Significance
We found that hundreds of years of selection by humans have produced sport-hunting breeds of superior speed and athleticism through strong selection on multiple genes relating to cardiovascular, muscle, and neuronal functions. We further substantiated these findings by showing that genes under selection significantly enhanced athleticism, as measured by racing speed and obstacle course success, using standardized measures from dogs competing in national competitions. Overall these results reveal both the evolutionary processes and the genetic pathways putatively involved in athletic success.
Keywords: positive selection, sport-hunting dogs, athletic ability, whole-genome sequencing
Abstract
Modern dogs are distinguished among domesticated species by the vast breadth of phenotypic variation produced by strong and consistent human-driven selective pressure. The resulting breeds reflect the development of closed populations with well-defined physical and behavioral attributes. The sport-hunting dog group has long been employed in assistance to hunters, reflecting strong behavioral pressures to locate and pursue quarry over great distances and variable terrain. Comparison of whole-genome sequence data between sport-hunting and terrier breeds, groups at the ends of a continuum in both form and function, reveals that genes underlying cardiovascular, muscular, and neuronal functions are under strong selection in sport-hunting breeds, including ADRB1, TRPM3, RYR3, UTRN, ASIC3, and ROBO1. We also identified an allele of TRPM3 that was significantly associated with increased racing speed in Whippets, accounting for 11.6% of the total variance in racing performance. Finally, we observed a significant association of ROBO1 with breed-specific accomplishments in competitive obstacle course events. These results provide strong evidence that sport-hunting breeds have been adapted to their occupations by improved endurance, cardiac function, blood flow, and cognitive performance, demonstrating how strong behavioral selection alters physiology to create breeds with distinct capabilities.
Extensive efforts have been made over the past three decades to understand the remarkable success and accomplishments of elite athletes (1). While environmental, psychological, and sociological factors are all important contributors, athletic performance is a complex trait to which genetic makeup contributes substantially (2). Performance-enhancing polymorphisms (PEPs) are germline variants that influence the outcome of athletic challenges (1). Over 200 PEPs have been identified (3), largely in humans, and they include genes which regulate blood pressure (4), muscle size, oxygen use (5), fatigue resistance (6), and blood lactate and ammonium ion accumulation (7).
While these findings are of interest, studies to date have focused on the nationally or internationally recognized elite athletes or individuals of similar geographic ancestry (8, 9), providing limited insight into performance variation in the general population. It is well established that genetic variants can confer differential athletic fitness for various sports, as each sport demands specific physical features, e.g., strong sprinters may also excel in the long jump but not in the marathon (10). This implies the presence of desirable and mappable morphologic traits for many sports, but previous studies fail to capture a holistic view of athletic performance. Perhaps more importantly, little is known about population selection for most athletic traits, including the targets of selection and historical forces that have shaped the history of athlete populations.
To address these issues, we assessed athletic ability in closed-breeding populations of domestic dogs (Canis lupus familiaris). Dogs are unique among domesticated mammals in that they display high levels of phenotypic and behavioral diversity across populations coupled with strong phenotypic and genotypic homogeneity within populations or breeds (11). There are over 450 breeds recognized worldwide, shaped by events such as an ancient bottleneck occurring during domestication, additional bottlenecks associated with breed formation (12–14), and continued population restructuring due to popular sires and shrinking or expanding population size. These factors affect both the phenotype and underlying genotypic profile of each dog breed (15), making domestic breeds an ideal system in which to disentangle complex phenotype–genotype associations (reviewed in refs. 16 and 17).
To date, several genes have been identified which define breed-specific differences. However, those differences are most often associated with physical attributes, including body size, leg length, skull shape, and fur color, among others (18–20). Previous studies have also described loci associated with disease susceptibility (reviewed in ref. 21) and anomalous behaviors patterns, such as those which mimic human obsessive–compulsive disorders (22, 23). However, none has successfully addressed the genetics of physiological traits concomitant with breed-specific behaviors or functional employment with the exception of the high-altitude adaptation of Tibetan Mastiffs (24).
In the United States, the American Kennel Club (AKC) is the foremost authority for purebred dog classification and registration. The AKC has used heritage, behavior, and physical attributes to assign each of 189 breeds to one of seven loosely defined groups (11). At one end of a behavioral continuum is the sport-hunting group, which includes breeds which aid sport hunters by pointing, retrieving, and flushing birds. These breeds are universally active and athletic (11). At the other end is the terrier group, which largely includes small breeds described as “feisty and energetic,” whose primary historical task was to locate and dispatch of vermin from both agricultural and urban settings. Although historically not bred for such, modern terriers may also serve as companions. We hypothesized that a whole-genome comparison of the sport-hunting to the terrier group will reveal genomic regions under selection for creating the component breeds or groups of breeds. The results highlight the utility of dog breed populations for advancing studies of complex traits in humans while illuminating how a small group of genes has been leveraged to create athletes of extraordinary ability.
Results
Population Differentiation.
Based on genetic similarities and definitions of breed groups (25), we leveraged the whole-genome sequencing (WGS) data from 21 sport-hunting and 27 terrier dogs (Fig. 1 A and B and SI Appendix, Table S1). The sport-hunting dataset includes Spaniels: Brittany (one individual), Clumber (one), American (six) and English Cocker (five), and English Springer (two); Setters: English (one), Gordon (one), and Irish (one); and Pointers: English (two), and German Wirehaired (one). The terrier varieties include Airedale (three), Border (three), Irish (one), Jack Russell (four), Kerry Blue (two), Scottish (three), Soft Coated Wheaten (four), West Highland White (six), and Yorkshire (one). Together these breeds comprise 10 of the 30 AKC-recognized sport-hunting breeds and 8 of the 31 recognized terrier breeds. We note that the Jack Russell Terrier breed is recognized by other kennel clubs such as the Fédération Cynologique Internationale and UK Kennel Club but not by the AKC. A summary of the characteristics of each breed is provided in SI Appendix, Table S2. WGS of 79 village dogs, which are generally unselected for any morphologic or behavior traits, from the Middle East, South America, Asia, and Africa was publicly available and is incorporated here (26), yielding data from a total of 127 dogs. Genome alignment indicated an average of 18.8-fold depth for each individual relative to the Boxer reference genome (SI Appendix, Table S1). A total of ∼14.9 million high-quality autosomal single-nucleotide variants (SNVs) were identified and investigated for this study (Methods).
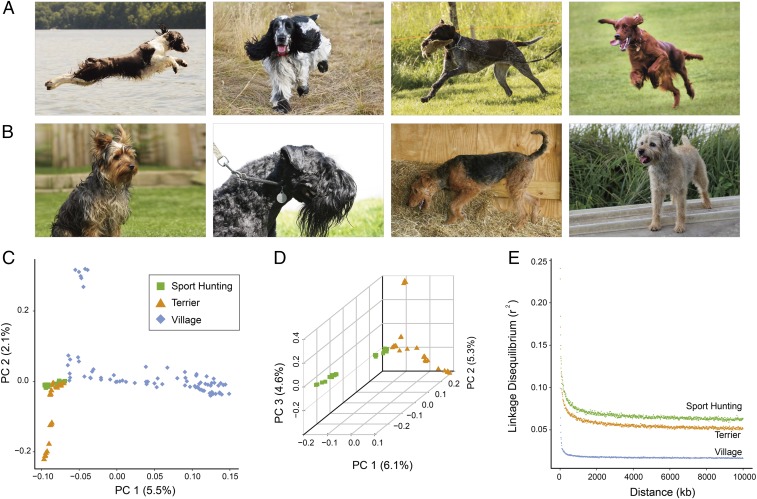
Fig. 1.
Population structure of sport-hunting, terrier, and village dog populations. (A) Sport-hunting breeds pictured include (Left to Right) English Springer Spaniel, English Cocker Spaniel, German Wirehaired Pointer, and Irish Setter. Images, from left to right, courtesy of Katrine Bremser (photographer), Jillian Mennie (photographer), Flickr/Tommi Valtanen, and Flickr/Rongem Boyo. (B) Terrier breeds pictured include (Left to Right) Yorkshire Terrier, Kerry Blue Terrier, Airedale Terrier, and Border Terrier. Images, from left to right, courtesy of Flickr/Michelle Dudley, Gerry Yeager (photographer), Kay Nellis (photographer), and Flickr/Sophie Lowe. (C and D) Data from each individual sample are plotted along the two main principal components (PC1 and PC2) on three populations (C) and the first three principal components (PC1, PC2, and PC3) on sport-hunting and terrier populations (D). (E) Genome-wide LD was estimated in each group by calculating r2 values between all pairs of SNVs with inter-SNV distances less than 10 Mb.
To examine genetic relationships among the three dog populations, we conducted principal component analysis (PCA) based on all SNVs. In an initial clustering analysis, the first eigenvector (5.5%) identified two distinct groups: the village dogs and a cluster consisting of domesticated dogs which included both sport-hunting and terrier breeds (Fig. 1C). The widely dispersed distribution of village dogs indicates the expected high level of heterogeneity in this population compared with the other two groups. An independent PCA incorporating three eigenvectors based on sport-hunting and terrier dogs provided evidence that these two groups are genetically distinct (Fig. 1D). In addition, both sport-hunting and terrier dogs exhibit higher levels of linkage disequilibrium (LD) than the village dogs (P < 2.2 × 10−16, Mann–Whitney u test), reflecting fewer recombination events associated with domestic breed formation (Fig. 1E).
Identification of Genomic Regions Under Selection.
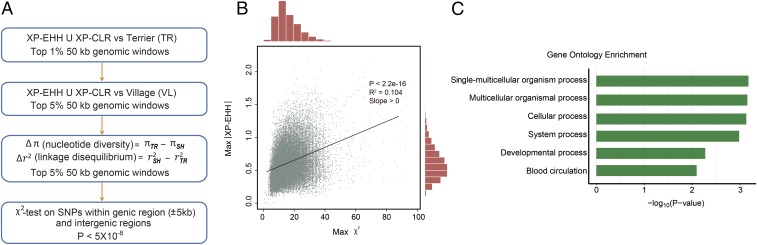
Dog breeds developed for use in hunting and field activities must naturally possess an active and alert demeanor and the athletic ability to fulfill the physically demanding roles specified in the AKC breed standards (11). Given that sport-hunting and terrier dogs have experienced unique selective pressure events over time to obtain desired phenotypes, particularly compared with the village dogs, we undertook a genome-wide pairwise comparison of each population to identify loci under selective pressure. To reduce the confounding effect of genetic drift and minimize the influence of breed-specific background, we combined the component breeds into their respective breed groups (sport-hunting, terrier, and village dogs). The fixation index (FST) was 0.047, 0.063, and 0.067 between the sport-hunting–terrier, terrier–village, and sport-hunting–village groups, respectively, indicating that each dataset was of sufficient sample size, with a minimum of 21 individuals, for this analysis (27). In addition, to identify genomic regions under extended scope of selection, we combined independent tests of selection based on nucleotide diversity, LD, and allele frequency. The overall study design of the positive selection analysis is depicted in Fig. 2A.
Fig. 2.
Evidence of positive selection in sport-hunting dogs (SH). (A) Flowchart of study design. (B) Maximum XP-EHH scores against maximum χ2 of each 50-kb window. (C) The significant GO terms (P < 0.01) enriched from positively selected genes in sport-hunting dogs.
The cross-population extended haplotype homozygosity (XP-EHH) and the cross-population composite likelihood ratio (XP-CLR) tests were used to identify 50-kb nonoverlapping genomic regions with significant LD and allele frequency differentiation between the sport-hunting and terrier groups. Using the empirical top 1% of genomic regions, a total of 457 genes and 396 intergenic regions (IRs) were identified as having potential selective sweeps in sport-hunting dogs. Although the overall distributions of the two metrics show a significant positive correlation (slope >0, P < 2.2 × 10−16) (SI Appendix, Fig. S1), these approaches complement each other, as ∼15% of the same candidate genes were detected by both tests.
To reduce the incidence of false positives and identify genomic regions with bona fide signals from among candidates identified in the initial scan, subsequent analyses were undertaken. Considering that village dogs were not subject to the same degree of selective pressure as sport-hunting dogs, we applied the same metrics (XP-EHH and XP-CLR) to the comparison of sport-hunting versus village dogs, which reduced the number of candidates to 280 genes and 126 IRs that remained within the top 5% of the distribution. The regions under selective sweeps also show significantly reduced diversity (π) and/or persist in strong LD (r2) in the selected population. Hence, the top 5% of empirical distributions of the relative nucleotide diversity, ∆π (πterrier − πsport-hunting), and LD ∆r2, (r2sport-hunting − r2terrier) (SI Appendix, Fig. S2), defined a further reduced set of windows (161 genes and 126 IRs) as outliers (Fig. 2A).
Positive Selection and Beneficial Alleles.
To finally define selected regions with the causal candidates, we performed a χ2 test on all SNVs within candidate genic regions (±5 kb) and IRs, determining the difference in allele frequency between sport-hunting and terrier groups. We found that stronger selective pressure leads to an increased incidence of highly differentiated allele frequencies between populations (Fig. 2B). Using the threshold of 5 × 10−8, this final scan retained a total of 59 genes and 51 IRs under strong selection in sport-hunting breeds (SI Appendix, Table S3). The median distance of positively selected IRs from the closest genes is 128 kb, ranging from 26 kb to 890 kb. We observed that alleles with low frequency within these positively selected regions were present in excess in sport-hunting group compared with nonselected groups (P < 2.2 × 10−16, Mann–Whitney u test) (SI Appendix, Fig. S3).
Signatures of Selection Associated with Athletic Performance in Sport-Hunting Dogs.
Fifty-nine genes were under positive selection in the sport-hunting breeds (SI Appendix, Table S3). Elite athletic performance is largely determined by integrative roles of muscular, cardiovascular, and neurological functions (28), and 11 of the 59 genes have biological functions consistent with an a priori hypothesis. Based on a manual review of the OMIM (Online Mendelian Inheritance in Man) and UniProt databases (SI Appendix, Table S4), positively selected genes are related to muscle contraction (RYR3), muscle development (ABLIM3 and CDH15), fatigue-enhanced muscle pain (ASIC3), vascular smooth muscle contraction (TRPM3), muscular dystrophy (UTRN), heart rate and hypertension (ADRB1 and GRK4), and neurological disorders including impaired learning (ROBO1 and RIMS1) and mental developmental delays and disabilities (KCNQ5 and CDH15). Analysis of gene ontology (GO) enrichment for positively selected genes indicated a significant overrepresentation for the category associated with blood circulation (GO: 0008015, P = 0.00803) (Fig. 2C and SI Appendix, Table S5). Other enriched biological functions include single-multicellular organism, multicellular organismal, cellular, system, and developmental processes (SI Appendix, Table S5). The selected genes also included those that are critical to neuronal functions such as neuronal migration, neurite outgrowth, and synapse formation (SI Appendix, Table S4), although the corresponding biological processes were not significantly overrepresented. Based on a χ2 test of the SNVs within each candidate gene, the variants with the strongest allele frequency difference between sport-hunting dogs and terriers at each gene were selected for further analyses.
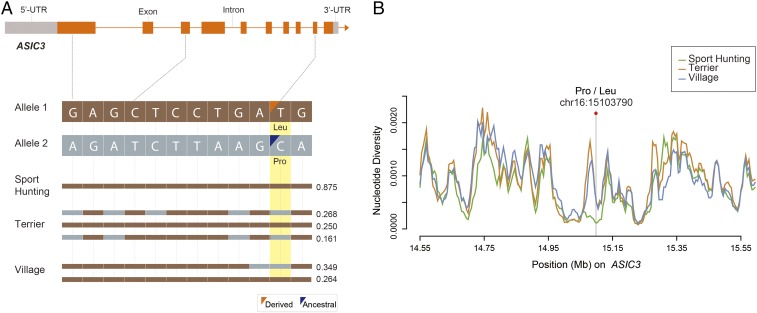
A Selective Sweep in the ASIC3 Gene.
We next examined the nonsynonymous mutations from each of the candidate genes, identifying one significant SNV (chr16:15103790, P = 4.9 × 10−11; χ2 test) in the ASIC3 gene that codes for a change at residue 512 (p.Leu512Pro) (Fig. 3A). The publicly available variants of 28 wolf samples were leveraged to infer the ancestral alleles (https://data.broadinstitute.org/vgb/435_dog_data/). The inferred ancestral allele from wolf (C allele) (SI Appendix, Table S1) suggests that a derived allele (T) experienced strong directional selection pressure within the sport-hunting group compared with other groups, including terrier and village dogs. This gene showed an apparent regional differentiation in terms of reduced nucleotide diversity in sport-hunting breeds (Fig. 3B). To confirm and extend these findings, we used Sanger sequencing to genotype this candidate SNV in the independent and larger dog population of 77 sport-hunting and 74 terrier samples from nine and six breeds, respectively (SI Appendix, Tables S6 and S7). This validation set additionally included two retriever breeds (Labrador and Golden) for sport hunting, which were not available in the initial WGS analysis. The allele frequency difference remained statistically significant between sport-hunting and terrier dogs (P = 8.8 × 10−9) (SI Appendix, Table S8).
Fig. 3.
Signatures of the selective sweep at the ASIC3 gene region. (A) Structure of the ASIC3 gene with exons indicated by orange bars. A nonsynonymous SNV (C: ancestral and T: derived) is highlighted in yellow. Different colors represent distinct alleles, and the frequency of each haplotype is indicated on the right. (B) Nucleotide diversity plot of three populations around the ASIC3 gene region.
Noise Sensitivity of Sport-Hunting Dogs.
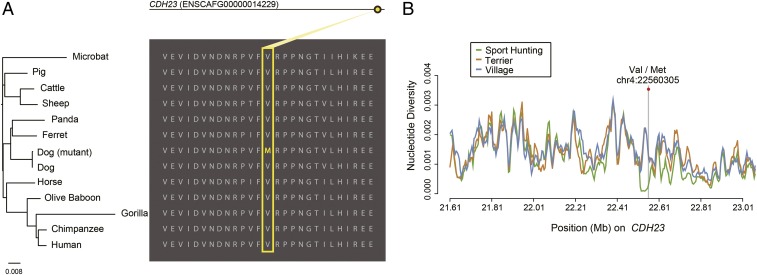
One strong result that was not overtly related to performance was in CDH23 and MSRB3; mutations in these genes are linked to sensory impairment (SI Appendix, Table S4). Previous studies demonstrate that terriers are among the breeds with the highest incidence of noise sensitivity, while popular sport-hunting breeds such as the Labrador, Cocker Spaniel, and Springer Spaniel are far less likely to show a startle response to noise, even in a comparison with crossbred dogs (29, 30).
To explore this result further, we screened for causal mutations in these genes segregating in sport-hunting dogs versus terriers and aligned the mutant genes with their orthologous proteins in other vertebrates to assess the functional impact of observed variants. This revealed a highly conserved mutation in CDH23 (chr4:22560305, p.Met2617Val), which is invariant among all 11 mammals closely related to dogs including horse, ferret, pig, sheep, and cattle (Fig. 4A). In addition, the pathogenicity assessment of this variant, assessed by using the Combined Annotation-Dependent Depletion (CADD) database (31), indicates that this change is predicted to be in the top 0.59% of the most deleterious single-nucleotide substitutions that can be generated from the human genome. Based on WGS, the mutant allele (M) was significantly more common in the sport-hunting dogs than in terriers (P = 3.7 × 10−6; χ2 test). This region shows a dramatic loss of nucleotide diversity compared with that observed in both terrier and village dogs, indicative of a strong selective sweep around a causal variant (Fig. 4B). Additional sequencing of this mutation on the same population used in a previous analysis of ASIC3 (SI Appendix, Tables S6 and S7) confirms the observation (P = 3.9 × 10−10; χ2 test), indicating that the nonsynonymous mutation may be responsible for the selective sweep at CDH23 (SI Appendix, Table S8).
Fig. 4.
Signatures of the selective sweep at the CDH23 gene region. (A, Right) Structural and evolutionary analysis of the amino acid variant in CDH23. The orthologous protein sequences from mammals are aligned with the mutant residues shown in yellow. (Left) The neighbor-joining tree derived from the multiple sequence alignment. (B) Nucleotide diversity plot of three populations around the CDH23 gene region.
ROBO1 Is Associated with Agility Performance.
To understand the relationship between the 59 genes suggested by this study and innate athletic ability across breeds, we leveraged the publicly available catalog of SNVs that we recently assembled, which includes an extended population of 298 dog samples representing 92 breeds, of which 67 (178 dogs) are not classified as either sport-hunting or terrier dogs (https://data.broadinstitute.org/vgb/435_dog_data/). We first assessed performance in agility, a popular canine sport which requires a dog directed by a human handler to navigate an obstacle course with a goal of achieving the fastest times. It thus provides an excellent test of canine athleticism and physical fitness (32). As the surrogate measure of breed-specific athletic ability, we used the total number of agility titles won by each breed, weighted by the total number of dogs registered for each breed (SI Appendix, Table S9), using data from the United States Dog Agility Association (USDAA).
We tested for differences in the allele frequency in the breed groups stratified according to the number of breed-specific agility titles after taking admixture and relatedness into account (33). The two groups included 57 individuals (18 breeds) and 44 individuals (18 breeds) in the top and bottom 20th percentiles, respectively (SI Appendix, Table S9). Of the 59 candidates tested, only an SNV within the ROBO1 gene (chr31:8305922, P = 3.05 × 10−4), which is related to dyslexia in humans (34), was significantly associated (threshold P = 8.47 × 10−3) with the breed-specific agility performance, even after correcting for height, weight, or height/weight ratio (SI Appendix, Table S10).
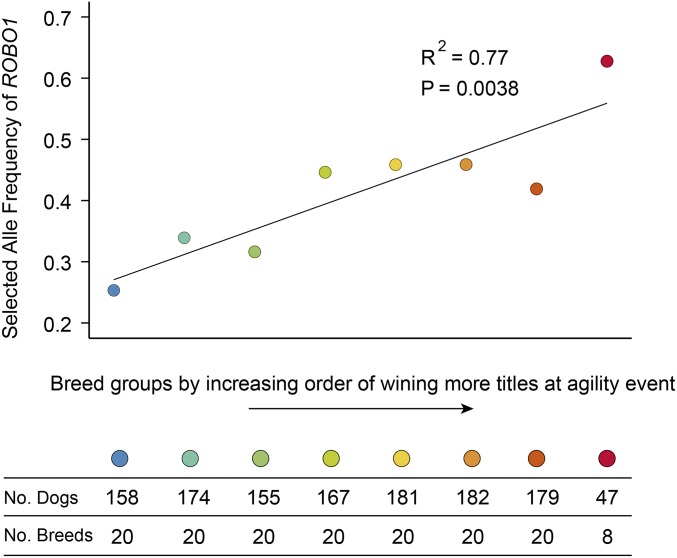
To examine whether this finding is relevant in a larger population, the genotypes of 1,243 dogs representing 148 breeds at the same position were retrieved from publicly available Canine HD Array data (25). Dog breeds are ordered and assigned to eight classes by ascending order of breed-specific agility titles (SI Appendix, Table S11), then plotted against the frequency of the advantageous A allele of ROBO1 (Fig. 5). The putatively advantageous allele was defined as that selected in sport-hunting dogs. We observed a significant trend (slope >0, R2 = 0.77) (Fig. 5), such that breeds with a higher frequency of the advantageous A allele were likely to earn more titles at agility events than those with a lower frequency of this allele.
Fig. 5.
Association of ROBO1 allele frequency with the breed’s agility titles. Shown on the x axis are the eight breed groups stratified according to the total number of agility titles won by each breed, weighted by the total number of dogs registered for each breed. The allele frequency on the y axis was calculated for each group based on the allele selected in sport-hunting dogs. The number of dogs and breeds within each group are indicated below the graph.
Racing Performance in Whippets.
Innate athletic performance was further investigated in a population of Whippets, an established racing breed for which we previously demonstrated an association between racing grade and a heterozygous knockout in myostatin (MSTN) (35). Such mutations cause an increase in muscle mass in humans, dogs, and mice (36, 37). To infer performance ability in the current study, we utilized data from 92 racing Whippets, which include the previously studied population of 85 Whippets (35). Each racing dog had previously been assigned a racing score: A, B, C, or D, in order from fastest to slowest, by the Whippet Racing Association, using their standard metrics (Methods).
Athletic success can stem from maximized delivery of oxygen and metabolic substrates due to increases in cardiac output (38), from improved skeletal muscle efficiency due to altered muscle fiber type and increased muscle mass (39), or from neurological factors related to the ability to learn (40). Hence, we further focused on the cardiovascular (ADRB1 and TRPM3) and muscle (ABLIM3, ASIC3, RYR3, CDH15, and UTRN) genes as well as genes involved in learning ability (KCNQ5, CDH15, and ROBO1) to unravel the complex mechanisms needed to attain elite racing performance. The variants of the selected genes were genotyped using Sanger sequencing (SI Appendix, Table S7). Note that the GRK4 and RIMS1 genes are excluded from this analysis as the candidate SNVs are located within the repeat-rich regions.
Our analysis revealed that a mutation in the TRPM3 gene (P = 1.59 × 10−3), in addition to the previously established variant in the MSTN gene (P = 1.71 × 10−4), showed strong evidence of association with racing performance even after Bonferroni multiple testing corrections for 10 independent tests (nine candidate genes and the MSTN gene) (Table 1). The advantageous allele (T, chr1:86,847,407) of TRPM3 significantly increased racing grade, thereby accounting for an additional 11.6% of the variance in racing performance beyond the effect of MSTN (15.3%). An excess of the T allele (75%) was noted within the racing Whippet population. As sex and height may affect racing performance, covariates are included in the regression analysis; however, the results remained significant (SI Appendix, Table S12). We found no evidence of interaction between markers. The allele distribution of TRPM3 in 28 Greyhound dogs with similar historical roles in racing was also tested but showed no signature of selection (52.5%).
Table 1.
Association of candidate genes with racing performance in Whippets
| Gene | Chromosome | POS | Selected allele | P | beta | h2 |
| ADRB1 | 28 | 24,904,824 | G | N.S. | — | — |
| ASIC3 | 16 | 15,103,790 | T | N.S. | — | — |
| TRPM3 | 1 | 86,847,407 | T | 1.59 × 10−3 | 0.556 | 0.116 |
| UTRN | 1 | 36,106,849 | A | N.S. | — | — |
| RYR3 | 30 | 1,235,869 | G | N.S. | — | — |
| ABLIM3 | 4 | 59,737,391 | T | N.S. | — | — |
| ROBO1 | 31 | 8,305,922 | A | N.S. | — | — |
| CDH15 | 5 | 64,274,090 | T | N.S. | — | — |
| KCNQ5 | 12 | 35,272,870 | A | N.S. | — | — |
| MSTN | 37 | 729,360–729,361* | Deletion | 1.72 × 10−4 | 1.000 | 0.153 |
After Bonferroni multiple testing corrections for 10 independent tests (threshold P = 5 × 10−3), TRPM3 and MSTN showed significant association with racing performance. Beta, regression coefficient; h2, narrow-sense heritability; N.S., nonsignificant; POS, candidate SNP position.
Two-base pair deletion.
Selective Sweep in Terriers.
Applying the same methodology to the terrier population, we identified 44 genes and 32 intergenic regions with significant signals of positive selection (SI Appendix, Tables S13 and S14). As expected, these include the gene associated with appearance, such as R-spondin-2 (RSPO2), which we previously showed is associated with the trait of “furnishings,” i.e., the characteristic moustache and eyebrows observed in the majority of terrier breeds (41). While the terrier group has a reputation for aggression (42), we observed no significant enrichment of GO categories related to behavior (SI Appendix, Table S15). However, the SHANK2 and OXR1 genes, which are involved in hyperactivity and panic responses, respectively (43, 44), were under selection. We also noted that NAALAD2, a member of the N-acetylated alpha-linked acidic dipeptidase (NAALADase) gene family whose inhibition results in a reduction in aggressive behavior (45), showed significant signatures of selection (XP-EHH, ∆π, ∆r2) in the terrier breeds tested, despite lacking a highly significant SNV (P < 5 × 10−8).
Discussion
In this study, we compared the genomes of sport-hunting and terrier breeds and village dog populations to identify genes that can unravel the genetic basis of complex traits which define terrier and sport-hunting breeds. Village dogs are nearly ubiquitous throughout the world, representing mixtures of regional indigenous and European-derived breed dogs in the absence of structured breeding (26), making them a valuable outgroup for comparative analyses. WGS enables accurate inferences of local adaptation, overcoming the limit of ascertainment bias that is inevitable with SNP genotyping arrays (46). However, challenges of positive selection analysis include a high false-positive rate and an inability to distinguish selection signatures from the effect of demographic history (47). To overcome this, we combined different population metrics (XP-EHH, XP-CLR, nucleotide diversity, extent of LD) and categorized 19 distinct breeds into their corresponding breed groups based on genetic similarity (25). The null distributions of these tests (XP-EHH and XP-CLR) demonstrated the robustness to variation in the diverse demographic scenarios simulated (48, 49), making them suitable metrics to define selective sweeps in dogs given that (i) the demographic history of each breed is expected to vary substantially (15) and (ii) demographic parameters and models of dog breeds are not explicitly defined. Finally, to effectively distinguish the effects of demographic history, which affects all loci with equal force from natural selection, we constructed empirical distributions of the test statistics on a dataset of ∼14.9 million SNVs and defined putative targets of selection based on outliers in the extreme tail of the distribution (i.e., outlier approach) (50).
A modest overlap in genes detected by XP-EHH and XP-CLR demonstrates that the two approaches are based on different patterns of genetic variation: levels of LD (XP-EHH) and allele frequency distribution (XP-CLR). As a result, the time frame of two approaches varies, as the XP-EHH statistic is designed to detect alleles that have increased in frequency to the point of fixation from recent selection, while XP-CLR has good power to detect regions affected by incomplete sweep or older selection (47).
The selective sweep mapping for sport hunting identified 59 candidate genes and 51 intergenic regions with significant divergence. The results were not sensitive to different definitions of window size (25, 50, 100, and 150 kb), as other window sizes yielded qualitatively similar results (SI Appendix, Table S16). The choice of 50 kb as a window size was driven by the intention to ensure a sufficient number of SNVs while detecting selection signatures with high resolution. We also showed by randomly sampling three individuals each from the six American and five English Cocker Spaniels, which represent the largest number of samples within the sport-hunting group while maintaining other breeds, that selection signatures are not driven solely by these particular breeds (SI Appendix, Table S17). We observed that positively selected regions tend to have SNVs with higher differentiation of allele frequencies in the selected population than in the nonselected population (Fig. 1D). This finding is concordant with a prior finding, which showed that variants within the previously defined selective sweeps for human population groups are more significantly associated with the tested phenotypes than variants located in the remainder of the genome (51). Further, as selection produces a skewed allele frequency toward rare alleles, positively selected regions demonstrate an excess of low-frequency variants compared with nonselected populations (Fig. 2B).
The positively selected genes are likely the manifestation of at least 300 y of intense selection that has altered the physiology of this breed group (43). We show that athletic breeds that excel at sports and hunting have experienced substantial selective pressure on the blood circulation system (GO: 0008015), possibly to maximize the delivery of oxygen and metabolic substrates to exercising muscle (1) by increasing cardiac output (ADRB1 and GRK4) and effectively regulating contractile response in vascular smooth muscle cells (TRPM3). Of these, ADRB1 (adrenoreceptor beta 1) showed evidence of association with maximum oxygen consumption during exercise and is one of the previously identified PEPs in humans (3). In addition, a ryanodine receptor (RYR3) likely mediates calcium ion release in the contracting skeletal muscles during field activity, and the contractile strength is mediated by the skeletal neuromuscular junction differentiation (UTRN) (52). The increased growth and improved function of skeletal muscle, which are controlled by ABLIM3 and CDH15, may have enhanced dogs’ physical fitness (53, 54). The ability of skeletal muscle to resist fatigue can be expressed as muscle endurance and is probably the result of adaptive response (ASIC3) (55). Neurological factors (ROBO1, RIMS1, KCNQ5, and CDH15) may have enhanced several aspects of canine behavior including motor control, skill learning, perceptual–cognitive skills, and ultimately athletic ability and success in the dogs (56).
Positive selection increases the power to detect association by driving the emergence of alleles with strong effect, which facilitates discovery of the causal (or tagging) variants (57). In this context, we scanned the coding variants in the candidate genes described above. Among the most compelling was a putative causal variant in the ASIC3 gene (p.Leu512Pro) (Fig. 3). Deletion of the ASIC3 gene prevents fatigue-enhanced muscle pain in a mouse model (58). This allele therefore may have increased in frequency with a loss-of-function role, owing to the burden of muscle pain after repetitive acute exercise. We note that only one of 28 wolf samples examined (SI Appendix, Table S1) carried this mutation, suggesting that the derived allele has reached near-fixation in sport-hunting breeds (82%) through strong positive selection in a short time frame.
Many sport-hunting dog breeds have a high incidence of congenital deafness (59, 60). We observe strong signatures of selection in sport-hunting breeds in the CDH23 gene, which is a member of the cadherin superfamily and is expressed in the neurosensory epithelium. Humans with missense mutations in CDH23 suffer sensory impairment, and the gene is located in the region associated with Usher syndrome type I in humans (61). Typical symptoms of Usher syndrome type I include peripheral vision and hearing loss as well as speech delays related to sensorineural hearing loss. The notion that any subset of sport-hunting dogs would have a selective sweep around the mutation (p.Met2617Val) in this gene was initially puzzling, as was the fact that it was not found in other closely related mammals (Fig. 4A). We also detected selection in the MSRB3 gene, whose deficiency is responsible for auditory hair cell loss, which ultimately results in profound deafness in mice (62). We hypothesize that partial loss of gene function may reduce the startle response in dogs, which, for sport-hunting dogs, would enhance their success as hunter companions. It also remains possible that other environmental factors such as conditioned training or acquired damage to the auditory system induced by exposure to gunshot noise may have contributed to their reduced startle reflex. Finally, considering that variants of CDH23 also confer susceptibility to age-related hearing loss in some inbred mouse strains (63), the mutation might cause an accelerated age-related hearing loss in these dogs after exposure to loud noise. Additional investigation is required to test these hypotheses, especially given that the crystal structure of this domain has not yet been determined and the cascade of molecular effects of this variant remains to be elucidated.
We observed a significant association of ROBO1 with breed-specific accomplishments in competitive agility events (Fig. 5). ROBO1 is a neuronal axon-guidance receptor gene involved in brain development, and disruption of this gene by translocation predisposes to the cognitive and learning disorder of dyslexia (34, 64, 65). Genetic variation in the ROBO1 gene may cause variability in cognitive plasticity that explains the marked differences in physical performance (56) and in the potential of adaptation for performance between dog breeds. More specifically, ROBO1 may have affected the ability to identify and acquire environmental information (e.g., an obstacle course) so that task-specific responses can be selected and executed at the agility events.
The TRPM3 gene shows a strong association with racing grades in Whippets. The gene product is a mammalian transient receptor potential channel and is expressed in smooth muscle cells of blood vessels, where the channel activity is related to contractile phenotypes (66). The significant association between the TRPM3 variant and racing speed suggests that the mutation may confer a selective advantage in regulating blood flow to skeletal muscle which can, by extension, be linked to enhanced athletic performance. Heritability estimates of performance-related traits range from 20–70% in humans (2) and 17–40% in racehorse populations (67, 68) but were generally unknown in dogs. We observed a significant genetic contribution from TRPM3 (11.6%), which, when combined with the 15.3% we observed previously in MSTN (35), accounts for 26.9% of the variation in racing speed in Whippets. Our previous study revealed that a 2-bp knockout mutation in MSTN is largely observed in the heterozygous state among racing Whippets (35). The homozygous state is not lethal but procures heavily muscled “bully” dogs that are typically removed from breeding stocks as they do not conform to breed standards and have health issues (69). Contrary to MSTN, the advantageous T allele in TRPM3 gene has a frequency of 75% in 92 Whippets and is present in both the homozygous (57%) and heterozygous states (43%). The high frequency of homozygous dogs implies that the allele does not confer health issues. Although Greyhounds and Whippets share a common ancestral gene pool, we found no evidence of selection for the T allele (52.5%) within Greyhound samples tested. This result is in agreement with our previous observation that Greyhounds do not carry the MSTN mutation (35). However, the Greyhounds tested were not solely selected as racing dogs. The allele frequency might have been shifted had they been from a cohort of racers. In thoroughbred racehorses, studies have revealed the relationship between polymorphisms in MSTN and the genetic potential to improve racing ability and stamina (70). However, no previous studies have demonstrated a role for TRPM3 in sports, suggesting additional avenues for improvement of athletic phenotypes in other species (e.g., marker-assisted selection).
Trade-offs frequently exist so that enhancements in one area of performance occur in conjunction with disadvantages in reciprocal performance areas (10). The superior performance is task specific and is dependent on advantages in cardiovascular (TRPM3 on racing speed) or neuronal (ROBO1 on agility sports) mechanisms. Thus, it is possible that variants in additional candidate genes may benefit performances in different canine sports which rely on abilities not directly measured in this study. For example, speed requires a high proportion of fast, fatigue-sensitive muscle fibers, while endurance relies on slower muscle fibers that are fatigue resistant (71). Consistent with this, the previous evaluation for breed composition of mixed-breed Alaskan sled dogs indicated a significant genomic contribution of Pointer to the sprint sled dog and of Alaskan Malamute to the distance sled dogs (72), as the two breeds represent strong short-distance racers and long-distance endurance competitors, respectively. We also note that it is likely that MSTN and TRPM3 are an unrepresentative sample of all genes associated with increased racing speed in Whippets; this limitation can be overcome by a more thorough and inclusive WGS analysis of a large Whippet population.
Terriers are described as courageous and tenacious dogs that have traditionally been employed to rid rural or urban landscapes of vermin (11). They were crossbred with Mastiffs and Bulldogs from 1860–1870 to create breeds that excel in dog fighting (25, 42). Our screening for regions under positive selection in terrier breeds revealed genes (SHANK2 and OXR1) that, when mutated, have been implicated in autism-like behaviors and hyperactivity and panic disorders, respectively. These human behavior complexes in the context of breed traits could explain the terriers’ distinctive responses to stimuli, territoriality, and confrontational attitudes that have resulted from generations of selective breeding. It is worth noting, however, that aggression is one of the most complex canine behavioral traits, and considerable work remains to be done to understand this phenotype in all breeds (45).
Intergenic regions also showed signatures of positive selection in each population, and some of these regions may reflect genetic hitchhiking effects near a selected locus (12) such as RYR3 in sport-hunting breeds (distance of 33 kb) and RSPO2 in terriers (distance of 35 kb). It is also possible that these regions highlight the adaptive regulatory divergence between populations within DNA methylation, transcription factor binding, DNase I hypersensitive sites, or other epigenetic factors (73). For instance, sport-hunting dogs exhibited a selection signature at 67 kb upstream of MSX2, which encodes a member of the muscle segment homeobox gene family. This gene is important in cardiac outflow tract morphogenesis and is also linked to the formation of the atrioventricular junction and valves (74). Although we did not find evidence of selection within this gene region, further functional analysis may provide evidence that this candidate regulatory region is physically associated with expressed MSX2.
Dog breeds illustrate extraordinary diversity in athletic prowess, providing a rare lens through which to view genetic variation and evolutionary forces that govern athletic success. Our study revealed positively selected genes associated with muscular, cardiovascular, and neuronal functions in sport-hunting versus terrier and village dogs that we now hypothesize played a role in enhancing their athletic ability. Indeed, sport-hunting breeds are truly unique in that they have achieved astounding genetic success in the course of their short history. As they continue to strive for greatness, selective pressures may lead to an even greater refinement of “the champion genome,” and their distinct and evolving genetic makeup is sure to further expand our knowledge about the genetics of athletic performance.
Materials and Methods
Sample Collection.
Blood samples were collected from purebred AKC-registered or pedigree-verified dogs after written consent was obtained from the dog owners as previously described (75). Genomic DNA was isolated using the proteinase-K/phenol-chloroform methods (76) and then was stored at −80 °C. All procedures were approved by the National Human Genome Research Institute Animal Care and Use Committee at the NIH before collection. Samples were collected from individuals that were unrelated to one another at the grandparent level.
Whole-Genome Sequencing Data.
Data from 127 individuals were obtained via the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) from previously published studies or were sequenced for this study by the NIH Intramural Sequencing Center using the Illumina TruSeq DNA PCR-Free Protocol (catalog no. FC-121-3001) (SI Appendix, Table S1). Previously unpublished data from six sequenced dogs have been deposited in the SRA (https://www.ncbi.nlm.nih.gov/sra), and a full list of accession numbers is provided in SI Appendix, Table S1. Together, the paired-end sequence reads were then mapped against the CanFam 3.1 reference genome (genome.ucsc.edu/cgi-bin/hgGateway?db=canFam3) using Burrows–Wheeler Aligner (BWA) 0.7.13 MEM (77), sorted with SAMtools 0.1.10 (78), and screened for putative PCR duplicate reads with PicardTools 1.119 (https://github.com/broadinstitute/picard). The Genome Analysis Toolkit 3.5 (GATK) (79) was used to perform local realignment of reads to correct misalignments due to the presence of indels using 714,278 variants (80) as the training set. SNVs were called per-individual in gVCF mode of HaplotypeCaller (81), with subsequent joint-calling across the entire population. GATK best practices and default parameters, together with the initial alignment training sets, were used for variant quality score recalibration of SNVs. A total of ∼14.9 million autosomal SNVs that were polymorphic in the population and passed our quality control criteria of a maximum missing rate <10% and quality score >20 were used for subsequent analyses. We used BEAGLE v.4.0 (82) to infer the haplotype phase for the entire population.
To infer the ancestral alleles from wolves and utilize the genotypes from expanded population of 92 breeds on candidate genes, we leveraged the publicly available variants of 435 dogs (SI Appendix, Table S1, https://data.broadinstitute.org/vgb/435_dog_data/).
Sanger Sequencing of Candidate Genes.
Candidate SNVs within the ASIC3 and CDH23 genes were first genotyped in 165 dogs representing 15 breeds using Sanger sequencing (SI Appendix, Table S6). We then genotyped additional candidate genes (ASIC3, ADRB1, TRPM3, UTRN, RYR3, ABLIM3, CDH15, ROBO1, and KCNQ5) in Whippets. Positions of the markers and the resulting allele frequency of each breed are provided in SI Appendix, Table S7. Primers were designed using Primer3 software (SI Appendix, Table S7) (83). The regions containing the tested SNV were amplified by PCR with AmpliTaq Gold (Applied Biosystems). PCR products were purified by ExoSap-It reaction (Affymetrix). The BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) was used for Sanger sequencing, and the products were run on an ABI 3730 DNA analyzer (Applied Biosystems). Sequence traces were analyzed using ApE software to make genotype calls.
Population Analysis.
PCA utilized Genome-Wide Complex Trait Analysis (GCTA) (84) to estimate the eigenvectors, incorporating genotype data from all samples. We used VCFtools 4.0 (85) to estimate nucleotide diversity (in windows of 50 kb) and minor allele frequency. LD between pairs of markers and allele frequency differentiation (χ2 test) were assessed using PLINK v.1.07 (86). The r2 value was calculated between all pairs of SNVs with inter-SNV distances of less than 50 kb (r2 and LD window parameters). Haploview software (87) was used to evaluate the haplotype structure and estimate haplotype frequencies in each dog population. The ortholog proteins of CDH23 in 11 species were retrieved from Ensembl (release 87), and Cluster Omega was used to perform multiple sequence alignment. The resulting tree was generated using FigTree v1.4.0.
Identification of Selective Sweeps.
The XP-EHH method was used to detect selective sweeps (hgdp.uchicago.edu/Software/) (88). An XP-EHH score is directional: In the current paper, an extreme positive score implies selection in sporting hunting dogs, whereas a negative score suggests selection in the terriers. We split the genome into nonoverlapping segments of 50 kb to use the maximum (sport hunting) or minimum (terrier) XP-EHH score of all SNVs within a window as a summary statistic for each window. To take into account the SNV density, we binned genomic windows according to their numbers of SNVs in increments of 200 SNVs (combining all windows with ≥600 SNVs into one bin). Within each bin, for each window i, the fraction of windows with a value of the statistic greater than that in i is defined as the empirical P value, following the method previously reported (48).
We also performed the XP-CLR test (https://reich.hms.harvard.edu/software) for detecting selective sweeps that involve jointly modeling the multilocus allele frequency between two populations (49). We used the following parameters: nonoverlapping sliding windows of 50 kb, a maximum of 600 SNVs within each window, and correlation level from which the SNVs contribution to XP-CLR result was down-weighted to 0.95. Independent XP-CLR tests were performed to identify selection signals in each population group separately. The genetic map was assumed to be 1 cM/Mb.
The regions with XP-EHH P values less than 0.01 (1%) and XP-CLR values in the top 1% of the empirical distribution were considered strong signals in sport-hunting versus terrier groups. (SI Appendix, Figs. S4 and S5). A more relaxed threshold of 5% was used in the comparison with the random-bred village dogs to consider the large genetic distance to the selected populations of sport-hunting and terrier dogs. Significant genomic regions identified from each step were annotated to the closest genes. The genes that overlapped the significant window regions were defined as candidate genes.
For gene enrichment analysis of candidate genes, PANTHER v.11 (89) was used to determine if there was any significant overrepresentation of genes with functional categories (GO-slim Biological Process). A P value of 0.01 (no correction for multiple testing) was used as the criterion for statistical significance.
Racing Grades of Whippets and Agility Titles of Breeds.
The racing grades of 92 Whippets were previously obtained from the Whippet Racing Association (WRA) website (www.whippetracing.org) (35). We followed the definition of WRA racing grades (www.whippetracing.org/Rules/2006/2006Chapter5.htm) as A, B, C, and D in order from fastest to slowest. Each dog was assigned a grade based on the highest grade achieved during that dog’s career. The racing grades were converted to numeric values (A, 4; B, 3; C, 2; D, 1) and were z-transformed for association analysis. The dog’s sex and height at the withers was obtained from our previous publication (35).
The total number of agility titles won by each breed and the number of dogs registered per breed was provided by the USDAA (https://www.usdaa.com/). The lm function in R version 3.2.2. was used to perform linear regression analyses. The linear mixed model implemented in GEMMA (33) was used to test for association between groups stratified by agility titles.
Data Availability.
The six newly sequenced dog genomes in this study are publicly available from GenBank (Bioproject accession no. PRJNA389682). NCBI-SRA accession numbers are available in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank the Intramural Program of the National Human Genome Research Institute of the NIH and the NIH Intramural Sequencing Center for support and dog owners and breeders for generously providing DNA samples and giving us permission to use their images for this study.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The six newly sequenced dog genomes in this study have been deposited in the GenBank database (Bioproject accession no. PRJNA389682).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800455115/-/DCSupplemental.
References
- 1.Ostrander EA, Huson HJ, Ostrander GK. Genetics of athletic performance. Annu Rev Genomics Hum Genet. 2009;10:407–429. doi: 10.1146/annurev-genom-082908-150058. [DOI] [PubMed] [Google Scholar]
- 2.Macarthur DG, North KN. Genes and human elite athletic performance. Hum Genet. 2005;116:331–339. doi: 10.1007/s00439-005-1261-8. [DOI] [PubMed] [Google Scholar]
- 3.Rankinen T, et al. The human gene map for performance and health-related fitness phenotypes: The 2005 update. Med Sci Sports Exerc. 2006;38:1863–1888. doi: 10.1249/01.mss.0000233789.01164.4f. [DOI] [PubMed] [Google Scholar]
- 4.Rankinen T, et al. Effect of endothelin 1 genotype on blood pressure is dependent on physical activity or fitness levels. Hypertension. 2007;50:1120–1125. doi: 10.1161/HYPERTENSIONAHA.107.093609. [DOI] [PubMed] [Google Scholar]
- 5.Saunders PU, Pyne DB, Telford RD, Hawley JA. Factors affecting running economy in trained distance runners. Sports Med. 2004;34:465–485. doi: 10.2165/00007256-200434070-00005. [DOI] [PubMed] [Google Scholar]
- 6.Coetzer P, et al. Superior fatigue resistance of elite black South African distance runners. J Appl Physiol (1985) 1993;75:1822–1827. doi: 10.1152/jappl.1993.75.4.1822. [DOI] [PubMed] [Google Scholar]
- 7.Weston AR, Karamizrak O, Smith A, Noakes TD, Myburgh KH. African runners exhibit greater fatigue resistance, lower lactate accumulation, and higher oxidative enzyme activity. J Appl Physiol (1985) 1999;86:915–923. doi: 10.1152/jappl.1999.86.3.915. [DOI] [PubMed] [Google Scholar]
- 8.Yang N, et al. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet. 2003;73:627–631. doi: 10.1086/377590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippi G, Longo UG, Maffulli N. Genetics and sports. Br Med Bull. 2010;93:27–47. doi: 10.1093/bmb/ldp007. [DOI] [PubMed] [Google Scholar]
- 10.Van Damme R, Wilson RS, Vanhooydonck B, Aerts P. Performance constraints in decathletes. Nature. 2002;415:755–756. doi: 10.1038/415755b. [DOI] [PubMed] [Google Scholar]
- 11.American Kennel Club . The Complete Dog Book. 20th Ed Ballantine Books; New York: 2006. [Google Scholar]
- 12.Marsden CD, et al. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci USA. 2016;113:152–157. doi: 10.1073/pnas.1512501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreger DL, et al. Whole-genome sequence, SNP chips and pedigree structure: Building demographic profiles in domestic dog breeds to optimize genetic-trait mapping. Dis Model Mech. 2016;9:1445–1460. doi: 10.1242/dmm.027037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 15.Vonholdt BM, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson EK, Lindblad-Toh K. Leader of the pack: Gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9:713–725. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- 17.Ostrander EA, Wayne RK, Freedman AH, Davis BW. Demographic history, selection and functional diversity of the canine genome. Nat Rev Genet. 2017;18:705–720. doi: 10.1038/nrg.2017.67. [DOI] [PubMed] [Google Scholar]
- 18.Boyko AR, et al. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaysse A, et al. LUPA Consortium Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011;7:e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayward JJ, et al. Complex disease and phenotype mapping in the domestic dog. Nat Commun. 2016;7:10460. doi: 10.1038/ncomms10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoenebeck JJ, Ostrander EA. Insights into morphology and disease from the dog genome project. Annu Rev Cell Dev Biol. 2014;30:535–560. doi: 10.1146/annurev-cellbio-100913-012927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang R, et al. Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biol. 2014;15:R25. doi: 10.1186/gb-2014-15-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dodman NH, et al. A canine chromosome 7 locus confers compulsive disorder susceptibility. Mol Psychiatry. 2010;15:8–10. doi: 10.1038/mp.2009.111. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, et al. Population variation revealed high-altitude adaptation of Tibetan mastiffs. Mol Biol Evol. 2014;31:1200–1205. doi: 10.1093/molbev/msu070. [DOI] [PubMed] [Google Scholar]
- 25.Parker HG, et al. Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 2017;19:697–708. doi: 10.1016/j.celrep.2017.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon LM, et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc Natl Acad Sci USA. 2015;112:13639–13644. doi: 10.1073/pnas.1516215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalinowski ST. Do polymorphic loci require large sample sizes to estimate genetic distances? Heredity (Edinb) 2005;94:33–36. doi: 10.1038/sj.hdy.6800548. [DOI] [PubMed] [Google Scholar]
- 28.Joyner MJ, Coyle EF. Endurance exercise performance: The physiology of champions. J Physiol. 2008;586:35–44. doi: 10.1113/jphysiol.2007.143834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storengen LM, Lingaas F. Noise sensitivity in 17 dog breeds: Prevalence, breed risk and correlation with fear in other situations. Appl Anim Behav Sci. 2015;171:152–160. [Google Scholar]
- 30.Blackwell EJ, Bradshaw JW, Casey RA. Fear responses to noises in domestic dogs: Prevalence, risk factors and co-occurrence with other fear related behaviour. Appl Anim Behav Sci. 2013;145:15–25. [Google Scholar]
- 31.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rovira S, Muñoz A, Benito M. Hematologic and biochemical changes during canine agility competitions. Vet Clin Pathol. 2007;36:30–35. doi: 10.1111/j.1939-165x.2007.tb00178.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannula-Jouppi K, et al. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:e50. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosher DS, et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007;3:e79. doi: 10.1371/journal.pgen.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuelke M, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 37.Szabó G, et al. A deletion in the myostatin gene causes the compact (Cmpt) hypermuscular mutation in mice. Mamm Genome. 1998;9:671–672. doi: 10.1007/s003359900843. [DOI] [PubMed] [Google Scholar]
- 38.Montgomery HE, et al. Human gene for physical performance. Nature. 1998;393:221–222. doi: 10.1038/30374. [DOI] [PubMed] [Google Scholar]
- 39.Bigard AX, Brunet A, Guezennec C-Y, Monod H. Effects of chronic hypoxia and endurance training on muscle capillarity in rats. Pflugers Arch. 1991;419:225–229. doi: 10.1007/BF00371099. [DOI] [PubMed] [Google Scholar]
- 40.Faubert J. Professional athletes have extraordinary skills for rapidly learning complex and neutral dynamic visual scenes. Sci Rep. 2013;3:1154. doi: 10.1038/srep01154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cadieu E, et al. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fogle B. The Encyclopedia of the Dog. Dorling Kindersley; New York: 1995. [Google Scholar]
- 43.Johnson PL, et al. A key role for orexin in panic anxiety. Nat Med. 2010;16:111–115. doi: 10.1038/nm.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmeisser MJ, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 45.Lumley LA, et al. Reduced isolation-induced aggressiveness in mice following NAALADase inhibition. Psychopharmacology (Berl) 2004;171:375–381. doi: 10.1007/s00213-003-1610-z. [DOI] [PubMed] [Google Scholar]
- 46.Lachance J, Tishkoff SA. SNP ascertainment bias in population genetic analyses: Why it is important, and how to correct it. BioEssays. 2013;35:780–786. doi: 10.1002/bies.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Lee H-J, et al. Deciphering the genetic blueprint behind Holstein milk proteins and production. Genome Biol Evol. 2014;6:1366–1374. doi: 10.1093/gbe/evu102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akey JM. Constructing genomic maps of positive selection in humans: Where do we go from here? Genome Res. 2009;19:711–722. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nat Rev Genet. 2014;15:379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grady RM, et al. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: A model for Duchenne muscular dystrophy. Cell. 1997;90:729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 53.Lamon S, Wallace MA, Russell AP. The STARS signaling pathway: A key regulator of skeletal muscle function. Pflugers Arch. 2014;466:1659–1671. doi: 10.1007/s00424-014-1475-5. [DOI] [PubMed] [Google Scholar]
- 54.Barrientos T, et al. Two novel members of the ABLIM protein family, ABLIM-2 and -3, associate with STARS and directly bind F-actin. J Biol Chem. 2007;282:8393–8403. doi: 10.1074/jbc.M607549200. [DOI] [PubMed] [Google Scholar]
- 55.Anderson WA, et al. Long-term neurostimulation of skeletal muscle: Its potential for a tether-free biologic cardiac assist device. Pacing Clin Electrophysiol. 1988;11:2128–2134. doi: 10.1111/j.1540-8159.1988.tb06361.x. [DOI] [PubMed] [Google Scholar]
- 56.Mann DT, Williams AM, Ward P, Janelle CM. Perceptual-cognitive expertise in sport: A meta-analysis. J Sport Exerc Psychol. 2007;29:457–478. doi: 10.1123/jsep.29.4.457. [DOI] [PubMed] [Google Scholar]
- 57.McCarthy MI, et al. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 58.Gregory NS, Brito RG, Fusaro MCGO, Sluka KA. ASIC3 is required for development of fatigue-induced hyperalgesia. Mol Neurobiol. 2016;53:1020–1030. doi: 10.1007/s12035-014-9055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cox C. Investigation of hearing loss in dogs. In Pract. 2002;24:494–501. [Google Scholar]
- 60.Strain GM. Congenital deafness and its recognition. Vet Clin North Am Small Anim Pract. 1999;29:895–907, vi. doi: 10.1016/s0195-5616(99)50079-x. [DOI] [PubMed] [Google Scholar]
- 61.Bork JM, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwon T-J, et al. Methionine sulfoxide reductase B3 deficiency causes hearing loss due to stereocilia degeneration and apoptotic cell death in cochlear hair cells. Hum Mol Genet. 2014;23:1591–1601. doi: 10.1093/hmg/ddt549. [DOI] [PubMed] [Google Scholar]
- 63.Noben-Trauth K, Zheng QY, Johnson KR. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat Genet. 2003;35:21–23. doi: 10.1038/ng1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kidd T, et al. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- 65.Bates TC, et al. Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav Genet. 2011;41:50–57. doi: 10.1007/s10519-010-9402-9. [DOI] [PubMed] [Google Scholar]
- 66.Naylor J, et al. Pregnenolone sulphate- and cholesterol-regulated TRPM3 channels coupled to vascular smooth muscle secretion and contraction. Circ Res. 2010;106:1507–1515. doi: 10.1161/CIRCRESAHA.110.219329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villela L, Mota M, Oliveira H. Genetic parameters of racing performance traits of Quarter horses in Brazil. J Anim Breed Genet. 2002;119:229–234. [Google Scholar]
- 68.Langlois B. Heritability of racing ability in Thoroughbreds—A review. Livest Prod Sci. 1980;7:591–605. [Google Scholar]
- 69.Casas E, Kehrli ME., Jr A review of selected genes with known effects on performance and health of cattle. Front Vet Sci. 2016;3:113. doi: 10.3389/fvets.2016.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hill EW, McGivney BA, Gu J, Whiston R, Machugh DE. A genome-wide SNP-association study confirms a sequence variant (g.66493737C>T) in the equine myostatin (MSTN) gene as the most powerful predictor of optimum racing distance for Thoroughbred racehorses. BMC Genomics. 2010;11:552. doi: 10.1186/1471-2164-11-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vanhooydonck B, Van Damme R, Aerts P. Speed and stamina trade-off in lacertid lizards. Evolution. 2001;55:1040–1048. doi: 10.1554/0014-3820(2001)055[1040:sastoi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 72.Huson HJ, Parker HG, Runstadler J, Ostrander EA. A genetic dissection of breed composition and performance enhancement in the Alaskan sled dog. BMC Genet. 2010;11:71. doi: 10.1186/1471-2156-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abdelwahid E, Rice D, Pelliniemi LJ, Jokinen E. Overlapping and differential localization of Bmp-2, Bmp-4, Msx-2 and apoptosis in the endocardial cushion and adjacent tissues of the developing mouse heart. Cell Tissue Res. 2001;305:67–78. doi: 10.1007/s004410100399. [DOI] [PubMed] [Google Scholar]
- 75.Parker HG, et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 77.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 81.Auwera GA, et al. From FastQ data to high‐confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.11–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 84.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 88.Sabeti PC, et al. International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thomas PD, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The six newly sequenced dog genomes in this study are publicly available from GenBank (Bioproject accession no. PRJNA389682). NCBI-SRA accession numbers are available in SI Appendix, Table S1.