Significance
Accurate cancer diagnostics is a prerequisite for optimal personalized cancer medicine. We propose an information-theoretic cancer diagnosis that identifies signatures comprising patient-specific oncogenic processes rather than cancer type-specific biomarkers. Such comprehensive transcriptional signatures should allow for more accurate classification of cancer patients and better patient-specific diagnostics. The approach that we describe herein allows decoding of large-scale molecular-level information and elucidating patient-specific transcriptional altered network structures. Thereby, we move from cancer type-associated biomarkers to unbiased patient-specific unbalanced oncogenic processes.
Keywords: information theory, surprisal analysis, cancer diagnostics, patient-specific gene expression signatures, intertumor heterogeneity
Abstract
Every individual cancer develops and grows in its own specific way, giving rise to a recognized need for the development of personalized cancer diagnostics. This suggested that the identification of patient-specific oncogene markers would be an effective diagnostics approach. However, tumors that are classified as similar according to the expression levels of certain oncogenes can eventually demonstrate divergent responses to treatment. This implies that the information gained from the identification of tumor-specific biomarkers is still not sufficient. We present a method to quantitatively transform heterogeneous big cancer data to patient-specific transcription networks. These networks characterize the unbalanced molecular processes that deviate the tissue from the normal state. We study a number of datasets spanning five different cancer types, aiming to capture the extensive interpatient heterogeneity that exists within a specific cancer type as well as between cancers of different origins. We show that a relatively small number of altered molecular processes suffices to accurately characterize over 500 tumors, showing extreme compaction of the data. Every patient is characterized by a small specific subset of unbalanced processes. We validate the result by verifying that the processes identified characterize other cancer patients as well. We show that different patients may display similar oncogene expression levels, albeit carrying biologically distinct tumors that harbor different sets of unbalanced molecular processes. Thus, tumors may be inaccurately classified and addressed as similar. These findings highlight the need to expand the notion of tumor-specific oncogenic biomarkers to patient-specific, comprehensive transcriptional networks for improved patient-tailored diagnostics.
Cancer results from the acquisition of genetic alterations, which in turn, lead to significant rewiring of molecular networks. Despite a diverse array of genetic mutations in tumors, there are typically fewer distinct phenotypes than the extent of genetic, epigenetic, and transcriptional heterogeneity would suggest (1). Many tumors eventually rely on a limited number of key proteins (oncogenes) that are responsible for cancer growth and survival, a phenomenon known as “oncogene addiction” (2). This has led to the idea that the identification of the key tumor-specific oncogene biomarkers would be an effective diagnostics strategy. However, extensive molecular variations between patients from the same cancer type, referred to as interpatient heterogeneity, render it difficult to find common gene markers that correlate well with drug sensitivity and patient survival. Indeed, it was recently proposed that reliable genomic markers should be identified and integrated into the pathology process to diagnose and treat each patient optimally (3). Moreover, different molecular processes may give rise to the same list of oncogenic biomarkers (1). Hence, tumors may be classified as similar for the purposes of diagnostics and treatment, despite being biologically different.
Our main goal in this study was to develop a method that classifies patients not only based on their tumor-specific list of oncogenic biomarkers but also, based on the molecular context that gave rise to this list of biomarkers. To this end, we investigated gene expression alterations in a cohort of 527 samples consisting of lymphoma, bladder cancer, gastric cancer, colorectal cancer, breast cancer, and normal gastric tissues, and we identified the set of ongoing molecular processes that make up each patient-specific transcriptional network. We show how the numerous gene expression alterations that occurred in the large cohort of tumors can be translated to a few altered molecular processes (4, 5) that repeat themselves in different combinations in every patient and accurately characterize the vast interpatient heterogeneity. These few unbalanced processes, identified by an information theoretic approach, achieved a significant compaction of the big dataset.
We show that similar expression levels of certain oncogenes in different patients can be attributed to different combinations of unbalanced processes. This suggests that, to accurately classify patients, transcriptional networks instead of lists of biomarkers should be identified.
The approach described herein provides an important additional step toward accurately decoding cancer information in a patient-specific manner. Our findings highlight the need to incorporate oncogene biomarkers into the context of transcriptional networks to accurately characterize patient-specific tumor biology and to improve patient-tailored diagnostics.
Using Information Theory to Identify Patient-Specific Ongoing Cancer Processes
Tumors are biological systems in which the balanced homeostatic state has been disturbed due to genomic and environmental factors or constraints (6–8). These constraints bring about an imbalance in the tissue and result in abnormal gene expression levels reflecting ongoing unbalanced molecular processes. To quantify the imbalance, we use a thermodynamically based information-theoretic strategy. Thermodynamic-based approaches (9–13) and/or information-theoretic approaches have been successfully applied to the analysis of biological systems in a number of cases (for example, refs. 14–17). In this study, we utilize the thermodynamic-motivated surprisal analysis (6, 7, 18, 19). We have previously applied this analysis to various biological systems (4, 20–22) and also showed its experimental validity (21, 23).
The equation used in the study represents the logarithm of the experimental transcript expression level, , of a measured transcript i in every patient k as (7)
| [1] |
where is the logarithm of the expression level of the transcript i at the balanced state and the sum, , represents the deviations in the logarithm of the expression level of this transcript from the balanced state level due to the environmental/genetic constraints that may operate in the system.
Surprisal analysis identifies which transcripts are at their balanced state level for every single tumor. The balanced state term can be represented as (7), allowing us to calculate an amplitude for the balanced state, , for every tumor k and the extent of the participation of each individual transcript i, , in the balanced state process . We have previously shown that this balanced state is robust, and it remains common to normal and cancer tissues and even to different organisms (i.e., the transcripts participating in this process do not show any dependence on the patients) (4, 6, 7, 20). The experimental data that we wished to analyze in this study originated from several different datasets. We expect that the expression level of every transcript i in the balanced state, , should be common to all patients and does not depend on the patient index, k.
The analysis further uncovers the complete set of constraints that operate in the system, including the transcripts that are affected by these constraints and thus, deviate from their balanced state levels. A constraint can result from any perturbation in a biological system. Each constraint significantly influences only a subset of transcripts in a similar way, causing collective deviations of the transcript levels (up or down) from their balanced levels. This group of covarying transcripts represents an altered transcript correlation subnetwork that we name an unbalanced process. The unbalanced processes are indexed by . Each unbalanced process can consist of several biological pathways. For example, proteins involved in aerobic glycolysis and MAPK signaling pathways can deviate in a coordinated manner from the balanced state and thus, participate in the same unbalanced process (20).
Several unbalanced processes may operate in each tumor, and each transcript can participate in several unbalanced processes due to the nonlinearity of biological networks (20).
Singular value decomposition (24–26) is used as a mathematical tool to determine the two sets of parameters that determine the unbalanced processes in surprisal analysis (7): (i) the values denoting the amplitude of unbalanced process in every tumor k and (ii) the values denoting the extent of the participation of each individual transcript i in the specific unbalanced process, (7). Transcripts with the highest/lowest values are used to determine the transcript composition of unbalanced processes (SI Appendix, Fig. S1). To assign a biological meaning for each process, transcripts with the most significant values are classified into biological categories according to Gene Ontology database (Dataset S1). Several biological categories appear in each process (Dataset S1). Note that the weight, , of transcript i in a process is the same for all patients (i.e., is independent of k). Hence, the network structure, composed of covarying transcripts participating in process , remains constant. The amplitude, , determines whether process is active in the patient k and to what extent.
Our goal was to utilize surprisal analysis to classify tumors according to the tumor-specific sets of constraints that deviate the cancer tissues from the stable, balanced state. We suggest that such a classification is essential to improve personalized cancer diagnostics.
Integrating Biological Datasets to Study Interpatient Heterogeneity
The field of personalized medicine has been accelerating, and a massive amount of gene expression data regarding different types of cancer is becoming available. Five different datasets were selected for analysis, each comprising samples from a different type of cancer: lymphoma, bladder cancer, gastric cancer, colorectal cancer, and breast cancer (527 in total: 506 tumor samples and 21 normal gastric samples). A concurrent analysis of different datasets will allow identification of the altered biological processes that characterize the interpatient heterogeneity. Additionally, a large-scale analysis should uncover the patient-specific sets of unbalanced processes with a better signal-to-noise ratio.
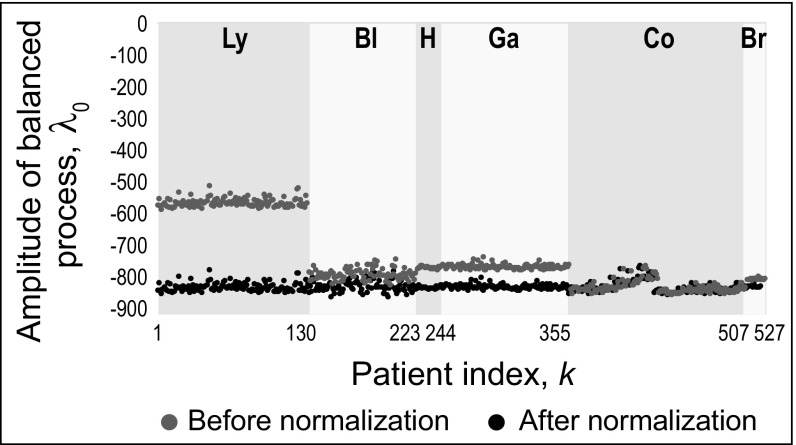
As expected, surprisal analysis of the five datasets identified a common balanced state for each type of cancer represented by an invariant amplitude of the balanced state for all patients, k, of a specific cancer type, including the normal gastric samples (Fig. 1, gray). This result corresponds to our previous findings showing the robustness of the balanced process (4, 6, 20). The levels of more than 470 transcript probes of ∼20,000 probes were well-fitted by the balanced term alone and were not influenced by any unbalanced process. These transcripts participate in the homeostatic functions of the cell, such as protein and RNA metabolism, and the cell cycle (Dataset S1).
Fig. 1.
Identifying the steady state in cancer patients and integrating biological datasets to study intertumor heterogeneity. Amplitudes of balanced process () for patients with lymphoma (Ly; patients 1–130), patients with bladder cancer (Bl; patients 131–223), healthy patients (H; patients 224–244), patients with gastric cancer (Ga; patients 245–355), patients with colorectal cancer (Co; patients 356–507), and patients with breast cancer (Br; patients 508–527) before and after normalization.
After determination of the balanced state term separately for each dataset, the intensities of the different sets were normalized and converted to a common scale, such that all five datasets shared a common balanced state term (Fig. 1). Importantly, the transcript composition of the steady state remained invariant before and after the normalization, suggesting that the intensity differences reflected experimental artifacts and not biological differences (SI Appendix, Fig. S2). The thermodynamic-based approach underlying surprisal analysis is what enables such a normalization. Importantly, we show that this normalization does not influence the amplitudes of the unbalanced processes or the weight of individual transcripts in these processes (SI Appendix, Integration of Different Datasets).
The notion that the balanced state is common to normal and cancerous tissues is highly significant, because it suggests that the search for the tumor gene markers should focus only on the unbalanced processes, greatly reducing the number of possible targets.
Interpatient Heterogeneity Among 506 Patients Is Characterized by 12 Unbalanced Processes
Our next step was to inspect the unbalanced processes that characterized the 506 tumors (not including the 21 normal gastric samples). The analysis revealed that 12 unbalanced processes sufficed to reproduce the deviations from the balanced state across the 506 tumors of five types (Datasets S1–S4). We used three different methods to identify the number of unbalanced processes that characterize the interpatient variability: (i) calculation of error limits that were based on the fluctuations in the expression levels of the most stable transcripts was used to determine which of the processes possesses an amplitude that exceeds the noise threshold; (ii) error bars for each patient were computed as described previously (27); and (iii) to validate that the number of significant unbalanced processes (only those having amplitude values exceeding error limits) is sufficient, we verify that these processes adequately reproduce the experimental data. These three methods are further discussed in SI Appendix, Calculation of Error Bars and Threshold Values, with results shown in SI Appendix, Figs. S3–S6.
To verify the robustness and accuracy of the analysis, we randomly picked 50% of the patients from each cancer type (264 patients total; representing about one-half of the complete dataset) and found that the unbalanced processes and patient-specific signatures remained the same (SI Appendix, Figs. S7 and S8).
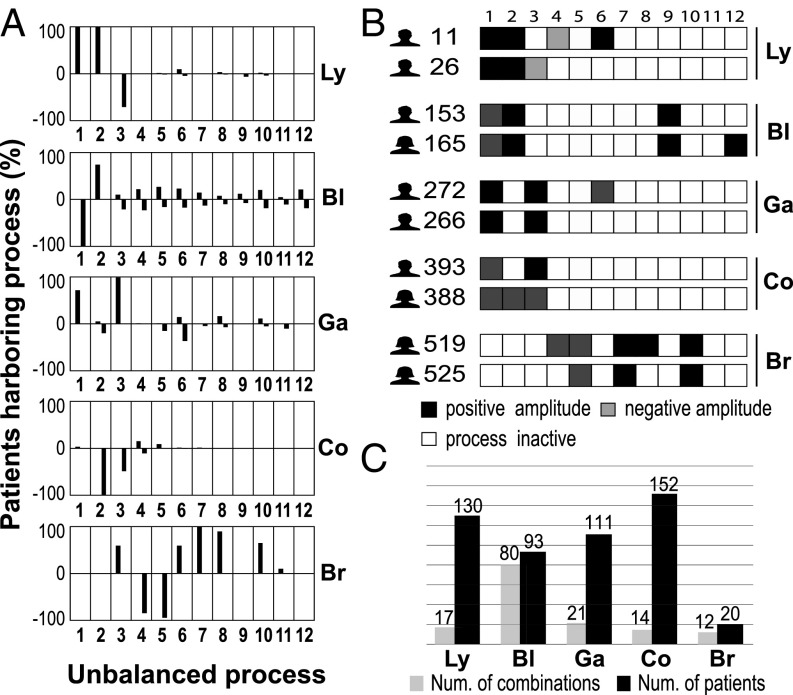
Transcripts can be involved in only one constraint (e.g., GRB2, PTK2B, and CALM3) (Dataset S3), whereas others participate in two or more unbalanced processes, such as EGF receptor (EGFR), programmed death ligand 1 (PD-L1; CD274), CD44, IRS2, EIF4E, and CDK1 (Dataset S3). Dataset S1 shows that each unbalanced process can include multiple (sometimes overlapping) biological categories. Importantly, we found that, in every cancer type, one or more unbalanced processes are shared by all of the patients with this cancer type (Fig. 2A). For example, all of the lymphoma patients were found to harbor the process with a positive amplitude [], which we define as process 1+ (Fig. 2A). Process 2+ was found in all patients with lymphoma as well (Fig. 2A). Genes involved in these processes were classified to multiple categories (for example, B-cell signaling, cell proliferation, platelet deregulation, and DNA repair) (Dataset S1). Recall that the weights, , are independent of the patient index, k, and that it is the amplitude, , that determines whether a process is active in the specific patient. The sign of determines the direction to which the process deviates from the transcripts. Thus, if all lymphoma patients harbor process 1+, it means that process 1 deviates the transcripts in the process in the same manner in all lymphoma patients (i.e., up-regulates or down-regulates them). Process 1− was found in all patients with bladder cancer (Fig. 2A) and includes genes involved, for example, in intracellular signal transduction and GTPase activation (Dataset S1). Process 3+ appeared in all patients with gastric cancer (Fig. 2A) and includes genes involved in angiogenesis and antiapoptosis (Dataset S1). Process 2− appeared in all patients with colorectal cancer (Fig. 2A) and includes genes involved in IL-4 and IL-10 production and NF-κB signaling (Dataset S1). The breast cancer patients were all found to harbor process 7+ (Fig. 2A), which includes VEGFR signaling and glucuronidation (a mechanism of intrinsic drug resistance) (Dataset S1). The finding that certain unbalanced processes are shared by all patients with a particular cancer type is consistent with our earlier findings that there is a dominant process that characterizes a particular type of cancer compared with normal samples (4, 6, 7). Note, however, that the same process may also appear in other cancer types, possibly less frequently. For example, process 3− is shared by lymphoma, bladder, and colorectal cancers (Fig. 2A). This constraint includes transcripts involved, for example, in PGDFR signaling pathway, mRNA processing, and splicing (Dataset S1). Process 5− appears in bladder, gastric, and breast cancers and comprises transcripts involved in, for example, Wnt signaling, cell–cell adhesion, and RNA splicing (Dataset S1). Processes of higher index appear in a smaller number of patients (Dataset S2).
Fig. 2.
Transcriptomic data from 506 patients were reduced to a 12D space of unbalanced processes. Each patient harbors a combination of 2–5 unbalanced processes of 12. (A) The frequency of the unbalanced processes in every cancer type: −100 denotes 100% with negative amplitude. Note that at least one unbalanced process is common to all patients with a given cancer type. For example, processes 1+ and 2+ were found in all patients with lymphoma (Ly); process 1− was found in all patients with bladder cancer (Bl). Furthermore, most of the processes each appear in at least two types of cancer. For example, process 3 (+ or −) was found in all cancer types [Ly, Bl, colorectal cancer (Co), gastric cancer (Ga), and breast cancer (Br)]. (B) Patient-specific combinations of unbalanced process. Two selected patients from each cancer type are shown. Although cancer patients can have the same type of cancer, they may harbor different sets of unbalanced processes. For example, patients 11 and 26 were diagnosed with Ly; they have process 1 and 2 in common and differ in the rest of the active unbalanced processes. Dataset S4 includes the full list of patient-specific combinations of unbalanced processes. Negative/positive amplitude denotes how the patients are correlated with respect to a particular process (SI Appendix, Signs of Giα, and λα(k)). For example, patient 165 harbors process 1−, whereas patient 272 harbors process 1+. Therefore, transcripts that participate in process 1 (Dataset S1) deviate from their balanced level in opposite directions in these patients. (C) A patient-specific combination of unbalanced processes was calculated for every patient. A total of 144 different combinations of two to five unbalanced process (Dataset S4) appeared in the entire dataset. Some cancer types possess a significantly higher degree of heterogeneity (e.g., Bl has 80 different combinations in the population of 93 patients), whereas others, such as Co, are less heterogeneous (14 combinations in the population of 152 patients).
From Unbalanced Processes to Patient-Specific Signatures
Twelve unbalanced processes repeat themselves across 506 tumors. However, not all processes are active in all tumors. Every individual tumor harbors a specific subset or signature of active unbalanced processes (Fig. 2 B and C and Dataset S4). Typically, every patient can be accurately represented by a combination of one to five ongoing processes (Fig. 2B and Dataset S4). Dataset S4 contains the entire list of 144 patient-specific sets of unbalanced processes that are repeated across 506 tumors.
Twelve unbalanced processes can be assembled into thousands of unique subsets of one to five processes. We found varying degrees of intertumor heterogeneity in each of the tumor types (Fig. 2C): 17 combinations of processes were found in the population of 130 lymphoma patients, 80 combinations were found in the population of 93 bladder cancer patients, 21 combinations were found the population of 111 gastric cancer patients, 14 unique combinations were found in the population of 152 colorectal cancer patients, and 12 combinations of processes were found in the population of 20 breast cancer patients. Thus, some cancer types possess a high degree of heterogeneity (e.g., bladder), whereas others, such as colorectal cancer, are significantly less heterogeneous (Fig. 2C).
Similar Gene Expression Levels Can Result from Different Combinations of Unbalanced Processes
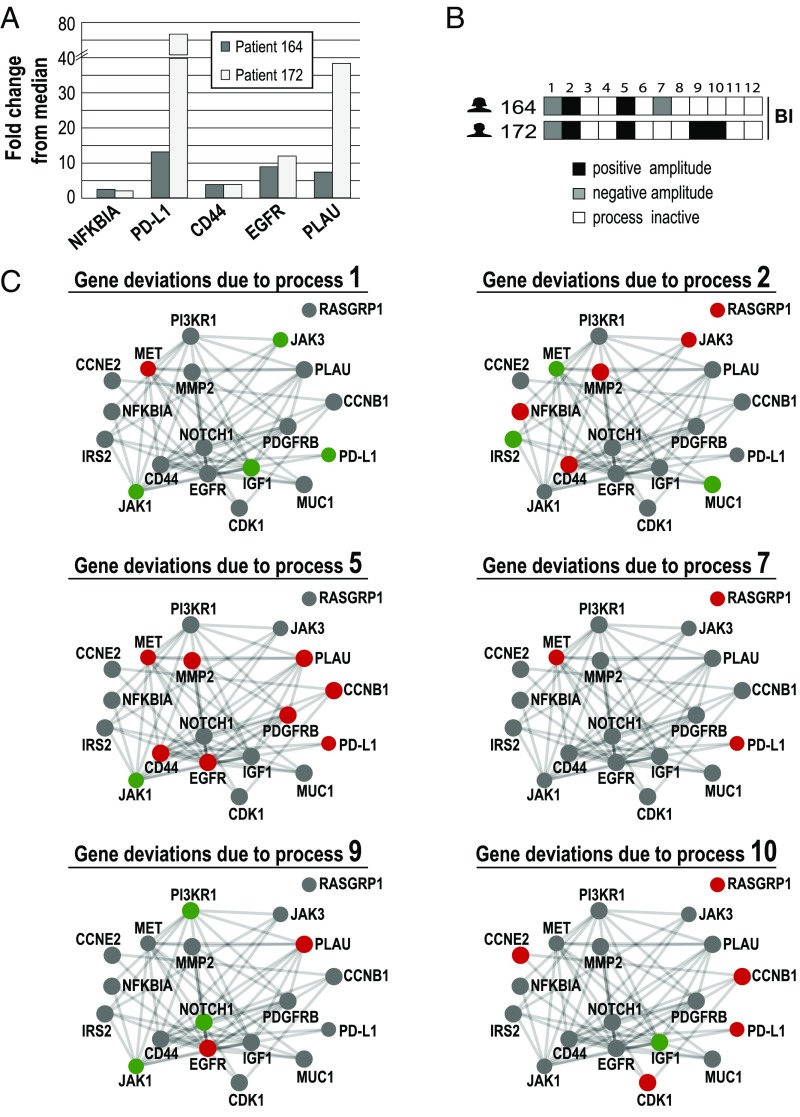
One of the main features of surprisal analysis is its ability to assign transcripts to more than one unbalanced process (see above) (7). For example, EGFR was found to independently participate in processes 4–6 and 9; PD-L1 (CD274; inhibitor of the immune system) participates in processes 5, 7, 8, and 10 (Dataset S3). Therefore, two patients can display similar gene expression levels, but their tumors may harbor different combinations of unbalanced processes. To demonstrate this point, we selected two bladder cancer patients, indexed 164 and 172, and inspected their tumor-specific experimental expression levels of five bladder cancer-associated genes: NF-κBIA (the inhibitor of NF-κB) (28, 29), PD-L1 (30), CD44 (31), EGFR (32), and PLAU (33) (Fig. 3A). In both tumors, these biomarkers were up-regulated relative to their median expression level (Fig. 3A). However, surprisal analysis revealed that the tumors are biologically different: patient 164 is characterized by a combination of processes 1, 2, 5, and 7, whereas patient 172 harbors a combination of processes 1, 2, 5, 9, and 10 (Fig. 3B and Dataset S4). Fig. 3C shows 19 selected genes and how they were affected by these unbalanced processes in the two patients. In both patients, the induction of NF-κBIA is associated with unbalanced process 2 (Fig. 3 B and C), and the induction of CD44 is associated with processes 2 and 5 (Fig. 3 B and C). However, the induction of other oncogenes was attributed to different processes. For example, in patient 164, PD-L1 induction was attributed to processes 5 and 7, whereas in patient 172, PD-L1 induction was attributed to processes 5 and 10 (Fig. 3 B and C). Similarly, EGFR was induced by process 5 in patient 164, whereas in patient 172, it was induced due to processes 5 and 9 (Fig. 3 B and C).
Fig. 3.
Similar gene expression levels in different patients may be attributed to different unbalanced processes. Two bladder cancer (Bl) patients were selected for the demonstration of the concept. (A) The fold changes of five selected Bl-associated oncogenes are shown. NF-κBIA, PD-L1, CD44, EGFR, and PLAU were up-regulated in both patients relative to their median expression levels across 506 patients. (B) Patient-specific combinations of unbalanced processes. Patient 164 harbors processes 1, 2, 5, and 7. Patient 172 harbors unbalanced processes 1, 2, 5, 9, and 10. Selected genes from these processes are shown in C. (C) The influences of these unbalanced processes on 19 selected oncogenes are shown. The complete list of transcripts that participate in each of the processes (∼370–1,500 transcripts for each process) can be found in Dataset S1. Red denotes up-regulation, green denotes down-regulation, and gray denotes no change due to the process. Functional connections are according to STRING database. For example, CD44 was up-regulated in both patients to a similar extent. This is attributed to unbalanced processes 2 and 5, which are active in both patients. The up-regulation of PD-L1 in patient 164 is attributed to processes 5 and 7, whereas in patient 172, the up-regulation in PD-L1 is attributed to processes 5 and 10. The up-regulation of PLAU in both patients is associated with process 5. However, in patient 172, the up-regulation of PLAU is also attributed to process 9, which is active in his tumor as well.
The full lists of values, representing the extent of the participation of each transcript in processes , are presented in Dataset S3.
Patients 164 and 172 serve as an example of two patients carrying tumors of the same type that may present with similar lists of oncogenic biomarkers, although their tumors are not the same. Classification of tumors according to similar biomarkers may lead to significant differences between cancer patients in terms of response to treatment, survival prediction, and more. Deciphering the complete altered transcriptional network in every tumor should enable more accurate diagnosis and classification of patients.
Twelve Unbalanced Processes Identified Are Active in Other Cancer Patients
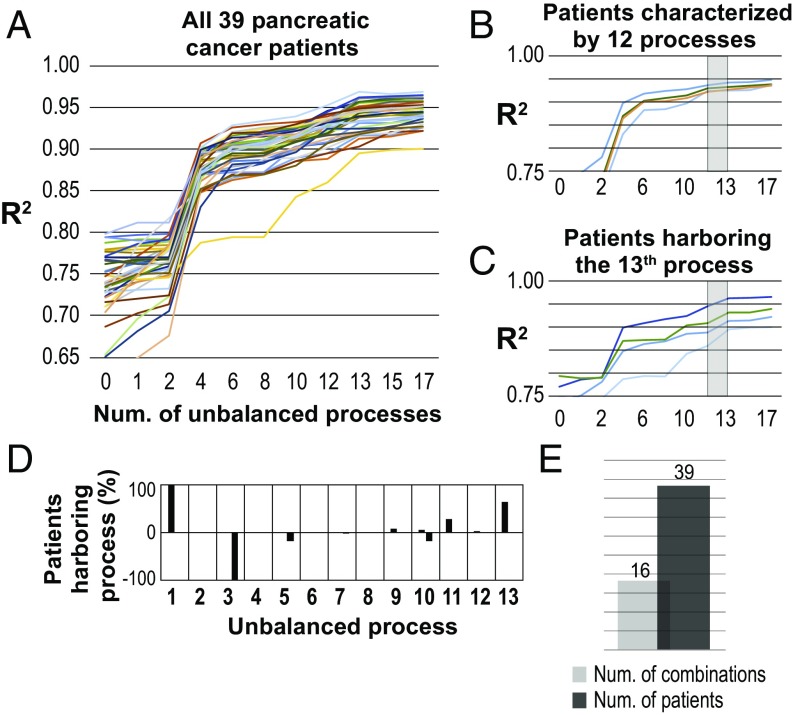
Our next step was to verify whether the 12 unbalanced processes that were identified in 506 tumors are relevant to other cancer patients as well. To answer this, we obtained an additional dataset, which consists of 39 pancreatic tumors. This additional dataset will be referred to as the validation set. The dataset was merged with the previously analyzed five datasets (utilizing the normalization method described above), and the combined dataset, comprising 566 patients, was analyzed using surprisal analysis (Fig. 4). Thirteen unbalanced processes were identified in this analysis. Strikingly, the first 12 unbalanced processes appeared to be the same 12 unbalanced processes that were identified in the analysis of the original 527 samples (SI Appendix, Figs. S9 and S10), and they could fully characterized 36% of the pancreatic patients (Fig. 4B and Dataset S5). The 13th process, which appeared only on addition of the validation set to the analysis, was essential for the characterization of the remaining ∼64% of pancreatic patients (Fig. 4C and Dataset S5). This process did not appear in the original dataset, suggesting that it is a pancreatic cancer-specific constraint. The transcripts involved in unbalanced process 13 were categorized, among others, to the Notch, IL-1, NF-κB, and EGFR signaling pathways (Dataset S5). These pathways were shown to be involved in pancreatic cancer (34–37).
Fig. 4.
The 12 original unbalanced processes and an additional pancreatic-specific unbalanced process fully characterize 39 pancreatic patients. (A) The 39 pancreatic cancer patients in the validation set were found to harbor 13 unbalanced processes. Processes 1–12 are the same processes found for 506 original patients. R2 values were calculated for all pancreatic patients with pancreatic cancer by plotting the natural logarithm of the experimental data vs. for different values of . The value of R2 approaches one as more unbalanced processes are added to the calculation. Mathematically, 506 unbalanced processes are calculated for each patient. However, not all of them are significant. The figure shows that all patients reach a plateau after 13 processes, suggesting that the first 13 unbalanced processes are significant and that the rest of the processes represent random noise in the biological system. (B) Thirty-six percent of the pancreatic patients were fully characterized by the first 12 unbalanced processes. Four selected patients are shown. The gray box highlights that the addition of the 13th constraint had no significant effect on the R2 value for these patients. (C) Sixty-four percent of the pancreatic patients were found to harbor an additional pancreatic-specific constraint, unbalanced process 13, which did not appear in the analysis of 506 original patients. The gray box highlights that the addition of the 13th constraint is significant for these patients. (D) The frequency of the unbalanced processes in the 39 pancreatic cancer patients; 64% of the pancreatic patients were found to harbor the additional pancreatic-specific unbalanced process 13. (E) Sixteen combinations of unbalanced processes were identified in the pancreatic cancer dataset.
Interestingly, unbalanced processes 1+ and 3− appeared active in all pancreatic patients (Fig. 4D). Unbalanced process 11, which was found in 14 patients with bladder cancer (Fig. 2A), was active in ∼28% of the validation pancreatic set (Fig. 4D). Process 12, which represented only bladder cancer patients previously (Fig. 2A), was found only in one pancreatic patient (Fig. 4D). Overall, 16 different combinations of unbalanced processes were found in 39 pancreatic patients, showing a relatively high degree of interpatient heterogeneity (Fig. 4E and Dataset S5).
Discussion
Personalized medicine aims to subdivide patients into different categories based on molecular-level information. Such a classification often uses biomarker lists to make more informed medical decisions regarding patient diagnosis or treatment. In this study, we expand the idea of gene biomarker profiling and show that integration of biomarkers into tumor ongoing unbalanced processes is critical for accurate identification of patient-specific cancer biology.
To obtain exhaustive patient signatures, we assembled a large-scale patient dataset of transcript expression levels comprising different cancer types. We showed that the notion of the balanced state allows us to integrate datasets from different experiments into one big dataset, thereby increasing the amount of information that can be extracted from the experimental data collected by different groups. The approach can, therefore, be used in a large spectrum of different studies that require large-scale analysis and integration of various molecular datasets.
Using 506 tumors from five different cancer types, we showed that this diverse collection of tumors can be altogether characterized by only 12 unbalanced processes. The majority of unbalanced processes spanned across different cancer types (Fig. 2A), suggesting that unbalanced processes can often be independent of cancer type.
The heterogeneity among the cancer patients was attributed to different patient-specific combinations of 1–5 unbalanced processes of 12, leading eventually to 144 different combinations that represent 144 types of disease. The fact that 144 different diseases can be characterized by only 12 processes makes our approach toward personalized diagnostics quite effective.
Each patient-specific signature is a set of a small number of unbalanced processes, thereby offering a considerable compaction of the data. The compaction means that there is a limited set of processes characterizing the entire dataset. Furthermore, each patient is represented by a subset of those processes.
The finding that each person usually harbors several processes and not only one unbalanced process may result from the coexistence of different intratumor cellular subpopulations in each patient. Each process may characterize a distinct cellular subpopulation. However, in some of the tumor cells, these processes may be active in parallel. Single-cell analysis of a large number of patients will be able to accurately address this topic.
We propose that our approach can provide guidance for patient-specific combined therapies, targeting distinct unbalanced signaling processes in each patient (this is an ongoing project in our laboratory). We validated the approach by adding an independent pancreatic cancer dataset. We show that the same set of 12 unbalanced processes remained valid; 36% of the pancreatic patients from the additional dataset were found to harbor different combinations of the previous 12 unbalance processes. However, the remaining 64% of the pancreatic patients were not fully characterized by the previous 12 unbalanced processes and were found to harbor an additional, pancreatic cancer-specific unbalanced process, indexed 13. This is consistent with our earlier finding that each cancer has a cancer type-specific dominant process.
The approach that we present herein enables extraction of significant signals from large datasets and gaining in-depth, unbiased, patient-specific information. Surprisal analysis efficiently uncovers the altered transcriptional networks in every individual patient, potentially allowing improved classification of cancer patients. Our finding that similar oncogene expression levels in different patients may stem from distinct sets of unbalanced processes underscores the need to extend the initial analysis of tumors and to increase the resolution of cancer patient diagnosis.
Methods
Surprisal Analysis.
All of the gene expression datasets used as an input in the study were obtained from Gene Expression Omnibus database and are publicly available: GSE17920 (lymphoma), GSE31684 (bladder cancer), GSE54129 (gastric cancer), GSE71222 (colorectal cancer), GSE82173 (breast cancer), and GSE15471 (pancreatic cancer). Surprisal analysis was carried out as described before (5–7) and in the text. The notion of the stable steady state, which was determined separately for each dataset, allowed us to integrate different datasets in one large matrix. This matrix was analyzed further to determine the unbalanced processes characterizing five different cancer types. More details are in the text and SI Appendix. SI Appendix includes sections comparing surprisal analysis with principal component analysis (PCA) and k-means clustering.
Calculation of Error Bars and Threshold Values.
To find significant unbalanced processes characterizing each patient and then, to calculate a patient-specific set of the processes, we calculated error bars for the amplitudes of the processes in each sample (27) as well as a threshold limit for each type of cancer using stable transcripts, representing baseline fluctuations in the population as described previously (27) and in SI Appendix, Calculation of Error Bars and Threshold Values.
Calculation of Patient-Specific Combinations of Unbalanced Processes.
Combinations presented in Dataset S4 were generated using () values that exceeded threshold limits and had error bars above 0 (SI Appendix, Calculation of Patient-Specific Combinations of Unbalanced Processes has more details).
Supplementary Material
Acknowledgments
The funding source for this work was the Abisch-Frenkel Foundation (N.K.-B.). F.R. thanks the Fonds National de la Recherche Scientifique for its support.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804214115/-/DCSupplemental.
References
- 1.Alizadeh AA, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinstein IB. Cancer. Addiction to oncogenes–The Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 3.Chibon F. Cancer gene expression signatures–The rise and fall? Eur J Cancer. 2013;49:2000–2009. doi: 10.1016/j.ejca.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Zadran S, Remacle F, Levine RD. miRNA and mRNA cancer signatures determined by analysis of expression levels in large cohorts of patients. Proc Natl Acad Sci USA. 2013;110:19160–19165. doi: 10.1073/pnas.1316991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kravchenko-Balasha N, et al. Convergence of logic of cellular regulation in different premalignant cells by an information theoretic approach. BMC Syst Biol. 2011;5:42. doi: 10.1186/1752-0509-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kravchenko-Balasha N, et al. On a fundamental structure of gene networks in living cells. Proc Natl Acad Sci USA. 2012;109:4702–4707. doi: 10.1073/pnas.1200790109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remacle F, Kravchenko-Balasha N, Levitzki A, Levine RD. Information-theoretic analysis of phenotype changes in early stages of carcinogenesis. Proc Natl Acad Sci USA. 2010;107:10324–10329. doi: 10.1073/pnas.1005283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poovathingal SK, Kravchenko-Balasha N, Shin YS, Levine RD, Heath JR. Critical points in tumorigenesis: A carcinogen-initiated phase transition analyzed via single-cell proteomics. Small. 2016;12:1425–1431. doi: 10.1002/smll.201501178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokolovski M, Bhattacherjee A, Kessler N, Levy Y, Horovitz A. Thermodynamic protein destabilization by GFP tagging: A case of interdomain allostery. Biophys J. 2015;109:1157–1162. doi: 10.1016/j.bpj.2015.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucia U. Thermodynamics and cancer stationary states. Phys A. 2013;392:3648–3653. [Google Scholar]
- 11.Haynie DT. Biological Thermodynamics. 2nd Ed Cambridge Univ Press; New York: 2008. [Google Scholar]
- 12.Salamon P, Wootton JC, Konopka AK, Hansen LK. On the robustness of maximum entropy relationships for complexity distributions of nucleotide sequences. Comput Chem. 1993;17:135–148. [Google Scholar]
- 13.Renken C, et al. A thermodynamic model describing the nature of the crista junction: A structural motif in the mitochondrion. J Struct Biol. 2002;138:137–144. doi: 10.1016/s1047-8477(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 14.Nykter M, et al. Critical networks exhibit maximal information diversity in structure-dynamics relationships. Phys Rev Lett. 2008;100:058702. doi: 10.1103/PhysRevLett.100.058702. [DOI] [PubMed] [Google Scholar]
- 15.Waltermann C, Klipp E. Information theory based approaches to cellular signaling. Biochim Biophys Acta. 2011;1810:924–932. doi: 10.1016/j.bbagen.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Levchenko A, Nemenman I. Cellular noise and information transmission. Curr Opin Biotechnol. 2014;28:156–164. doi: 10.1016/j.copbio.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atwal GS, et al. An information-theoretic analysis of genetics, gender and age in cancer patients. PLoS One. 2008;3:e1951. doi: 10.1371/journal.pone.0001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine RD. Molecular Reaction Dynamics. Cambridge Univ Press; Cambridge, UK: 2005. [Google Scholar]
- 19.Levine RD, Bernstein RB. Energy disposal and energy consumption in elementary chemical reactions. Information theoretic approach. Acc Chem Res. 1974;7:393–400. [Google Scholar]
- 20.Kravchenko-Balasha N, Johnson H, White FM, Heath JR, Levine RD. A thermodynamic-based interpretation of protein expression heterogeneity in different glioblastoma multiforme tumors identifies tumor-specific unbalanced processes. J Phys Chem B. 2016;120:5990–5997. doi: 10.1021/acs.jpcb.6b01692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kravchenko-Balasha N, Shin YS, Sutherland A, Levine RD, Heath JR. Intercellular signaling through secreted proteins induces free-energy gradient-directed cell movement. Proc Natl Acad Sci USA. 2016;113:5520–5525. doi: 10.1073/pnas.1602171113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Facciotti MT. Thermodynamically inspired classifier for molecular phenotypes of health and disease. Proc Natl Acad Sci USA. 2013;110:19181–19182. doi: 10.1073/pnas.1317876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kravchenko-Balasha N, Wang J, Remacle F, Levine RDD, Heath JRR. Glioblastoma cellular architectures are predicted through the characterization of two-cell interactions. Proc Natl Acad Sci USA. 2014;111:6521–6526. doi: 10.1073/pnas.1404462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aiello KA, Alter O. Platform-independent genome-wide pattern of DNA copy-number alterations predicting astrocytoma survival and response to treatment revealed by the GSVD formulated as a comparative spectral decomposition. PLoS One. 2016;11:e0164546. doi: 10.1371/journal.pone.0164546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sankaranarayanan P, Schomay TE, Aiello KA, Alter O. Tensor GSVD of patient- and platform-matched tumor and normal DNA copy-number profiles uncovers chromosome arm-wide patterns of tumor-exclusive platform-consistent alterations encoding for cell transformation and predicting ovarian cancer survival. PLoS One. 2015;10:e0121396. doi: 10.1371/journal.pone.0121396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alter O, Brown PO, Botstein D. Generalized singular value decomposition for comparative analysis of genome-scale expression data sets of two different organisms. Proc Natl Acad Sci USA. 2003;100:3351–3356. doi: 10.1073/pnas.0530258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross A, Levine RD. Surprisal analysis of transcripts expression levels in the presence of noise: A reliable determination of the onset of a tumor phenotype. PLoS One. 2013;8:e61554. doi: 10.1371/journal.pone.0061554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu J, et al. NF-κB p65 overexpression promotes bladder cancer cell migration via FBW7-mediated degradation of RhoGDIα protein. Neoplasia. 2017;19:672–683. doi: 10.1016/j.neo.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito Y, et al. Down-regulation of NF kappa B activation is an effective therapeutic modality in acquired platinum-resistant bladder cancer. BMC Cancer. 2015;15:324. doi: 10.1186/s12885-015-1315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellmunt J, Powles T, Vogelzang NJ. A review on the evolution of PD-1/PD-L1 immunotherapy for bladder cancer: The future is now. Cancer Treat Rev. 2017;54:58–67. doi: 10.1016/j.ctrv.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Wu C-T, Lin W-Y, Chang Y-H, Chen W-C, Chen M-F. Impact of CD44 expression on radiation response for bladder cancer. J Cancer. 2017;8:1137–1144. doi: 10.7150/jca.18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colquhoun AJ, Mellon JK. Epidermal growth factor receptor and bladder cancer. Postgrad Med J. 2002;78:584–589. doi: 10.1136/pmj.78.924.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasui Y, Nishi S, Kitada S, Osada Y. [Urokinase-type plasminogen activator antigen as a prognostic factor in bladder cancer] Nippon Hinyokika Gakkai Zasshi. 1993;84:1624–1628. doi: 10.5980/jpnjurol1989.84.1624. [DOI] [PubMed] [Google Scholar]
- 34.Avila JL, Kissil JL. Notch signaling in pancreatic cancer: Oncogene or tumor suppressor? Trends Mol Med. 2013;19:320–327. doi: 10.1016/j.molmed.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prabhu L, Mundade R, Korc M, Loehrer PJ, Lu T. Critical role of NF-κB in pancreatic cancer. Oncotarget. 2014;5:10969–10975. doi: 10.18632/oncotarget.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuang Z, et al. IL1 receptor antagonist inhibits pancreatic cancer growth by abrogating NF-κB activation. Clin Cancer Res. 2016;22:1432–1444, erratum (2017) 23:868. doi: 10.1158/1078-0432.CCR-14-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troiani T, et al. Targeting EGFR in pancreatic cancer treatment. Curr Drug Targets. 2012;13:802–810. doi: 10.2174/138945012800564158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.