphorylation sites within IBMs in cells, we employed mass spectrometry. FLAG-tagged JPO2, POGZ1117–1410, MLL1-ELL (an oncogenic MLL-fusion), and IWS1 were expressed in HEK293T cells, immunoprecipitated using anti-FLAG antibodies (SI Appendix, Fig. S4), and analyzed by mass spectrometry. We detected phosphorylation events on all Ser and Thr residues in the IBMs examined, confirming that these residues are subject to posttranslational modification by kinases in cells. Together with previous reports (36–39), our proteomic study reveals that each IBM harbors at least one amino acid residue that can be phosphorylated in cells (SI Appendix, Tables S2 and S3). Interestingly, the investigated IBMs had very high abundance of peptides carrying at least one phospho-group in the linker region (Fig. 3B). Other posttranslational modifications were not detected in studied IBM regions. A detailed overview of specific peptides identified by mass spectrometry is provided in SI Appendix, Table S3.
To visualize the dynamic effects of IBM phosphorylation, we performed 500 ns of molecular dynamics (MD) simulations for all of the experimentally confirmed phosphorylated variants of binding partners based on our solution structures shown in Fig. 1C. Comprehensive networks of electrostatic interactions were readily established between every tested phospho-group and the positively charged side chains from the IBD within 200 ns of MD simulation (SI Appendix, Fig. S5). Hence, the addition of negative charge density arising from phosphorylation of the IBM results in rapid establishment of new ionic interactions with the LEDGF/p75 IBD.
To validate the effect of IBM phosphorylation on the interaction with the IBD, we synthesized a comprehensive set of IBM peptide variants including phosphorylated Ser/Thr residues or phosphomimetic mutations (Asp or Glu) (Fig. 3A and SI Appendix, Table S4). We used NMR to follow changes in positions of the LEDGF/p75 IBD backbone amide signals induced upon titration with IBM peptide and determined dissociation constants for all synthetic peptides (Fig. 3B and SI Appendix, Fig. S6). Of all WT IBMs, IWS1 contains the highest number of charged residues in the linker region and also has the highest affinity for the IBD (Kd = 7.6 ± 0.8 μM). This observation is consistent with the notion that elevated negative charge density in the linker region stabilizes the LEDGF/p75 IBD–IBM interface.
Affinity for the LEDGF/p75 IBD was significantly increased (reflected by reduced dissociation constant Kd) upon addition of negative charge by phosphorylation or phosphomimetic mutation in the IBM linker region in all tested variants (Fig. 3C). Phosphorylation of the motif typically increased binding affinity to the IBD 2- to 10-fold. The partial effects of phosphomimetic mutation observed in some cases can be attributed to the lower negative charge (−1) carried by aspartate or glutamate compared with the phosphate group (−1 to −2) at physiological pH (40).
AlphaScreen experiments confirmed the positive effect of additional negative charges in the IBM on the interaction with the IBD (Fig. 3 D–F). WT and/or mutant recombinant MBP-tagged POGZ1117–1410 was titrated against FLAG-tagged full-length LEDGF/p75 (Fig. 3D). Introduction of a S1392D phosphomimetic mutation (POGZ1117–1410 S1392D) in the IBM increased the AlphaScreen signal twofold, while mutation of both Ser residues to Asp (POGZ1117–1410 S1392 D/S1396D) led to a 16-fold increase (Fig. 3D). Likewise, introduction of S136D or S136D/S142D double mutation in GST-tagged MLL11–160 (MLL-GST) resulted in increased AlphaScreen counts (Fig. 3E). Mutation of the sole Ser residue in JPO2 M1 (S21D) or all phosphorylatable residues in the second motif (T77D, S79D, and T81D) yielded no discernible differences in the AlphaScreen signal, whereas the combination of both mutants did increase affinity (Fig. 3F).
To confirm these results in a cellular setting we performed coimmunoprecipitation with FLAG-tagged JPO2 expressed in HEK293T cells in the absence or presence of phosphatase inhibitors. Following expression of FLAG-tagged JPO2, we observed increased coimmunoprecipitation of LEDGF/p75 using anti-FLAG antibodies in the presence of phosphatase inhibitors (Fig. 3 G and H). Furthermore, the interaction between LEDGF/p75 and FLAG-tagged POGZ could not be observed using anti-FLAG immunoprecipitation in the absence of phosphatase inhibitors; however, coimmunoprecipitation of LEDGF/p75 was detected upon addition of phosphatase inhibitors (Fig. 3 G and I). To confirm that the serine residues in the POGZ IBM mediate this interaction, we introduced a double S1392A/S1396A mutation. Because the double mutant did not interact with LEDGF/p75 in the presence or absence of phosphatase inhibitors, we conclude that phosphorylation at these positions acts as a switch to regulate the affinity between POGZ and LEDGF/p75.
Phosphorylation of MED1 Drives Interaction with LEDGF/p75.
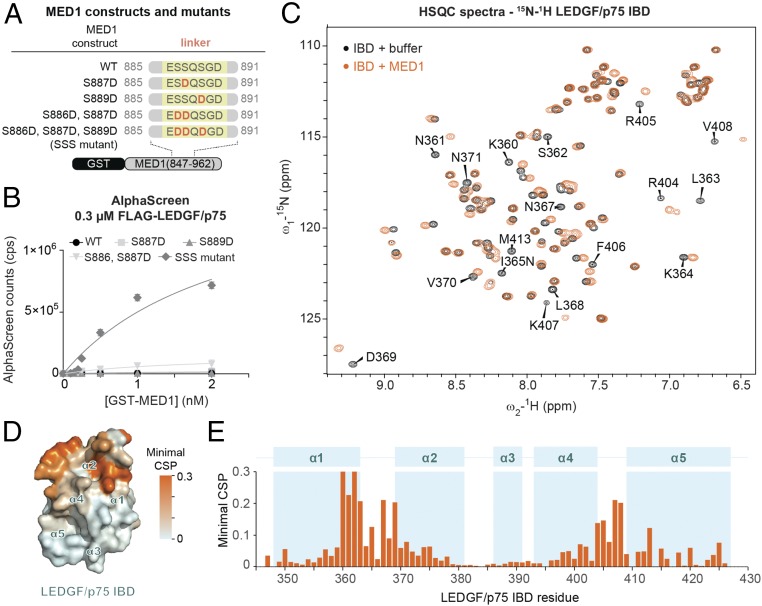
MED1 (Mediator subunit 1, DRIP205, Trap220) is a component of the Mediator complex, a coactivator involved in transcription of nearly all RNA polymerase II-dependent genes (41). MED1 harbors an evolutionarily conserved IBM (Fig. 1A) (17). The WT MED1 IBM synthetic peptide had low-to-modest affinity for the IBD in NMR titrations (269 ± 15 μM) (Fig. 3B). Interestingly, the MED1 IBM contains three putative phosphorylation sites (S886, S887, and S889), of which one has been experimentally detected in vivo (SI Appendix, Table S2) (42). To examine the effects of phosphorylation on the MED1–LEDGF/p75 interaction, we introduced phosphomimetic mutations in the MED1 IBM upstream of the FxGF motif and assayed them with AlphaScreen (Fig. 4A). While single substitution of Ser to Glu at positions 887 or 889 did not stimulate interaction in AlphaScreen, the combination mutants S886D/S887D and S886D/S887D/S889D (“SSS mutant”) increased the affinity of MED1 for LEDGF/p75 (Fig. 4B). The interaction was also confirmed by anti-FLAG coimmunoprecipitation in HEK293T cells expressing FLAG-tagged MED1 (SI Appendix, Fig. S7).
Fig. 4.
Validation of MED1 as an LEDGF/p75 IBD binding partner. (A) Diagrams of MED1 constructs and mutants used in B. (B) WT and/or mutated recombinant GST-MED1 was titrated against FLAG-tagged LEDGF/p75 in AlphaScreen. Error bars represent SDs calculated from independent measurements (n = 4). (C) Comparison of the 2D 15N/1H HSQC NMR spectra of the IBD in the absence (black) or presence (orange) of MED1847–930. Spectra were obtained from the 15N-labeled recombinant IBD and an unlabeled recombinant MED1 fragment. The residues with the strongest shifts are annotated. (D) IBD amino acid residues that are significantly perturbed upon addition of MED1847–930 (as determined by NMR spectroscopy) are highlighted in orange on the surface of the IBD structure. (E) Representation of the minimal CSP in backbone amide signals of the IBD upon addition of MED1847–930.
For a detailed characterization of the LEDGF/p75–MED1 interface, we used NMR spectroscopy. Specifically, we compared the changes in positions of backbone amide signals in the spectra of 15N-labeled LEDGF/p75 IBD (amino acids 345–426) in the presence or absence of a recombinant MED1 fragment (amino acids 847–930) (Fig. 4 C–E). Analysis of the chemical shift perturbations (CSPs) in the IBD backbone induced by binding of MED1 revealed a pattern remarkably similar to that induced upon addition of MLL1, POGZ, JPO2, and IWS1 (SI Appendix, Fig. S9). Together, these results establish MED1 as a genuine LEDGF/p75 IBD interactor and confirm that the MED1 fragment binds in the same conserved structural mode as other factors in the LEDGF/p75 IBD interaction network.
ASK Bears LEDGF/p75 Interaction Sites with Differential Kinase-Regulated Affinities.
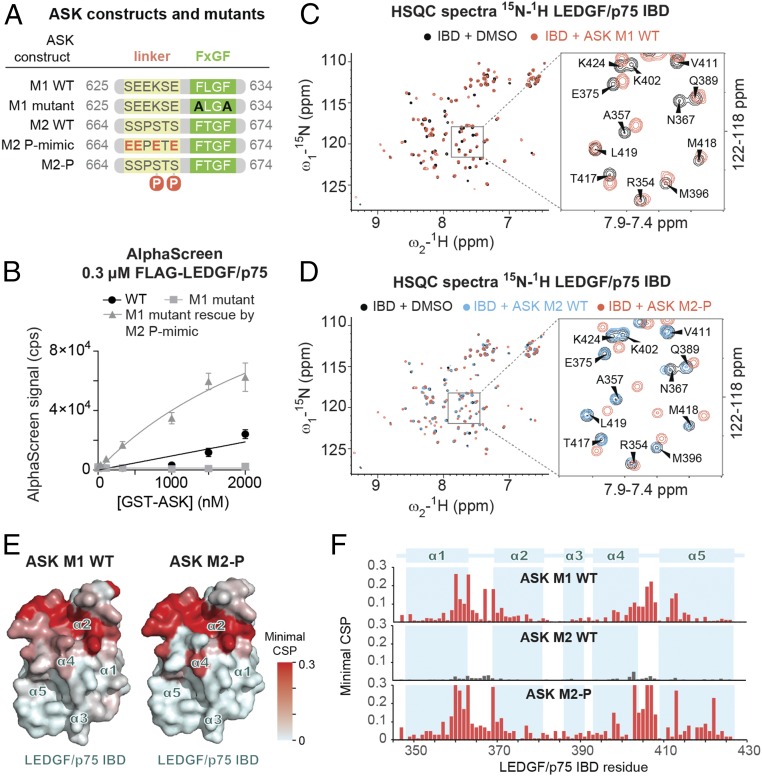
All known physiological LEDGF/p75 IBD interaction partners harbor several negatively charged residues in the linker between helix-1 and the FxGF motif of the IBM. Since negative charge can also be provided through phosphorylation, we performed a new search for proteins containing helix-1 and FxGF motifs interspersed by phosphorylatable Ser/Thr residues. Surprisingly, ASK showed such a region at its extreme C terminus (ASK M2), right behind the first motif (ASK M1). To establish whether ASK M2 is able to interact with LEDGF/p75, we purified a GST-tagged C-terminal fragment of ASK (amino acids 598–674, GST-ASK) and evaluated the interaction of WT and/or mutated ASK constructs (Fig. 5A). Using an AlphaScreen assay with FLAG-tagged LEDGF/p75, we found that WT ASK598–674 readily interacts with LEDGF/p75, and that double mutation of FLGF to ALGA in the FxGF motif of ASK M1 abolished the interaction (Fig. 5B). However, phosphomimetic mutation of the other motif, ASK M2 (SSPSTS to EEPETE in the linker, “P-mimic”), restored binding to LEDGF/p75, suggesting that ASK M2 may interact with LEDGF/p75 upon phosphorylation (Fig. 5B). Indeed, previous studies detected phosphorylation of at least two of the Ser residues in the ASK M2 linker in vivo (SI Appendix, Table S2) (36).
Fig. 5.
ASK has two IBMs whose affinities for the LEDGF/p75 IBD are differentially regulated by phosphorylation. (A) Diagrams of WT and modified constructs of ASK motif 1 and 2 (M1, M2) used in B–E. (B) The WT and/or mutated recombinant ASK protein constructs were titrated against FLAG-tagged LEDGF/p75 in AlphaScreen. Error bars represent SDs calculated from independent measurements (n = 4). (C and D) Comparison of the NMR 2D 15N/1H HSQC spectra of the 15N-labeled IBD in the absence and presence of different ASK synthetic peptides (see also SI Appendix, Fig. S8). (E) IBD amino acid residues that are significantly perturbed upon addition of ASK M1 and phosphorylated ASK M2 (as determined by NMR spectroscopy) are highlighted in red on the surface of the IBD structure. (F) Representation of the minimal CSP in backbone amide signals of the IBD upon addition of ASK-derived peptides.
To confirm this observation and characterize the ASK–IBD interaction in more detail, we compared the changes in positions of backbone amide signals of 15N-labeled LEDGF/p75 IBD in the 15N/1H HSQC spectra before and after addition of ASK M1 and ASK M2 peptides as well as a phosphorylated ASK M2 (pS667 and pS669, “M2-P”) (Fig. 5 C and D and SI Appendix, Fig. S8). The CSPs in the IBD backbone induced by binding of ASK M1 are comparable to those observed earlier for MED1 (Fig. 4 D and E), as well as the other binding partners (SI Appendix, Fig. S9) (17). ASK M2 induced only minor changes even when present at a 16-fold molar excess over the IBD. However, we could clearly detect interaction with IBD upon titration of the double-phosphorylated ASK M2-P (Fig. 5F). We obtained dissociation constants for these interactions from NMR titration experiments (Fig. 3B), with the exception of WT ASK M2, whose very low affinity (≥500 μΜ) precluded accurate characterization by NMR due to the peptide’s limited solubility. The modest affinity observed for ASK M1 (233 ± 16 μM) was comparable to that obtained for MED1 (269 ± 15 μM) and consistent with the lack of negatively charged residues within the linker region (Fig. 1A). In summary, we find that phosphorylation or phosphomimetic mutation at Ser624 (ASK M1) or Ser667/S669 (ASK M2) led to a 5- to 10-fold increase in binding affinity compared with the unmodified motifs (Fig. 3B).
Kinase-Dependent Phosphorylation of IBMs in LEDGF/p75-Binding Partners.
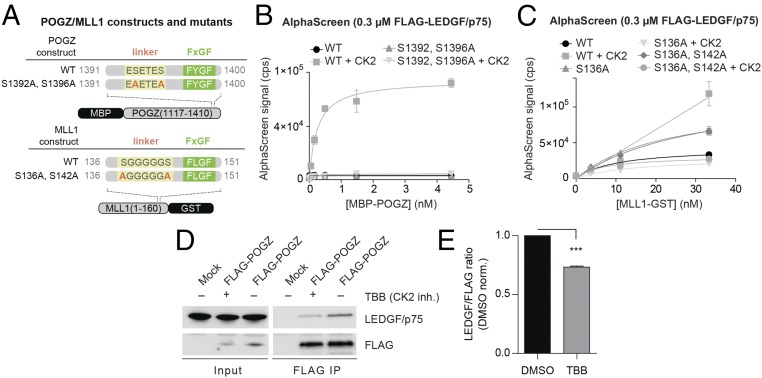
Using mass spectrometry following anti-FLAG immunoprecipitation of either FLAG-tagged MLL1-ELL or JPO2 in HEK293T cells, we identified several potential binding partners, including several kinases (SI Appendix, Table S5). Interestingly, multiple peptides of CK2 subunits were detected as JPO2 and MLL1 interacting proteins. Furthermore, the general CK2 recognition motifs for phosphorylation are SxxE/D, SxxpS/pT, and SxE/D (43). All known LEDGF/p75 IBD binders contain at least one of these CK2 consensus motifs in their IBMs. Because we identified CK2 as a recurrent binding partner, and because of the presence of CK2 motifs in each IBM, we sought to test whether CK2 could mediate phosphorylation of the IBM motifs. To investigate the effect of CK2 treatment on the interaction of LEDGF/p75 with POGZ, we designed constructs bearing Ser/Thr to Ala substitutions, blocking phosphorylation of the linker (Fig. 6A). We then performed AlphaScreen assays of MBP-tagged POGZ with or without CK2 preincubation (Fig. 6B). CK2 treatment significantly increased the affinity of WT POGZ for LEDGF/p75, whereas S1392/1396A POGZ mutation (which cannot be phosphorylated in the IBM region) was insensitive to the effects of CK2 preincubation. Similar but less-pronounced results were obtained for MLL1 (Fig. 6C), where binding of a S136A/S142A mutant was unaffected by CK2 phosphorylation.
Fig. 6.
IBM phosphorylation by CK2 increases the affinity of POGZ and MLL1 for the LEDGF/p75 IBD. (A) Diagrams of POGZ and MLL1 constructs and mutants used in B and C. (B and C) WT or mutants of POGZ (B) and MLL1 (C) were titrated against FLAG-tagged LEDGF/p75 in AlphaScreen after preincubation with or without CK2. Error bars represent SDs calculated from independent measurements (n = 4). (D) HEK 293T cells were transfected with FLAG-tagged POGZ. The proteins were immunoprecipitated with anti-FLAG antibody in the absence (−) or presence (+) of selective CK2 inhibitor (TBB). The presence of FLAG-tagged POGZ and endogenous LEDGF/p75 was confirmed using Western blot. (E) Densitometry of Western blot bands. Data are plotted as the ratio of LEDGF/p75 over FLAG-POGZ, normalized by DMSO control (mean ± SD from n = 3 independent replicates). P values obtained from Student’s t test; ***P < 0.0001.
To confirm that LEDGF/p75 binding partners can be phosphorylated by CK2 in a cellular context, we expressed FLAG-tagged POGZ in HEK293T cells and performed coimmunoprecipitation in the presence or absence of 4,5,6,7-tetrabromobenzotriazole (TBB) (44), a selective inhibitor of CK2 (Fig. 6D). We reproducibly observed decreased coimmunoprecipitation of LEDGF/p75 using anti-FLAG antibodies in the presence of TBB (Fig. 6 D and E). While we do not rule out a role for other kinases, our results confirm that CK2-dependent phosphorylation of IBMs can act as an affinity switch regulating association with LEDGF/p75.
Helix-1 and Phosphorylation of the Linker Contribute to Leukemic Transformation by MLL1 Fusion Proteins.
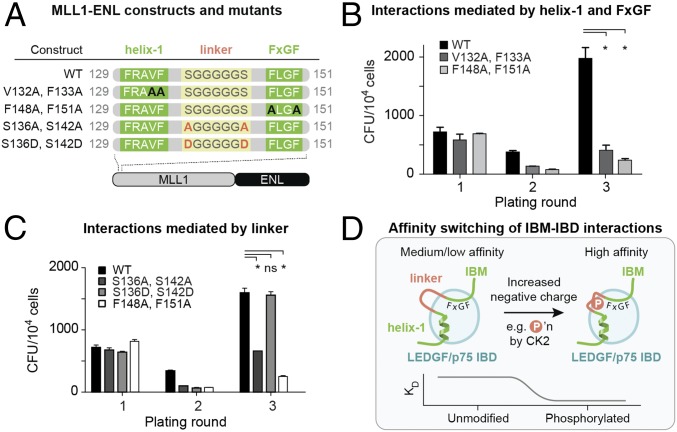
Our bioinformatic and experimental results above show that the IBM consists of three distinct conserved regions: helix-1, the acidic linker, and the hydrophobic FxGF motif. We also demonstrated that mutations in both the helix-1 and acidic linker regions of IBMs abolish association with the IBD. Since we previously reported that mutations at the FxGF–IBD interface interfere with leukemic transformation (18), we sought to assess the contributions of IBM helix-1 and linker phosphorylation to MLL1-rearranged leukemia. To address this, we measured the colony-forming potential of MLL1 fusions following mutation of these regions in primary murine lineage-depleted hematopoietic stem cells. We generated the V132A, F133A double mutation (at helix-1) of the MLL1-ENL oncogenic fusion (Fig. 7A) and measured the number of colony forming units (CFU) using serial plating. As a positive control, we mutated the phenylalanine residues in the FxGF motif to alanine. Like the FxGF mutant, the helix-1 mutant significantly reduced the transforming capacity of MLL1-ENL fusion after three plating rounds (Fig. 7B).
Fig. 7.
Helix-1 and phosphorylation of flexible linker in MLL1 IBM are both crucial for MLL1-fusion-driven transformation. (A) Diagrams of MLL1 constructs and mutants used in B and C. (B and C) The colony-forming capacity of murine stem cells transformed with an MLL1-ENL fusion was compared with those transformed with MLL1-ENL mutants in helix-1 (B) or in the flexible linker and FxGF motif (C). CFU of lineage-depleted murine hematopoietic stem cells transduced with vectors carrying the WT or mutated MLL1-ENL fusions were determined in three rounds. Errors bars indicate SDs determined from independent replicates (n = 3). Statistical differences were determined using Student’s t test; *P < 0.05; ns, nonsignificant compared with WT. (D) Summary and conceptual model of IBM affinity switching.
To assess the effect of phosphorylation of the MLL1 IBM on MLL1-rearranged leukemia, we introduced several mutations that blocked phosphorylation in the linker of MLL1 (Fig. 7A) and expressed them in an MLL1-ENL fusion. Substitution of the phosphorylatable serines S136 and S142 to alanine significantly reduced the transformation capacity of the fusions (Fig. 7C). In contrast, introduction of the phosphomimetic mutations S136D and S142D rescued this loss, almost fully recovering the oncogenic activity of the fusion. Overall, these results validate our structural findings and demonstrate an essential role of helix-1 and linker phosphorylation for in vivo, with particular importance for the MLL1 IBD during leukemogenesis.
Discussion
IBMs present in all LEDGF/p75 IBD interaction partners are typical examples of SLiMs, conserved amino acid sequences found in natively disordered parts of a protein. Protein sequence motifs lacking well-defined structure are surprisingly common in eukaryotes. However, the characterization of SLiM-mediated interactions represents a major challenge due to their modest affinity as well as the transient and dynamic nature of their interactions. An ideal tool to overcome these issues is NMR spectroscopy, which allowed us to examine the structures of IBMs in the context of their native protein-binding scaffold. Our approach has permitted characterizing the complete binding interfaces of these complexes and revealed a well-defined tertiary structure of the IBM upon docking to its cognate binding site on the IBD of LEDGF/p75. Extensive structural analysis of the LEDGF/p75 IBD interactome revealed that IBMs recognize the IBD through three distinct structurally conserved regions: helix-1, the flexible acidic linker, and the FxGF motif. Our mutational analysis of various IBMs underscored the importance of each of these regions for stabilization of the IBM–IBD complex. In addition, the conservation of the IBM–IBD binding mode extends beyond the LEDGF/p75 interactome. We found that other TNDs are recognized by their binding partners in a structurally conserved manner preserved throughout evolution.
MLL1 is the only known IBD binding partner that forms a ternary complex with tumor suppressor MENIN (16, 29). In comparison with the other interaction partners of LEDGF/p75, the MLL1 IBM shows greater flexibility of the linker, likely mediated through poly-G repeats and some flexibility (∼40° rotation) in the positioning of helix-1 in the absence of MENIN (Fig. 1C and SI Appendix, Fig. S3A). Comparison of our solution structure with an available crystal structure that includes MENIN, the IBD, and an MLL1-derived peptide, but which did not resolve the full IBM-IBD interface (29), suggests that MENIN binding induces a ∼50–90° rotation of helix-1 (SI Appendix, Fig. S3C), likely necessitating the elevated flexibility of the MLL1 IBM. Because this flexibility precluded resolution in previous X-ray crystallography studies, our NMR experiments provide important insights into the highly dynamic interaction surface between MLL1 and LEDGF/p75.
While most IBD binding partners have a single IBM, ASK and JPO2 each have two copies of the motif, leaving open the possibility that these factors may participate in higher-order structures such as oligomers, or nucleate higher concentrations of LEDGF/p75 at biological active sites. Such a model would, for example, permit the accumulation of LEDGF/p75 in gene bodies. Future studies are needed to test and confirm this notion.
Despite LEDGF/p75’s only containing a single PWWP domain capable of binding methylated H3K36, the genome-wide chromatin binding profile of LEDGF/p75 does not simply follow H3K36 methylation marks. Instead, LEDGF/p75 is enriched at sites containing additional marks of active transcription like H3 and H4 acetylation, H3K4me1, and RNA polymerase II binding (45). Since several LEDGF/p75 binding partners have diverse roles on chromatin, it is appealing to speculate that physiological binding partners may contribute to the specificity of the LEDGF/p75–chromatin interactions.
Through motif conservation and biochemical characterization, we validated MED1 as a LEDGF/p75 interaction partner that recognizes the IBD through its IBM. Mediator is a large transcriptional coactivator complex composed of ∼30 distinct subunits that acts as a bridge between transcriptional activators and RNA polymerase II (46). It remains to be investigated under which conditions MED1 interacts with LEDGF/p75 and how LEDGF/p75 may influence the association of the Mediator complex at transcription start sites and/or gene bodies.
In our studies, we have found that the flexible linker connecting helix-1 and the FxGF motif of IBMs spans 8–16 residues, and negatively charged residues contribute >50% of its sequence content (Fig. 1A). Furthermore, we found that addition of negative charges through phosphorylation increases the affinity of binding partners for LEDGF/p75. The increased charge density in the acidic linker results in stabilization of the complexes by formation of new electrostatic interactions between the linker and a positively charged surface on the LEDGF/p75 IBD (Fig. 1B and SI Appendix, Fig. S2). Phosphorylation provides a reversible mechanism for affinity switching (Fig. 7D), which can respond to intrinsic and extrinsic signaling cues. This property makes IBMs versatile elements whose interactions with LEDGF/p75 can be tuned in a modular differential fashion through posttranslational modification. Because IBM–IBD interactions are stabilized through a small number of intermolecular contacts, the addition of a single electrostatic contact can shift the steady-state equilibrium, dramatically affecting the interaction landscape between individual IBMs and LEDGF/p75. The strong changes we observe are reminiscent of the strong sensitivity of SH2-binding proteins to tyrosine phosphorylation (31).
HIV integrase is the only known LEDGF/p75 IBD interaction partner which does not harbor an IBM. Although the interaction mechanism differs significantly, the HIV integrase binding site on the IBD partially overlaps the surface recognized by IBMs. The N-terminal domain of HIV integrase interacts with the positively charged surface recognized by the IBM acidic linker, and the integrase catalytic core domain binds to part of the hydrophobic pockets occupied by the FxGF motif (SI Appendix, Fig. S10) (47). Compared with the IBMs, HIV integrase buries considerably more hydrophobic surface area upon interaction with the IBD, allowing it to compete with physiological binding partners and ensure efficient viral replication.
In our studies, we demonstrated that IBMs undergo phosphorylation in vitro as well as in cellulo, altering their affinity for the LEDGF/p75 IBD. Using proteomic approaches, we identified kinases CK2, DNA-dependent PKC, TPK-JAK1 and P38 associated with JPO2 and CK2 and Rho-PK2 associated with MLL-ELL. Interestingly, JPO2 and its target monoamine oxidase A are involved in serum starvation-induced p38 kinase-mediated apoptosis downstream of p38 kinase activity (48). Additionally, both LEDGF/p75 and DNA-dependent PKC regulate DNA end resection and are components of the homologous recombination DNA repair machinery (49–52). The exact relationship between the LEDGF/p75 interactome and these kinases remains to be investigated.
Since CK2 was found in both the JPO2 and MLL-ELL mass spectrometry experiments and all IBMs possess at least one general recognition motif for phosphorylation by CK2, this kinase represents a prime candidate for phosphorylation of LEDGF/p75 interactors. However, we do not exclude that the IBMs may be modulated by other kinases as well. CK2 is overexpressed in numerous types of cancer and associates with poor prognosis (48, 53–56). CK2 promotes cell proliferation, cell growth, and survival and is involved in the modification of cell morphology and angiogenesis (57–59). The ability of CK2 to act as an oncogenic factor is largely based on its role in several signal transduction pathways including PI3K/PTEN/Akt/mTOR, JAK/STAT, Wnt, and Ras/MEK/ERK signaling (53, 60, 61). By phosphorylation of specific residues in components of these pathways, CK2 contributes to the up-regulation of genes conferring a survival advantage (62–64). An example of CK2-mediated regulation in the Ras/MEK/ERK signaling cascade is its direct phosphorylation of ERK1/2 S244, S245, and S246 within the nuclear localization signal with subsequent enhancement of nuclear translocation and activity (65). In addition, CK2 is known to inhibit both intrinsic and extrinsic apoptotic pathways by targeting caspase signaling (66). These findings make CK2 an attractive target for anticancer therapy, with several inhibitors currently in clinical trials (67–69).
Interestingly, CK2 was recently shown to play a role in regulating transcriptional elongation in both gene bodies and enhancer regions through phosphorylation of Y57 of histone H2A (70). Genome-wide binding analysis revealed that CK2α, the catalytic subunit of CK2, binds across RNAP2-transcribed coding genes and active enhancers. Kinase-dependent regulation of the LEDGF/p75 interactome therefore remains an exciting frontier with great potential for therapeutic application. Our results reveal that phosphorylation of IBMs plays a crucial role in the transformation of MLL-rearranged leukemic cells. Hence phosphorylation of IBMs (in particular on MLL1) represents an oncogenic dependency that may be exploited for development of precision therapies to treat MLL-rearranged leukemia.
Materials and Methods
Cloning, Protein Expression, and Purification.
For NMR experiments, the expression plasmids coding for the IBD (LEDGF/p75345–426), JPO21–130, POGZ1117–1410, IWS1352–548, or MLL1111–160, were described earlier (17, 18). The IBD–IBM chimeras described in this study consist of the C-terminally extended IBD (LEDGF/p75345–442) fused to JPO21–32, JPO256–91, POGZ1370–1404, IWS1447–548, and MLL1111–160, respectively. In case of the IBD-MLL1 tandem protein chimera, the flexible linker SG(GS)4SG was introduced between the IBD and MLL parts. Detailed information about plasmid construction for either prokaryotic or eukaryotic expression is provided in SI Appendix, Supplementary Methods. Expression and purification of FLAG-tagged LEDGF/p75, MBP-JPO2, MLL1–160-GST, and MBP-POGZ1117–1410 or mutants thereof was described earlier (7, 11, 14, 26). GST-tagged MED1847–962, ASK598–674, and IWS1446–496 plasmids were expressed in Escherichia coli cells and purified using Glutathione-Sepharose beads (GE Life Sciences) using standard protocol.
NMR Spectroscopy and MD.
NMR spectra were acquired at 25 °C on 600-MHz and 850-MHz Bruker Avance spectrometers equipped with 15N/13C/1H cryoprobes. A series of double- and triple-resonance spectra (71, 72) were recorded to determine essentially complete sequence-specific resonance backbone and side-chain assignments. Restraints for 1H–1H distance required for structural calculations were derived from 3D 15N/1H NOESY-heteronuclear single quantum coherence (HSQC) and 13C/1H NOESY-heteronuclear multiple quantum coherence, which were acquired using a NOE mixing time of 100 ms.
The families of converged structures were initially calculated using Cyana (73). Subsequently, five cycles of simulated annealing combined with redundant dihedral angle restraints were used to produce sets of converged structures with no significant restraint violations, which were further refined in explicit solvent. The statistics for the resulting structures are summarized in SI Appendix, Table S1. The structures, NMR restraints, and resonance assignments were deposited in the Protein Data Bank (PDB ID codes 6EMO, 6EMP, 6EMQ, 6EMR, and 5YI9) and the Biological Magnetic Resonance Data Bank (accession nos. 34179, 34180, 34181, 34182, and 36120).
To follow the changes in the chemical shifts of a protein when a ligand is bound and to determine the location of the ligand interaction site, we calculated minimal CSPs. Minimal CSP of each assigned resonance in the 2D 15N/1H HSQC spectra of the protein in free state is calculated as the geometrical distance in parts per million to the closest peak in the 2D 15N/1H HSQC spectra of the same protein upon ligand binding using the formula , where α is a weighing factor of 0.2 accounting for differences in the proton and nitrogen spectral widths (74).
The representative models from the experimentally derived NMR structures for the IBD complexes with JPO2 M1 and M2, POGZ, IWS1, and MLL1 were changed into phosphorylated variants by automatic building in the LEaP module of AMBER14 (75). The production run was 500 ns long, executed on graphics processing units. The resulting trajectories were analyzed using CPPTRAJ program (76) of AMBER14 and hydrogen bonds formed between the IBD and the residues from the flexible linker of binding partners are summarized in SI Appendix, Table S6.
Cell Culture and Isolation of Primary Murine Hematopoietic Stem Cells.
All cells were grown in a humidified atmosphere containing 5% CO2 at 37 °C. The 293T cells were grown in DMEM (Gibco-BRL) supplemented with 8% vol/vol heat inactivated FBS (Sigma-Aldrich) and 50 µg/ml gentamicin (Gibco). To isolate lineage-depleted murine hematopoietic stem cells, femurs and tibias of 8- to 10-wk-old C57BL/6J mice (Janvier) were harvested to extract crude bone marrow cells. Lineage-depleted (Lin-) progenitors were isolated from the crude extracts with the EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (STEMCELL Technologies) and cultured in RPMI-1640 (10% FCS and 50 µg/mL gentamicin), supplemented with mIL-6 (10 ng/mL), mIL-3 (6 ng/mL), and mSCF (100 ng/mL; PeproTech).
Coimmunoprecipitations.
HEK 293T cells were transfected with the plasmids using branched polyethylenimine. On the second day, cells were washed with PBS and lysed. Nuclei were pelleted and lysed. The lysate was cleared by centrifugation and incubated with ANTI-FLAG M2-agarose affinity resin (Sigma-Aldrich) and DNase (Roche). In the phosphatase inhibitor condition, all buffers contained PhosStop inhibitor (Roche). Immunoprecipitated protein was eluted and visualized by Western blotting (SI Appendix, Supplementary Methods).
AlphaScreen.
AlphaScreen measurements were performed in a total volume of 25 μL in 384-well Optiwell microtiter plates (PerkinElmer). The optimal protein concentrations for each experiment were determined by cross-titration to avoid binding curve perturbation while still yielding a high signal-to-noise ratio. The concentrations of proteins used in these titrations were selected to optimally distinguish the WT and mutant curves. At these concentrations, WT proteins remain close to background, while higher-affinity mutants yield high AlphaScreen counts. Each titration was performed in duplicate, and assays were independently repeated three times.
Mass Spectrometry.
For detection of JPO2 and MLL1 interactors, HEK 293T cells were transfected with the respective FLAG-tagged expression constructs, nuclear extracts were prepared and proteins were immunoprecipitated and digested using trypsin or chymotrypsin. All samples were analyzed on UltiMate 3000 RSLCnano system (Dionex; Thermo Scientific) coupled to a TripleTOF 5600 mass spectrometer with a NanoSpray III source (Sciex). TOF MS mass range was set to 350–1500 m/z; in MS/MS mode the instrument acquired fragmentation spectra within m/z range 100–2000. For analysis of phosphopeptides, immunoprecipitated proteins were separated by SDS/PAGE. Peptides generated by trypsin digestion were subjected to mass spectrometric analysis using MALDI-FTMS solariX (Bruker Daltonics) equipped with a 15-T superconducting magnet and SmartBeam laser.
Generation of Retroviral Vectors.
For retroviral vector production, 6 × 106 HEK293T cells were seeded in 8.5-cm Petri dishes. On the next day, 5 μg of the respective pMSCV transfer plasmid and 5 μg ecotropic packaging plasmid (iPak), a kind gift from J. Schwaller, University Children’s Hospital Basel, Basel, Switzerland, were incubated for 15 min at room temperature in the presence of 2 mL serum-free medium and 20 μL turbofect (Thermo Scientific). The transfection mix was added to the cells in a final volume of 5 mL DMEM supplemented with 10% FCS (Thermo Scientific) and 50 μg/mL gentamicin (Thermo Scientific). After 24 h, the medium was refreshed. Vector-containing supernatant was harvested 48 h posttransfection, filtered through a 0.45-µM filter, and stored at −80 °C.
Clonogenic Growth Assays in Vitro.
Murine lineage-depleted stem cells were transduced on two consecutive days with the respective pMSCV-based vectors using spinoculation (2,000 × g, 90 min) in the presence of 4 μg/mL polybrene. After 24 h, the murine myeloid CFU assay was initiated. Transduced lineage-depleted cells were plated in murine methylcellulose containing mIL-3, mIL-6, and mSCF (Methocult M3534; STEMCELL Technologies). The number of colonies was scored after 6–7 d. Colonies were harvested and washed twice with RPMI-1640 (containing 10% FCS and 50 µg/mL gentamicin). Cells were counted in a Neubauer chamber using trypan blue for dead-cell exclusion and plated in fresh medium for the subsequent round.
Supplementary Material
Acknowledgments
We thank Magdalena Hořejší, Marcela Mádlíková, Martine Michiels, Lukáš Vrzal, and Rozálie Hexnerová for technical assistance. This work was supported by KU Leuven Interdisciplinair onderzoeksprogramma (IDO) Program Grant IDO/12/008-3E130241, Czech Science Foundation Grant 16-06357S (to V.V.), Research Foundation - Flanders Grant G.0595.13 (to Z.D.), Ministry of Education of the Czech Republic Grants LO1304, LM2015070, LM2015043, and LK11205, the European Regional Development Fund, Operational Programme Research, Development and Education, the project Chemical Biology for Drugging Undruggable Targets (ChemBioDrug) Grant CZ.02.1.01/0.0/0.0/16_019/0000729 (to M.L., P.Ř., and V.V.), Czech Academy of Sciences Grants RVO61388963, RVO61388971, and RVO68378050, Agency for Innovation by Science and Technology Grant SBO 140038 (ChromaTarget), and the Belgian National Foundation Against Cancer. S.E.A. is a postdoctoral fellow supported by the IDO program. F.C. is supported by the KU Leuven Industrial Research Fund. The laboratory of H.C.H. is supported by National Cancer Institute Grant R00CA187565, Cancer Prevention and Research Institute of Texas Grant RR170036, and Gabrielle’s Angel Foundation for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6EMO–6EMR and 5YI9) and the NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 34179–34182 and 36120).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803909115/-/DCSupplemental.
References
- 1.Dhayalan A, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eidahl JO, et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okuda H, et al. MLL fusion proteins link transcriptional coactivators to previously active CpG-rich promoters. Nucleic Acids Res. 2014;42:4241–4256. doi: 10.1093/nar/gkt1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 6.Ciuffi A, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 7.Maertens G, et al. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 8.Schrijvers R, et al. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog. 2012;8:e1002558. doi: 10.1371/journal.ppat.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shun MC, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christ F, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 11.Bartholomeeusen K, et al. Differential interaction of HIV-1 integrase and JPO2 with the C terminus of LEDGF/p75. J Mol Biol. 2007;372:407–421. doi: 10.1016/j.jmb.2007.06.090. [DOI] [PubMed] [Google Scholar]
- 12.Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- 13.Chan TS, Hawkins C, Krieger JR, McGlade CJ, Huang A. JPO2/CDCA7L and LEDGF/p75 are novel mediators of PI3K/AKT signaling and aggressive phenotypes in medulloblastoma. Cancer Res. 2016;76:2802–2812. doi: 10.1158/0008-5472.CAN-15-2194. [DOI] [PubMed] [Google Scholar]
- 14.Bartholomeeusen K, et al. Lens epithelium-derived growth factor/p75 interacts with the transposase-derived DDE domain of PogZ. J Biol Chem. 2009;284:11467–11477. doi: 10.1074/jbc.M807781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes S, Jenkins V, Dar MJ, Engelman A, Cherepanov P. Transcriptional co-activator LEDGF interacts with Cdc7-activator of S-phase kinase (ASK) and stimulates its enzymatic activity. J Biol Chem. 2010;285:541–554. doi: 10.1074/jbc.M109.036491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tesina P, et al. Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif. Nat Commun. 2015;6:7968. doi: 10.1038/ncomms8968. [DOI] [PubMed] [Google Scholar]
- 18.Cermáková K, et al. Validation and structural characterization of the LEDGF/p75-MLL interface as a new target for the treatment of MLL-dependent leukemia. Cancer Res. 2014;74:5139–5151. doi: 10.1158/0008-5472.CAN-13-3602. [DOI] [PubMed] [Google Scholar]
- 19.Stessman HAF, et al. Disruption of POGZ is associated with intellectual disability and autism spectrum disorders. Am J Hum Genet. 2016;98:541–552. doi: 10.1016/j.ajhg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homsy J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang A, et al. Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res. 2005;65:5607–5619. doi: 10.1158/0008-5472.CAN-05-0500. [DOI] [PubMed] [Google Scholar]
- 22.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 23.Yu M, et al. MLL tandem duplication and multiple splicing in adult acute myeloid leukemia with normal karyotype. Leukemia. 1996;10:774–780. [PubMed] [Google Scholar]
- 24.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 25.Zeisig BB, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Méreau H, et al. Impairing MLL-fusion gene-mediated transformation by dissecting critical interactions with the lens epithelium-derived growth factor (LEDGF/p75) Leukemia. 2013;27:1245–1253. doi: 10.1038/leu.2013.10. [DOI] [PubMed] [Google Scholar]
- 27.Cherepanov P, et al. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 28.Booth V, Koth CM, Edwards AM, Arrowsmith CH. Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70. J Biol Chem. 2000;275:31266–31268. doi: 10.1074/jbc.M002595200. [DOI] [PubMed] [Google Scholar]
- 29.Huang J, et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482:542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu BA, et al. The SH2 domain-containing proteins in 21 species establish the provenance and scope of phosphotyrosine signaling in eukaryotes. Sci Signal. 2011;4:ra83. doi: 10.1126/scisignal.2002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch CA, Anderson D, Moran MF, Ellis C, Pawson T. SH2 and SH3 domains: Elements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- 32.Adey NB, et al. Threonine phosphorylation of the MMAC1/PTEN PDZ binding domain both inhibits and stimulates PDZ binding. Cancer Res. 2000;60:35–37. [PubMed] [Google Scholar]
- 33.Tzivion G, Avruch J. 14-3-3 proteins: Active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 34.Diebold M-L, et al. The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26. EMBO J. 2010;29:3979–3991. doi: 10.1038/emboj.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald SM, Close D, Xin H, Formosa T, Hill CP. Structure and biological importance of the Spn1-Spt6 interaction, and its regulatory role in nucleosome binding. Mol Cell. 2010;40:725–735. doi: 10.1016/j.molcel.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornbeck PV, et al. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mertins P, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mertins P, et al. NCI CPTAC Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci. 2012;367:2513–2516. doi: 10.1098/rstb.2012.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Mu JJ, et al. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 43.Pinna LA. Protein kinase CK2: A challenge to canons. J Cell Sci. 2002;115:3873–3878. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- 44.Sarno S, et al. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (‘casein kinase-2’) FEBS Lett. 2001;496:44–48. doi: 10.1016/s0014-5793(01)02404-8. [DOI] [PubMed] [Google Scholar]
- 45.De Rijck J, Bartholomeeusen K, Ceulemans H, Debyser Z, Gijsbers R. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 2010;38:6135–6147. doi: 10.1093/nar/gkq410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeronimo C, Robert F. The mediator complex: At the nexus of RNA polymerase II transcription. Trends Cell Biol. 2017;27:765–783. doi: 10.1016/j.tcb.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortega CE, Seidner Y, Dominguez I. Mining CK2 in cancer. PLoS One. 2014;9:e115609. doi: 10.1371/journal.pone.0115609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daugaard M, et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat Struct Mol Biol. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y, Paull TT. DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM) J Biol Chem. 2013;288:37112–37125. doi: 10.1074/jbc.M113.514398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neal JA, et al. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol Cell Biol. 2011;31:1719–1733. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Convery E, et al. Inhibition of homologous recombination by variants of the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) Proc Natl Acad Sci USA. 2005;102:1345–1350. doi: 10.1073/pnas.0406466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seldin DC, et al. CK2 as a positive regulator of Wnt signalling and tumourigenesis. Mol Cell Biochem. 2005;274:63–67. doi: 10.1007/s11010-005-3078-0. [DOI] [PubMed] [Google Scholar]
- 54.Dominguez I, Sonenshein GE, Seldin DC. Protein kinase CK2 in health and disease: CK2 and its role in Wnt and NF-kappaB signaling: Linking development and cancer. Cell Mol Life Sci. 2009;66:1850–1857. doi: 10.1007/s00018-009-9153-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trembley JH, Wang G, Unger G, Slaton J, Ahmed K. Protein kinase CK2 in health and disease: CK2: A key player in cancer biology. Cell Mol Life Sci. 2009;66:1858–1867. doi: 10.1007/s00018-009-9154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruzzene M, Pinna LA. Addiction to protein kinase CK2: A common denominator of diverse cancer cells? Biochim Biophys Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 57.Montenarh M. Protein kinase CK2 and angiogenesis. Adv Clin Exp Med. 2014;23:153–158. doi: 10.17219/acem/37040. [DOI] [PubMed] [Google Scholar]
- 58.Feng D, et al. Protein kinase CK2 is a regulator of angiogenesis in endometriotic lesions. Angiogenesis. 2012;15:243–252. doi: 10.1007/s10456-012-9256-2. [DOI] [PubMed] [Google Scholar]
- 59.Kramerov AA, et al. Inhibition of protein kinase CK2 suppresses angiogenesis and hematopoietic stem cell recruitment to retinal neovascularization sites. Mol Cell Biochem. 2008;316:177–186. doi: 10.1007/s11010-008-9831-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCubrey JA, et al. Targeting survival cascades induced by activation of Ras/Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways for effective leukemia therapy. Leukemia. 2008;22:708–722. doi: 10.1038/leu.2008.27. [DOI] [PubMed] [Google Scholar]
- 61.Steelman LS, et al. Contributions of the Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways to leukemia. Leukemia. 2008;22:686–707. [Google Scholar]
- 62.Gray GK, McFarland BC, Rowse AL, Gibson SA, Benveniste EN. Therapeutic CK2 inhibition attenuates diverse prosurvival signaling cascades and decreases cell viability in human breast cancer cells. Oncotarget. 2014;5:6484–6496. doi: 10.18632/oncotarget.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Y, et al. Targeting protein kinase CK2 suppresses prosurvival signaling pathways and growth of glioblastoma. Clinical Cancer Res. 2013;19:6484–6494. doi: 10.1158/1078-0432.CCR-13-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng Y, et al. A CK2-dependent mechanism for activation of the JAK-STAT signaling pathway. Blood. 2011;118:156–166. doi: 10.1182/blood-2010-01-266320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schevzov G, et al. Regulation of cell proliferation by ERK and signal-dependent nuclear translocation of ERK is dependent on Tm5NM1-containing actin filaments. Mol Biol Cell. 2015;26:2475–2490. doi: 10.1091/mbc.E14-10-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duncan JS, et al. A peptide-based target screen implicates the protein kinase CK2 in the global regulation of caspase signaling. Sci Signal. 2011;4:ra30. doi: 10.1126/scisignal.2001682. [DOI] [PubMed] [Google Scholar]
- 67.Siddiqui-Jain A, et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010;70:10288–10298. doi: 10.1158/0008-5472.CAN-10-1893. [DOI] [PubMed] [Google Scholar]
- 68.Perea SE, et al. CIGB-300, a novel proapoptotic peptide that impairs the CK2 phosphorylation and exhibits anticancer properties both in vitro and in vivo. Mol Cell Biochem. 2008;316:163–167. doi: 10.1007/s11010-008-9814-5. [DOI] [PubMed] [Google Scholar]
- 69.Buontempo F, et al. Therapeutic targeting of CK2 in acute and chronic leukemias. Leukemia. 2018;32:1–10. doi: 10.1038/leu.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Basnet H, et al. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature. 2014;516:267–271. doi: 10.1038/nature13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Renshaw PS, et al. Sequence-specific assignment and secondary structure determination of the 195-residue complex formed by the Mycobacterium tuberculosis proteins CFP-10 and ESAT-6. J Biomol NMR. 2004;30:225–226. doi: 10.1023/B:JNMR.0000048852.40853.5c. [DOI] [PubMed] [Google Scholar]
- 72.Veverka V, et al. NMR assignment of the mTOR domain responsible for rapamycin binding. J Biomol NMR. 2006;36(Suppl 1):3. doi: 10.1007/s10858-005-4324-1. [DOI] [PubMed] [Google Scholar]
- 73.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 74.Veverka V, et al. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: Compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene. 2008;27:585–595. doi: 10.1038/sj.onc.1210693. [DOI] [PubMed] [Google Scholar]
- 75.Case DA, et al. 2014 Amber 14 reference manual. Available at ambermd.org/doc12/Amber14.pdf.
- 76.Roe DR, Cheatham TE., 3rd PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.